Abstract
Primate lentivirus Vif proteins function by suppressing the antiviral activity of the cell-encoded apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like (APOBEC) proteins APOBEC3G and APOBEC3F. It has been hypothesized that species-specific susceptibilities of APOBEC proteins to Vif proteins may help govern the transmission of primate lentiviruses to new host species. Consistent with this view and with previous results, we report that the Vif proteins of several diverse simian immunodeficiency viruses (SIVs) that are not known to infect humans are not effective inhibitors of human APOBEC3G or APOBEC3F when assessed in transient-transfection experiments. Unexpectedly, this lack of SIV Vif function did not prevent the replication of two vif-deficient SIVs (SIVtan and SIVmnd1; isolated from tantalus monkeys and mandrills, respectively) in a human T-cell line, HUT78, that expresses both APOBEC 3G and APOBEC3F, a finding which demonstrates that some SIVs are partially resistant to the antiretroviral effects of these enzymes irrespective of Vif function. Additional virus replication studies also revealed that the Vif protein of SIVtan is, in fact, active in human T cells, as it substantially enhanced the replication of its cognate virus and human immunodeficiency virus type 1. In sum, we now consider it improbable that species-specific restrictions to SIV Vif function can explain the lack of human infection with certain SIVs. Instead, our data reveal that the species-specific modulation of Vif function is more complex than previously envisioned and that additional (as-yet-unidentified) viral or host factors may be involved in regulating this dynamic interaction between host and pathogen.
The establishment of human immunodeficiency virus type 1 (HIV-1) and HIV-2 infections in humans is the consequence of multiple cross-species (zoonotic) transmissions of simian immunodeficiency viruses (SIVs) from chimpanzees (SIVcpz) and sooty mangabeys (SIVsmm) (15). Interestingly, however, the majority of SIVs now known to infect over 30 different primate species in sub-Saharan Africa have no known human counterparts. Physical barriers, such as a lack of geographic coincidence of the host species with humans or a lack of exposure to infectious SIV, could account for the apparent absence of additional SIV transmissions to humans. Alternatively, it is possible that species-specific molecular restrictions to virus replication may also prevent replication of these SIVs in human cells.
The regulatory gene vif is well conserved among lentiviruses, with only equine infectious anemia virus not appearing to carry it. Vif has long been recognized as being essential for HIV-1 replication, most notably in cultures of primary blood mononuclear cells and in a limited number of immortalized T-cell lines (collectively known as nonpermissive because of their ability to restrict the growth of vif-deficient [Δvif] viruses) (9, 10, 24, 33, 35, 36, 41, 43). In recent years, the human APOBEC3G gene (hA3G, formerly known as CEM15) has been pinpointed as being responsible for the nonpermissive phenotype (33). hA3G is a member of the apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like (APOBEC) family of cytidine deaminases (19) and is incorporated into HIV-1 particles (16, 26, 33), allowing it to catalyze the deamination of deoxycytidine to deoxyuridine in (mostly) nascent minus-strand reverse transcript DNA during the early stages of viral infection (16, 20, 26, 46, 48). This process results in the hypermutation of viral sequences as well as in the premature degradation of viral cDNA (26, 36), effects which together account for the profound loss of infectivity seen for Δvif viruses. The Vif protein protects HIV-1 from hA3G-mediated inhibition by recruiting an E3-ubiquitin ligase to hA3G (47), thereby leading to its polyubiquitination, proteasomal degradation, and exclusion from assembling virus particles (7, 23, 29, 30, 34, 40, 47). Thus, the cellular enzyme hA3G confers innate resistance to HIV-1 infection but is counteracted by the viral Vif protein.
Recent studies have extended not only the spectrum of human APOBEC proteins that can inhibit retroviral infection but also the range of retroviruses that are susceptible to APOBEC action. Thus, hAPOBEC3B (hA3B) and hAPOBEC3F (hA3F) also inhibit HIV-1 and HIV-2 infection (5, 22, 44, 49), and murine leukemia virus, equine infectious anemia virus, SIVtan, and an SIV isolated from experimentally infected rhesus macaques (SIVmac) are susceptible to hA3G (16, 26, 28). Interestingly, however, the suppression of hA3G by Vif has been described as being constrained in a species-specific manner, leading to the view that this interaction may be a predictor of the cross-species transmission potential of primate lentiviruses (37). For instance, it has been shown that SIVtan Vif does not inhibit the anti-HIV or -SIV effects of hA3G in single-cycle infection assays (6, 23, 27, 28, 32). Replacement of the aspartic acid residue at position 128 of hA3G with the lysine residue that naturally occurs at this position in the A3G proteins of various African green monkey species (a grouping that includes tantalus, vervet, grivet, and green monkeys) confers susceptibility to SIVtan Vif, suggesting that this region of A3G may be contacted by Vif directly (6, 27, 32). Accordingly, it has been proposed that the resistance of hA3G to SIV Vif could have played a key role in preventing the transmission of SIV strains such as SIVtan or SIVver (isolated from vervet monkeys) to humans.
Analysis of HIV and SIV Vif function in transfected H9 cells.
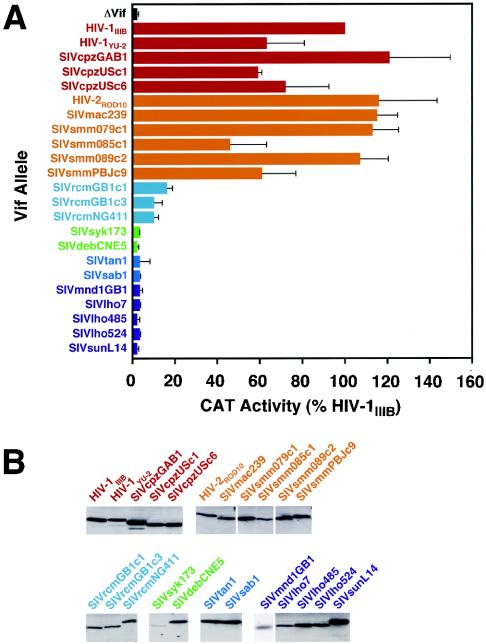
To probe further the potential role of SIV Vif as a determinant of SIV cross-species transmission, we first surveyed a broad array of HIV and SIV Vif proteins for their abilities to regulate HIV-1 infectivity in human T cells. Specifically, we subcloned assorted vif alleles representing six phylogenetic lineages of primate immunodeficiency viruses between the XbaI and BamHI sites of the previously described vector pgVif:T7/HIV-1IIIB (2-4, 8, 12, 13, 17, 18, 21, 37). The lineages that were represented were HIV-1/SIVcpz, HIV-2/SIVmac/SIVsmm, SIVrcm (rcm, red-capped mangabeys), SIVsyk/SIVdeb (syk, Sykes' monkeys; deb, DeBrazza's monkeys), SIVtan/SIVsab (sab, green monkeys), and SIVmnd1/SIVlho/SIVsun (lho, L'Hoest's monkeys; sun, sun-tailed monkeys). The resulting vectors expressed T7 epitope-tagged versions of the various Vif proteins in a Rev-dependent manner. We then used a previously described complementation assay whereby H9 T cells were cotransfected by electroporation with a vif-deficient HIV-1 provirus (HIV-1/Δvif) and either the control vector pgΔVif or one of the Vif expression vectors (36, 37). Virus particles were harvested ∼24 h later, normalized according to p24Gag antigen levels, and used in single-cycle challenges of C8166-CCR5/HIV-CAT indicator cells (Fig. 1A). Since H9 cells express hA3G and hA3F, and therefore closely mimic primary human T cells in terms of their APOBEC protein expression profile (5, 44), HIV-1/Δvif particles that were produced in the absence of any Vif protein had negligible infectivity compared to that of the positive control sample, Vif-IIIB (set at 100%).
FIG. 1.
trans complementation of vif-deficient HIV-1 by HIV and SIV Vif proteins in H9 cells. (A) Virus infectivity. Ten million H9 cells were electroporated with 4 μg of pIIIB/Δvif and 10 μg of pgΔVif or a single pgVif:T7 vector. At ∼24 h, culture supernatants were clarified at 500 × g for 5 min, filtered, and stored. Viruses were normalized according to p24Gag concentration, and levels of infectivity were determined using C8166-CCR5/HIV-CAT cells. Values taken from three experiments are presented as percentages of infectivity relative to that of virus produced in the presence of HIV-1IIIB Vif, and error bars show standard deviations. Similar results were also obtained using HUT78 cells (data not shown). CAT, chloramphenicol acetyltransferase. (B) Protein expression. Whole-cell lysates of 293T cells cotransfected with the indicated pgVif:T7 expression vectors and pcREV (25) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and hybridized with a T7-specific monoclonal antibody (Novagen). Bound antibody was detected using a horseradish peroxidase-conjugated secondary antibody, enhanced chemiluminescence, and autoradiography. All lanes were loaded with equal amounts of lysate derived from the same set of transfections, and the exposure times for all gels were equivalent. The sole exception is the gel for SIVmnd1GB1 Vif, which is from a different experiment.
All Vif proteins that were tested from any single lineage displayed a remarkably consistent phenotype (Fig. 1A). Specifically, all HIV-1/SIVcpz and HIV-2/SIVmac/SIVsmm Vif proteins were >50% active relative to HIV-1IIIB Vif, all SIVrcm proteins displayed 10 to 20% activity, and all remaining Vif proteins from the SIV-only SIVsyk/SIVdeb, SIVtan/SIVsab, or SIVmnd1/SIVlho/SIVsun lineages were inactive (i.e., indistinguishable from the ΔVif sample). Immunoblot analysis of transfected 293T cells suggested that the inactivity of certain SIV Vif proteins was not due to the lack of expression (Fig. 1B) (note that Vif expression levels in the transfected H9 cells were too low to detect). Also, based on the fact that similar results were obtained with analogous constructs lacking the epitope tag (data not shown), we conclude that use of the T7 tag was unlikely to be skewing our results. Thus, all tested Vif proteins from SIV lineages that have been transmitted to humans were functional in human cells, whereas those derived from SIVs not known to infect humans appeared to be inactive. The capacity of any given Vif protein to function in this H9-based single-cycle complementation assay therefore seemed to be a strong predictor of the parental virus' potential to become established in exposed human populations.
The fact that the Vif proteins of SIVrcm (which itself is not a human pathogen) displayed partial activity is also consistent with this hypothesis, since it is now known that SIVcpz (the immediate precursor of HIV-1) is a recombinant of ancestral viruses infecting red-capped mangabeys and a subset of Cercopithecus species (greater spot-nosed, mona, and mustached monkeys) (1). Since the vif region of SIVcpz is most likely derived from SIVrcm (P. M. Sharp, personal communication), it would be predicted that Vif proteins derived from a shared ancestor would also have some activity in human cells.
Regulation of HIV and SIV infection by single APOBEC genes in transfected cells.
In addition to Vif's capacity to inhibit APOBEC function, the effectiveness of the APOBEC proteins of a potential primate host in inhibiting the replication of a nonnative challenge SIV could also represent a zoonotic risk factor and help predict the likelihood of successful cross-species transmission. For instance, should APOBEC protein-mediated inhibition be partial, it is conceivable that suboptimal Vif function would still be sufficient to facilitate cross-species transmission. To begin to address this possibility, we performed a series of titration experiments using hA3G or hA3F cDNA expression vectors and full-length proviruses (Fig. 2). The proviruses used were HIV-1 (pIIIB and pIIIB/Δvif) (38), SIVtan (pTAN and pTAN/Δvif) (37, 39), and SIVmnd1 (pMND and pMND/Δvif). The wild-type pMND vector was modified from pMD121 (31) by repairing the vif and vpr genes on the basis of comparison with the pSMH103 clone of SIVmnd1GB1 (42) and the vif genes of SIVlho and SIVsun (codon 12, TGA→TGG; codon 168, GGΔ→GGG; codon 173, TGA→CGA). pMND/Δvif contains an engineered deletion between nucleotides 66 and 79 of vif that introduces both nonsense and frameshift mutations.
FIG. 2.
Inhibition of HIV-1, SIVmnd1, and SIVtan infection by expression of hA3G (A) or hA3F (B) in 293T cells. Subconfluent 293T monolayers (35 mm in diameter) were cotransfected with 3 μg of wild-type (wt) or Δvif provirus and 0, 3, or 9 μg of pAPOBEC3G or pAPOBEC3F; the DNA content of each transfection cocktail was increased to 12 μg, using a control vector. At ∼24 h, culture supernatants were harvested and normalized by RT content, and virus infectivities were determined using C8166-CCR5/HIV-CAT cells. Values are presented as percentages of infectivity relative to that of virus produced in the absence of hA3G or hA3F, and error bars show standard deviations.
Constant levels of each provirus were cotransfected into 293T cells with a control vector or differing doses of the pAPOBEC3G or pAPOBEC3F expression vectors (5, 33). Virus-containing supernatants were then harvested at ∼24 h, normalized on the basis of reverse transcriptase (RT) activity, and used in challenges of C8166-CCR5/HIV-CAT cells as described above. Consistent with earlier findings from the use of similar experimental set-ups (5, 26, 28, 33), hA3G was a potent inhibitor of all three Δvif viruses (Fig. 2A). Similarly high levels of inhibition were also seen for each wild-type SIV, whereas HIV-1 Vif afforded efficient (but not absolute) protection. It was noticeable, however, that SIVtan/Δvif seemed to be slightly less affected than the other two viruses by hA3G (see below).
As recently demonstrated, hA3F has an hA3G-like phenotype with respect to HIV-1 infection, though the suppressive effects tend to be somewhat less dramatic (Fig. 2B) (5, 22, 44, 49). Unexpectedly, SIVtan was found to be remarkably resistant to hA3F; the infectivity of the wild-type virus was unaffected by any dose tested, and infection by SIVtan/Δvif was, at best, suppressed approximately twofold at the higher dose of transfected plasmid. Taken together, these experiments suggest that SIVtan is less sensitive to restriction by hA3G and hA3F. Interestingly, the drastically different sensitivities of SIVtan to the antiviral effects of hA3G and hA3F mirror similar observations recently made concerning the effects of these APOBEC proteins on infection by the distantly related gammaretrovirus, murine leukemia virus (5). Precisely how the virus substrate range for any given APOBEC protein is determined remains to be defined.
Regulation of HIV and SIV replication in human T cells by SIVtan Vif.
Despite the perceived concordance between the potential of a given SIV to infect humans and the function of its Vif protein in transfected H9 cells (Fig. 1A), we suspected that these matters were more complicated. First, hA3G and more evidently hA3F are not as effective at inhibiting SIVtan as they are at inhibiting HIV-1 (Fig. 2). Second, certain SIVs, most notably SIVlho, SIVmnd1, and some SIVs isolated from African green monkeys, replicate in human peripheral blood mononuclear cells (14, 17). Based on many previous analyses of HIV-1 replication, virus growth would be anticipated to be dependent on Vif function, yet the Vif proteins we tested from these SIV lineages appeared inactive in transfected H9 cells (Fig. 1A). Accordingly, we next conducted a series of experiments that employed multiple rounds of virus replication, rather than single-cycle infectivity assays, to monitor APOBEC protein or Vif function (Fig. 3).
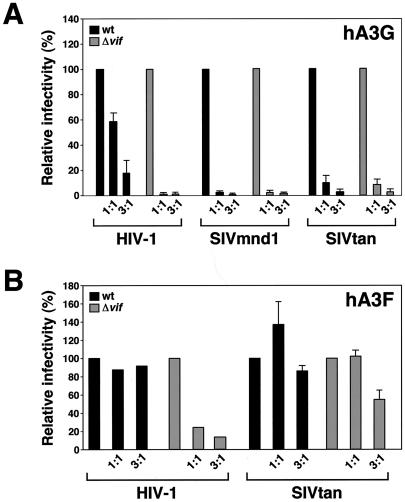
FIG. 3.
Effect of HIV and SIV Vif proteins on replication of HIV-1, SIVmnd1, or SIVtan in T cells. (A to D) HUT78-CCR5 or Jurkat-CCR5 cells stably transduced with HIV-1IIIB vif, HIV-1IIIB Δvif, or SIVtan1 vif were infected with normalized wild-type (wt) or Δvif virus stocks and maintained for up to 4 weeks at 0.25 × 106 to 2 × 106 cells per ml. Virus replication was monitored by measuring supernatant levels of p24Gag or RT. (E) Jurkat cells express hA3F. An ethidium bromide-stained agarose gel shows the products of PCRs performed using primers specific for hA3G or hA3F (5). The templates were either cDNA generated from mRNA isolated from the indicated T cell lines (lanes 3 to 5), water as a negative control (lane 2), or plasmids encoding hA3G or hA3F as positive controls (lane 1).
In each case, the vif genes tested were either carried by the virus under examination or were expressed in trans (i.e., stably expressed by the cell) following standard retrovirus vector-mediated gene transfer (33); similarly, all cells were rendered CCR5 positive to avoid any restrictions on virus entry (11). All infections were initiated with cell-free viral inocula derived from transfections of 293T cells with full-length proviruses, and replication was then monitored over time by observing the accumulation of p24Gag (for HIV-1) or RT activity (for SIVs) in culture supernatants (37).
As previously established, nonpermissive HUT78 cells (which express both hA3F and hA3G [5]) support the growth of wild-type HIV-1, but replication of a Δvif counterpart was completely blocked (Fig. 3A) (38). Consistent with data shown in Fig. 1A, replication of the Δvif virus was rescued in trans by HIV-1 Vif but not by the Vif protein of SIVtan. In contrast, this block was not observed with vif-deficient derivatives of SIVmnd1 or SIVtan, as both viruses were able to replicate to significant extents (Fig. 3B and C). In each case, SIV replication and, by inference, infectivity were further improved by the presence of HIV-1 Vif, presumably through the inactivation and degradation of hA3F and hA3G. Most importantly, and in conflict with results from many experiments carried out using single-cycle measurements of infectivity and viruses generated by transient transfection (Fig. 1A and 2A) (6, 23, 27, 28, 32, 37, 45), the presence of SIVtan Vif clearly stimulated replication of its cognate virus by ∼2 orders of magnitude (Fig. 3C). Indeed, the extent of enhancement was equivalent to that achieved with HIV-1 Vif.
Confirming that this instance of “species-mismatched” SIV Vif function was not a curiosity limited to this combination of cell line and virus, we also found that SIVtan Vif enhanced the replication of a second virus, HIV-1/Δvif, in Jurkat T cells (Fig. 3D). This particular cell line is regarded as semipermissive, as it supports attenuated growth of HIV-1/Δvif (38); this is most likely due to its expression of hA3F but not hA3G, as judged by RT-PCR analysis of extracted mRNA (Fig. 3E, lane 4). Thus, not only are some SIVs partially resistant to the antiviral effects of hA3F and hA3G, but SIVtan Vif can overcome the inhibitory effects of these human proteins to a certain degree in the context of spreading viral infections.
Conclusions.
The data presented in this report reveal that the regulation of primate lentivirus replication by the human APOBEC proteins is more complex than was previously recognized. Specifically, we propose that the potential for cross-species infections by these viruses cannot be assessed in cultured-cell experiments using currently available methodologies and reagents. For instance, even though extensive testing of diverse SIV Vif proteins in transfected H9 cells suggested that their activities correlate closely with the abilities of their cognate viruses to establish infections in humans (Fig. 1), analyses of the replicative potential of some of these same viruses in HUT78 and Jurkat cells yielded contradictory results (Fig. 3). Most notably, the finding that SIVtan Vif can substantially improve the replication of two vif-deficient viruses (Fig. 3), HIV-1/Δvif and SIVtan/Δvif, demonstrates that this particular SIV Vif protein is active (albeit at a lower level than HIV-1 Vif) against the APOBEC proteins naturally expressed in human T cells. Thus, the fact that humans do not appear to be susceptible to infection with SIVs from African green monkeys can no longer be explained by invoking a species-specific barrier to Vif function in human cells. We do not know why transient-transfection-based experiments fail to show the effect of SIVtan Vif on hA3G (Fig. 1A and 2A) and show only a marginal effect on hA3F (Fig. 2B), but this shortcoming presumably reflects the limitations of our understanding of the regulatory interface between APOBEC proteins and HIV or SIV Vif.
It will be of considerable interest to determine why certain SIVs, unlike HIV-1 or HIV-2, are able to replicate in nonpermissive T cells such as HUT78 in the absence of an active vif gene (Fig. 3B and C). We outline a couple of potential contributory factors. First, other elements carried by these SIVs may confer a level of anti-APOBEC activity that is independent of Vif; in other words, these viruses may be inherently more resistant to human APOBEC proteins. Transient-transfection experiments using full-length proviruses provide some hints that this may be the case for SIVtan (Fig. 2), though more extensive analyses will be required to demonstrate a broad trend. Second, coexpression of hA3G and hA3F (5, 22, 44) may result in diminished antiviral activity relative to hA3G alone. Indeed, these two proteins are known to heteromultimerize (44), and recent transfection-based studies of HIV-1 infection have shown that hA3F can interfere with hA3G function (22). Thus, in the case of SIVtan, where the virus is quite refractory to inhibition by hA3F (Fig. 2B), hA3F may act as a significant dominant inhibitor of hA3G and thereby facilitate low levels of Vif-independent replication (Fig. 3B and C).
Importantly, the noted growth of some vif-deficient SIVs in human T cells further emphasizes the notion that the Vif-APOBEC protein interaction is unlikely to underlie the apparent lack of transmission of certain SIVs to humans. SIVtan and SIVmnd1 are less sensitive to inhibition by human APOBEC proteins, yet neither has crossed this species barrier; had the Vif-APOBEC protein interaction been a key selective factor, one might have predicted that diminished restriction would have increased the likelihood of cross-species transmission. Whether the Vif-APOBEC protein interaction proves to be the critical barrier for other potential transmissions of lentiviruses between species remains to be determined.
Nucleotide sequence accession numbers.
The sequences of the previously unpublished vif alleles in this study have been submitted to GenBank under accession numbers AY336731 (SIVcpzUSc1), AY336732 (SIVcpzUSc6), AY336733 (SIVrcmGB1c1), AY336734 (SIVrcmGB1c3), AY336735 (SIVrcmNG411), AY336736 (SIVsmm079c1), AY336737 (SIVsmm085c1), AY336738 (SIVsmm089c2), and AY336739 (SIVdebCNE5).
Acknowledgments
We thank Vanessa Hirsch for supplying the SIVrcmNG411, SIVlho7, SIVlho485, SIVlho524, and SIVsun vif clones. We also thank Paul Sharp for his helpful insights. In addition, we express our appreciation to Bryan Cullen and Bob Doms for providing lab space and other resources for this investigation.
This work was supported by NIH research grants AI46246 (M.H.M.), AI10460 (A.M.S.), AI44596 (P.A.M.), and AI50529 (B.H.H.), NIH training grant AI07325, and grants from the United Kingdom Medical Research Council (M.H.M.) and the Royal Society (A.M.S.). N.C.G. received support as a National Science Foundation predoctoral fellow. M.H.M. is an Elizabeth Glaser Scientist supported by the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 2.Beer, B. E., E. Bailes, G. Dapolito, B. J. Campbell, R. M. Goeken, M. K. Axthelm, P. D. Markham, J. Bernard, D. Zagury, G. Franchini, P. M. Sharp, and V. M. Hirsch. 2000. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J. Virol. 74:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer, B. E., E. Bailes, R. Goeken, G. Dapolito, C. Coulibaly, S. G. Norley, R. Kurth, J.-P. Gautier, A. Gautier-Hion, D. Vallet, P. M. Sharp, and V. M. Hirsch. 1999. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73:7734-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. St. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S.-J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 8.Dewhurst, S., J. E. Embretson, D. C. Anderson, J. I. Mullins, and P. N. Fultz. 1990. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature 345:636-640. [DOI] [PubMed] [Google Scholar]
- 9.Fisher, A. G., B. Ensoli, L. Ivanoff, M. Chamberlain, S. Petteway, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237:888-893. [DOI] [PubMed] [Google Scholar]
- 10.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaddis, N. C., E. Chertova, A. M. Sheehy, L. E. Henderson, and M. H. Malim. 2003. Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J. Virol. 77:5810-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 13.Georges-Courbot, M. C., C. Y. Lu, M. Makuwa, P. Telfer, R. Onanga, G. Dubreuil, Z. Chen, S. M. Smith, A. Georges, F. Gao, B. H. Hahn, and P. A. Marx. 1998. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J. Virol. 72:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm, T. A., B. E. Beer, V. M. Hirsch, and K. A. Clouse. 2003. Simian immunodeficiency viruses from multiple lineages infect human macrophages: implications for cross-species transmission. J. Acquir. Immune Defic. Syndr. 32:362-369. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 16.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch, V. M., B. J. Campbell, E. Bailes, R. Goeken, C. Brown, W. R. Elkins, M. Axthelm, M. Murphey-Corb, and P. M. Sharp. 1999. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J. Virol. 73:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 19.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 20.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 21.Li, Y., H. Hui, C. J. Burgess, R. W. Price, P. M. Sharp, B. H. Hahn, and G. M. Shaw. 1992. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J. Virol. 66:6587-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liddament, M. T., W. L. Brown, A. J. ASchumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 23.Liu, B., X. Yu, K. Luo, Y. Yu, and X.-F. Yu. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 78:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 26.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 27.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 28.Mariani, R., D. Chen, B. Schröfelbauer, F. Navarro, R. König, B. Bollman, C. Münk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 29.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 30.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 31.Sakai, H., J. Sakuragi, S. Sakuragi, R. Shibata, M. Hayami, A. Ishimoto, and A. Adachi. 1992. Genetic characterization of simian immunodeficiency virus isolated from an African mandrill. Arch. Virol. 125:1-14. [DOI] [PubMed] [Google Scholar]
- 32.Schröfelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 34.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 35.Simon, J. H. M., N. C. Gaddis, R. A. M. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 36.Simon, J. H. M., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon, J. H. M., D. L. Miller, R. A. M. Fouchier, M. A. Soares, K. W. C. Peden, and M. H. Malim. 1998. The regulation of primate immunodeficiency virus infectivity by Vif is cell-species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, J. H. M., T. E. Southerling, J. C. Peterson, B. E. Meyer, and M. H. Malim. 1995. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J. Virol. 69:4166-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soares, M. A., D. L. Robertson, H. Hui, J. S. Allan, G. M. Shaw, and B. H. Hahn. 1997. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228:394-399. [DOI] [PubMed] [Google Scholar]
- 40.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 41.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV “A ” (sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 42.Tsujimoto, H., A. Hasegawa, N. Maki, M. Fukasawa, T. Miura, S. Speidel, R. W. Cooper, E. N. Moriyama, T. Gojobori, and M. Hayami. 1989. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature 341:539-541. [DOI] [PubMed] [Google Scholar]
- 43.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 47.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, Y.-H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]