Abstract
In addition to their antibacterial activities, certain antimicrobial peptides inactivate enveloped viruses, including the human immunodeficiency virus (HIV). To determine whether peptide bactericidal activities are predictive of antiviral activity, the anti-HIV properties of recombinant human α-defensin 5, mouse α-defensins, cryptdins (Crp) 3 and 4, and rhesus macaque myeloid α-defensins (RMADs) 3 and 4 were determined in vitro. The peptides, purified to homogeneity, had equivalent bactericidal activities that were similar to those of the native molecules. Nuclear magnetic resonance spectroscopy showed RMAD-4 and Crp3 had characteristic α-defensin tridisulfide arrays. Of the peptides analyzed, only RMAD-4 inhibited HIV infectivity at 150 μg/ml, and Crp3 unexpectedly increased HIV replication. Quantitative real-time PCRs for minus-strand strong stop DNA and complete viral cDNA synthesis were used to distinguish between preentry and postentry anti-HIV effects by RMAD-4. Viral exposure to RMAD-4 for 1 h prior to infection reduced HIV minus-strand strong stop DNA and HIV cDNA by 4- to 20-fold during the first round of replication, showing that RMAD-4-exposed virions were not entering cells during the first 24 h. On the other hand, when RMAD-4 was added coincident with HIV inoculation, no anti-HIV activity was detected. Viral exposure to Crp3 resulted in a threefold increase in both HIV minus-strand strong stop DNA and HIV cDNA over the first round of replication. Therefore, two α-defensins, RMAD-4 and Crp3, inhibit or augment HIV replication, respectively, by mechanisms that precede reverse transcription.
Antimicrobial peptides (AMPs) exist in all species that have been investigated, and they occur in diverse host defense settings. AMP primary structures vary greatly, but they tend to be ≤5 kDa in size, cationic at neutral pH, and amphipathic, and they have broad-spectrum microbicidal activities in vitro at low micromolar concentrations. In mammals, AMPs occur in two peptide families: cathelicidins and defensins, both of which have been reviewed recently (15, 44, 45). The α-defensins, characterized by a tridisulfide array (34), are major constituents of neutrophil azurophilic granules (9) and are also secreted as abundant granule components by Paneth cells, an epithelial cell lineage that is specific to small intestinal crypts. Many α-defensins are bactericidal at low micromolar concentrations (≤10 μg/ml); furthermore, they occur at millimolar concentrations in phagolysosomes and at the point of secretion into the crypt lumen in vivo (1, 9, 10). The general features of AMP biology have been reviewed comprehensively (45).
Three mechanisms of AMP-mediated anti-human immunodeficiency virus (HIV) activity are apparent from the literature. The first, direct virolysis (28), is similar to the mechanism of bactericidal activity by many AMPs. The second, suppression of transcription from the HIV long terminal repeat, has been described for human α-defensins 1 to 3 (46) and the α-helix class of AMPs, such as cecropin and melittin (39). The third, a lectin-like effect, whereby the AMP binds to cell surface glycoproteins and blocks HIV entry, has been described for the synthetic θ-defensin, retrocyclin (3, 21, 40). Although the bactericidal activities of α-defensins have been studied extensively, their antiviral effects are not well characterized. For example, human neutrophil α-defensins (HNPs) 1, 2, and 3 have anti-HIV activity (46), but there are inconsistencies in the literature concerning their antiviral mechanism (2). The bovine neutrophil cathelicidin, indolicidin, has potent anti-HIV activity, nearly sterilizing HIV-containing culture fluid at a concentration of 333 μg/ml (28). Indolicidin treatment induces 51Cr release from H9 cells, the producer cell line for the HIV used in the experiments, and the lytic activity is temperature dependent: indolicidin becomes less effective at inactivating HIV as temperatures approach 4°C (28), consistent with HIV inactivation by viral membrane disruption.
In this study, the anti-HIV, antibacterial, and cytostatic/cytolytic activities of five mouse and primate α-defensin peptides were determined. The peptides chosen for this study, cryptdins (Crp) 3 and 4, rhesus myeloid α-defensins (RMADs) 3 and 4, and human Paneth cell α-defensin 5 (HD5), differ in primary structure but have identical disulfide arrays (Fig. 1A) and topologies and also have similar bactericidal activities. Therefore, one objective was to determine whether peptides with similar overall structures and bactericidal activities would exhibit similar anti-HIV relationships.
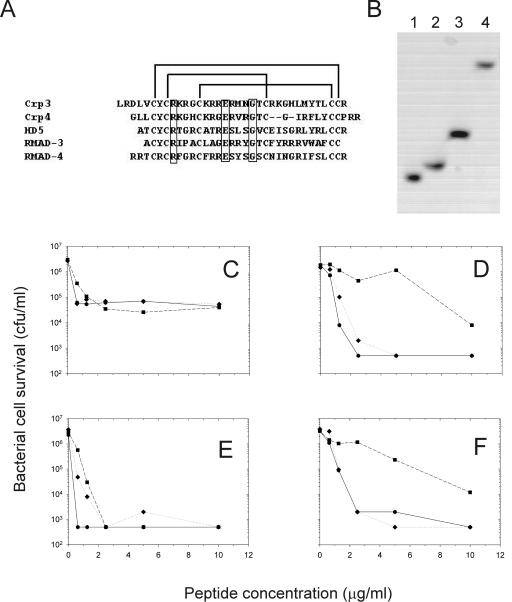
FIG. 1.
Bactericidal activity of recombinant α-defensins. (A) Primary sequence alignment for each of the peptides showing the tridisulfide array and the position of three invariant residues (boxed region). (B) AU-PAGE of recombinant α-defensins. Lane 1, Crp4; lane 2, Crp3; lane 3, RMAD-3; and lane 4, RMAD-4. Representative bactericidal activities of recombinant RMAD-3 (hexagon), RMAD-4 (diamond), and HD5 (square) against E. coli (C), V. cholerae (D), L. monocytogenes (E), and S. aureus (F) are illustrated.
MATERIALS AND METHODS
Cells and virus.
H9 and MT-2 cells are CD4+ lymphoblastoid cells and were grown in RPMI-1640 containing 25 mM HEPES supplemented with 11.5% fetal bovine serum and 2 mM l-glutamine. HIVLAI was obtained from the NIH AIDS Research and Reference Reagent program. HIVLAI was produced in H9 cells; the supernatant fluids were clarified of cells by low-speed centrifugation followed by filtration through 0.45-μm-pore-size cellulose acetate filters. Conditioned culture fluid was used immediately.
Preparation of recombinant peptides.
RMAD-3, RMAD-4, and Crp4 were prepared using the pET28a expression system (Novagen, Madison, Wis.) as previously reported (31, 36). Forward primers included a 5′ EcoRI site and a Met codon at the peptide N terminus; reverse primers contained consecutive stop codons and a 5′ SalI site. HD5 was prepared by amplifying exon 2 of the HD5 gene from human genomic DNA as template with primers HD5-F227 (5′-GCCAC CTGCT ATTGC CGAA) and HD5-R325 (5′-CAGCG ACAGC AGAGT CTG) and then was adapted for cloning into pET-28a by amplification with pET-HD5-F (5′-ATATA TGAAT TCATG GCCAC CTGCT ATTGC) and pET-HD5-R (5′-ACACA CGTCG ACTCA TCAGC GACAG CAGAG TCT) and handled as described above. The Crp3 peptide used in these studies was synthesized in solid phase and has been shown previously to be biochemically and biologically equivalent to natural Crp3 (16). Following PCR amplification, samples (25 μl) of individual reactions were gel purified using 2% agarose gels and were extracted using QIAEX II (QIAGEN Inc., Valencia, Calif.). Amplification products were cloned in pCR2.1-TOPO, sequenced, and digested sequentially with SalI and EcoRI. Gel-purified inserts were then ligated into EcoRI- and SalI-digested pET-28a plasmid DNA and transformed into both Escherichia coli XL-2 Blue and BL21 (DE3) Codon Plus cells (Stratagene Cloning Systems, Inc., La Jolla, Calif.).
To prepare recombinant α-defensins, His-tagged fusion proteins induced at 37°C for 6 h were purified by nickel nitrilotriacetic acid resin affinity chromatography (Ni-NTA Superflow; QIAGEN) with 100 or 200 mM imidazole in 6 M guanidine-HCl and 100 mM Tris-HCl, pH 8.0 (31, 36). Eluted fusion proteins were cleaved with 5 mg of CNBr/ml in 6 M guanidine and 0.1 N HCl for 24 h at room temperature in the dark, samples were acidified with 0.01% trifluoroacetic acid, and they were purified by C18 reverse-phase high performance liquid chromatography (RP-HPLC) using a 15 to 45% acetonitrile gradient developed over 90 min (31, 36). Peptides were purified to homogeneity by rechromatography using shallower acetonitrile gradients as assessed by RP-HPLC chromatograms (data not shown) and comigration by acid urea-polyacrylamide gel electrophoresis (AU-PAGE). Peptide molecular masses were confirmed by matrix-assisted laser desorption-ionization-time-of-flight mass spectrometry (MALDI-TOF MS), and peptide concentrations were determined by absorbance at 280 nm, using extinction coefficients obtained at the EXPASY web site (http://us.expasy.org/). Peptides were lyophilized and stored at room temperature.
Bactericidal peptide activity assays.
E. coli ML35, Staphylococcus aureus 710a, Listeria monocytogenes 10403S, and Vibrio cholerae 0395 were target organisms for bactericidal assays. Quantities of individual peptides were incubated with 1 × 106 to 5 × 106 CFU of log phase bacterial cells per ml in 50 μl of buffer consisting of 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid), pH 7.4] supplemented with 1% (vol/vol) trypticase soy broth at 37°C for 1 h. After 100-fold dilution, samples of incubation mixtures were plated on semisolid media using a Spiral Biotech Autoplater 4000 (Spiral Biotech, Bethesda, Md.). Surviving bacterial cells were determined as bacterial CFU by counting after overnight growth (32).
Cell toxicity.
Lyophilized peptides were dissolved in H2O at 300 μg/ml. Cell toxicity studies were performed as described previously (20, 28). Briefly, peptides were diluted serially in 96-well plates in triplicate. Approximately 2.0 × 105 MT-2 cells were added to each well, and the peptide-cell mixture was incubated at 37°C for 3 days. The cells were transferred to poly-l-lysine-coated plates and stained with Finter's Neutral Red vital dye. The A540 was determined on a microcolorimeter, and the percentage of viable cells was calculated relative to eight replicate control wells that were not exposed to peptide. Because the assay is performed over 3 days, it measures both cytotoxic and cytostatic peptide activities. This assay has been extensively discussed (13, 20, 26, 27, 30) and is highly reproducible, with 1 standard deviation less than or equal to one twofold dilution, as shown recently for 15 anti-HIV agents (7).
Anti-HIV activity.
HIV, approximately 1.5 × 105 tissue culture infectious doses, was incubated with each peptide at 150 μg/ml in a final volume of 200 μl, a concentration that is substantially below α-defensin levels in phagolysosomes and in small intestinal crypts. Total fetal bovine serum concentration was 5.75%. Due to its greater cytotoxicity, Crp3 was incubated with HIV at 50 μg/ml. The anti-HIV and cell toxicity concentrations of indolicidin were previously reported to be nearly equal (28). In the case of infections in triplicate, three separate aliquots of virus were incubated with each peptide. After incubation of peptide-HIV mixtures for 1 h at 37°C, samples were diluted fivefold to 1 ml and then added to 9 ml of MT-2 cells at a density of 7.5 × 105 cells per ml. This 50-fold dilution reduced peptide levels to well below their cytotoxic concentrations. The final multiplicity of infection was approximately 0.02. In each case, infections were performed in separate tissue culture flasks, thereby ensuring that each treatment was indeed a unique infection. Conditions differed for Crp3, which, due to its greater toxicity (Fig. 2), was diluted 300-fold, although the virus itself was diluted the same as for the other peptides. Cells were harvested at 4, 8, 12, 24, 48, 72, and 96 h postinoculation for real-time PCR to measure viral replication and at 24, 48, 72, 96, and 144 h for indirect immunofluorescence assay (IFA) to monitor HIV spread through tissue culture.
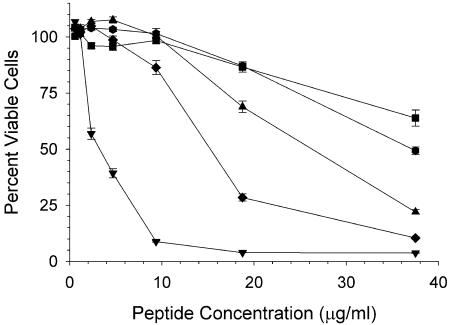
FIG. 2.
Cytostatic/cytotoxic activities of α-defensins against MT-2 cells. Crp3 (inverted triangle), Crp 4 (triangle), HD5 (square), RMAD-3 (hexagon), and RMAD-4 (diamond) were incubated for 72 h with MT-2 cells. Cells were harvested for viability, and the percentage of viable cells was calculated based on eight cell replicates (100% viable) and eight blank wells (0% viable). Each point is the mean of triplicates; error bars are 1 standard deviation.
Indirect immunofluorescence assay.
Cells were fixed on glass slides and stained with pooled human immune globulins containing antibodies to all HIV proteins and fluorescein-conjugated goat anti-human immunoglobulin G as described previously (29). The percentage of HIV antigen-positive cells was calculated on a Nikon epifluorescence microscope.
Real-time PCR.
Quantitative real-time PCR using SYBR green-I was performed as described previously (38). Briefly, 106 cells were harvested and dissolved in Tris lysis buffer, which includes Tween-20, Nonidet P40, and Proteinase K (12). Minus-strand strong stop DNA was quantified using the AA55/M667 primer pair (38, 43), and completely reverse transcribed cDNA was quantified using the M661/M667 primer pair (38, 43). Each PCR contained internal controls. Controls, chronically HIVLAI-infected H9 cells ranging from 20,000 to 2 infected cells, were lysed. These controls were used to construct the standard curves. Any individual reactions in which the standard curves had r2 below 0.98 were repeated, although no PCR had to be repeated in these studies. All data reported herein were calculated from standard curves constructed from the mean of each individual standard curve; this eliminates run-to-run variability in the calculations of infected cell equivalents (38, 43).
In addition to quantifying viral replication, real-time PCR can reveal the site of replication defects. For example, the AA55/M667 product is lowered over the first 24 h when entry is inhibited (38), leading to a subsequent decrease in complete cDNA synthesis. Postentry events, such as inhibition of reverse transcriptase, lead to a different profile. For example, when reverse transcriptase is inhibited, minus-strand strong stop DNA is near the levels present in control infections, but cDNA is reduced by more than 1 log10 (38). Later events, such as inhibition of the HIV-specific protease, demonstrate their effects principally on the second round of HIV spread (after 24 h) (38).
NMR spectroscopy.
Nuclear magnetic resonance (NMR) samples were prepared by dissolving 0.5 to 1 mg of peptide in 25 mM sodium phosphate buffer (pH 6.0) or H2O and adjusting the pH to 5.0. Experiments were performed on a Varian Inova 800 MHz NMR spectrometer. Data was collected on the sample in 90% H2O-10% D2O, and then data was collected again after the sample was lyophilized and resuspended in an equal volume of 100% D2O. Nuclear Overhauser effect spectroscopy (NOESY) (τm = 200 and 300 ms), total correlation spectroscopy (TOCSY) (τm = 45 ms), and double quantum-filtered correlation spectroscopy (DQF-COSY) experiments were performed; for the 90% H2O sample, the water peak was eliminated using WATERGATE. Data were processed using NMRPipe (6) and were analyzed using Sparky (http://www.cgl.ucsf.edu/home/sparky/).
RESULTS
Purification of recombinant α-defensins.
Efficient expression of recombinant Crp4, HD5, RMAD-3, and RMAD-4 (Fig. 1B) was obtained using the pET-28 vector system (31, 36). All peptides were purified to homogeneity by RP-HPLC as verified using analytical RP-HPLC (data not shown) and AU-PAGE analyses (Fig. 1B). The molecular masses of individual recombinant peptides were determined by MALDI-TOF MS, and they matched the respective theoretical values exactly. Thus, the purified recombinant peptides were homogeneous and their biochemical features were consistent with direct comparisons (see Fig. 8) or with reports of the corresponding natural molecules.
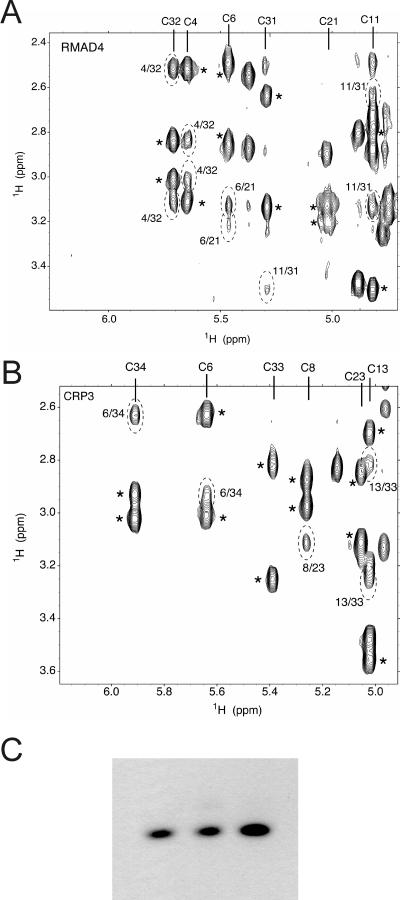
FIG. 8.
Disulfide connectivities of RMAD-4 and Crp3. (A) RMAD-4; (B) Crp3. Sections of NOESY (τm = 300 ms) spectra, 100% D2O. Intraresidue NOEs (H-alpha to H-beta) are marked with asterisks. Interresidue NOEs that define Cys-Cys bonds are highlighted with hatched ovals. Disulfides confirmed in these panels were RMAD-4, C4-C32, C6-C21, C11-C31; CRP3, C6-C34, C8-C23, C13-C33. (C) Lane 1, AU-PAGE of native RMAD-4; lane 2, preparation 1 of recombinant RMAD-4; lane 3, preparation 2 of recombinant RMAD-4.
The bactericidal activities of each peptide were tested against E. coli, V. cholerae, L. monocytogenes, and S. aureus (Fig. 1C to F). All peptides tested displayed bactericidal activities in the low micromolar range, again consistent with published values for the native peptides (22-24, 35, 37). Of the peptides tested, RMAD-3 and RMAD-4 were consistently the most bactericidal against all species, achieving 99.9% killing at 1 to 2 μM concentrations. Thus, under these conditions, the peptides had equivalent and potent antibacterial activities, enabling a test of whether relative bactericidal and anti-HIV activities would correlate.
Toxicity and anti-HIV activities of recombinant antimicrobial peptides.
To determine the toxicity of the peptides against eukaryotic cells, MT-2 cells were cultured in the presence of individual peptides. All peptides were cytotoxic, with Crp3 having the greatest toxicity (Fig. 2). Peptide levels that inhibited MT-2 cell growth by 50%, or the 50% cytotoxic dose (CT50), ranged from 3.0 to 43.8 μg/ml. The CT50s for the peptides were the following: Crp3, 3.0 μg/ml; Crp4, 26.0 μg/ml; HD5, 43.8 μg/ml; RMAD-3, 35.8 μg/ml; and RMAD-4, 16.9 μg/ml. The high reproducibility of the assay is evident from repeat analyses of Crp4, which had a CT50 of 20.7 μg/ml with 95% confidence intervals between 16 and 27 μg/ml. This degree of reproducibility is a hallmark of this assay and indicates that these measurements of AMP toxicity are as reproducible as toxicities measured for other anti-HIV agents (7).
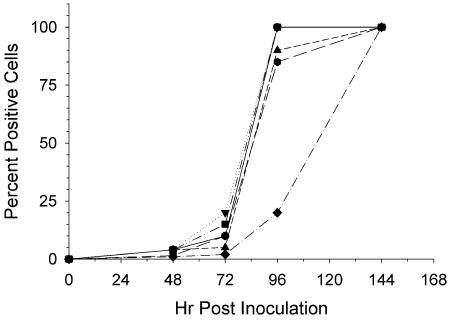
For the anti-HIV assays, the 1:1 peptide-virus mixtures were diluted 50-fold prior to addition of target MT-2 cells. Because HIV is an intracellular parasite, inhibition of cell growth can suppress HIV replication, even if the inhibitory compound has no direct effect on HIV replication. Researchers have previously described that the CT5, or 5% cytotoxic dose, is truly nontoxic (20, 26, 30). The CT5s for mouse peptides were the following: Crp3, 0.6 μg/ml (0.3 to 1.1 μg/ml); Crp4, 12.9 μg/ml (10.3 to 16.2 μg/ml), with repeat analysis of 7.1 μg/ml (5.3 to 10 μg/ml) and the calculated 95% confidence intervals noted parenthetically. For HD5, the CT5 was 14.1 μg/ml (11.5 to 17.2 μg/ml), and for RMAD-3 and -4 the CT5s were 14.8 μg/ml (13.3 to 16.6 μg/ml) and 6.8 μg/ml (5.5 to 8.4 μg/ml), respectively. Except for the more cytotoxic Crp3, all peptides had very similar CT5s between 7 and 15 μg/ml. Because infected cells were exposed only to 3-μg/ml concentrations of peptides, 0.17 μg/ml in the case of Crp3, anti-HIV peptide assays were performed at concentrations well below the CT5. Under the conditions of these assays, RMAD-4 delayed the spread of HIV through the culture significantly, Crp4 and RMAD-3 showed slight anti-HIV activity by IFA at 72 and 96 h (Fig. 3), and Crp3 modestly increased the spread of HIV through the culture.
FIG. 3.
Spread of HIVLAI in MT-2 cells by IFA. HIV was incubated with H2O (circle), Crp3 (inverted triangle), Crp4 (triangle), HD5 (square), RMAD-3 (hexagon), or RMAD-4 (diamond) for 1 h at 37°C. Next, each mixture was diluted 50-fold in growth medium and was added to MT-2 cells. At each time point the cells were harvested for IFA, and the percentage of cells expressing HIV antigens are indicated.
α-Defensins exhibit different effects on HIV replication.
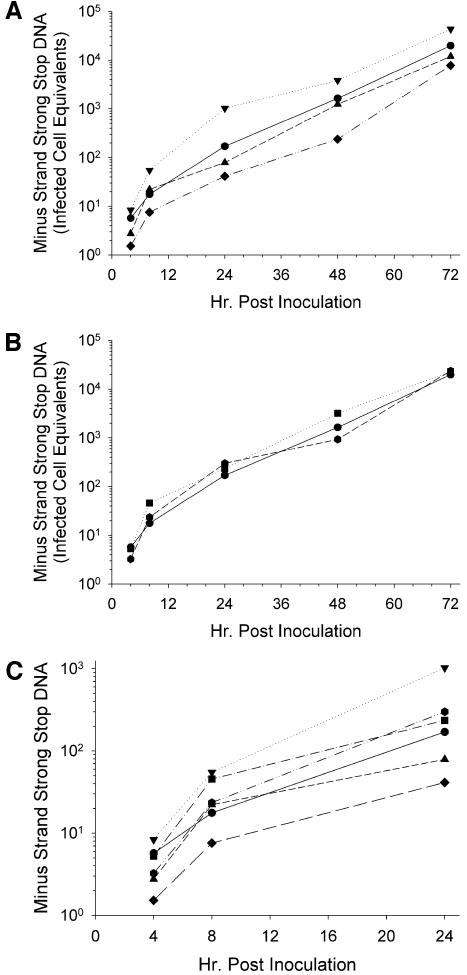
To distinguish the mechanism of viral inhibition by RMAD-4, MT-2 cells were infected with peptide-treated HIV, and the syntheses of both HIV minus-strand strong stop DNA and full-length cDNA were monitored by quantitative real-time PCR. Inhibitors of viral entry, such as dextran sulfate, block minus-strand strong stop DNA synthesis, while inhibitors of later steps in the viral life cycle, including reverse transcription, primarily reduce synthesis of full-length cDNA (38). Previous work had suggested that indolicidin, a cathelicidin, inhibited HIV replication by interacting with the viral membrane and lysing the virions (28). As shown in Fig. 4A, Crp4 had no effect on minus-strand strong stop DNA production over the 72-h time course of infection. In contrast, Crp3 increased minus-strand strong stop DNA levels (Fig. 4A), consistent with the IFA results (Fig. 3). RMAD-4, on the other hand, decreased production of minus-strand strong stop DNA by approximately fourfold during the first 24 h after infection (Fig. 4B), also consistent with results of IFA (Fig. 3). After the first 24 h postinfection, the slope of the minus-strand strong stop DNA accumulation curve was parallel to that of the virus control, showing that viral inhibition by RMAD-4 occurred during the first round of replication, perhaps prior to entry. Viral exposure to HD5 and RMAD-3 had no significant effect on minus-strand strong stop DNA synthesis over the 72-h course of infection (Fig. 4B), showing that the RMAD-4- and Crp3-induced effects were peptide specific. Focusing on α-defensin-mediated effects on HIV entry and the first round of viral replication, HIV treatment with Crp3 increased levels of minus-strand strong stop DNA by nearly 1 log10, but RMAD-4 exposure decreased minus-strand strong stop DNA by nearly 1 log10. The remaining peptides had little effect on the first round of viral replication (Fig. 4).
FIG. 4.
α-Defensins affect the synthesis of minus-strand strong stop DNA during the first round of HIV replication. HIVLAI was incubated for 1 h at 37°C with (A) H2O (circle), Crp3 (inverted triangle), Crp4 (triangle), or RMAD-4 (diamond) or (B) H2O (circle), HD5 (square), or RMAD-3 (hexagon). Virus-peptide mixtures were subsequently diluted 50-fold and added to MT-2 cells. At each time point shown, the cells were harvested for real-time PCR using the AA55/M667 primer pair to quantify minus-strand strong stop DNA. Infected cell equivalents were calculated using HIVLAI chronically infected H9 cells from a single standard curve (for an example, see Fig. 10A). Each reaction mixture contained an internal standard curve from 20,000 to 2 infected cells. (C) A compilation of all the data from the first round of replication. Standard curve for calculations: n = 15, r2 = 0.9989.
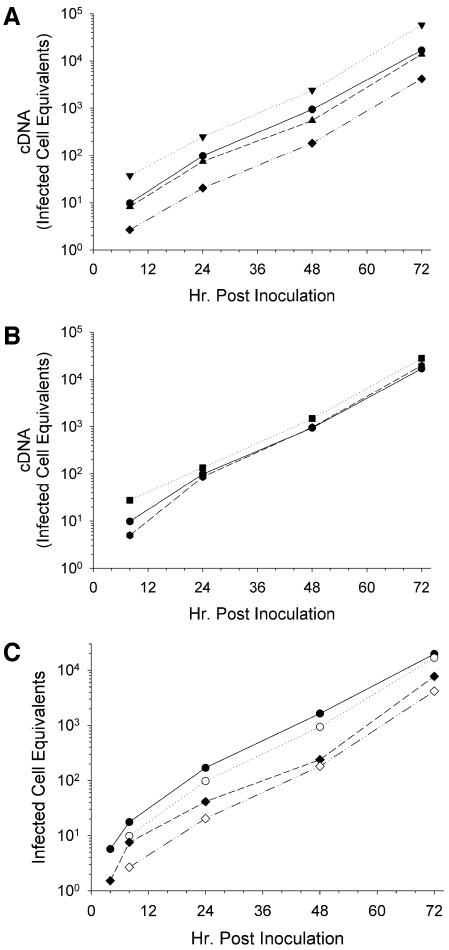
If AMPs suppress transcription from the HIV long terminal repeat, the synthesis of both minus-strand strong stop DNA and viral cDNA should diminish after 24 h, indicating that second-round spread was delayed. As indicated, none of the AMPs tested affected minus-strand strong stop DNA production after the first 24 h postinoculation. To determine whether reverse transcription was being affected, full-length HIV cDNA was quantified. As shown in Fig. 5A and B, accumulation of full-length cDNA resembled that of minus-strand strong stop DNA synthesis. Only RMAD-4 and Crp3 had significant, albeit opposite, effects on HIV replication. Figure 5C compares the levels of minus-strand strong stop DNA to cDNA for the two samples relevant to HIV inactivation: the virus control and RMAD-4-treated HIV. The kinetics of replication were identical between the samples, with only the level of minus-strand strong stop DNA production in the first 24 h being inhibited by RMAD-4. Therefore, the inhibitory effects of RMAD-4 occur early in viral infection and prior to reverse transcription (Fig. 5C).
FIG. 5.
α-Defensins do not affect reverse transcription as measured by complete HIV cDNA synthesis. HIVLAI was incubated for 1 h at 37°C with (A) H2O (circle), Crp3 (inverted triangle), Crp4 (triangle), or RMAD-4 (diamond) or (B) H2O (circle), HD5 (square), or RMAD-3 (hexagon). Virus-peptide mixtures were diluted 50-fold and added to MT-2 cells. At each indicated time point the cells were harvested for real-time PCR using the M661/M667 primer pair to quantify full-length HIV cDNA. Infected cell equivalents were calculated using HIVLAI chronically infected H9 cells from a single standard curve (for an example, see Fig. 10B). Each reaction mixture contained an internal standard curve from 20,000 to 2 infected cells. (C) Direct comparison of minus-strand strong stop DNA (closed symbols) and full-length cDNA (open symbols). Standard curve for calculations: n = 15, r2 = 0.9992.
Crp3 enhances HIV entry.
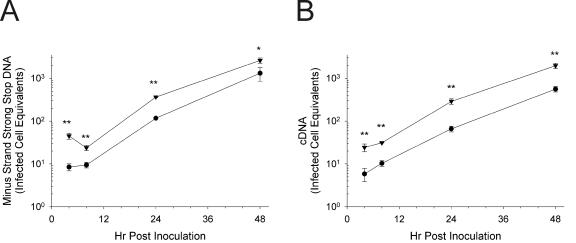
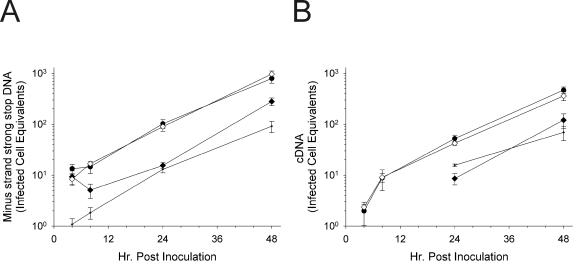
To confirm the infection-enhancing effects of Crp3, an additional time course was performed. As illustrated in Fig. 6, Crp3 treatment of HIV caused a statistically significant increase in levels of both minus-strand strong stop DNA (Fig. 6A) and completely synthesized HIV cDNA (Fig. 6B). The increase was approximately threefold at all time points. As seen in the previous experiment (Fig. 4), the effects were manifest during the first 24 h postinoculation, indicative of an effect on HIV entry.
FIG. 6.
Infections of MT-2 cells by HIVLAI treated with Crp3 results in significantly increased levels of HIV throughout infection. (A) Minus-strand strong stop DNA; (B) full-length HIV cDNA. HIVLAI was preincubated in triplicate with either H2O (circles) or Crp3 (inverted triangles) for 1 h at 37°C. The virus peptide mixture was diluted 50-fold and added to MT-2 cells. Cells were lysed for real-time PCR at the indicated times. Infected cell equivalents were determined using a mean standard curve. Each point is the mean of triplicate infections; the error bars are 1 standard deviation. Significance was determined at each point using Student's two-tailed t test assuming equal variances. *, P < 0.02; **, P < 0.002. Standard curves for calculations: (A) n = 14, r2 = 0.9996; (B) n = 13, r2 = 0.9997.
RMAD-4 inhibits HIV entry.
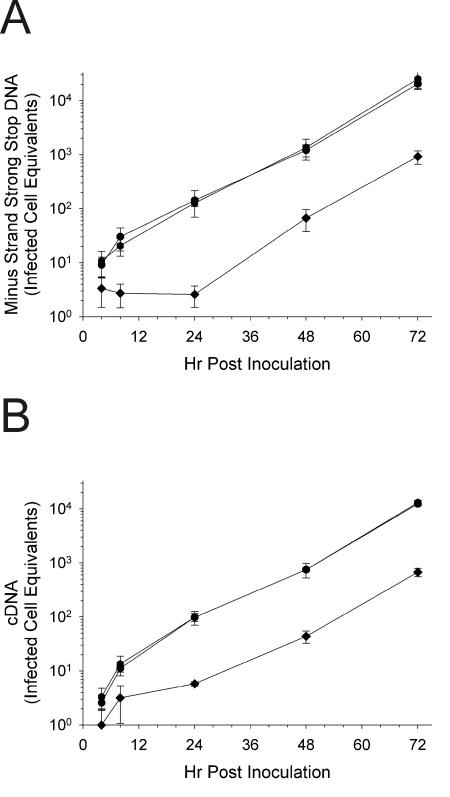
The effects of RMAD-4 treatment on the replication kinetics of HIV were confirmed by performing additional courses of infection in triplicate and comparing rates of viral replication relative to triplicate RMAD-3 and control HIV infections. As shown in Fig. 7A, RMAD-4 had statistically significant effects on HIV entry, as shown by replicate measurements of minus-strand strong stop DNA levels during the first 24 h after infection. Likewise, the RMAD-4 effect on minus-strand strong stop DNA synthesis resulted in a comparable, approximately 20-fold, decrease in full-length HIV cDNA synthesis (Fig. 7B). However, the effect of RMAD-4 was limited to the first round of replication, as shown by the identical slopes of viral production curves for the control and peptide treated HIV infections after the first 24 h (Fig. 7A and B). The kinetics of both minus-strand strong stop DNA (Fig. 7A) and completely synthesized HIV cDNA (Fig. 7B) for RMAD-3, on the other hand, were superimposable with those of the virus controls. To confirm the difference between RMAD-3 and -4, both peptides were prepared a second time, and the relative anti-HIV activities of the two new peptide lots were indistinguishable from those of the previous preparations (data not shown).
FIG. 7.
Infections of MT-2 cells by HIVLAI treated with RMAD-4 results in significantly decreased levels of HIV throughout infection but no evidence of an effect other than entry. (A) Minus-strand strong stop DNA; (B) full-length HIV cDNA. In triplicate reactions HIVLAI was incubated with H2O (circles), RMAD-3 (hexagon), or RMAD-4 (diamond) for 1 h at 37°C. The virus peptide mixture was diluted 50-fold and added to MT-2 cells. Cells were lysed for real-time PCR at the indicated times. Each point is the mean of triplicate infections; the error bars are 1 standard deviation. Statistical significance was calculated using Student's two-tailed t test assuming equal variances. For both minus-strand strong stop DNA and completely synthesized HIV cDNA, P < 0.05 for RMAD-4 pretreatment at all points except 4 h, and P > 0.05 for RMAD-3 at all points compared to H2O-treated HIVLAI. Standard curves for calculations: (A) n = 14, r2 = 0.9990; (B) n = 16, r2 = 0.9992.
Recombinant and synthetic peptides maintain canonical disulfide arrays. Electrophoretic mobility and high-resolution NMR studies show that the synthetic and recombinant α-defensins investigated have the canonical tridisulfide array. The migration of AMPs in AU-PAGE is a sensitive index of correct folding and disulfide bond formation in α-defensins (33). Focusing on the peptides with disparate anti-HIV activities, preliminary NMR data showed that Crp3 and RMAD-4 are similar in beta-sheet content, topology, and cysteine pairings in their disulfide arrays. Sequential assignments were made through each strand of the beta sheet using NOE and TOCSY data to determine the identity of each cysteine spin system. Sections of NOESY spectra that include the cysteine side chain alpha and beta protons for RMAD-4 and Crp3 (Fig. 8A and B, respectively) confirmed that the disulfide connectivities correspond to those of native α-defensins. In addition, the AU-PAGE mobilities of both pure RMAD-4 peptide preparations were identical to that of native RMAD-4 (Fig. 8C). Also, Crp4 produced in E. coli is properly folded (23), and the Crp4 disulfide array has been verified structurally by NMR (17a). These findings show that the differential anti-HIV activities of Crp3, Crp4, RMAD-4, and other α-defensins cannot be attributed to an artifact of misfolding.
Replication profile of HIV is similar for RMAD-4 and dextran sulfate treatment.
To confirm that a decrease in minus-strand strong stop DNA synthesis over the first 24 h of HIV replication is indicative of inhibition of entry (38), HIVLAI was treated with either H2O, RMAD-4, or dextran sulfate for 1 h. Alternatively, at the time of inoculation onto H9 cells RMAD-4 was diluted to the same final concentration. As shown in Fig. 9A, both RMAD-4 and dextran sulfate had a statistically significant effect on HIV replication. During the first 24 h, minus-strand strong stop DNA was significantly lower than that in the control infection. There was a subsequent decrease in later products of reverse transcription as well: RMAD-4 and dextran sulfate both lowered the levels of HIV cDNA to below detectable levels until 24 h postinoculation (Fig. 9B). RMAD-4 added at the same time as HIV infection had no effect; thus, the low level of RMAD-4 present throughout the infection was not a factor in HIV replication (Fig. 9).
FIG. 9.
Anti-HIV activity of RMAD-4 is consistent with virolysis. In triplicate, HIV was preincubated at 37°C with H2O (circles), RMAD-4 (closed diamond), or dextran sulfate (dots). The virus-peptide mixture was diluted 50-fold and added to MT-2 cells, or RMAD-4 was diluted 50-fold and added to MT-2 cells with untreated HIVLAI (open diamond). Cells were lysed for real-time PCR at the indicated times. (A) Minus-strand strong stop DNA; (B) completely reverse transcribed HIV cDNA. Each point is the mean of triplicate infections; the error bars are 1 standard deviation. Statistical significance was calculated using Student's two-tailed t test assuming equal variances. For both panels, P < 0.05 for RMAD-4 pretreatment at all points except 4 h, while P < 0.01 for dextran sulfate at all time points and P > 0.05 for RMAD-4 posttreatment at all time points compared to those of H2O-treated HIVLAI. Infected cell equivalents were calculated from the standard curves illustrated in Fig. 10.
Calculations for real-time PCR.
The infected cell equivalents for all of the real-time PCR data were calculated using mean standard curves constructed from internal standards that were run with each PCR as reported previously (38). Experiments that generated nonlinear standard curves or standards that varied by more than 10% would have been repeated, although no PCR met these exclusion criteria and, thus, no data were excluded. Examples of standard curves used in these studies are illustrated in Fig. 10.
FIG. 10.
Representative standard curves for real-time PCR. In each PCR an internal standard curve from 20,000 infected cells to 2 infected cells was used. The standards were constructed by diluting HIVLAI chronically infected H9 cells into uninfected H9 cell DNA to maintain a constant level of DNA (equivalent to that from 20,000 cells). Each point is the mean, the error bars are 1 standard deviation, the solid line is the linear regression analysis, and the dashed curves are the 95% confidence intervals. Linear regression analysis was performed for minus-strand strong stop DNA (A) and completely synthesized HIV cDNA (B).
DISCUSSION
The findings reported here show that α-defensin bactericidal activity and eukaryotic cell toxicity do not correlate with peptide anti-HIV activity. Although RMAD-3 and RMAD-4 have equivalent bactericidal activities against E. coli, V. cholerae, L. monocytogenes, and S. aureus (Fig. 1), only RMAD-4 is virucidal, and the data suggest that RMAD-4 may be virolytic. While RMAD-4 was more toxic to H9 cells than RMAD-3, the most toxic molecule, Crp3, had no anti-HIV activity. Indeed, Crp3 augmented HIV replication. This observation supports the conclusion that AMP anti-HIV activities cannot be predicted reliably from microbicidal activity but require that anti-HIV activity be assayed directly.
In theory, differences in AMP biological activity could result from artifacts of peptide misfolding, particularly for small peptides with high cysteine content. Nevertheless, the adoption of correct tertiary structures appears to be favorable for α-defensins prepared by solid-phase synthesis or recombinant expression. In the studies reported here, the correct folding and tridisulfide arrays of Crp3 and RMAD-4 were confirmed, dispelling the possibility that their differential anti-HIV effects were the result of inconsistent folding. In fact, there is precedent for this class of peptide to fold correctly regardless of source (5, 18).
The antiviral activities of certain AMPs have been studied in detail. For example, MCP-1 and MCP-2, found in rabbit lung macrophages, directly inactivate enveloped viruses, including herpes simplex viruses 1 and 2 but not nonenveloped viruses such as echovirus (14). This direct virolysis is temperature sensitive, as cooling on ice stops the inactivation; temperature dependence is consistent with a membrane effect (14). Nearly identical results were observed for HNP defensins HNP1, -2, and -3 against herpes simplex virus type 1 (4). Indolicidin at 333 μg/ml inactivated 99.99% of HIV in culture fluid in a temperature-dependent manner (28). Based on measurements of viral cDNA during the first round of HIV replication, RMAD-4 at 150 μg/ml inactivated 94 to 98% of HIV in culture fluid. While hypotonicity might play a role in the anti-HIV effect of RMAD-4 (see Material and Methods), of the five peptides tested, only RMAD-4 had detectable anti-HIV effects. Moreover, when HIV was incubated with RMAD-4 at 150 μg/ml under isotonic conditions and in the presence of 11.5% fetal bovine serum, anti-HIV activity equivalent to that present under hypotonic conditions was observed by both IFA and real-time PCR analyses (data not shown).
Direct virolysis of HIV by RMAD-4 would be consistent with the general understanding of AMP microbicidal mechanisms. Studies with model membranes support the view that α-defensins kill their targets by permeabilizing the cell envelope, leading to dissipation of electrochemical gradients. However, certain α-defensin peptides operate by apparently different mechanisms. For example, the crystal structure of HNP-3 is a noncovalent dimer. HNP-3 forms stable, ∼20-Å multimeric pores in large unilamellar vesicles (LUV) (41). In contrast, rabbit neutrophil α-defensins are monomeric in solution and cause large, transient defects in LUV (11). Mouse Crp4 resembles the neutrophil α-defensin mode of action, inducing graded leakage from quenched fluorophore-loaded LUV (32). Indolicidin, a cathelicidin, is also capable of forming voltage-dependent ion channels (8). HNP-1-mediated lysis of the eukaryotic cell line K562 occurs rapidly, results in release of 51Cr, and is eliminated by CaCl2. The cytolytic effects have been confirmed by vital dye exclusion (17), consistent with the effect of indolicidin on H9 cells (28).
The anti-HIV activity of RMAD-4 resembles that of indolicidin more than that of retrocyclin or HNP-1 to -3. When retrocyclin was preincubated with HIV and diluted prior to infection, no anti-HIV activity was detected, even at concentrations of 200 μg/ml (3). On the other hand, HIV exposure to 150 μg of RMAD-4/ml for 1 h followed by 50-fold dilution before infection inhibited HIV replication significantly via a prereverse transcription mechanism (Fig. 4, 5, 7, and 9). Recently, the affinity of retrocyclin for the HIV envelope glycoprotein gp120 as well as for CD4 and galactosylceramide suggested that its anti-HIV activity may be mediated through binding to carbohydrates on CD4 or galactosylceramide, thereby blocking HIV entry (40). Although RMAD-4 has not been tested for either lectin-like or membrane-disruptive activities, several observations support the conclusion that RMAD-4 and retrocyclin antiviral activities differ in mechanism. First, RMAD-4 is cytotoxic at 17 μg/ml (Fig. 2), while retrocyclin has little cytotoxicity at peptide concentrations of 100 μg/ml (3). Second, RMAD-4, but not retrocyclin, inactivates HIV when peptide and virus are incubated before infection. Finally, if RMAD-4 functioned as a lectin in the culture fluid, its presence throughout the time course would have diminished viral production in subsequent rounds of replication. However, virus in RMAD-4-treated cultures was produced at a rate parallel to that of cells infected by control virus as measured by real-time PCR. Moreover, when diluted RMAD-4 was added to cells at the time of HIV addition, it had no effect on HIV replication (Fig. 9). Thus, only the first round of viral replication was affected by RMAD-4, in contrast to retrocyclin, which inhibited HIV production for 9 days when incubated with cells (3).
The mechanism of anti-HIV activity for human α-defensins 1, 2, and 3 has been described as suppression of viral transcription (2, 46). This mechanism of action is clearly not the mechanism by which RMAD-4 inhibits HIV replication, as RMAD-4 treatment led to decreased levels of minus-strand strong stop DNA in the first round of replication, which can only be due to virolysis, decreased entry of HIV, or an effect on reverse transcription. Indeed, the potent reverse transcriptase inhibitors zidovudine and nevirapine have little effect on minus-strand strong stop DNA levels during the first 8 h postinoculation (38). As shown in Fig. 7 and 9, the principal effect of RMAD-4 was on the level of minus-strand strong stop DNA over the first 24 h postinoculation. The profile of RMAD-4 was similar, but not identical, to that of the entry inhibitor dextran sulfate (38 and Fig. 9). Dextran sulfate inhibits entry, but, as shown in Fig. 9, viral entry and reverse transcription continue in the presence of dextran sulfate over the first 24 h. RMAD-4, on the contrary, exhibited no change in either minus-strand strong stop DNA or completely synthesized HIV cDNA from 4 to 24 h. This finding would be consistent with virolysis or viral inactivation rather than inhibition of entry: if the virions were lysed, no particle core, including the RNA genome and reverse transcriptase, would be available for entry. Thus, no increase in genome would be detectable from 4 to 24 h postinoculation.
The mechanism of enhanced HIV entry mediated by Crp3 is not known. Possibly, at nonlytic concentrations, the cationic Crp3 molecule may be juxtaposed between electronegative viral and cell membrane phospholipids and thus stabilize the virus-cell interactions. This may facilitate HIV gp120 interactions with the CD4 receptor and its chemokine coreceptor. Alternatively, if Crp3 is behaving like a lectin, similar to retrocyclin, it is feasible that a carbohydrate-carbohydrate interaction between either gp120 and CD4 or gp120 and a secondary receptor might be stabilized, again resulting in increased binding and uptake of HIV by the susceptible cells. These interpretations seem improbable, however, because Crp4, HD5, and RMAD-3 did not exhibit similar activities. Crp3 also is known to induce the formation of anion-selective pores in mammalian cells (16, 19, 42). We speculate that the induction of such pores could facilitate HIV entry into cells in culture and thus increase the effective infectious dose of the virus. Perhaps such pores increase the rate of uncoating of the reverse transcription complex. Because α-defensins are secreted at epithelial surfaces (25) and AMPs are inducible in response to pathogens (reviewed in reference 45), the role of low doses of Crp3 on HIV entry should be studied further to determine whether certain endogenous AMPs may be implicated in the pathogenesis of HIV infection.
In summary, RMAD-4 is a potent bactericidal α-defensin that also inhibits HIV replication. Its mechanism of anti-HIV activity is consistent with virolysis, similar to the reported anti-HIV effect of indolicidin (28). AMP-mediated anti-HIV activity is dissociated from the tridisulfide array, overall topology, cytotoxicity, and cytolytic activities on mammalian cells as well as microbicidal activity. Thus, AMP antiviral activities must be determined empirically for each molecule. Indeed, AMPs with equivalent bactericidal activities may be virucidal (RMAD-4), enhance viral infection (Crp3), or lack anti-HIV properties (RMAD-3, Crp4, and HD5).
Acknowledgments
We thank Brenda McDougall for excellent technical assistance. Dat Tran and Michael E. Selsted generously provided mouse Crp3 and native RMAD-4. Bao Nguyen acquired some of the NMR data.
This work was supported in part by a grant from the Public Health Service DK044632 (A.J.O.). W. Edward Robinson, Jr., is a Burroughs-Wellcome Fund Clinical Scientist in Translational Research.
REFERENCES
- 1.Ayabe, T., D. P. Satchell, C. L. Wilson, W. C. Parks, M. E. Selsted, and A. J. Ouellette. 2000. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 1:113-118. [DOI] [PubMed] [Google Scholar]
- 2.Chang, T. L.-Y., F. Francois, A. Mosoian, and M. E. Klotman. 2003. CAF-mediated human immunodeficiency virus (HIV) type 1 transcriptional inhibition is distinct from α-defensin-1 HIV inhibition. J. Virol. 77:6777-6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, A. M., T. Hong, L. M. Boo, T. Nguyen, C. Zhao, G. Bristol, J. Zack, A. J. Waring, O. O. Yang, and R. I. Lehrer. 2002. Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc. Natl. Acad. Sci. USA 99:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daher, K. A., M. E. Selsted, and R. I. Lehrer. 1986. Direct inactivation of viruses by human granulocyte defensins. J. Virol. 60:1068-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson, N. F., D. J. Craik, A. M. McManus, S. G. Dashper, E. C. Reynolds, G. W. Tregear, L. Otvos, Jr., and J. D. Wade. 2000. Chemical synthesis, characterization and activity of RK-1, a novel alpha-defensin-related peptide. J. Pept. Sci. 6:19-25. [DOI] [PubMed] [Google Scholar]
- 6.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 7.Essey, R. J., B. R. McDougall, and W. E. Robinson, Jr. 2001. Mismatched double-stranded RNA (polyI-polyC12U) is synergistic with multiple anti-HIV drugs and is active against drug-sensitive and drug-resistant HIV-1 in vitro. Antivir. Res. 52:189-202. [DOI] [PubMed] [Google Scholar]
- 8.Falla, T. J., D. N. Karunaratne, and R. E. W. Hancock. 1996. Mode of action of the antimicrobial peptide indolicidin. J. Biol. Chem. 271:19298-19303. [DOI] [PubMed] [Google Scholar]
- 9.Ganz, T., M. E. Selsted, D. Szklarek, S. S. Harwig, K. Daher, D. F. Bainton, and R. I. Lehrer. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh, D., E. Porter, B. Shen, S. K. Lee, D. Wilk, J. Drazba, S. P. Yadav, J. W. Crabb, T. Ganz, and C. L. Bevins. 2002. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat. Immunol. 3:583-590. [DOI] [PubMed] [Google Scholar]
- 11.Hristova, K., M. E. Selsted, and S. H. White. 1996. Interactions of monomeric rabbit neutrophil defensins with bilayers: comparison with dimeric human defensin HNP-2. Biochemistry 35:11888-11894. [DOI] [PubMed] [Google Scholar]
- 12.Kellogg, D. E., and S. Kwok. 1990. Detection of human immunodeficiency virus, p. 337-347. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 13.King, P. J., G. Ma, W. Miao, Q. Jia, B. R. McDougall, M. G. Reinecke, C. Cornell, J. Kuan, T. R. Kim, and W. E. Robinson, Jr. 1999. Structure-activity relationships: analogues of the dicaffeoylquinic and dicaffeoyltartaric acids as potent inhibitors of human immunodeficiency virus type 1 integrase and replication. J. Med. Chem. 42:497-509. [DOI] [PubMed] [Google Scholar]
- 14.Lehrer, R. I., K. Daher, T. Ganz, and M. E. Selsted. 1985. Direct inactivation of viruses by MCP-1 and MCP-2, natural peptide antibiotics from rabbit leukocytes. J. Virol. 54:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehrer, R. I., and T. Ganz. 2002. Cathelicidins: a family of endogenous antimicrobial peptides. Curr. Opin. Hematol. 9:18-22. [DOI] [PubMed] [Google Scholar]
- 16.Lencer, W. I., G. Cheung, G. R. Strohmeier, M. G. Currie, A. J. Ouellette, M. E. Selsted, and J. L. Madara. 1997. Induction of epithelial chloride secretion by channel-forming cryptdins 2 and 3. Proc. Natl. Acad. Sci. USA 94:8585-8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichtenstein, A. 1991. Mechanism of mammalian cell lysis mediated by peptide defensins: evidence for an initial alteration of the plasma membrane. J. Clin. Investig. 88:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Maemoto, A., X. Qu, K. J. Rosengren, H. Tanabe, A. Henschen-Edman, D. J. Craik, and A. J. Ouellette. Functional analysis of the alpha-defensin disulfide array in mouse cryptdin-4. J. Biol. Chem., in press. [DOI] [PubMed]
- 18.McManus, A. M., N. F. Dawson, J. D. Wade, L. E. Carrington, D. J. Winzor, and D. J. Craik. 2000. Three-dimensional structure of RK-1: a novel alpha-defensin peptide. Biochemistry 26:15757-15764. [DOI] [PubMed] [Google Scholar]
- 19.Merlin, D., G. Yue, W. I. Lencer, M. E. Selsted, and J. L. Madara. 2001. Cryptdin-3 induces novel apical conductance(s) in Cl-secretory, including cystic fibrosis, epithelia. Am. J. Physiol. Cell. Physiol. 280:C296-302. [DOI] [PubMed] [Google Scholar]
- 20.Montefiori, D. C., J. W. E. Robinson, S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies against human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munk, C., G. Wei, O. O. Yang, A. J. Waring, W. Wang, T. Hong, R. I. Lehrer, N. R. Landau, and A. M. Cole. 2003. The θ-defensin, retrocyclin, inhibits HIV-1 entry. AIDS Res. Hum. Retrovir. 19:875-881. [DOI] [PubMed] [Google Scholar]
- 22.Ouellette, A. J., M. M. Hsieh, M. T. Nosek, D. F. Cano-Gauci, K. M. Huttner, R. N. Buick, and M. E. Selsted. 1994. Mouse Paneth cell defensins: primary structures and antibacterial activities of numerous cryptdin isoforms. Infect. Immun. 62:5040-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouellette, A. J., D. P. Satchell, M. M. Hsieh, S. J. Hagen, and M. E. Selsted. 2000. Characterization of luminal Paneth cell alpha-defensins in mouse small intestine. Attenuated antimicrobial activities of peptides with truncated amino termini. J. Biol. Chem. 275:33969-33973. [DOI] [PubMed] [Google Scholar]
- 24.Porter, E. M., E. van Dam, E. V. Valore, and T. Ganz. 1997. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect. Immun. 65:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quayle, A. J., E. M. Porter, A. A. Nussbaum, Y. M. Wang, C. Brabec, K. P. Yip, and S. C. Mok. 1998. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am. J. Pathol. 152:1247-1258. [PMC free article] [PubMed] [Google Scholar]
- 26.Reinke, R. A., P. J. King, J. G. Victoria, B. R. McDougall, G. Ma, Y. Mao, M. G. Reinecke, and W. E. Robinson, Jr. 2002. Dicaffeoyltartaric acid analogues inhibit human immunodeficiency virus type 1 (HIV-1) integrase and HIV-1 replication at non-toxic concentrations. J. Med. Chem. 45:3669-3683. [DOI] [PubMed] [Google Scholar]
- 27.Robinson, W. E., Jr., M. Cordeiro, S. Abdel-Malek, Q. Jia, S. A. Chow, M. G. Reinecke, and W. M. Mitchell. 1996. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus integrase: iInhibition of the core catalytic domain of human immunodeficiency virus integrase. Mol. Pharmacol. 50:846-855. [PubMed] [Google Scholar]
- 28.Robinson, W. E., Jr., B. McDougall, D. Tran, and M. E. Selsted. 1998. Anti-HIV-1 activity of indolicidin, an antimicrobial peptide from neutrophils. J. Leukoc. Biol. 63:94-100. [DOI] [PubMed] [Google Scholar]
- 29.Robinson, W. E., Jr., D. C. Montefiori, D. H. Gillespie, and W. M. Mitchell. 1989. Complement-mediated, antibody-dependent enhancement of HIV-1 infection in vitro is characterized by increased protein and RNA syntheses and infectious virus release. J. Acquir. Immune Defic. Syndr. 2:33-42. [PubMed] [Google Scholar]
- 30.Robinson, W. E., Jr., M. G. Reinecke, S. Abdel-Malek, Q. Jia, and S. A. Chow. 1996. Inhibitors of HIV-1 replication that inhibit HIV integrase. Proc. Natl. Acad. Sci. USA 93:6326-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satchell, D. P., T. Sheynis, J. E. Cummings, T. K. Vanderlick, R. Jelinek, M. E. Selsted, and A. J. Ouellette. 2003. Quantitative interactions between cryptdin-4 amino terminal variants and membranes. Peptides 24:1793-1803. [DOI] [PubMed] [Google Scholar]
- 32.Satchell, D. P., T. Sheynis, Y. Shirafuji, S. Kolusheva, A. J. Ouellette, and R. Jelinek. 2003. Interactions of mouse paneth cell alpha-defensins and alpha-defensin precursors with membranes: prosegment inhibition of peptide association with biomimetic membranes. J. Biol. Chem. 278:13838-13846. [DOI] [PubMed] [Google Scholar]
- 33.Selsted, M. E. 1993. Investigational approaches for studying the structures and biological functions of myeloid antimicrobial peptides. Genet. Eng. 15:131-147. [DOI] [PubMed] [Google Scholar]
- 34.Selsted, M. E., and S. S. Harwig. 1989. Determination of the disulfide array in the human defensin HNP-2. A covalently cyclized peptide. J. Biol. Chem. 264:4003-4007. [PubMed] [Google Scholar]
- 35.Selsted, M. E., S. I. Miller, A. H. Henschen, and A. J. Ouellette. 1992. Enteric defensins: antibiotic peptide components of intestinal host defense. J. Cell Biol. 118:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirafuji, Y., H. Tanabe, D. P. Satchell, A. Henschen-Edman, C. L. Wilson, and A. J. Ouellette. 2003. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J. Biol. Chem. 278:7910-7919. [DOI] [PubMed] [Google Scholar]
- 37.Tang, Y. Q., J. Yuan, C. J. Miller, and M. E. Selsted. 1999. Isolation, characterization, cDNA cloning, and antimicrobial properties of two distinct subfamilies of alpha-defensins from rhesus macaque leukocytes. Infect. Immun. 67:6139-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Victoria, J. G., D. J. Lee, B. R. McDougall, and W. E. Robinson, Jr. 2003. Replication kinetics for divergent type 1 human immunodeficiency viruses using quantitative SYBR green I real-time polymerase chain reaction. AIDS Res. Hum. Retrovir. 19:865-874. [DOI] [PubMed] [Google Scholar]
- 39.Wachinger, M., A. Kleinschmidt, D. Winder, N. von Pechmann, A. Ludvidsen, M. Neumann, R. Holle, B. Salmons, V. Erfle, and R. Brack-Werner. 1998. Antimicrobial peptides melittin and cecropin inhibit replication of human immunodeficiency virus 1 by suppressing viral gene expression. J. Gen. Virol. 79:731-740. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W., A. M. Cole, T. Hong, A. J. Waring, and R. I. Lehrer. 2003. Retrocyclin, an antiretroviral θ-defensin, is a lectin. J. Immunol. 170:4708-4716. [DOI] [PubMed] [Google Scholar]
- 41.White, S. H., W. C. Wimley, and M. E. Selsted. 1995. Structure, function, and membrane integration of defensins. Curr. Opin. Struct. Biol. 5:521-527. [DOI] [PubMed] [Google Scholar]
- 42.Yue, G., D. Merlin, M. E. Selsted, W. I. Lencer, J. L. Madara, and D. C. Eaton. 2002. Cryptdin 3 forms anion selective channels in cytoplasmic membranes of human embryonic kidney cells. Am. J. Physiol. Gastrointest. Liver Physiol. 282:G757-765. [DOI] [PubMed] [Google Scholar]
- 43.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Y. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 44.Zanetti, M., R. Gennaro, M. Scocchi, and B. Skerlavaj. 2000. Structure and biology of cathelicidins. Adv. Exp. Med. Biol. 479:203-218. [DOI] [PubMed] [Google Scholar]
- 45.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, L., W. Yu, T. He, J. Yu, R. E. Caffrey, E. A. Dalmasso, S. Fu, T. Pham, J. Mei, J. J. Ho, W. Zhang, P. Lopez, and D. D. Ho. 2002. Contribution of human α-defensin 1, 2, and to the anti-HIV-1 activity of CD8 antiviral factor. Science 298:995-1000. [DOI] [PubMed] [Google Scholar]