CONCEPT OF THE IONOME
Living systems are supported and sustained by the genome through the action of the transcriptome, proteome, metabolome, and ionome, the four basic biochemical pillars of functional genomics. These pillars represent the sum of all the expressed genes, proteins, metabolites, and elements within an organism. The dynamic response and interaction of these biochemical “omes” defines how a living system functions, and its study, systems biology, is now one of the biggest challenges in the life sciences. Studies on the functional connections between the genome and the transcriptome (Martzivanou and Hampp, 2003; Becher et al., 2004; Leonhardt et al., 2004), proteome (Koller et al., 2002), and metabolome (Fiehn et al., 2000) are well under way; however, the study of the ionome, in contrast, is still in its infancy (Lahner et al., 2003; for review, see Hirschi, 2003; Rea, 2003), with the majority of genes and gene networks involved in its regulation still unknown. Moreover, because the ionome is involved in such a broad range of important biological phenomena, including electrophysiology, signaling, enzymology, osmoregulation, and transport, its study promises to yield new and significant biological insight. An understanding of the ionome and how it interacts with other cellular systems such as the genome, proteome, metabolome, and environment are integral to our full understanding of how plants integrate their organic and inorganic metabolisms.
Lahner and colleagues first described the ionome to include all the metals, metalloids, and nonmetals present in an organism (Lahner et al., 2003), extending the term metallome (Outten and O'Halloran, 2001; Williams, 2001; Szpunar, 2004) to include biologically significant nonmetals such as nitrogen, phosphorus, sulfur, selenium, chlorine, and iodine. It is important to note here that the boundaries between the ionome, metabolome, and proteome are blurred. Compounds containing the nonmetals phosphorus, sulfur, or nitrogen, for example, would fall within both the ionome and metabolome, and metals such as zinc, copper, manganese, and iron in metalloproteins would fall within the proteome, or metalloproteome as it has been described (Szpunar, 2004). The elements to be measured in the ionome will be determined by their biological importance or environmental relevance, in conjunction with their amenability to quantitation. However, each element measured must be present in sufficient concentrations in the plant tissue so as to be well above the Limit of Quantitation, defined as the concentration equal to the signal from a blank plus 10 sds of the blank signal.
CHARACTERIZATION OF THE PLANT IONOME: A SINGLE ION AT A TIME
Remarkable progress has been made in describing and understanding the basic biology of nutrient ion homeostasis in plants since its establishment as a scientific disciple in the 19th century (Marschner, 1995). The development and application of modern molecular genetic techniques and completion of the Arabidopsis and rice genomes has accelerated progress; however, much remains to be discovered. During evolution, the first proto cells faced a major obstacle. The outer membrane, while needed to keep the cellular contents organized as a functional unit, created a barrier that prevented the uptake of nutrient ions. As multicellular and terrestrial organisms evolved, the challenges of moving solute ions from the environment to the appropriate tissues further increased. Thus, one of the key advances enabling organisms to survive was the evolution of ion transport systems. Because of their central importance, ion transporters have been the primary focus of most work involved in characterizing the ionome in plants. In the past few years, transporters for many different ions have been characterized (Mäser et al., 2001). Multiple genes, and even multiple gene families, appear to be responsible for transport. This is not surprising considering that transport across different membranes is required in plant tissues with diverse nutritional and energy requirements. In addition, multiple membrane proteins may be needed for ion uptake from the soil to adapt to varying extracellular conditions and nutrient availability. Such paradigms are exemplified in our current understanding of the regulation of numerous mineral ions in plants, including iron, zinc, sodium, phosphorus, potassium, and calcium (Rausch and Bucher, 2002; Sanders et al., 2002; Curie and Briat, 2003; Véry and Sentenac, 2003; Zhu, 2003). Though extensive progress has clearly been made, a careful analysis of the Arabidopsis genome reveals the existence of approximately 1,000 ion transporters, most of which have not yet been characterized. Furthermore, it is estimated that 5% of the approximately 25,000 genes in the Arabidopsis genome are involved in regulating the ionome (Lahner et al., 2003). Clearly, a major challenge to understanding the genes and gene networks involved in ion homeostasis in plants is to design ways to probe gene function on a genomic scale.
CHARACTERIZATION OF THE PLANT IONOME: MULTIPLE IONS AT A TIME
Progress in our understanding of plant mineral nutrition has been closely related to the development of chemical quantification methods and availability of pure reagents. This relationship is clearly reflected in the timeline of discovery for the essentiality of various micronutrients in plants from iron in 1860 to nickel in 1987. More recent developments in parallel and massively parallel analytical techniques have played a critical role in the explosive development of genomic biology. The advent of DNA microarray technology has certainly accelerated the pace at which genes regulated by ionic changes can be identified. Not surprisingly, many genes are transcriptionally responsive to changes in nutrient availability, including transporters, transcription factors, and signaling factors (Thimm et al., 2001; Negishi et al., 2002; Maathuis et al., 2003; Wang et al., 2003; Wintz et al., 2003). It is clear from these and other studies that plants can respond specifically to availability of individual nutrients and nutrient deficiencies, suggesting that many regulatory pathways exist. The challenge is how to integrate alterations in transcription with a functional understanding of how ion homeostasis networks operate. As part of this challenge, we have developed a high throughput ion-profiling strategy based on the use of recently developed robust inductively coupled plasma mass spectroscopy (ICP-MS) technology for genomic scale profiling of nutrient and trace elements (Lahner et al., 2003; Fig. 1). Using this system, we have analyzed shoot tissue from more than 40,000 plant samples to date, averaging 1,000 per month, including various ecotypes and both fast neutron and T-DNA insertional mutants. We have also analyzed the seed ionome in numerous mutants and ecotypes. During this project, we have also collected data on the shoot and seed ionome of numerous wild-type plants, providing a picture of both the shoot and seed ionome of Arabidopsis, under our standard soil growth conditions (Fig. 1).
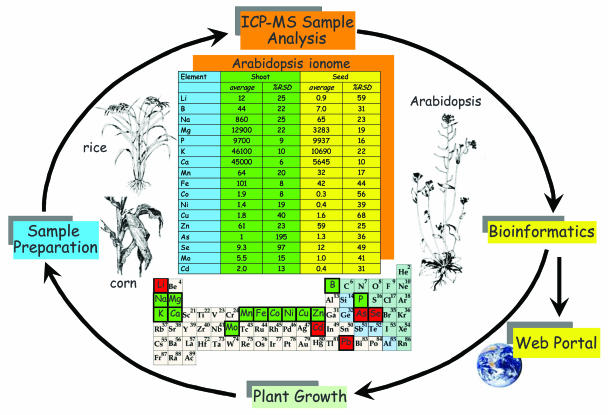
Figure 1.
High-throughput ionomics. Putative mutants and wild-type Arabidopsis plants are grown together with known ionomic mutants, used as positive controls, under standardized conditions. Plants are uniformly sampled, digested in concentrated nitric acid, diluted, and analyzed for numerous elements using ICP-MS. Raw ICP-MS data are normalized using analytical standards and calculated weights based on wild-type plants (Lahner et al., 2003). Data are processed using custom tools and stored in a searchable, World Wide Web-accessible database. Ionomic analysis can also be applied to other plants with available genetic resources, including rice and maize. Elements in the Periodic Table highlighted in black boxes represent those elements analyzed during our ionomic analyses using ICP-MS, elements highlighted in green are essential for plant growth, and those in red represent nonessential trace elements. The table represents Arabidopsis (Col 0) shoot and seed ionomes, all elements presented as μg g−1 dry weight. Data represent the average shoot concentrations from 60 individual plants and seed from 12 individuals ± sd as percentage of average (%RSD), all plants grown as described by Lahner et al. (2003).
HIGH-THROUGHPUT ION PROFILING
For comparative ionomics on a genome-wide scale, wild-type and mutant plants are ideally grown side-by-side under identical conditions, in a uniform growth media. They all grow to the same size and are harvested at the same time, sampling equivalent amounts and parts of tissue in every case. They are grown and harvested under clean room conditions, using tools that won't impart any measured elements to the samples, washed of any surface contaminants, dried, weighed, and analyzed accurately and precisely. The data are processed efficiently, summarized in an easily understood format, and made widely accessible. However, in the imperfect world, a screen of a significant portion of the genome takes months or years. Conditions change, personnel change. Samples get contaminated with growth media, growth media varies from batch to batch. Plants grown on soil are under- or overwatered. Sample sizes vary and include different tissues from different plants. Instruments vary with the maintenance cycle and operating conditions. Data are mixed up, lost, or misinterpreted, and programs have bugs. The success or failure of such an ionomic project is determined by which of these two scenarios we stand closer to. Within these boundaries lies another dimension to consider: sample throughput versus the quality and breadth of the data collected. The best approach to this key tradeoff is by no mean universally agreed upon. Nearer to one end of this dimension lie scientists such as Sydney Brenner, who feel that “…data that goes into a database should be complete, accurate and permanent, so you never have to do it again” (Duncan, 2004, p. 21). At the other end lie scientists who emphasize speed to maximize the number of mutants found, hoping to find the lower-hanging fruit by screening larger portions of genomes, and tend to rely on various statistical tools to extract good information from noisy and incomplete data.
For ionomic analysis samples are typically digested in concentrated acid and diluted before analyzing. Nitric acid digests most plant material easily and, of the common inorganic acids, interferes with ICP-MS analysis the least. Open-air digestion below the boiling point works well, while microwave digestion is becoming more common, especially where loss of an analyte of interest is a concern. Digestion time is much shorter under the higher pressure and temperatures of the microwave digestion apparatus; however, capacity issues shift the overall speed equation toward open-air digestion, where several hundred samples can be run in the same hood that would vent the microwave, and with far less sample handling. The three most common methods for elemental analysis are atomic absorption spectroscopy, ICP-optical emission spectroscopy (ICP-OES), and ICP-MS. Atomic absorption spectroscopy quantitates one element at a time or a few elements in rapid succession. This method is very well established and quite precise, but not nearly as sensitive as ICP-MS and with a dynamic range of only 3 or 4 orders of magnitude. ICP-OES, often referred to as simply ICP, is less sensitive than ICP-MS; however, some of this sensitivity is won back by the robustness of ICP in more concentrated matrices. ICP is a reasonable choice for an ionomics screen, at the possible expense of some of the trace elements due to its lower sensitivity. Both ICP and ICP-MS can measure multiple elements essentially simultaneously in the same sample, a very important property in an ionomics project. One critical advantage of ICP-MS is that it allows for a smaller sample size due to its greater sensitivity, making nondestructive sampling of small plants possible, a prerequisite in a random forward genetic screen—interesting mutants need to be saved not destroyed by the ICP. ICP-MS also has a dynamic range of 6 or 7 orders of magnitude, making it possible to simultaneously measure both macronutrients and micronutrient concentrations in the same sample. Effective genome-scale ionomics requires that many thousands of plant samples will need to be grown, harvested, dried, weighed, digested, and analyzed over long periods of time. Such an effort means that hundreds of samples will need to be processed in a consistent manner, weekly, for several years. Such an effort continuously generates thousands of pieces of data, and the final critical task in ionomics is data handling, including everything from getting the data off the analytical instrument, through date tracking, data storage, data processing, analyses, and publication. Analysis of raw data generated in an ionomics project can be carried out in a number of valid ways. The key objective is identification of plants with disturbed ionomes. This goal does not require that actual concentrations of any of the elements be determined. Instead, interesting mutants can be identified by their overall elemental profiles using average signal normalization and various supervised and unsupervised clustering systems such as discriminant analysis, principle component analysis, and neural net trolling. Clearly, an efficient and robust process containing all these components needs to be designed and implemented before any successful ionomic project can be performed (Lahner et al., 2003).
LINKING THE IONOME AND GENOME
By considering the ionome as a whole, the concept of ion homeostasis networks arises, in which various ions within an organism are coordinately regulated. The observation that only 11% of the 50 Arabidopsis ion-profile mutants recently identified (Lahner et al., 2003) showed changes in only one element strongly supports the existence of such regulatory networks in the ionome of plants. Characterization and mapping of these ion homeostasis networks should help uncover not only their genetic basis but also how they interact with both the proteome and the metabolome. Identification of genes underlying any biological phenomena can take the two different but complementary approaches of forward and reverse genetics. Forward genetics is the more traditional approach of mapping genotypic variation to a specific phenotype. Genotypic variation can be either naturally occurring, such as between different ecotypes of Arabidopsis (Alonso-Blanco and Koornneef, 2000), or induced using various mutagens including ethyl methanesulfonate (EMS), x-rays, fast neutrons (FN; Koornneef et al., 1982), T-DNA (Azpiroz-Leehan and Feldmann, 1997; Krysan et al., 1999), and transposable elements such as Dissociation (Parinov et al., 1999). Once a plant population has been established with significant genotypic variation, a suitable screen needs to be developed to identify plants with the phenotype of interest. The probability of identifying a plant harboring a mutation in a gene that affects the trait of interest, in this case the ionome, is dependent on both the mutation frequency and size of the gene(s). Mutation frequency varies between mutagens, with FN and EMS producing on average 30 to 60 mutations per diploid genome (Koornneef et al., 1982), compared to 1.4 mutations for T-DNA (Feldmann, 1991). To perform a saturation screen using an EMS or FN mutagenized population would therefore require phenotyping of approximately 10,000 to 20,000 M2 plants, whereas the same screen with T-DNA would require 200,000 to 400,000 M2 plants. Clearly, even when using an EMS or FN mutagenized population, the screening system used to identify plants with an altered ionome needs to be relatively high throughput in order to achieve saturation.
Achieving high-throughput ICP-MS analysis at the high precision needed to produce a viable screen of the ionome is challenging and requires both good analytical techniques and data handling. We have performed such a screen on shoot tissue from an Arabidopsis FN mutagenized population of approximately 6,000 M2 plants and identified 51 mutants with altered shoot ionomes (Lahner et al., 2003). Of these mutants, only one mutant, frd3-5, appeared to show dominance after analysis of the high manganese phenotype in the M3 generation. However, to confirm the recessive nature of the majority of these mutations backcrossing to the wild type (Col 0), selfing of the hybrid F1 and scoring of the ionomic phenotype in the F2 is required. Such analyses have been performed on three of these ionomics mutants (145:01, 121:33, and 132:31) that show alterations in various elements, including boron, calcium, potassium, molybdenum, sodium, and nickel, and all were found to be recessive. A similar ICP-MS based forward genetic screen is also under way in the laboratory of Mark Tester (http://www.designscene.com.au/clients/nn/30_group/tester_mark.htm). This project plans to screen activation tagged lines of Arabidopsis to identify genes involved in regulating the ionome; however, few details are yet available. Activation tag mutagenesis involves transformation with T-DNA vectors that contain a multimerized transcription enhancer (Weigel et al., 2000). Such a strategy allows for the identification of both insertional mutants and dominant gain-of-function mutations, in which the transcription enhancer activates expression of genes adjacent to the insertion of the T-DNA. This can be contrasted with other forms of mutagenesis, including EMS and FN, in which loss-of-function mutations are more common.
The availability of Arabidopsis high-density gene arrays now makes it possible to simultaneously genotype plants for several hundred thousand loci. By using total genomic DNA instead of mRNA for hybridization and pooling DNA from only 15 homozygous recombinants displaying the mutant phenotype, it is possible to map a locus to approximately 12 cM (Borevitz et al., 2003). Simulations suggest that using DNA from a pool of >200 plants would allow mapping down to <0.5 cM (Hazen and Kay, 2003). Because FN mutagenesis produces deletions of between 2 to 4 kb (Li et al., 2001) and assuming the deletion is within an exon, bulk segregant analysis of pooled F2 plants using genomic DNA as a probe should also provide a rapid strategy for the identification of mutant genes. The emerging high-density Arabidopsis genome tiling arrays should greatly facilitate the identification of genomic polymorphisms, allowing the rapid identification of deletions produced by FN mutagenesis in a single F2 bulk segregant analysis experiment. Such an approach could revolutionize forward genetics in Arabidopsis, making it possible to identify a mutation within 3 to 4 months. Positional cloning of the genes responsible for ion-profile changes in several of the ionomic mutants (Lahner et al., 2003) is under way in our laboratory. Outcrosses to Landsberg erecta (Ler) have been performed, hybrid F1 allowed to self, and the F2 progeny screened for homozygous mutants. These F2 mapping populations are now being screened for cosegragating genetic polymorphisms.
As an alternative to laboratory-induced mutations, genetic variation occurring among and within natural populations of Arabidopsis can be used (Alonso-Blanco and Koornneef, 2000). Since Arabidopsis shows a wide geographic distribution, many Arabidopsis ecotypes or accessions have been collected and are available from the Arabidopsis Biological Resource Center (ABRC). Considerable variation for such traits as resistance to biotic and abiotic stress, development, and metabolic traits has been described (for review, see Alonso-Blanco and Koornneef, 2000). Observed variation between accessions can either be qualitative, defined by phenotypic distributions that fall into discrete classes and is caused by one or two major loci, or quantitative, defined by a continuous phenotypic distribution and caused by the combined effect of multiple loci. Natural variation in Arabidopsis seed and shoot phosphate accumulation is known to exist between the Ler and Cvi accessions (Bentsink et al., 2003), and for potassium, sodium, calcium, magnesium, iron, manganese, zinc, and phosphorus in seeds of numerous ecotypes (Vreugdenhil et al., 2004). Analyses of Ler/Cvi recombinant inbred lines revealed quantitative trait loci (QTL) that explain between 10% and 79% of this variation for the different elements (Vreugdenhil et al., 2004). Natural variation in several Arabidopsis ecotypes has also been observed for shoot caesium. Four QTL accounting for >80% of the genetic contribution governing this variability have been identified in Ler × Col recombinant inbred lines, in which this variability has been fixed (Payne et al., 2004). The robust and precise quantification of the ionomic variation in various ecotypes holds the promise of Mendelian or QTL mapping to identify genes involved in regulating the ionome in Arabidopsis.
An alternative to the forward genetic approach described is the opposite strategy of starting with a mutation in a known gene and asking the question, “Does this mutation have an ionomic phenotype?” Such an approach switches the focus from one of high-throughput screening and mapping to an approach that requires in-depth biochemical and physiological analyses of the mutant. A close-to-saturation collection of Arabidopsis T-DNA insertional mutants with sequenced boarders is curated at the Salk Institute Genomic Analysis Laboratory (SIGnAL), making this approach attractive in Arabidopsis. High-throughput screening for induced point mutations (TILLING) also makes it possible to identify alternative alleles in genes of interest in numerous species, including maize (Zea mays), black cottonwood (Populus balsamifera subsp. trichocarpa), Brassica oleracea, and Lotus japonicus. The use of genomic DNA pooling and PCR also makes the identification of mutants in specific genes possible in other species using fast neutron deletion mutagenesis (Li et al., 2001). As part of the ionomics project in our laboratory, we are in the process of quantifying the shoot ionome in approximately 1,000 homozygous T-DNA insertional lines of Arabidopsis. Mutants have been chosen with insertions in exons of genes thought to be involved in regulating the ionome, including ion transporters and various regulatory proteins. To date, we have analyzed the shoot ionome in 12,000 plants, representing insertions in approximately 600 genes.
FUTURE DIRECTIONS
To date, ionomics has been applied to bulk tissue samples. Such analyses only provide a very limited view of the tissue, and cellular and subcellular complexities of ion homeostasis mechanisms. The ability to profile the elemental content of different plant tissues such as meristematic and vascular tissue requires a 10- to 50-μm spatial sampling resolution. Such imaging has been achieved in vivo for individual elements such as selenium in plants using x-ray spectroscopy (Pickering et al., 2000, 2003), though not for multielement analyses using ICP-MS. However, use of laser ablation sampling coupled to ICP-MS holds promise for development of high resolution ionomic imaging (Narewski et al., 2000; Kang et al., 2004). If developed in living plant tissue, such technology would open up a completely new window onto the ionome, allowing changes in the total shoot, seed, or root ionomes, for example, to be mapped to specific tissues and cell types. Such ionomic imaging would also allow colocalization of in vivo gene expression and protein localization patterns with ionomic changes, providing spatial linkage between gene, protein, and ionomic function.
COMMUNITY IONOMIC RESOURCES
To maximize the value of any large-scale genomics effort, it is critical that the data be made available to as wide a selection of people as possible. To facilitate such a process, we have developed a searchable online database containing ionomic information on more than 22,000 plants. The database can be searched for fast neutron mutagenized mutants altered in a specific or set of elements, as well as on gene name and AGI gene codes for reverse genetics in T-DNA insertional lines. This ionomics database can be found at http://hort.agriculture.purdue.edu/Ionomics/database.asp. Ionomic mutants reported by Lahner et al. (2003) are detailed in the ionomics database and available through the ABRC (http://www.biosci.ohio-state.edu/ approximately plantbio/Facilities/abrc/abrchome.htm). Seeds of other M3 fast neutron mutant lines identified in the database are available on request (dsalt@purdue.edu), dependent on availability. Genes that appear to alter the ionome, identified in the T-DNA insertional ionomics database, can be further characterized using T-DNA insertional lines curated at the SIGnAL database (http://signal.salk.edu/cgi-bin/tdnaexpress). The Purdue Ionomics Database will be periodically updated and expanded, and our ultimate goal is to provide a biologist-friendly synthetic laboratory that integrates ionomic data with other types of genomic, biochemical, and physiological datasets. Such an environment will allow in silico experiments to be performed that will facilitate the development of sophisticated hypotheses that can be tested with wet experiments. It is hoped that such an environment will accelerate the speed at which researchers design and perform experiments that deliver new and significant biological insight.
Acknowledgments
The development of ionomics as a functional genomics approach has been supported in part by the NSF Plant Functional Genomics program (0077378–DBI) through a collaborative grant to Mary Lou Guerinot, David Eide, Jeff Harper, David E. Salt, Julian Schroeder, and John Ward. We also thank Brett Lahner for his insightful and sustained input on the design, implementation, and running of our ionomics system at Purdue and help with the content of this update.
This work was supported in part by the National Science Foundation (NSF) Plant Functional Genomics program (grant no. 0077378–DBI).
References
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22–29 [DOI] [PubMed] [Google Scholar]
- Azpiroz-Leehan R, Feldmann KA (1997) T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet 13: 152–156 [DOI] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Krämer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 73: 251–268 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Yuan K, Koorneef M, Vreugdenhil D (2003) The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor Appl Genet 106: 1234–1243 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J (2003) Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res 13: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat J-F (2003) Iron transport and signaling in plants. Annu Rev Plant Biol 54: 183–206 [DOI] [PubMed] [Google Scholar]
- Duncan DE (2004) The man who made worms the workhorses of genetics. Discover 25: 20–23 [Google Scholar]
- Feldmann KA (1991) T-DNA insertional mutagenesis in Arabidopsis—mutational spectrum. Plant J 1: 71–82 [Google Scholar]
- Fiehn O, Kopka J, Dömann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Hazen SP, Kay SA (2003) Gene arrays are not just for measuring gene expression. Trends Plant Sci 8: 413–416 [DOI] [PubMed] [Google Scholar]
- Hirschi KD (2003) Striking while the ionome id hot: making the most of plant genomic advances. Trends Biotechnol 21: 520–521 [DOI] [PubMed] [Google Scholar]
- Kang D, Amarasiriwardena D, Goodman AH (2004) Application of laser ablation-inductively coupled plasma-mass spectroscopy (LA-ICP-MS) to investigate trace metal distributions in human tooth enamel and dentine growth layers and pulp. Anal Bioanal Chem 378: 1608–1615 [DOI] [PubMed] [Google Scholar]
- Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schielts D, Ulaszek R, Wei J, et al (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99: 11969–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LW, van der Veen JH (1982) EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat Res 93: 109–123 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L, et al (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol 21: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analysis of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Song Y, Century K, Straight S, Ronald P, Dong X, Lassner M, Zhang Y (2001) A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J 27: 235–242 [DOI] [PubMed] [Google Scholar]
- Maathuis FJ, Filatov V, Herzyk P, Krijger GC, Axelsen KB, Chen S, Green BJ, Li Y, Madagan KL, Sanchez-Fernandez R, et al (2003) Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J 35: 675–692 [DOI] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D, et al (2001) Phylogenetic relationships within cation-transporter families of Arabidopsis thaliana. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London
- Martzivanou M, Hampp R (2003) Hyper-gravity effects on the Arabidopsis transcriptome. Physiol Plant 118: 221–231 [DOI] [PubMed] [Google Scholar]
- Narewski U, Werner G, Schulz H, Vogt C (2000) Application of laser ablation inductively coupled mass spectrometry (LA-ICP-MS) for the determination of major, minor, and trace elements in bark samples. Fresenius J Anal Chem 366: 167–170 [DOI] [PubMed] [Google Scholar]
- Negishi T, Nakanishi H, Yazaki J, Kishimoto N, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T, Kikuchi S, et al (2002) cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots. Plant J 30: 83–94 [DOI] [PubMed] [Google Scholar]
- Outten CE, O'Halloran TV (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292: 2488–2492 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang W-C, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KA, Bowen HC, Hammond JP, Hampton CR, Lynn JR, Mead A, Swarup K, Bennett MJ, White PJ, Broadley MR (2004) Natural genetic variation in caesium (Cs) accumulation by Arabidopsis thaliana. New Phytol 162: 535–548 [Google Scholar]
- Pickering IJ, Hirsch G, Prince RC, Yu EY, Salt DE, George GN (2003) Imaging of selenium in plants using tapered metal capillary optics. J Synchrotron Radiat 10: 289–290 [DOI] [PubMed] [Google Scholar]
- Pickering IJ, Prince RC, Salt DE, George GN (2000) Quantitative chemically-specific imaging of selenium transformation in plants. Proc Natl Acad Sci USA 97: 10717–10722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Rea PA (2003) Ion genomics. Nat Biotechnol 21: 1149–1151 [DOI] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownless C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14: S410–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpunar J (2004) Metallomics: a new frontier in analytical chemistry. Anal Bioanal Chem 378: 54–56 [DOI] [PubMed] [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ (2001) Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol 127: 1030–1043 [PMC free article] [PubMed] [Google Scholar]
- Véry A-A, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54: 575–603 [DOI] [PubMed] [Google Scholar]
- Vreugdenhil D, Aarts MGM, Koornneef M, Nelissen H, Ernst WHO (2004) Natural variation and QTL analysis for cationic mineral content in seeds of Arabidopsis thaliana. Plant Cell Environ 27: 828–839 [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RJP (2001) Chemical selection of elements by cells. Coordin Chem Rev 216-217: 583–595 [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C (2003) Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
- Zhu J-K (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6: 441–445 [DOI] [PubMed] [Google Scholar]