Soil salinity represents an increasing threat to agricultural production. High sodium (Na+) concentrations in soils are toxic to most higher plants. More than 40% of irrigated lands worldwide show increased salt levels. Several studies have shown that under saline conditions, Na+ influx into root cells occurs via Na+ permeable transporters (Amtmann et al., 1997; Roberts and Tester, 1997; Tyerman et al., 1997), which in turn elevates the cytoplasmic sodium concentration and causes toxicity (Kingsbury and Epstein, 1986). Channels and transporters for nutrient cations such as Ca2+ and K+ allow sodium influx. Furthermore, other Na+ transporters that are more specific for sodium transport play diverse roles in protecting plant cells and whole tissues from Na+ toxicity. Important mechanisms that contribute to Na+ tolerance are Na+ efflux transporters (DuPont, 1992; Shi et al., 2002) and Na+ transporters that mediate Na+ sequestration into vacuoles (Blumwald and Poole, 1985, 1987). Moreover, recent research also has shown that plasma membrane Na+ influx can mediate Na+ tolerance by reducing vascular long-distance Na+ transfer to leaves, thus protecting photosynthetically active tissues from salinity stress (Mäser et al., 2002a; Berthomieu et al., 2003). Here we review findings of plant Na+ transporter genes that have been shown to contribute to important and specific mechanisms during plant salinity stress.
SEQUESTRATION OF SODIUM INTO VACUOLES
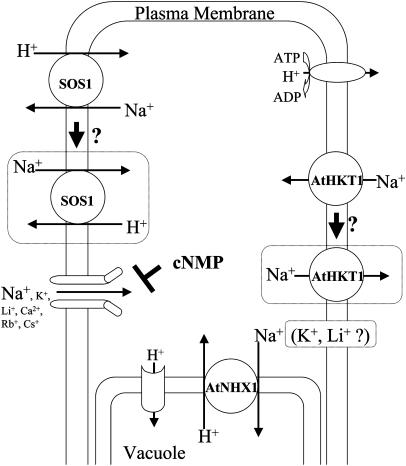
One distinctive structural feature of plant cells is the presence of large membrane-bound compartments, the vacuoles. Early biochemical and tonoplast transport analyses led to the model that the sequestration of excessive Na+ under salt stress is mediated by Na+/H+ antiporters localized in the vacuolar membrane (Blumwald and Poole, 1985, 1987). These Na+/H+ antiporters mediate Na+ uptake into vacuoles, which is driven by the vacuolar proton gradient established by the vacuolar (V-type) proton ATPase that acidifies the vacuolar lumen (Fig. 1). The resulting vacuolar Na+ sequestration protects essential enzymatic reactions in the cytoplasm from excess Na+ levels while maintaining turgor (Glenn et al., 1999).
Figure 1.
Schematic drawing of a simplified plant cell model representing Na+ transport proteins discussed in this review. Regions surrounded by dotted lines indicate hypotheses and subjects that remain to be shown. Plasma membrane Na+/H+ antiporter SOS1 and Na+ transporter AtHKT1 have been hypothesized to transport in either direction depending on conditions (Shi et al., 2002; Berthomieu et al., 2003). Vacuolar Na+/H+ antiporter AtNHX1 is hypothesized to transport K+ (Zhang and Blumwald, 2001; Venema et al., 2002) and Li+ (Qiu et al., 2004) in addition to Na+.
The Arabidopsis genome-sequencing project led to the identification of a plant Na+/H+ antiporter gene, AtNHX1, and subsequently five additional Arabidopsis homologs of AtNHX1 (Apse et al., 1999; Gaxiola et al., 1999; Yokoi et al., 2002). AtNHX1 is highly homologous to the Na+/H+ antiporters from Saccharomyces cerevisiae (Nhx1) and the NHX family of Caenorhabditis elegans and humans. Expression of the AtNHX1 cDNA in the yeast nhx1 mutant complemented the salt-sensitive phenotype (Gaxiola et al., 1999). Apse et al. (1999) biochemically demonstrated tonoplast localization of AtNHX1 and Na+-dependent tonoplast proton transport via AtNHX1 with AtNHX1-overexpressing Arabidopsis plants. AtNHX1-overexpressing Arabidopsis showed a salt-tolerant phenotype compared to wild type and sequestered more Na+ in plants under salt stress conditions, providing molecular evidence that Na+/H+ exchange in vacuoles is an important factor for salt tolerance. In support of these findings, T-DNA tagged atnhx1 plants showed an increase in salt sensitivity compared to wild type (Apse et al., 2003). Improvement in salt tolerance evoked by AtNHX1 overexpression was also observed in Brassica and tomato (Lycopersicon esculentum; Zhang and Blumwald, 2001; Zhang et al., 2001). An Nhx1 homolog was also identified in rice (Oryza sativa), and overexpression of OsNHX1 in rice also conferred an increase in salt tolerance (Fukuda et al., 2004). These studies show that overexpression of vacuolar NHX transporters provides an approach that can contribute to the molecular breeding of salt-tolerant plants.
Several studies have appeared that analyze the ion selectivity and activity regulation of AtNHX1. Potassium-proton exchange activity mediated by AtNHX1 was found in addition to Na+/H+ exchange in tonoplast vesicles isolated from AtNHX1-overexpressing tomato plants and yeast (Zhang and Blumwald, 2001), and in liposomes reconstituted with purified AtNHX1 (Venema et al., 2002; Fig. 1). Tonoplast vesicles isolated from mutant atnhx1 showed reduced H+-coupled cation transport activity not only for Na+ but also for K+ (Apse et al., 2003). These data suggest that AtNHX1 contributes to vacuolar K+ sequestration. Yamaguchi et al. (2003) reported that deletion of the C terminus of AtNHX1 causes enhanced Na+/H+ but lower K+/H+ exchange rates. By contrast, Qiu et al. (2004) reported that cation/H+ exchange in tonoplast vesicles isolated from Arabidopsis cultured cells shows Na+ and, to a lesser extent, Li+ transport but not K+ transport.
PLASMA MEMBRANE SODIUM EXTRUSION
Salt-sensitive SOS mutant loci were identified as essential factors for salt tolerance in Arabidopsis by screening for reduced root bending of seedlings grown in the presence of sodium (Wu et al., 1996). The sos1, sos2, and sos3 loci have been shown to affect Na+ transport and Na+ transport regulation. Detailed phenotyping of sos1 plants revealed two features. Firstly, hypersensitivity of sos1 plants is specific to Na+ and Li+. Secondly, sos1 plants show growth deficiencies on media containing low potassium concentrations. The SOS1 gene encodes a Na+/H+ antiporter that has 12 transmembrane domains in the N-terminal half and a long hydrophilic C-terminal tail (Shi et al., 2000). Based on imaging experiments of SOS1-GFP fusions SOS1 was predicted to be targeted to the plasma membrane of cells (Shi et al., 2002; Fig. 1). Plasma membrane Na+/H+ antiporters mediate Na+ extrusion that is energized by proton influx. The SOS2 gene encodes a protein kinase (Liu et al., 2000) that is activated by SOS3 (Halfter et al., 2000). SOS3 encodes a Ca2+ binding protein that interacts with SOS2 (Liu and Zhu 1998). Qiu et al. (2002) biochemically demonstrated that plasma membrane Na+/H+ exchange activity of sos1, sos2, and sos3 plants are reduced compared to wild type. However, addition of a constitutively active SOS2 kinase mutant in vitro stimulated and recovered Na+/H+ exchange in vesicles isolated from sos2 and sos3 plants but not in those from sos1 plants (Guo et al., 2001; Qiu et al., 2002). Furthermore, Quintero et al. (2002) reconstituted the SOS system in yeast and demonstrated that SOS2 and SOS3 regulate Na+ transport by SOS1. These results support a model in which SOS1 activity is regulated by a complex composed of the SOS2 kinase and the SOS3 Ca2+ binding protein in vivo.
SOS2 has been reported to also regulate tonoplast Na+/H+ exchange activity. The sodium/proton exchange activity in tonoplast vesicles of sos2 plants was reported to be greatly reduced in comparison to that of wild type. But interestingly, Na+/H+ exchange activity in tonoplast vesicles of sos3 plants was not affected (Qiu et al., 2004). These data therefore indicate that the SOS signaling pathway is branched, such that vacuolar Na+/H+ antiport activity is regulated in an SOS3-independent manner (Qiu et al., 2004).
Transgenic plants expressing the β-glucuronidase (GUS) gene under the control of the SOS1 promoter showed GUS activity predominantly in xylem parenchyma cells (Shi et al., 2002). Furthermore, xylem sap extracted from salt-stressed sos1 plants contained more Na+ than wild type. Additional GUS activity was detected in epidermal cells of root tips. Reduced Na+/H+ transport activity in whole leaf plasma membrane vesicles isolated from sos1, however, indicates that SOS1 may be more widely expressed in plants (Qiu et al., 2002). sos1 plants show a strong growth defect under severe salt stress (e.g. 100 mm NaCl) and accumulate more Na+ in both roots and shoots than wild type. However, under moderate stress conditions, such as 25 mm NaCl, sos1 seedlings and plants accumulate less Na+ than wild type. From these results, Shi et al. (2002) hypothesized a long-distance Na+ transport model, in which under moderate stress, SOS1 functions to load Na+ into the xylem in roots for Na+ transfer and storage in leaf mesophyll vacuoles. Whereas under severe Na+ stress, SOS1 is proposed to function in unloading Na+ from the root xylem to reduce Na+ damage of leaves that might be caused by exceeding the capacity of Na+ sequestration in leaf cell vacuoles. This model would require reversibility in the Na+ transport direction of SOS1 in xylem parenchyma cells.
SODIUM EXTRUDING ATPASES ARE FOUND IN SOME PLANT SPECIES
In fungi, such as the yeast S. cerevisiae, plasma membrane sodium extrusion is mediated by Na+-ATPases named ENA. ENA is derived from the Latin, exitus natrus, for sodium efflux. ENAs function in addition to the Na+/H+ antiport system (Haro et al., 1991). Recently, ENA1 homologs were isolated from the moss, Physcomitrella patens (PpENA1, PpENA2A; Benito and Rodríguez-Navarro, 2003). The expression of the PpENA1 cDNA in the highly sodium sensitive yeast mutant, ena1-4 nha1, complemented the salt-sensitive phenotype, whereas PpENA2A did not. PpENA2 transcripts seem to be subject to differential splicing in Physcomitrella, yielding three different transcripts, PpENA 2A, 2B, and 2C. Functions of the PpENA2B and 2C transcripts have not yet been tested. ENA homologs have not been identified in the Arabidopsis and rice genomes or in expressed sequence tags of other higher plants. Findings in Physcomitrella suggest that Na+-extruding pumps existed in primitive land plants but these genes may have been lost in higher plants during evolution (Benito and Rodríguez-Navarro, 2003).
HIGH-AFFINITY POTASSIUM UPTAKE AND SODIUM TRANSPORT
Potassium influx transporters have long been proposed to mediate sodium influx (Epstein et al., 1963). The HKT1 cDNA was isolated from a wheat (Triticum aestivum) root cDNA library by complementation screening of a K+ uptake deficient yeast mutant strain, trk1 trk2 (Schachtman and Schroeder, 1994). HKT1 mediates saturable high-affinity K+ uptake activity in yeast (Km approximately 3 μm K+ and 29 μm Rb+; Schachtman and Schroeder, 1994; Rubio et al., 1995). Detailed analyses of HKT1 expressed in yeast and Xenopus oocytes showed that HKT1 shows two transport modes, saturable high-affinity K+-Na+ couptake (symport) and low affinity Na+ transport (Rubio et al., 1995). The transport properties of the KtrAB K+ uptake system have been analyzed in Vibrio alginolyticus, a marine bacterium that can cause food poisoning. The KtrB subunit gene in V. alginolyticus is homologous to HKT1 (Durell et al., 1999) and was also shown to mediate Na+-coupled K+ uptake (Tholema et al., 1999). For wheat HKT1, high-affinity K+ uptake is stimulated by micromolar extracellular Na+ concentrations. However, at high toxic extracellular Na+ concentrations, K+ uptake mediated by HKT1 is blocked and selective low-affinity channel-like Na+ uptake occurs (Rubio et al., 1995; Gassmann et al., 1996). Another class of K+ uptake transporters, the KUP/HAK/KT family, has also been shown to mediate low-affinity Na+ influx at high Na+ concentrations (Santa-María et al., 1997).
Na+-coupled high-affinity K+ uptake activity was shown to be the predominant form of high-affinity potassium uptake in certain higher plant species, including Egeria, Elodea, and Vallisneria and charophyte algae such as Chara australis and Nitella translucens (Smith and Walker, 1989; Walker and Sanders, 1991; Maathuis et al., 1996), showing that this activity has physiological relevance in plants. However, in roots of wheat, barley (Hordeum vulgare), and maize (Zea mays), Na+-coupled high affinity K+ uptake could not be detected, indicating that HKT1-type transport provides a minor component to total K+ uptake in these plants (Maathuis et al., 1996). Similarly proton-stimulated high-affinity K+ uptake could also not be resolved in maize roots (Newman et al., 1987; Kochian et al., 1989). Together, these studies indicate that many transporter types may have overlapping K+ uptake activities in these plants (Schroeder et al., 1994), thus rendering individual activities difficult to resolve in wild-type backgrounds (Maathuis et al., 1996; Rubio et al., 1996; Mäser et al., 2001).
HKT1 homologs have been isolated or detected from many species, including Arabidopsis, eucalyptus, rice, ice plant (Mesembryanthemum), and poplar (Populus spp.; Fairbairn et al., 2000; Uozumi et al., 2000; Horie et al., 2001; Golldack et al., 2002; Garciadeblás et al., 2003; Sul et al., 2003). The completed sequence of the japonica rice genome shows the presence of up to nine OsHKT genes (Garciadeblás et al., 2003). Interestingly, HKT homologs fall into two different classes of transport properties in heterologous expression systems. For instance, K+ uptake was not observed for the Arabidopsis homolog AtHKT1 in both yeast and Xenopus oocytes, although a large Na+ uptake activity was detected (Uozumi et al., 2000; Fig. 1). In the case of eucalyptus two HKT1 homologs, EcHKT1 and EcHKT2, were isolated, and both transporters showed K+ and Na+ currents in oocytes (Fairbairn et al., 2000). In rice two highly homologous proteins, OsHKT1 and OsHKT2 (91% identity), were identified in the indica variety Pokkali. Interestingly, OsHKT1 showed an AtHKT1-like Na+-specific transport activity, whereas OsHKT2 showed a wheat HKT1 like K+-Na+ transport activity (Horie et al., 2001).
Through a combination of functional chimeric HKT analyses and sequence analyses an amino acid was identified in HKT transporters that plays an important role in determining the transport mode of HKT transporters (K+-Na+ symporter or Na+ transporter; Mäser et al., 2002b). This amino acid lies within a predicted “pore-loop” domain. The presence of a Gly residue resulted in K+-Na+ transport, whereas a Ser residue at this position caused more Na+ selective transport (Mäser et al., 2002b). Interestingly, Gly residues also form the narrowest portion of the selectivity filter in K+ channel pore-loop domains (Mackinnon and Yellen, 1990; Nakamura et al., 1997; Doyle et al., 1998). Thus, HKT analyses indicate that Gly residues in P-loops may provide a more general mechanism for producing K+ selectivity in transport proteins. These data also provide support for the hypothesis that HKT transporters have an ion channel type transport mode (Gassmann et al., 1996).
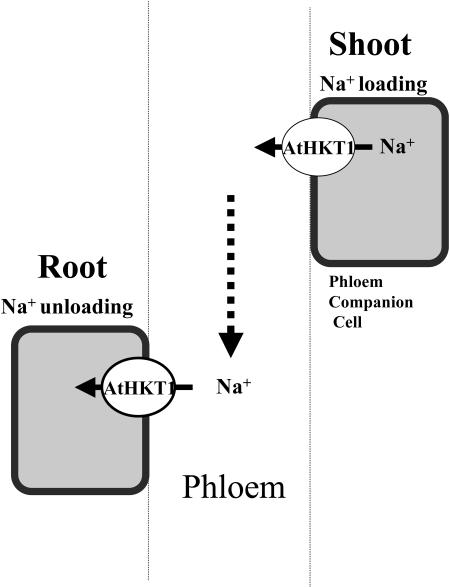
Several reports have analyzed the physiological roles of HKT transporters in vivo. Laurie et al. (2002) found that transgenic wheat plants expressing an HKT1 antisense construct showed Na+ tolerance under saline conditions, and Na+ uptake activity and accumulation were reduced. Consistent with these results RNA in situ hybridizations in wheat showed that HKT1 is expressed in root cortex cells (Schachtman and Schroeder, 1994). The Arabidopsis genome includes only one HKT1 gene, AtHKT1. T-DNA insertions in AtHKT1 gene suppressed the Na+ hypersensitivity of sos3 plants, leading to the proposal that AtHKT1 mediates Na+ uptake into Arabidopsis roots (Rus et al., 2001). However, Mäser et al. (2002a) and Berthomieu et al. (2003) showed that loss-of-function mutations in the AtHKT1 gene lead to over accumulation of Na+ in shoots and rendered plant leaves Na+ hypersensitive. Transgenic plants harboring an AtHKT1 promoter-GUS construct showed AtHKT1 expression in vascular tissues (Mäser et al., 2002a; Berthomieu et al., 2003). Further in situ hybridizations showed expression in phloem tissues (Berthomieu et al., 2003). Taken together with the previous reports showing the Na+ selectivity of AtHKT1, a model was proposed in which AtHKT1 would mediate Na+ loading into the phloem in leaves, and also Na+ unloading from the phloem sap in roots (Berthomieu et al., 2003; Figs. 1 and 2). Na+ selectivity and HKT mutant studies show that members of this gene family play important roles in long-distance Na+ transport and plant salt tolerance.
Figure 2.
Model for role of AtHKT1 in long-distance transport in Arabidopsis (Mäser et al., 2002a; Berthomieu et al., 2003). Na+ recirculation is proposed to be mediated by AtHKT1 at the phloem (Berthomieu et al., 2003). Overaccumulated Na+ in shoots is loaded into the phloem and unloaded from the phloem in roots.
TOXIC SODIUM INFLUX INTO ROOTS
One of the important questions to be addressed with respect to salinity stress in plants is the identification of channels and transporters responsible for toxic Na+ influx into root cells. Classical 22Na+ influx studies showed multiple kinetic components of Na+ influx into barley roots (Rains and Epstein, 1965, 1967). Furthermore, single locus mutations that greatly diminish Na+ influx into plants have not been found, suggesting that several parallel redundant pathways exist (Schroeder et al., 1994). Na+ influx currents have been characterized in electrophysiological studies in root cortex cells of wheat (Tyerman et al., 1997), maize (Roberts and Tester, 1997), and barley suspension-cultured cells (Amtmann et al., 1997). These studies suggest that Na+ influx is mediated by voltage-independent, nonselective cation channels (named VIC or NSC; Tyerman and Skerrett, 1999). Calcium inhibition of Na+ influx in wheat was observed (Tyerman et al., 1997; Buschmann et al., 2000; Davenport and Tester, 2000). The NSC current in wheat cortex cells is weakly voltage dependent and nonselective among monovalent cations (Davenport and Tester, 2000). Furthermore, in wheat root cortex cells K+ deprivation was shown to enhance Na+ influx currents, providing evidence that K+ starvation-induced transporters contribute to Na+ influx (Buschmann et al., 2000). However, the molecular identity of VIC/NSC remains unknown and more than one transporter gene may contribute to this activity.
A cDNA was isolated from wheat that mediates low-affinity K+ and cation transport in yeast and was named LCT1 (Schachtman et al., 1997). Analysis of the secondary structure of LCT1 predicts the presence of 8 to 10 hydrophobic domains with a hydrophilic N terminus. The hydrophobic region is distinctive and novel compared to other transporter genes. LCT1 mRNA is detected at low levels in wheat roots and leaves. LCT1 functions as a nonselective cation permeable transporter in yeast mediating not only K+ influx but also Rb+, Na+, Cd2+, and Ca2+ transport (Schachtman et al., 1997; Clemens et al., 1998). LCT1 expression rendered yeast more salt sensitive (Amtmann et al., 2001). However, further analyses will be required to determine to which plant membrane and cell types LCT1 is targeted and physiological roles of LCT1 remain to be identified.
VIC/NSC currents in Arabidopsis are down-regulated by the addition of cAMP and cGMP (Maathuis and Sanders, 2001; Fig. 1). Furthermore, 22Na+ tracer influx experiments show reduction in Na+ influx in the presence of cyclic nucleotides and salt tolerance of Arabidopsis plants was improved (Maathuis and Sanders, 2001). These results support the hypothesis that cyclic nucleotide inhibited channels may contribute to VIC/NSC currents (Fig. 1). The Arabidopsis genome includes 20 cyclic nucleotide gated channel-like genes (Mäser et al., 2001), and their roles in root Na+ influx remain to be determined.
CONCLUSIONS
Salt stress is a major problem threatening agricultural productivity and yields in the 21st century. Salinity threatens many arid and heavily populated regions of the world. The combination of physiological, biochemical, genomic, genetic, and molecular biological analyses has led to the identification and characterization of important Na+ transporter genes and proteins. Interestingly, the genes that have been analyzed via mutagenesis in plants to date show important and distinct roles in controlling salinity stress. These findings have led to the formulation of novel hypotheses on Na+ sequestration, long-distance transport, and influx that point to mechanisms mediating plant salt tolerance and demonstrate that salt tolerance can be manipulated by molecular engineering of plants using these genes. Further research and development using these genes and diverse promoters, which are tissue and condition dependent, will likely contribute to the future engineering of crops with enhanced salinity resistance. Genome-wide analyses indicate that additional classes of Na+ transporters are likely to exist and characterization of further complexities and interesting functions of Na+ transporters are on the horizon.
This work was supported by the U.S. Department of Energy (grant no. DOE–DE–FG02–03ER15449 to J.I.S.).
References
- Amtmann A, Fischer M, Marsh EL, Stefanovic A, Sanders D, Schachtman DP (2001) The wheat cDNA LCT1 generates hypersensitivity to sodium in a salt-sensitive yeast strain. Plant Physiol 126: 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Laurie S, Leigh R, Sanders D (1997) Multiple inward channels provide flexibility in K+/Na+ discrimination at the plasma membrane of barley suspension culture cells. J Exp Bot 48: 431–440 [DOI] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Apse MP, Sottosanto JB, Blumwald E (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J 36: 229–239 [DOI] [PubMed] [Google Scholar]
- Benito B, Rodríguez-Navarro A (2003) Molecular cloning and characterization of a sodium-pump ATPase of the moss Physcomitrella patens. Plant J 36: 382–389 [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Poole R (1985) Na+/H+-antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol 78: 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Poole R (1987) Salt-tolerance in suspension cultures of sugar beet. I. Induction of Na+/H+-antiport activity at the tonoplast by grown in salt. Plant Physiol 83: 884–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann PH, Vaidyanathan R, Gassmann W, Schroeder JI (2000) Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol 122: 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Antosiewicz DM, Ward JM, Schachtman DP, Schroeder JI (1998) The plant cDNA LCT1 mediates the uptake of calcium and cadmium un yeast. Proc Natl Acad Sci USA 95: 12043–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Tester M (2000) A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiol 122: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, Mackinnon R (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280: 69–77 [DOI] [PubMed] [Google Scholar]
- Dupont F (1992) Salt-induced changes in ion transport: regulation of primary pumps and secondary transporters. In D Cooke, D Clarkson, eds, Transport and Receptor Proteins of Plant Membranes. Plenum Press, New York, pp 91–100
- Durell SR, Hao Y, Nakamura T, Bakker EP, Guy HR (1999) Evolutionary relationship between K+ channels and symporters. Biophys J 77: 775–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E, Rains DW, Elzam OE (1963) Resolution of dual mechanism of potassium absorption by barley roots. Proc Natl Acad Sci USA 49: 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn DJ, Liu W, Schachtman DP, Gomez-Gallego S, Day SR, Teasdale RD (2000) Characterization of two distinct HKT-like potassium transporters from Eucalyptus camaldulensis. Plant Mol Biol 43: 515–525 [DOI] [PubMed] [Google Scholar]
- Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45: 149–159 [DOI] [PubMed] [Google Scholar]
- Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34: 1–14 [DOI] [PubMed] [Google Scholar]
- Gassmann W, Rubio F, Schroeder JI (1996) Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J 10: 869–882 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96: 1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn EP, Brown JJ, Blumwald E (1999) Salt tolerance and crop potential of halophytes. Crit Rev Plant Sci 18: 227–256 [Google Scholar]
- Golldack D, Su H, Quigley F, Kamasani UR, Muñoz-Garay C, Balderas E, Popova OV, Bennett J, Bohnert HJ, Pantoja O (2002) Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J 31: 529–542 [DOI] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu J-K (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro R, Garciadeblás B, Rodríguez-Navarro A (1991) A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett 291: 189–191 [DOI] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A (2001) Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J 27: 129–138 [DOI] [PubMed] [Google Scholar]
- Kingsbury RW, Epstein E (1986) Salt sensitivity in wheat. Plant Physiol 80: 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Shaff JE, Lucas WJ (1989) High affinity K+ uptake in maize roots: a lack of coupling with H+ efflux. Plant Physiol 91: 1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJM, Heard PJ, Brown SJ, Leigh RA (2002) A role for HKT1 in sodium uptake by wheat roots. Plant J 32: 139–149 [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K (2000) The Arabidopsis thaliana SOS2 gene encodes protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Sanders D (2001) Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol 127: 1617–1625 [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM, Verlin D, Smith FA, Sanders D, Fernández JA, Walker NA (1996) The physiological relevance of Na+-coupled K+-transport. Plant Physiol 112: 1609–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon R, Yellen G (1990) Mutations affecting TEA blockage and ion permeation in voltage-activated K+ channel. Science 250: 276–279 [DOI] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, et al (2002. a) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Mäser P, Hosoo Y, Goshima S, Horie T, Eckelman B, Yamada K, Yoshida K, Bakker EP, Shinmyo A, Oiki S, et al (2002. b) Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci USA 99: 6428–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D, et al (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura RL, Anderson JA, Gaber RF (1997) Determination of key structural requirements of a K+ channel pore. J Biol Chem 272: 1011–1018 [DOI] [PubMed] [Google Scholar]
- Newman IA, Kochian LV, Grusak MA, Lucas WJ (1987) Fluxes of H+ and K+ in corn roots: characterization and stoichiometries using ion-selective microelectrodes. Plant Physiol 84: 1177–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem 279: 207–215 [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rains DW, Epstein E (1965) Transport of sodium in plant tissue. Science 148: 1611. [DOI] [PubMed] [Google Scholar]
- Rains DW, Epstein E (1967) Sodium absorption by barley roots: the role of the dual mechanisms of alkali cation transport. Plant Physiol 42: 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SK, Tester M (1997) A patch clamp study of Na+ transport in maize roots. J Exp Bot 48: 431–440 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660–1663 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI (1996) High-affinity potassium uptake in plants. Science 273: 977–979 [DOI] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9: 2281–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Kumar R, Schroeder JI, Marsh EL (1997) Molecular and functional characterization of a novel low-affinity cation transporter (LCT1) in higher plants. Proc Natl Acad Sci USA 94: 11079–11084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370: 655–658 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Ward JM, Gassmann W (1994) Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu Rev Biophys Biomol Struct 23: 441–471 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FA, Walker NA (1989) Transport of potassium in Chara australis. I. A symport with sodium. J Membr Biol 108: 125–137 [DOI] [PubMed] [Google Scholar]
- Sul H, Balderas E, Vera-Estrella R, Golldack D, Quigley F, Zhao C, Pantoja O, Bohnert HJ (2003) Expression of the cation transporter McHKT1 in a halophyte. Plant Mol Biol 52: 967–980 [DOI] [PubMed] [Google Scholar]
- Tholema N, Bakker EP, Suzuki A, Nakamura T (1999) Change to alanine of one out of four selectivity filter glycines in KtrB causes a two orders of magnitude decrease in the affinities for both K+ and Na+ of the Na+ dependent K+ uptake system KtrAB from Vibrio alginolyticus. FEBS Lett 450: 217–220 [DOI] [PubMed] [Google Scholar]
- Tyerman SD, Skerrett M (1999) Root ion channels and salinity. Sci Hortic (Amst) 78: 175–235 [Google Scholar]
- Tyerman SD, Skerrett M, Garrill A, Findlay GP, Leigh RA (1997) Pathways for the permeation of Na+ and Cl− into protoplasts derived from the cortex of wheat roots. J Exp Bot 48: 459–480 [DOI] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema K, Quintero FJ, Pardo JM, Donaire JP (2002) The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low-affinity Na+ and K+ transport in reconstituted liposomes. J Biol Chem 277: 2413–2418 [DOI] [PubMed] [Google Scholar]
- Walker NA, Sanders D (1991) Sodium-coupled solute transport I charophyte algae: a general mechanism for transport energization in plant cells? Planta 185: 443–445 [DOI] [PubMed] [Google Scholar]
- Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Apse MP, Shi H, Blumwald E (2003) Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci USA 100: 12510–12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30: 529–539 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Blumwald E (2001) Transgenic salt tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang HX, Hodson J, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98: 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]