Much has been learned about the energization of nutrient transport since Hoagland in 1944 gave his famous series of lectures on plant nutrition. Already at that time it was speculated that energy for transport of solutes into cells was provided by compounds containing energy-rich phosphate bonds (Hoagland, 1944). We now know that ATP-consuming proton pumps drive nutrient transport at several entry points in the plant body. In this Update, we will focus on those entry points within the plant body where nutrient transport is intense, and we will discuss their energization and regulation by proton pumps.
TRANSPORT BARRIERS SEPARATE THE SYMPLAST FROM THE APOPLAST
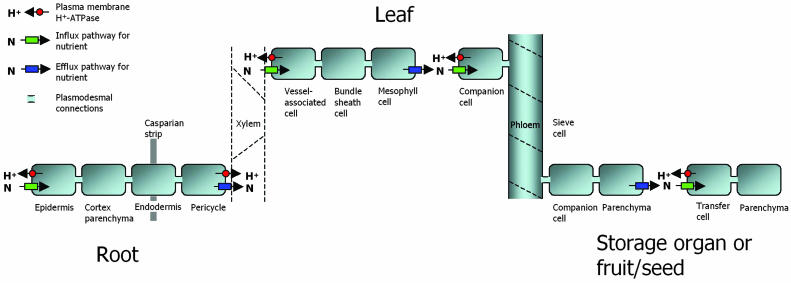
Any nutrient taken up by any cell must, at some stage, pass the plasma membrane (Fig. 1). The plasma membrane is a lipid bilayer structure that surrounds the cell and, in principle, is impermeable to solutes, such as ions and polar molecules. It is quite obvious why the membrane must be tight for nutrients. A root is always enriched in nutrients compared to the surrounding soil and without a tight wrapping most of its contents would leak out of the plant and back into the growth medium. Thus, there is only one way that solutes can be transported from the outside medium, the apoplast, into a cell, and this is by specific transport proteins that span the plasma membrane. As cells tend to accumulate nutrients, this transport is most often uphill, i.e. against a concentration gradient and/or an electrical gradient, and needs to be energized.
Figure 1.
Overview of main transport barriers in the plant body and the energization of cellular nutrient uptake by plasma membrane H+-ATPase.
Once inside a plant cell, a given solute can diffuse from cell to cell via cellular bridges called plasmodesmata, forming a cellular continuum, the symplast. Plasmodesmal transport by diffusion is not very effective, and for long-distance transport the nutrient in question might have to leave the symplast to enter a new from the apoplast in a neighboring cell or in another part of the plant. Here again, uptake needs to be energized and occurs through specialized transport proteins. Specialized cells throughout the plant body serve as transport interfaces between symplast and apoplast, and intense transport occurs across the plasma membrane of these cells.
COMMON CHARACTERISTICS OF CELLS SPECIALIZED TO ACCOMMODATE MASSIVE SOLUTE TRANSPORT
Cells with large fluxes of solutes across the plasma membrane share a number of important characteristics in common. With regard to structure, cells specialized for transport are often characterized by (1) exposing a large surface area toward the uptake interface, e.g. the plasma membrane exhibits many protrusions (e.g. epidermal cells) or invaginations (e.g. transfer cells); and (2) having a great number of mitochondria, the role of which is to supply ATP for active transport. Biophysically, transport competent cells are characteristic by exhibiting (1) a large membrane potential difference between the internal and external face of the membrane, typically ranging from −150 mV to −200 mV, negative on the inside; and (2) an acidic exterior, where apoplastic pH is typically between pH 4 and 5. Thus, across the plasma membrane of these cells, we observe a gradient of electric charge and chemical matter, which is termed the electrochemical gradient (Fisher, 2000).
Although uphill transport of solutes into cells is an energy-consuming process, it is rarely or never driven directly by metabolic energy, i.e. ATP hydrolysis. Rather, nutrient uptake systems are energized indirectly. Thus, ATP is primarily consumed by pumps, which export H+ in order to generate an electrochemical proton gradient across the plasma membrane. This electrochemical gradient of protons in turn energizes nutrient uptake by channel proteins and carriers.
Accordingly, transport-competent cells have at the molecular level (1) a large number of channel proteins and carriers (symporters, if nutrients get cotransported with H+ in the same direction; antiporters, if nutrients and H+ are cotransported in opposite directions; and uniporters, if nutrients are transported as such without being accompanied by H+) and (2) large amounts of the plasma membrane H+-ATPase, a proton pump. The proton pump exports H+ from the cytoplasm into the apoplast at the expense of ATP. It is this pump that is responsible for formation of the trans-plasma membrane electrochemical gradient (Palmgren, 2001; Arango et al., 2002). Transfer cells are considered to be the most specialized cell type for membrane transport, and, therefore, it is not surprising that H+-ATPase is abundant in the various transfer cell types of these cells (Bouche-Pillon et al., 1994; Harrington et al., 1997; Schikora and Schmidt, 2002).
The Plasma Membrane H+-ATPase
The major ion pumps in plants and fungi are plasma membrane H+-ATPases. Similar pumps are not found in animals, in which the equivalent enzyme is the Na+/K+-ATPase, which in turn is absent from plants. However, both types of pumps are evolutionarily related and belong to the superfamily of P-type ATPases (Axelsen and Palmgren, 1998; Kuhlbrandt, 2004). P-type pumps are characterized by forming a phosphorylated reaction-cycle intermediate during catalysis. In this way, plasma membrane H+-ATPases differ from all other proton pumps in the plant cell, including the vacuolar membrane H+-ATPase, another major proton pump which energizes the vacuolar membrane. The vacuolar membrane H+-ATPase has more than 10 different subunits, whereas, in contrast, the functional unit of plasma membrane H+-ATPase is a monomer, although it might be organized in the membrane as a dimer or in oligomers.
The yeast Saccharomyces cerevisiae is equipped with two plasma membrane H+-ATPase genes, of which one, PMA1, is essential for growth (Serrano et al., 1986). In plants, there are many more isoforms. The genomes of the dicot Arabidopsis and the monocot rice (Oryza sativa) have been sequenced, resulting in identification of 11 and 10 plasma membrane H+-ATPase isoforms, respectively (Baxter et al., 2003). The high number of isoforms in plants might indicate that some pumps have redundant functions. Isoform diversity may also be related to the multicellular nature of plants. Individual isoforms exhibit tissue- and developmental-specific expression patterns and have slightly different biochemical and regulatory properties (Palmgren, 2001; Arango et al., 2002). Thus, it is likely that duplication of pump genes has provided a means for assuring appropriate activities of pump protein in any given cell at any given time of development.
CELLS SPECIALIZED FOR NUTRIENT TRANSPORT
Nutrient Uptake into the Root
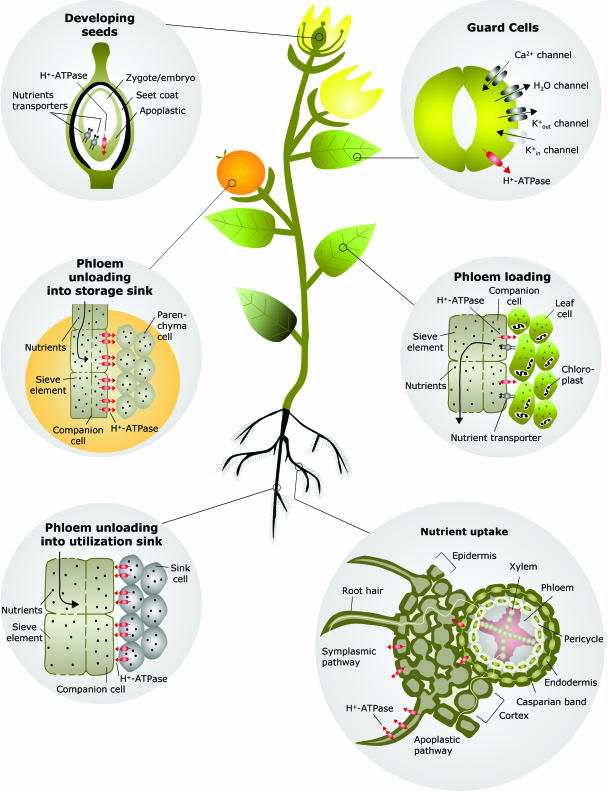
Any nutrient in the soil that is to enter into the plant will need to make contact with the root. The periphery of the root is therefore a crucial transport barrier (Kochian and Lucas, 1983). Nutrients are transferred from the surface of the roots toward the long-distance transport system for water, the xylem, either by apoplastic or symplastic pathways (Fig. 2). Either way, the nutrients have to pass at least one plasma membrane in order to be transported into the xylem. When transferred via the symplastic pathway, nutrients have to pass the plasma membrane of epidermal cells. When transferred via the apoplastic pathway, nutrients have to cross the plasma membrane of cortex or endodermis cells in order to pass the Casparian strip, which is an ion permeability barrier surrounding the endodermal layer. Membrane transport is thus a decisive tool for the selective uptake of nutrients and rejection of toxic ions from the environment. Measurements by x-ray microanalysis of the ionic composition of cells in the cortex, the endodermis, and the stele have shown that the endodermis is indeed an effective barrier, e.g. to Na+ (Pitman et al., 1981; Karahara et al., 2004). The key function of the endodermis in selective uptake of nutrients is indicated by the presence of high amounts of plasma membrane H+-ATPase as detected by immunological methods (Parets-Soler et al., 1990). At least one H+-ATPase isoform is specific for these cells as shown by promoter-GUS (β-glucuronidase) reporter expression of the plasma membrane H+-ATPase isoform AHA4 (Vitart et al., 2001).
Figure 2.
Overview of cells and organs in the plant body where intense nutrient transport takes place. Plasma membrane H+-ATPase is always active at interfaces between the living symplast and the dead apoplast. Nutrient transporters can be either channel proteins or carrier proteins.
The Role of Root Hairs for Nutrient Uptake
The epidermis constitutes the outermost layer of root cells, and the individual cells are equipped with thin protrusions, root hairs, that serve to increase the area of the plant-soil interface. Root hairs may constitute more than 60% of the surface area of the root. Using vibrating microelectrodes, it has been possible to detect large H+ currents around root hairs (for review, see Palmgren, 2001). These are most likely generated by the plasma membrane H+-ATPase. In support of this hypothesis, it has been shown that root epidermal cells show a high concentration of immunodetectable plasma membrane H+-ATPase (Samuels et al., 1992; Jahn et al., 1998).
Many genes encoding transport proteins are expressed in the root hairs, and root hairs of the model plant Arabidopsis have been shown to influence the uptake of nutrients such as phosphate (Bates and Lynch, 2000) and K+ (Jungk, 2001). These lines of evidence indicate that root hairs are central entry pathways for nutrients into the plant. A number of Arabidopsis mutants have been described that exhibit altered root hair morphology. Examples are the trh1-1 mutant (a knockout of the AtKT3/KUP4 K+ transporter), which has a “tiny root hairs” phenotype (Rigas et al., 2001), and rhd6, which is nearly devoid of root hairs (Masucci and Schiefelbein, 1994). By comparing these mutants with wild-type plants, it has been demonstrated that there is a positive correlation between the number of root hairs and growth on low K+ (Ahn et al., 2004). However, since the poor growth of these mutants on low K+ might also be correlated with a lack of specific K+ transport systems in the root periphery, more experiments are required to establish the role of root hairs for plant nutrition.
Xylem Loading: The Role of Pericycle and Xylem Parenchyma Cells
Once taken up into the root symplast, nutrients must be released into the dead xylem elements for their long-distance transport, with the transpiration stream to the shoot. David Clarkson proposed that ion secretion into the xylem vessels occurs across the plasma membrane of the living cells bordering the xylem (Clarkson, 1993), consisting of pericycle cells and xylem parenchyma (Fig. 1).
Many transporters have been localized to the root pericycle, and in several cases it has been shown that they are involved in xylem loading of nutrients. Some examples, such as the SKOR, BOR1, and SOS1 transporters, are illustrative. The SKOR gene encodes an outward-rectifying K+ channel in the plasma membrane that is expressed in the root pericycle and stelar parenchyma cells (Gaymard et al., 1998). Its disruption by a transferred DNA results in strongly decreased K+ translocation toward the shoots. The Arabidopsis BOR1 gene encodes a boron transporter that, when fused to the fluorescent reporter green fluorescent protein, can be detected in the pericycle and at the inner side of the endodermis (Takano et al., 2002). Mutant plants deficient in BOR1 have a strongly changed root-to-shoot ratio of boron. The Na+/H+ antiporter SOS1 is expressed in the stelar parenchyma cells, and mutants are affected in long-distance transport of Na+, presumably via the xylem (Shi et al., 2002).
The pH of the xylem sap is acidic (pH 5.0–6.5; Fisher, 2000), which is in accordance with proton extrusion from xylem parenchyma and pericycle cells. The membrane potential of these cells has not been measured so far. However, assuming that the membrane potential is negative on the inside due to the activity of the plasma membrane H+-ATPase, this raises the question as to how positively charged cations, such as K+, can leave the cells by channel-mediated passive transport. Xylem K+ concentrations are typically around 5 mm in contrast to cytoplasmic K+ concentrations, which vary between 50 and 150 mm. Thus, the electrochemical gradient for K+ might still be in favor of K+ efflux into the xylem (Wegner and De Boer, 1997). For divalent cations, however, it might be required to energize xylem loading. The plasma membrane-localized Zn2+ pumps HMA2 and HMA4 are present in vascular tissues (Hussain et al., 2004). In a double mutant, where both genes encoding these pumps are disrupted, Zn2+ accumulate in the root, whereas shoots show Zn2+ deficiency symptoms. This would suggest that these pumps are indeed involved in xylem loading of Zn2+ in the root.
Xylem Unloading: The Role of Vessel-Associated Cells
When nutrients have arrived via the apoplastic transpiration stream to the aerial parts of the plant, they can again be taken up into the symplast by leaf cells. En route, the xylem parenchyma cells, bordering the dead tracheary elements of the stem xylem, seem to play a function in the reabsorption of minerals and nutrients from the xylem sap. In some species, xylem parenchyma cells can reabsorb minerals, such as nitrate, potassium, and sodium, when the root supply is abundant, and the same cells can release the minerals back into the xylem sap in periods of mineral deficiency. A Mg2+/H+ cotransporter has been localized to the vacuolar membrane of Arabidopsis xylem parenchyma cells and may play a role in such homeostatic processes (Shaul et al., 1999). Reabsorption of nutrients is structurally reflected in many plants by xylem transfer cells showing numerous plasma membrane invaginations toward the tracheary elements.
Interestingly, at least in walnut, vessel-associated cells (VACs) seem to be very rich in plasma membrane H+-ATPase (Alves et al., 2004). This enzyme might thus play a role in energizing nutrient reabsorption from the xylem sap via VACs. VACs are a special class of xylem parenchyma cells surrounding xylem vessels present in many species. They are typically small cells characterized by a dense cytoplasm, numerous small vacuoles, and many mitochondria and are associated with numerous large pits to neighboring xylem vessels. On the other hand, VACs are connected to ordinary xylem parenchyma cells by numerous plasmodesmata, which constitute a pathway for reabsorbed nutrients into neighboring cells.
Phloem Loading: The Role of Companion Cells
Products of photosynthesis and metabolism have to be transported to other parts of the plants. This transport, which takes place from source tissues, such as leaves, to sink tissues, such as other leaves, fruits, and roots, utilizes the other long-distance transport pathway, the phloem. Phloem transport is dependent on the osmotic generation of pressure in leaves. This pressure is affected by active uptake of sugars across the plasma membrane of phloem cells in many plant species. Not only metabolites, such as sugars and amino acids, but also minerals and salts use the phloem pathway. For example, under phosphate deficiency, phosphate is redistributed from the old, fully expanded source leaves toward young, expanding sink leaves, a process requiring phosphate export to phloem sieve tubes.
The phloem is strongly decorated when probed with antibodies against proton pumps (Parets-Soler et al., 1990). In Arabidopsis, an epitope-tagged AHA3 plasma membrane H+-ATPase has been detected in the phloem (Dewitt and Sussman 1995) in the same cell type where an AHA3 promoter-GUS fusion is active (DeWitt et al., 1991).
Two modes of phloem loading have been identified in the minor veins of leaves of dicotyledonous plants. The mechanism of phloem loading is either symplastic or apoplastic. The two mechanisms differ with respect to (1) the plasmodesmal connections between phloem and surrounding cells and with (2) the morphology of companion cells. Apoplastic loaders have virtually no or only a few plasmodesmata between sieve-element/companion cell complexes and surrounding cells and often have companion cells in minor veins that resemble transfer cells. The transfer cells possess cell wall ingrowths, varying in surface area with the transit of photosynthate, and unfragmented vacuoles. Companion cells in minor veins of symplastic loaders have no cell wall protrusions and are connected with the surrounding cells by numerous plasmodesmata (for review, see Schulz, 1998).
Transfer cells of minor veins are rich in plasma membrane H+-ATPase (Bouche-Pillon et al., 1994). This finding supports the notion that this enzyme is energizing the loading of nutrients into these cells. However, localization of specific H+-ATPase isoforms to transfer cells has not been demonstrated so far.
Phloem Unloading and Postphloem Transport in Fruits and Developing Seeds: The Role of Transfer Cells
Fruits and seeds accumulate massive amounts of salts and organic compounds, and this process is tightly controlled and has to be energized. Nutrients to supply seed growth arrive through the phloem and leave the sieve tubes through plasmodesmata. Postphloem transport is mostly symplastic up to the interface between maternal and filial tissues. There are never plasmodesmal contacts across this interface. Nutrients are accordingly secreted into the seed apoplast and actively taken up by filial cells, which are enriched in transporters and H+-ATPases. In some species, both the cells releasing the nutrient and those taking them up are modified to a transfer cell morphology (Patrick and Offler, 2001; Thompson et al., 2001). Increase in membrane surface of the seed transfer cells was calculated to be up to 20-fold, compared to a typical cell of the same size. The membrane potential of seed coat transfer cells is in the range of −150 to −200 mV, as expected for transport-specialized cells (Offler et al., 2003). An Arabidopsis plasma membrane H+-ATPase isoform, AHA10, has been localized to developing seeds as the result of a promoter-GUS fusion analysis (Harper et al., 1994). The cell-specific localization of this isoform, however, remains unclear.
In fruits, such as apples and berries, the phloem unloading process is less studied, but in several cases it seems clear that an apoplastic step is involved (Wang et al., 2003; Zhang et al., 2004). The unloading monosaccharides from the phloem to the fruit flesh of apple pass through the sieve element-companion cell complex (Fig. 2) and into the parenchyma cells. Several plasmodesmata were observed in the sieve element-companion cell complex and between parenchyma cells but not between the companion and parenchyma cells, indicating an apoplastic pathway for loading of sugars into apple fruit (Zhang et al., 2004). The plasma membrane H+-ATPase has been detected in the sieve element-companion cell complex of vascular bundles in sepals feeding fruit flesh (Zhang et al., 2004). This would suggest that the phloem unloading process requires energy that is supplied by this pump.
Seed Germination: Mobilization of Stored Energy
When seeds germinate, they mobilize stored energy (fats and proteins) in the endosperm and release it in the form of sugars (Suc) and amino acids to the apoplast. From the apoplast it has to be transported into the phloem of the young cotyledon. Plasma membrane H+-ATPase has been shown to be involved in this process by supplying the electrochemical gradient used by the H+/Suc cotransporter (for review, see Williams et al., 2000). Recent studies in castor bean (Ricinus communis) cotyledons show that plasma membrane H+-ATPase is strongly expressed in the epidermis and the phloem, indicating that uptake of nutrients at this location takes place during initial development (Williams and Gregory, 2004).
OSMOREGULATION OF CELL SIZE
In many specialized cells, the primary role of active transport is not to allow nutrient uptake but rather to control water fluxes. These cells are small osmotic machines used to force plant movements. When absorbing water, they swell and shrink when water leaves the cells. To control water uptake and release, these cells use active transport to control their salt concentration. When the concentration of salt inside is high, the water concentration is lowered accordingly and will permit entry of water by water channels. This mechanism applies to stomatal guard cells and to pulvinar motor cells, which allow movement of leaves in various plants.
Opening of the Stomatal Pore: The Role of Guard Cells
The opening of stomata is mediated by an accumulation of K+ in guard cells. K+ accumulation is driven by an inside-negative electrical potential across the plasma membrane (for review, see Dietrich et al., 2001). This electrical potential is created by H+-ATPases in the plasma membrane. A fungal phytotoxin fusicoccin, an activator of plasma membrane H+-ATPase, produced by Fusicoccum amygdali, causes irreversible activation of H+ pump in guard cells and irreversible stomatal opening, which results in wilting of leaves and, eventually, the death of trees. The plasma membrane H+-ATPase in guard cells is regulated physiologically by blue light. Blue light is absorbed by PHOT1 and PHOT2, which are blue-light receptors with protein kinase activity (Kinoshita et al., 2001). Once activated, these proteins, most likely indirectly, cause phosphorylation of the plasma membrane H+-ATPase at its penultimate residue. Phosphorylation of the H+-ATPase alone is not enough to activate H+ pumping, as subsequent binding of 14-3-3 proteins is also needed (Kinoshita and Shimazaki, 2002; see below).
REGULATION OF PLASMA MEMBRANE H+-ATPase ACTIVITY
Small changes in pump activity are thought to be important for many aspects of plant growth and development. For example, many studies have found changes in pump activity in response to a variety of environmental conditions, including salt stress, hormones, light, and pathogens (for review, see Palmgren, 2001; Arango et al., 2002). Although blue light activates the H+-ATPase in guard cells, it has been shown to inactivate plasma membrane H+-ATPase activity of pulvinar motor cells of common bean (Phaseolus vulgaris; Okazaki, 2002). Auxin is involved in cell growth, but the mechanism is not entirely resolved. Plant cells treated with auxin excrete proton resulting in apoplastic acidification. This acidification provides a favorable condition for cell wall loosening, which could be an early step in auxin-induced cell expansion. Different results have been obtained in solving how auxin activates the proton pump at the transcriptional level, but the fact that plant cells respond to auxin within minutes suggests that H+-ATPases are directly activated at the posttranslational level. In rice, ABP57 is able to bind auxin and activate purified plasma membrane H+-ATPase, at least in vitro (Kim et al., 2001).
The C terminus of H+-ATPases acts as an autoinhibitory domain regulating enzyme activity (for review, see Palmgren, 2001). Removal of the C terminus leads to an activation of H+-ATPase activity, and, on this basis, it was hypothesized that the C terminus is a target for physiological factors activating or inactivating proton pumps (Palmgren et al., 1991). The role of 14-3-3 proteins in regulation of plant plasma membrane H+-ATPases is well studied. The 14-3-3 proteins are regulatory proteins found in all eukaryotic systems and generally bind to sequence motifs, including a phosphorylated Ser or Thr residue. In plasma membrane H+-ATPase, 14-3-3 proteins bind to a phosphorylated Thr, the penultimate residue in the C-terminal regulatory domain. The effect of 14-3-3 protein binding might be to displace the C-terminal domain leading to activation of enzyme activity.
The fungal toxin fusicoccin stabilizes complex formation between H+-ATPase and 14-3-3 protein even in the absence of regulatory phosphorylation. A crystal structure has been solved for the protein complex consisting of 14-3-3 proteins, fusicoccin, and a peptide of the consensus extreme C terminus of plant H+-ATPase (Wurtele et al., 2003). The structure reveals that fusicoccin binding to 14-3-3 protein strongly increases the binding affinity for the peptide.
Phosphorylation and dephosphorylation of Ser/Thr residue in the C terminus of H+-ATPases have in several experiments been shown to affect the activity of the pump. Oat plasma membrane H+-ATPase is activated when phosphorylated at Ser/Thr residues by a Ca2+-dependent plasma membrane kinase (Schaller et al., 1992), and in tomato, the activity of the plasma membrane H+-ATPase increases when dephosphorylated by a membrane-bound phosphatase (Vera-Estrella et al., 1994). Recently, other phosphorylation sites have been identified in a screening for phosphorylated membrane proteins in planta (Nuhse et al., 2003), indicating that regulation through the C terminus of H+-ATPase is a correlation between numerous kinases and phosphatases that still remains to be identified.
FUTURE PERSPECTIVES
We are beginning to get a picture of how the flow of nutrients into and within the plant body is energized and regulated by plasma membrane H+-ATPases. However, we know the tissue-specific localization of only a few H+-ATPase isoforms and their physiological roles have proven difficult to analyze, as no phenotypes of H+-ATPase knockouts have been reported so far. A rigorous analysis of each individual member of the plasma membrane H+-ATPase family in a given organism is required before we can conclude about their physiological role in nutrient uptake and translocation.
This work was supported by the European Union Framework 6 program.
References
- Ahn SJ, Shin R, Schachtman DP (2004) Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 134: 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves G, Ameglio T, Guilliot A, Fleurat-Lessard P, Lacointe A, Sakr S, Petel G, Julien JL (2004) Winter variation in xylem sap pH of walnut trees: involvement of plasma membrane H+-ATPase of vessel-associated cells. Tree Physiol 24: 99–105 [DOI] [PubMed] [Google Scholar]
- Arango M, Gevaudant F, Oufattole M, Boutry M (2002) The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216: 355–365 [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46: 84–101 [DOI] [PubMed] [Google Scholar]
- Bates TR, Lynch JP (2000) Plant growth and phosphorus accumulation of wild type and two root hair mutants of Arabidopsis thaliana (Brassicaceae). Am J Bot 87: 958–963 [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132: 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche-Pillon S, Fleurat-Lessard P, Fromont JC, Serrano R, Bonnemain JL (1994) Immunolocalization of the plasma membrane H+-ATPase in minor veins of Vicia faba in relation to phloem loading. Plant Physiol 105: 691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT (1993) Roots and the delivery of solutes to the xylem. Philos Trans R Soc Lond B Biol Sci 341: 5–17 [Google Scholar]
- DeWitt ND, Harper JF, Sussman MR (1991) Evidence for a plasma membrane proton pump in phloem cells of higher plants. Plant J 1: 121–128 [DOI] [PubMed] [Google Scholar]
- DeWitt ND, Sussman MR (1995) Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell 7: 2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich P, Sanders D, Hedrich R (2001) The role of ion channels in light-dependent stomatal opening. J Exp Bot 52: 1959–1967 [DOI] [PubMed] [Google Scholar]
- Fisher DB (2000) Long-distance transport. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants, Ed 1. American Society of Plant Physiologists, Rockville, MD, pp 730–784
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferriere N, Thibaud JB, Sentenac H (1998) Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Harper JF, Manney L, Sussman MR (1994) The plasma membrane H+-ATPase gene family in Arabidopsis: genomic sequence of AHA10 which is expressed primarily in developing seeds. Mol Gen Genet 244: 572–587 [DOI] [PubMed] [Google Scholar]
- Harrington GN, Franceschi VR, Offler CE, Patrick JW, Tegeder M, Frommer WB, Harper JF, Hitz WD (1997) Cell specific expression of three genes involved in plasma membrane sucrose transport in developing Vicia faba seed. Protoplasma 197: 160–173 [Google Scholar]
- Hoagland DR (1944) Lectures on the Inorganic Nutrition of Plants. Chronica Botanica, Waltham, MA
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T, Baluska F, Michalke W, Harper JF, Volkmann D (1998) Plasma membrane H+-ATPase in the root apex: evidence for strong expression in xylem parenchyma and asymmetric localization within cortical and epidermal cells. Physiol Plant 104: 311–316 [Google Scholar]
- Jungk A (2001) Root hairs and the acquisition of plant nutrients from soil. J Plant Nutr Soil Sci 164: 121–129 [Google Scholar]
- Karahara I, Ikeda A, Kondo T, Uetake Y (2004) Development of the Casparian strip in primary roots of maize under salt stress. Planta 219: 41–47 [DOI] [PubMed] [Google Scholar]
- Kim YS, Min JK, Kim D, Jung J (2001) A soluble auxin-binding protein, ABP57. Purification with anti-bovine serum albumin antibody and characterization of its mechanistic role in the auxin effect on plant plasma membrane H+-ATPase. J Biol Chem 276: 10730–10736 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (2002) Biochemical evidence for the requirement of 14-3-3 protein binding in activation of the guard-cell plasma membrane H+-ATPase by blue light. Plant Cell Physiol 43: 1359–1365 [DOI] [PubMed] [Google Scholar]
- Kochian LV, Lucas WJ (1983) Potassium-transport in corn roots. The significance of the root periphery. Plant Physiol 73: 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W (2004) Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol 5: 282–295 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhse TS, Stensballe A, Jensen ON, Peck SC (2003) Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics 2: 1234–1243 [DOI] [PubMed] [Google Scholar]
- Offler CE, McCurdy DW, Patrick JW, Talbot MJ (2003) Transfer cells: cells specialized for a special purpose. Annu Rev Plant Physiol Plant Mol Biol 54: 431–454 [DOI] [PubMed] [Google Scholar]
- Okazaki Y (2002) Blue light inactivates plasma membrane H+-ATPase in pulvinar motor cells of Phaseolus vulgaris L. Plant Cell Physiol 43: 860–868 [DOI] [PubMed] [Google Scholar]
- Palmgren MG (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C (1991) Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem 266: 20470–20475 [PubMed] [Google Scholar]
- Parets-Soler A, Pardo JM, Serrano R (1990) Immunocytolocalization of plasma membrane H+-ATPase. Plant Physiol 93: 1654–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JW, Offler CE (2001) Compartmentation of transport and transfer events in developing seeds. J Exp Bot 52: 551–564 [PubMed] [Google Scholar]
- Pitman MG, Lauchli A, Stelzer R (1981) Ion distribution in roots of barley seedlings measured by electron-probe X-ray-microanalysis. Plant Physiol 68: 673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, Dolan L, Hatzopoulos P (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels AL, Fernando M, Glass ADM (1992) Immunofluorescent localization of plasma membrane H+-ATPase in barley roots and effects of K nutrition. Plant Physiol 99: 1509–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Harmon AC, Sussman MR (1992) Characterization of a calcium- and lipid-dependent protein kinase associated with the plasma membrane of oat. Biochemistry 31: 1721–1727 [DOI] [PubMed] [Google Scholar]
- Schikora A, Schmidt W (2002) Formation of transfer cells and H+-ATPase expression in tomato roots under P and Fe deficiency. Planta 215: 304–311 [DOI] [PubMed] [Google Scholar]
- Schulz A (1998) The phloem. Structure related to function. Prog Bot 59: 429–475 [Google Scholar]
- Serrano R, Kielland-Brandt MC, Fink GR (1986) Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 319: 689–693 [DOI] [PubMed] [Google Scholar]
- Shaul O, Hilgemann DW, de-Almeida-Engler J, Van Montagu M, Inz D, Galili G (1999) Cloning and characterization of a novel Mg2+/H+ exchanger. EMBO J 18: 3973–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, Hayashi H, Yoneyama T, Fujiwara T (2002) Arabidopsis boron transporter for xylem loading. Nature 420: 337–340 [DOI] [PubMed] [Google Scholar]
- Thompson RD, Hueros G, Becker H, Maitz M (2001) Development and functions of seed transfer cells. Plant Sci 160: 775–783 [DOI] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, Higgins VJ, Blumwald E (1994) Plant defense response to fungal pathogens (activation of host-plasma membrane H+-ATPase by elicitor-induced enzyme dephosphorylation). Plant Physiol 104: 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitart V, Baxter I, Doerner P, Harper JF (2001) Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis. Plant J 27: 191–201 [DOI] [PubMed] [Google Scholar]
- Wang ZP, Deloire A, Carbonneau A, Federspiel B, Lopez F (2003) An in vivo experimental system to study sugar phloem unloading in ripening grape berries during water deficiency stress. Ann Bot (Lond) 92: 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner LH, De Boer AH (1997) Properties of two outward-rectifying channels in root xylem parenchyma cells suggest a role in K+ homeostasis and long-distance signaling. Plant Physiol 115: 1707–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LE, Gregory A (2004) Changes in the expression pattern of the plasma membrane H+-ATPase in developing Ricinus communis cotyledons undergoing the sink/source transition. Planta 218: 562–568 [DOI] [PubMed] [Google Scholar]
- Williams LE, Lemoine R, Sauer N (2000) Sugar transporters in higher plants—a diversity of roles and complex regulation. Trends Plant Sci 5: 283–290 [DOI] [PubMed] [Google Scholar]
- Wurtele M, Jelich-Ottmann C, Wittinghofer A, Oecking C (2003) Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J 22: 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Peng YB, Pelleschi-Travier S, Fan Y, Lu YF, Lu YM, Gao XP, Shen YY, Delrot S, Zhang DP (2004) Evidence for apoplasmic phloem unloading in developing apple fruit. Plant Physiol 135: 574–586 [DOI] [PMC free article] [PubMed] [Google Scholar]