Abstract
Transcriptome analysis, using Affymetrix ATH1 arrays and a real-time reverse transcription-PCR platform for >1,400 transcription factors, was performed to identify processes affected by long-term nitrogen-deprivation or short-term nitrate nutrition in Arabidopsis. Two days of nitrogen deprivation led to coordinate repression of the majority of the genes assigned to photosynthesis, chlorophyll synthesis, plastid protein synthesis, induction of many genes for secondary metabolism, and reprogramming of mitochondrial electron transport. Nitrate readdition led to rapid, widespread, and coordinated changes. Multiple genes for the uptake and reduction of nitrate, the generation of reducing equivalents, and organic acid skeletons were induced within 30 min, before primary metabolites changed significantly. By 3 h, most genes assigned to amino acid and nucleotide biosynthesis and scavenging were induced, while most genes assigned to amino acid and nucleotide breakdown were repressed. There was coordinate induction of many genes assigned to RNA synthesis and processing and most of the genes assigned to amino acid activation and protein synthesis. Although amino acids involved in central metabolism increased, minor amino acids decreased, providing independent evidence for the activation of protein synthesis. Specific genes encoding expansin and tonoplast intrinsic proteins were induced, indicating activation of cell expansion and growth in response to nitrate nutrition. There were rapid responses in the expression of many genes potentially involved in regulation, including genes for trehalose metabolism and hormone metabolism, protein kinases and phosphatases, receptor kinases, and transcription factors.
Nitrogen (N) is the most important inorganic nutrient in plants and a major constituent of proteins, nucleic acids, many cofactors, and secondary metabolites (Marschner, 1995). N affects all levels of plant function, from metabolism to resource allocation, growth, and development (Crawford, 1995; Marschner, 1995; Stitt and Krapp, 1999). Addition of nitrate (NO3−) induces genes involved in NO3− uptake and reduction, and the production of organic acids to act as acceptors and counter anions (Scheible et al., 1997a, 2000; Amarasinghe et al., 1998; Wang et al., 2000, 2001, 2003). Genes are induced in the oxidative pentose phosphate pathway to provide reducing equivalents for NO3− assimilation (Scheible et al., 1997a; Wang et al., 2000, 2003). NO3− addition also modifies resource allocation, growth, and development, by modulating shoot-root allocation (Scheible et al., 1997b; Stitt and Krapp, 1999) and lateral root growth (Zhang and Forde, 1998; Zhang et al., 1999), accelerating senescence, and promoting flowering and tuber initiation (Marschner, 1995).
Little is known about the molecular basis of NO3− sensing and signaling or, more generally, regulatory responses triggered by metabolites further downstream in N metabolism (Stitt, 1999; Stitt and Krapp, 1999; Stitt et al., 2002). Some responses, including those affecting NO3− uptake and reduction, organic acid metabolism (Crawford, 1995; Scheible et al., 1997a, 2000), and root architecture (Scheible et al., 1997b; Zhang and Forde, 1998; Zhang et al., 1999), may be triggered by NO3− itself. Plants contain putative homologs to components of microbial N signaling components, including the PII protein (Hsieh et al., 1998; Smith et al., 2003) and NIT2 (see Stitt and Krapp 1999), but their precise role has not been clarified. Several plant-specific genes, including ANR1 (Zhang and Forde, 1998; Zhang et al., 1999) as well as ABI4 and ABI5 (Signora et al., 2001), have been implicated in the response of lateral roots to NO3−. NO3− induction of IPT3 in the roots leads to increased synthesis and export of cytokinins to the shoot (Sakakibara et al., 1998). Transcript profiling has identified several uncharacterized transcription factors, protein kinases, and protein phosphatases that are induced or repressed by NO3− (Wang et al., 2003).
The breadth of response to NO3− makes it a rich but challenging area for the application of post-genomic strategies. Although platforms like expression profiling have the potential to provide a comprehensive overview of system responses, several factors currently limit their interpretation. First, genome annotation and software tools to analyze large datasets are lagging behind technical advances in the hardware for profiling. Results are typically presented as a statistical analysis of large numbers of genes where the individual players remain anonymous, or as lists of individual genes that are difficult to interpret because they do not provide an overview of the responses of all the genes in a particular functional area, which is needed to put them into a biological context. Second, some important classes of genes may not be reliably measured on conventional arrays (Czechowski et al., 2004). Third, changes in transcript levels do not, on their own, provide evidence that the encoded proteins have changed, let alone that the relevant metabolic pathway or biological process has been affected.
A recent analysis using ATH1 arrays revealed numerous genes and processes that respond rapidly to NO3− in Arabidopsis (Wang et al., 2003). We have extended the analysis by combining ATH1 arrays with a novel resource for real-time reverse transcription (RT)-PCR profiling of >1,400 putative transcription factor genes (Czechowski et al., 2004) and metabolite analyses to analyze the response to N deprivation and the response to readdition of NO3−. The results were interpreted using MapMan (Thimm et al., 2004), a new software tool that displays responses of genes onto diagrams of biological function, and by detailed searches for specific correlations and associations.
RESULTS AND DISCUSSION
Physiological and Metabolic Responses to N Deprivation and Nitrate Readdition
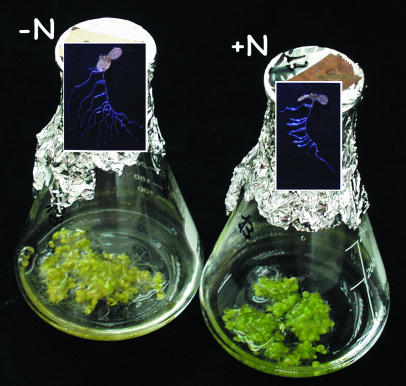
Arabidopsis seedlings were grown in liquid culture with low levels of Suc in the medium and continuous light to minimize diurnal changes in carbohydrate and N metabolism (Scheible et al., 2000; Matt et al., 2001a, 2001b), which would otherwise complicate interpretation of experimental data (see “Materials and Methods”). After 7 d on full N, some cultures were shifted to low N by changing the medium. Other cultures remained in N-replete conditions. On day 9, all seedlings (+N and −N) had developed cotyledons and first leaves (Fig. 1). N-starved seedlings exhibited the typical phenology of N-limited plants, including reduced chlorophyll, accumulation of anthocyanins in the leaves, and pronounced root and especially lateral root growth (Fig. 1; data not shown). N-replete seedlings and some batches of N-starved seedlings were harvested to investigate transcript and metabolite levels in full nutrition and N depletion. Some batches of N-starved seedlings received 3 mm KNO3 and were harvested 30 min and 3 h later. At both time points, two controls were harvested corresponding to N-starved seedlings that received 3 mm KCl and N-starved seedlings where the flask was opened and reclosed without making an addition.
Figure 1.
Phenology of 9-d-old N-limited and N-replete Arabidopsis seedlings grown in sterile liquid culture. Seedlings were grown for 7 d in full nutrients and were then transferred to low N (−N) or maintained in full nutrients (+N) for another 2 d.
After 2 d, NO3− was below the detection limit in the medium and the seedlings in the low-N cultures. The seedlings contained high levels of sugars and starch (Supplemental Fig. 1, available at www.plantphysiol.org), slightly increased malate (data not shown), 2- to 3-fold higher 2-oxoglutarate, 50-fold lower Gln, and 6-fold lower Glu compared to seedlings in full nutrient medium. 2-Oxoglutarate is the immediate acceptor, and Gln and Glu are the two first amino acids produced after assimilation of NO3−. After adding 3 mm NO3−, NO3− increased in the seedlings within 10 min (data not shown), reaching >1 μmol g fresh weight (FW)−1 after 30 min (Supplemental Fig. 1). Gln, Glu, starch, sugars, 2-oxoglutarate, and medium pH remained unchanged after 30 min. Changes of transcripts at this time will reveal rapid responses to NO3−. After 3 h, Gln and Glu increased by 6- and 2-fold, respectively, 2-oxoglutarate decreased by approximately 40%, and medium pH increased by approximately 0.2 units (Supplemental Fig. 1), as a consequence of NO3− uptake. Changes in expression at this time will include slower responses to NO3− and changes triggered by secondary events. Suc, reducing sugars, and starch were unaltered after 3 h (Supplemental Fig. 1) but started to decline from 8 h onward (data not shown).
Two independent experiments were carried out at an interval of 2 months. There was very good agreement between the two experiments, with only a small number of genes showing fluctuations. In Supplemental Figure 2, A and B, the expression values and the linear regression (R2 = 0.923; with all signals called absent included) are shown for the two biological replicates of N-starved seedlings resupplied with 3 mm KNO3 for 30 min. Of the 22,750 ATH1 probe sets, 98.6% yielded gene expression ratios between 0.5 and 2, and only 17 probe sets gave ratios >5 or <0.2. Expression levels in controls that were provided with 3 mm KCl for 30 min and 3 h were virtually identical with those in N-starved seedlings (Supplemental Fig. 2, C and D). In a comparison of N-starved seedlings and N-starved seedlings supplied with 3 mm KCl for 3 h, 98.4% of all ATH1 probe sets yielded gene expression ratios between 0.5 and 2, and only 22 probe sets gave ratios >5 or <0.2 (Supplemental Fig. 2, C and D), which resembles the variance between two biological replicates (see above). There were also hardly any changes of expression between N-starved seedlings at the beginning and end of the 3-h treatment (Supplemental Fig. 2, E and F). Furthermore, gene-by-gene comparisons showed that 3 mm KCl addition had virtually no effect on the genes that responded to 3 mm KNO3 (Supplemental Fig. 2, G–I). For simplicity, we present gene expression levels in full nutrition or after NO3− addition relative to the level in N-starved seedlings. Whole plants were harvested to provide an overview of all the responses in the seedlings. Organ-, tissue-, or cell-specific responses can be subsequently extracted from such datasets using published information (see below for examples).
Display in the MapMan Software
The results are presented using the data visualization tool MapMan (Thimm et al., 2004; http://gabi.rzpd.de/projects/MapMan/). The reader is encouraged to download the software package and use it to explore the data. A contact address is provided on the Web site, in case users experience difficulties in downloading or installing the package, or uploading the data files. The changes are expressed relative to those in N-deficient seedlings; the ratios in the biological replicates were averaged, converted to a log2 scale, and imported into MapMan, which converts the data values to a false color scale. Transcripts called not present are shown as gray, transcripts that change by less than a given threshold are white, transcripts that increase are blue, and transcripts that decrease are red. In the scale used for the hard-copy figures, a 2-fold change is required to produce a visible coloration, and the scale saturates at an 8-fold (=3 on a log2 scale) change. The user can change the scale and call up the Affymetrix code number, the Arabidopsis gene identifier (AGI), the gene annotation, and the extent of the change in a display box at the bottom of the screen. All the results are also deposited as an Excel spreadsheet (Supplemental Table II), to allow readers to explore the data with tools of their own choice.
Rapid Coordinated Induction of Genes for Nitrate Uptake and Assimilation
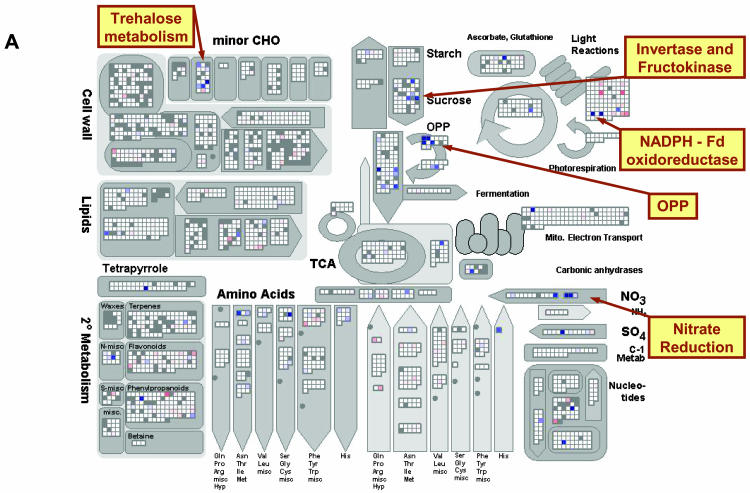
The results will first be presented for the response after NO3− addition to N-starved seedlings. There were marked changes for many genes directly involved in NO3− transport and assimilation (Fig. 2, A and B; see Supplemental Fig. 3 for BIN description; Supplemental Fig. 4). Several NO3− transporter genes were strongly induced, including NRT2.1 and NRT2.2 (At1g08090 and At1g08100, respectively), NRT2.4 (At5g60770), and to a weaker extent NRT1.1 (At1g12110). A homolog (At5g50200; Maathuis et al., 2003) of the Chlamydomonas reinhardtii NAR2 gene was also induced (see Supplemental Fig. 4A). NAR2 is required for high-affinity NO3− transport in C. reinhardtii (Zhou et al., 2000). Our results point to an analogous role in higher plants. All genes known to be directly required for NO3− assimilation were also strongly induced, including NIA1, NIA2, and NII (Fig. 3A; see also Scheible et al., 1997a; Wang et al., 2003).
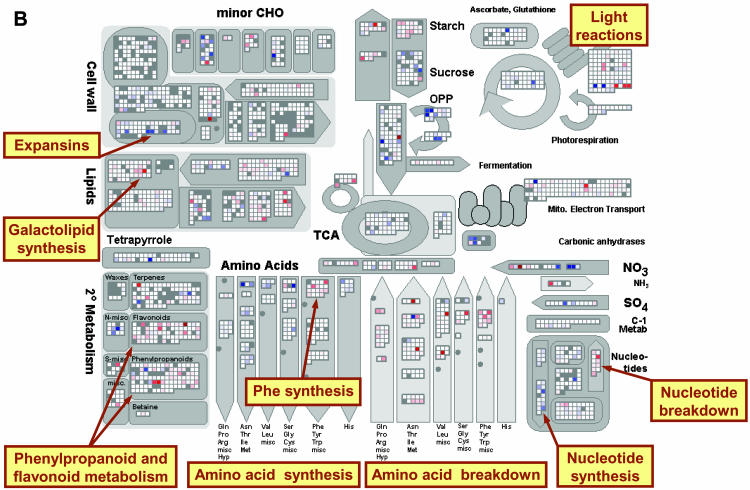
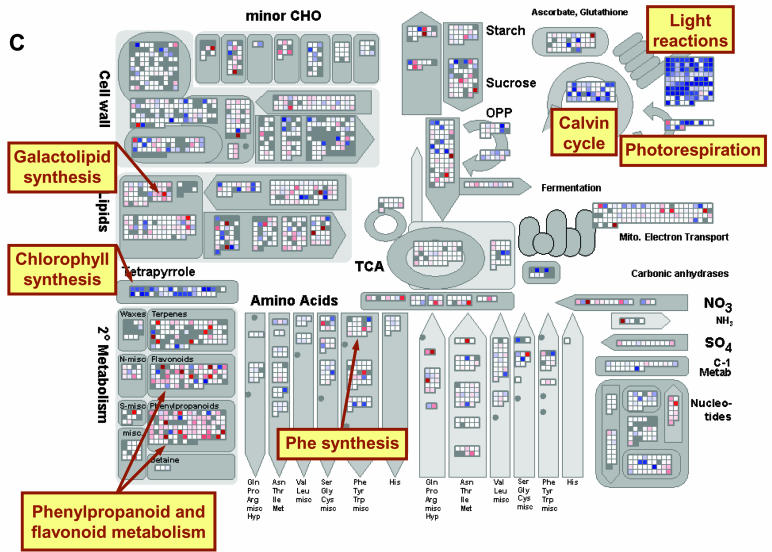
Figure 2.
Expression of genes involved in metabolism after addition of NO3−. A, Transcript levels in N-deficient seedlings 30 min after NO3− addition relative to the level in N-deficient seedlings. B, Transcript levels in N-deficient seedlings 3 h after NO3− addition relative to the level in N-deficient seedlings. C, Transcript levels in N-sufficient seedlings relative to the level in N-deficient seedlings. The experiment was carried out as described in Supplemental Figure 1 and in “Materials and Methods.” The results are the mean of two biological replicates. All results are shown on a log2 scale. The results are displayed using the MapMan software (Thimm et al., 2004), and a full description of the bins for the metabolism plot is shown in Supplemental Figure 3. Genes that are called absent by Affymetrix software are shown as gray, genes that do not change by more than a threshold value as white, and genes that increase and decrease by a increasingly intense blue and red coloration, respectively. A scale was selected in which values of 0.6 and 3 on a log2 scale gave faint and full saturation, respectively. The reader is encouraged to visit http://gabi.rzpd.de/projects/MapMan/ and download the experimental data files and MapMan software in order to explore these data interactively (see “Materials and Methods”).
Figure 3.
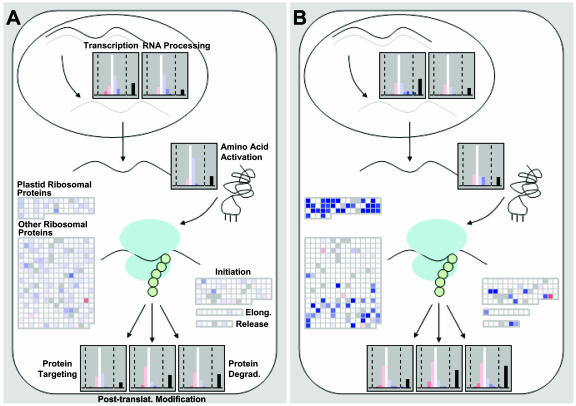
Expression of genes involved in RNA and protein synthesis. A, Three hours after adding NO3− to N-starved seedlings. B, In N-replete seedlings. The experiment was carried out and analyzed as described in the legend of Figure 2. An additional MapMan display mode was introduced that treats all the genes in a BIN as a population and shows their response as a frequency histogram. The results are shown as changes relative to the level in N-deficient seedlings. Genes whose expression changes by less than a filter value (<0.33 and >−0.33 on a log2 scale) are grouped in the central white bar. Genes that decrease or increase by 0.33 to 0.99, 0.99 to 1.66, 1.66 to 2.33, 2.33 to 3.0, and >3.0 on a log2 scale are grouped in a series of red bars at left side and blue bars at right side, respectively. Genes called absent by the Affymetrix software are grouped in a black bar on the far right side. The y axis gives the number of genes in each group (see online version). A relative scale is used on the y axis, to allow a uniformly sized plot. The display shows the responses of all genes involved in RNA synthesis (transcription), RNA processing, and protein synthesis, with the latter being subdivided into plastid ribosomal proteins, other ribosomal proteins, and initiation, elongation, and release factors. The response of gene populations implicated in protein targeting, posttranslational modification, and protein degradation is shown at the bottom of each section. Corresponding datasets for seedlings 30 min after NO3− addition are shown in Supplemental Figure 6.
There was no coordinated induction of genes for ammonium transport or assimilation. No genes assigned to ammonium transport were induced in 30 min, and one was repressed after 3 h (Supplemental Fig. 4, B and C). No members of the Gln synthetase family were induced after 30 min, and some were repressed by 3 h (Fig. 2, A and B). A NADH-dependent GOGAT was induced, but genes annotated as ferredoxin (Fd)-dependent GOGAT were unaffected (Fig. 2, A and B).
Rapid Coordinated Induction of Genes Required To Provide Reducing Equivalents
Reduction of NO3− and nitrite consumes NADH in the cytosol and reduced Fd in the plastid. In leaves in the light, photosynthesis provides the reducing equivalents. In respiratory tissues, NADH from the mitochondria is used to reduce Fd via NADPH from the oxidative pentose phosphate (OPP) pathway. NO3− rapidly repressed several genes that reduce NADH in the mitochondria (see below for details), and induced genes that are required to generate NADPH and use it to reduce Fd (Fig. 2, A and B). Within 30 min, one member of the Fd family was weakly induced, and two members of the Fd-NADPH oxidoreductase family were strongly induced. Two genes in the small gene families for Glc-6-P dehydrogenase (GPDH) and 6-phosphogluconate dehydrogenase (PGDH) were strongly induced. Two genes encoding transketolase and transaldolase, which are required in the regenerative part of the OPP pathway, were weakly induced. Their expression increased further after 3 h, along with other genes encoding OPP pathway enzymes. One member of the small gene family for phospho-Glc isomerase (PGI) was induced (Fig. 2, A and B; see also Wang et al., 2003). PGI is usually considered a glycolytic enzyme. However, this was the only gene involved in the first part of glycolysis that was induced by NO3− (Fig. 2, A and B). PGI is required in the OPP pathway when it operates at a high rate relative to the flux through glycolysis, and Fru-6-P is recycled to Glc-6-P and reenters the OPP pathway.
Rapid Coordinated Induction of Genes Required for the Synthesis of Organic Acids
NO3− assimilation requires the coordinated synthesis of organic acids, which act as acceptors for the reduced carbon and as counter anions to replace NO3− and maintain the pH balance (Scheible et al., 1997a). NO3− rapidly induced several invertases (Fig. 2, A and B; see Supplemental Fig. 3 for BIN description), two sugar transporters (Supplemental Fig. 4B), and a fructokinase (Fig. 2, A and B). None of the six-member Suc synthase family was induced. Preferential induction of invertases may reflect the high levels of sugars in N-starved seedlings (Supplemental Fig. 1). Many genes assigned to Suc synthesis were repressed following NO3− addition. Likewise, many genes involved in starch degradation were also repressed, including both genes for starch phosphorylase and several amylases and isoamylases (Fig. 2, A and B).
A set of genes encoding enzymes in the latter part of glycolysis and organic acid metabolism, which are required to synthesize malate and 2-oxoglutarate, were rapidly induced by NO3−. This was apparent after 30 min (Fig. 2A), and more marked at 3 h (Fig. 2B; see Supplemental Fig. 5 for resolution to the enzyme level). This included single members of the phosphoglycerate mutase and enolase families, two of the four members of the PPC family for phosphoenolpyruvate carboxylases, one member of the large pyruvate kinase family, and several members of the families encoding pyruvate dehydrogenases, aconitases, and isocitrate dehydrogenases. A plastid envelope 2-oxoglutarate/malate exchanger was strongly induced by NO3−, while several transporters on the mitochondrial membrane, including one annotated as a dicarboxylate carrier, were weakly induced (Supplemental Fig. 4, B and C). Both isoforms of pyruvate phosphate dikinase were repressed. In C3 plants, this enzyme is thought to shuttle carbon skeletons during N deficiency.
The response was specific for the steps leading to malate and to 2-oxoglutarate. Other genes involved in TCA cycle and in mitochondrial electron transport were not induced. Several genes encoding complex I components were weakly repressed, and a nonphosphorylating NADH dehydrogenase was strongly repressed after 3 h of exposure to NO3−. Respiration rose rapidly from 35 ± 3 to 57 ± 8 μmol O2 g FW−1 h−1 after adding 3 mm KNO3. This increase, which presumably reflects the metabolic cost of uptake and assimilation of NO3−, occurred independently of changes in transcription. The overview provided by MapMan suggests that there is a coordinate reprogramming of respiratory carbon metabolism to increase the synthesis but not the respiration of organic acids. This is achieved by specific changes in the expression of individual members of gene families.
Coordinated Induction of Synthesis and Repression of Breakdown of Amino Acids and Nucleotides
NO3− addition induced many of the genes assigned to amino acid synthesis. Some changes were apparent within 30 min (Fig. 2A) and others after 3 h (Fig. 2B). The only exception was Phe synthesis, where several genes were repressed (see below for further discussion). A reciprocal response occurred for amino acid breakdown, where many genes were repressed. Many genes annotated as amino acid transporters were repressed, indicating a role in N recycling rather than the transport of amino acids made using newly assimilated N (Supplemental Fig. 4, B and C). A specific isoform of Glu dehydrogenase (GDH3) was repressed, implicating it in the recycling of ammonium during amino acid catabolism. This isoform is normally expressed at very low levels and is not induced by sugar deprivation, which induces the other two members of the GDH family (Thimm et al., 2004).
An analogous picture emerged for nucleotide metabolism (Fig. 2, A and B). NO3− induced most of the genes required for de novo purine and pyrimidine synthesis, nucleotide salvage, and deoxynucleotide synthesis, and repressed almost all the genes assigned to nucleotide degradation. For both amino acids and nucleotides, the coordinated switch from degradation to synthesis would not be apparent from inspection of lists of affected genes because many of the changes are relatively small. It becomes striking when genes are systematically collected in functional categories.
Coordinate Induction of Genes Required for Protein Synthesis
NO3− addition had far-reaching consequences for the expression of genes involved further downstream in N use, for example, RNA and protein synthesis (Fig. 3). After 30 min, only minor changes were seen in the BINS corresponding to RNA synthesis, RNA processing, amino acid activation, and protein synthesis (Supplemental Fig. 6). After 3 h, a substantial proportion of the genes assigned to the first three categories and the vast majority of the genes assigned to protein synthesis were induced (Fig. 3A). This included genes annotated as plastid and cytosolic ribosomal proteins, and initiation, elongation, and termination (release) factors. A S6 kinase gene (At3g08720) was repressed, indicating a role in the regulation of translation in response to N depletion. Expression of genes assigned to protein targeting changed only slightly. This striking and coordinated induction of >100 genes required for RNA and protein synthesis would not be readily apparent from analyses that identify the most strongly induced genes because the individual changes in expression are not large.
Protein Synthesis Is Stimulated Independently of Changes in Minor Amino Acid Levels
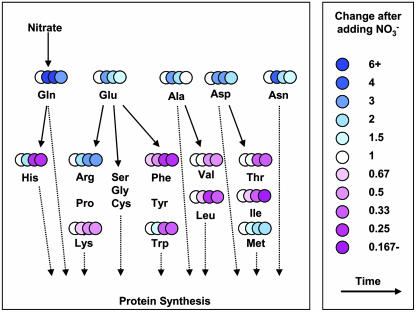
Amino acid levels were measured to provide information about the accompanying metabolic events (Fig. 4). There were no changes after 30 min, except for a slight increase of Phe. After 3 h, central amino acids like Gln, Glu, Asp, and Ala increased, Asn, Arg, and His increased, but the other minor amino acids decreased. At later time points, this decrease becomes even more marked, showing that the overall rate of protein synthesis was stimulated more strongly than the rate of minor amino acid synthesis. The transcript and metabolite data provide evidence for a highly coordinated stimulation of all processes between NO3− uptake and the use of the N for protein synthesis.
Figure 4.
Addition of NO3− leads to decreased levels of minor amino acid. The figure uses a false color scale to depict the relative level of individual amino acids. The symbols running from left to right show, for each amino acid, the change 30 min, 3 h, 8 h, and 24 h after adding NO3−, relative to the level in controls that received 3 mm KCl. The results are the mean of five biological replicates.
Cell Wall and Lipid Metabolism
There were relatively few changes in the expression of genes involved in cell wall polysaccharide synthesis in the first 3 h, but a substantial proportion of the genes for cell wall proteins were induced. Interestingly, a subset of genes for cell wall-modifying enzymes was also induced, including three members of the expansin family (Fig. 2B). A tonoplast integral protein (TIP) gene was strongly induced, and several TIPs and plasma membrane intrinsic protein genes were weakly induced (Supplemental Fig. 4C). More detailed studies and refinements of gene annotation could lead to further important insights into the regulation of cell expansion by N.
In lipid metabolism, a small subset of genes involved in fatty acid elongation and desaturation in the plastid were induced (see below for further discussion), and three genes required for galactolipid synthesis are repressed. Replacement of phospholipids by galactolipids saves phosphate during P starvation (Härtel et al., 2000; Kelly et al., 2003). Our results suggest there is an analogous adaptation in N starvation, which will save the N contained in the polar group.
Coordinated Repression of the Shikimate Pathway, and Phenylpropanoid and Flavonoid Metabolism
NO3− led to marked changes in phenylpropanoid and flavonoid metabolism, where a large number of genes were repressed and a small number induced. The changes were visible within 30 min and marked within 3 h (Fig. 2, A and B). Although the changes of individual genes were often small, the overview provided by MapMan reveals a clear coordinated response. As already noted, while NO3− led to a general induction of genes for amino acid biosynthesis, it repressed the shikimate pathway. The latter is responsible for the synthesis of Phe (Fig. 2B), which is the precursor for phenylpropanoids and contributes to the carbon skeleton of flavonoids. After NO3− addition, its expression is coordinated with that of phenylpropanoid and flavonoid metabolism rather than protein synthesis.
Plants possess large gene families for cytochrome P450s, UDP-glucosyl transferases, alcohol dehydrogenases, glucosidases, O-methyl transferases, nitrilases, cyanohydrinlyases, berberine bridge enzymes/reticuline oxidases, troponine reductase-like proteins, acetyltransferases, β-1,3-glucan hydrolases, and peroxidases. The individual members are involved in various biosynthetic and secondary pathways, but their precise function is seldom known. There are widespread changes of expression in the families (Supplemental Fig. 7). Several cytochrome P450s, a glutathione S-transferase, and a nitrilase are induced within 30 min of adding NO3− (Supplemental Fig. 7A) and remain high after 3 h (Supplemental Fig. 7B). In some families there are marked trends, for example, a substantial proportion of the UDP-glucosyl transferases are repressed, and several glucosidases are induced by 3 h. Intriguingly, NO3− also induced a small set of genes annotated as homologous to enzymes of alkaloid synthesis (Fig. 2B). Our results suggest there is broad reprogramming of secondary metabolism, including a shift away from carbon-rich secondary metabolites.
Changes of Metabolites in Phenylpropanoid and Flavonoid Metabolism
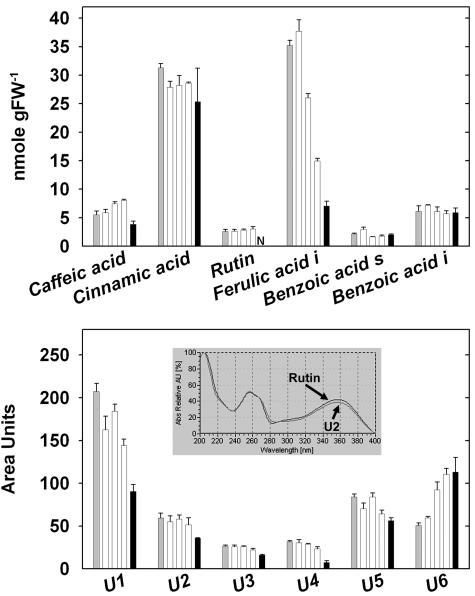
Figure 5 summarizes the impact of NO3− on phenylpropanoid and flavonoid metabolism. Metabolites located early in the pathway, like cinnamic and caffeic acid, showed only slight changes. This is consistent with the relatively small changes of transcripts of PAL, 4CL, and other genes in the initial stages of phenylpropanoid metabolism. Compared to N-replete seedlings, N-deficient seedlings contained high levels of several products, including rutin, bound ferulic acid, and several unidentified peaks, which on the basis of their elution profile and absorption spectra (e.g. Fig. 5B, insert; data not shown) are likely to be flavonoids and phenylpropanoids. After adding NO3−, relatively small changes were seen from 8 h onward. This may reflect a delay between changes of transcripts and enzyme activity, or the rate of turnover of the metabolites. Significant decreases were measured for insoluble (bound) ferulic acid, rutin, and several unidentified metabolites. A small number of metabolites increased, for example, an unidentified peak that, based on its spectrum (data not shown), probably represents a derivative of sinapinic acid (Fig. 5B, U6). This reflects the patterns of gene expression (see above) where many genes involved in phenylpropanoid and flavonoid metabolism were repressed but a few specific genes were induced.
Figure 5.
Levels of selected phenylpropanoid and flavonoid metabolites. A, Levels of known metabolites. B, Levels of unknown metabolites. The samples were collected from the same materials analyzed in Figure 2. Metabolite levels in N-starved seedlings prior to NO3− addition are shown as gray bars, those of seedlings harvested 3, 8, and 24 h (second, third, and fourth columns in each block) after addition of 3 mm KNO3 as white bars, and those of N-replete seedlings as black bars. The results are the mean ± se (n = 4 biological replicates). The levels are expressed either in nmol g FW−1 (A) or as peak area units (B). I, Insoluble; S, soluble; N, not detectable. The insert in B shows that unknown metabolite U2 has a spectrum with high similarity to the one of rutin.
Limited Response of Genes Involved in the Assimilation of Other Nutrients
There has been considerable interest in the coordination of NO3− and sulfate metabolism (e.g. Vidmar et al., 1999; Prosser et al., 2001). Several genes annotated as sulfate transporters and one gene encoding adenosyl phosphosulfate transferase were induced by NO3−. However, genes for the other three steps of the sulfate assimilation pathway were not induced, and Cys and Met biosynthesis was not preferentially induced compared to other amino acid biosynthesis pathways (see above; Fig. 2, A and B). The overview provided by MapMan highlights the danger of overinterpreting responses of individual genes. Genes annotated as phosphate transporters were unaffected by NO3− addition (Supplemental Fig. 4), but many genes involved in iron utilization, including nicotianamine synthase 1 and nicotianamine synthase 2, were strongly induced.
Redox Processes
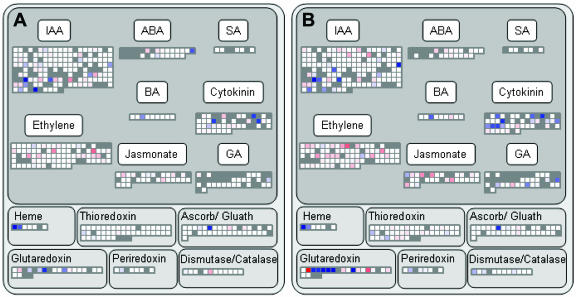
NO3− addition resulted in unexpectedly large changes in the expression of specific genes involved in redox status. Several genes encoding glutaredoxin were induced, and others repressed (Fig. 6). This was one of the most strongly responding functional categories identified by the MapMan program. These changes are of interest in view of the recent report that reactive oxygen species are involved in responses to nutrient deficiency (Shin and Schachtman, 2004). NO3− rapidly and strongly induced AtHB1 and weakly induced AtHB2, encoding nonsymbiotic hemoglobins (Fig. 6; Wang et al., 2000, 2003). Other redox-related genes were less strongly affected, with only restricted and small changes in the expression of thioredoxins, genes for ascorbate and glutathione metabolism, periredoxins, catalases, and dismutases in the first 3 h after adding NO3− (Fig. 6).
Figure 6.
Overview of changes in redox processes, hormone synthesis, and sensing after NO3− addition. A, Transcript levels in N-deficient seedlings 30 min after NO3− addition. B, Transcript levels in N-deficient seedlings 3 h after NO3− addition. The expression levels are shown relative to the one in untreated N-deficient seedlings. The experiment was carried out and analyzed as described in the legend of Figure 2. Corresponding datasets for N-replete seedlings are shown in Supplemental Figure 8.
Concerted Responses to N
Our results also reveal the difference between N-starved and N-replete seedlings. Changes in transcript levels of other genes reflected a maintained induction of nucleotide and amino acid synthesis, and repression of the respective degradative pathways in N-replete plants. These changes were associated with increased transcript levels for much of the machinery of protein synthesis in the cytoplasm and (especially) in chloroplasts (Figs. 2C and 3B).
Photosynthesis was profoundly affected. N-replete plants had substantially increased transcript levels for proteins involved in chlorophyll synthesis, photosynthetic lights reactions, the Calvin cycle, and photorespiration (Fig. 2C). This is apparent as several striking blue blocks in Figure 2C. Together with the increases in transcripts for the chloroplast protein synthesis machinery, these results point to a coordinated induction of photosynthesis in N-replete plants. Expression of other sets of genes required for photosynthesis or chloroplast biogenesis was also strongly increased. One set includes the triose phosphate translocator, the cytosolic FBPase, and individual members of the gene families for enzymes like phosphoglucoisomerase, phosphoglucomutase, and UDP-Glc pyrophosphorylase (Fig. 2C; see Supplemental Fig. 5C for a more detailed analysis at the pathway level). These genes are required for the export of photosynthate and its conversion to the immediate precursors of Suc synthesis. Another set includes several fatty acid elongases and four fatty acid desaturases involved in the plastid fatty acid synthesis pathway (Fig. 2C).
Transcript levels for many proteins involved in growth-related processes increased in N-replete seedlings, including cell wall-modification enzymes like xyloglucan endotransglycosylases, expansins, pectinesterases, and polygalacturonases (Fig. 2C). Other notable changes in the N-replete seedlings included widespread changes of transcripts for secondary metabolism (Fig. 2C) and redox metabolism (Fig. 6). There was a marked shift in the expression of genes encoding components of the mitochondrial electron transport pathway, with a shift to increased expression of complex I compared to complexes II to IV and genes involved in nonphosphorylating electron transport pathways, which is consistent with increased coupling of respiration to ATP production.
Trehalose Metabolism
The results were analyzed to uncover possible regulatory mechanisms. Ectopic overexpression of trehalose-phosphate synthase (TPS) and trehalose-phosphate phosphatase (TPP) has strong but opposite effects on starch levels and plant growth, implicating trehalose-6-P as a novel signal molecule (Leyman et al., 2001; Schluepmann et al., 2003). Of all the functional areas delimited by MapMan, trehalose metabolism is one of the most strongly affected by NO3− addition. TPS and TPP are each encoded by a small gene family. One TPS gene was induced by NO3− after 30 min and several after 3 h (Fig. 2, A and B), while another was weakly repressed. Two genes annotated as TPP were repressed (see also Wang et al., 2003). The same subset of TPS genes is induced by sugar deprivation, and the same outlier is repressed (Thimm et al., 2004). However, the magnitude of the responses differs, and the two members of the TPP family that are induced by NO3− are unaffected during sugar deprivation. These results indicate some of the changes after NO3− addition are a direct response, and others may be indirect due to changes in sugars.
Hormone Synthesis and Sensing
There were marked changes in the expression of genes involved in hormone synthesis and sensing (Fig. 6; Supplemental Fig. 8). Several genes involved in cytokinin synthesis, including IPT3 and several downstream response elements involved in cytokinin signaling, were induced within 30 min (Fig. 6A), and further genes involved in cytokinin signaling were induced by 3 h (Fig. 6B). One gene (DF4) involved in brassinosteroid synthesis and several genes involved in GA synthesis were also induced by 3 h. Many genes involved in ethylene synthesis and sensing were repressed, including a large number of 1-aminocyclopropane-1-carboxylate oxidases. These results identify transcriptional reprogramming of hormone metabolism and sensing as one of the early responses to NO3− addition.
Transcriptional Regulators
The Arabidopsis genome probably encodes >2,000 transcription factors or transcriptional regulators (Riechmann, 2002; http://www.arabidopsis.org; http://Arabidopsis.med.ohio-state.edu; Davuluri et al., 2003; http://genetics.mgh.harvard.edu/sheenweb/AraTRs.html), and new classes of transcriptional regulators (TFs) are still being discovered (e.g. LBD; Lin et al., 2003). TFs are of special interest since they are capable of coordinating the expression of several or many downstream target genes and, hence, entire metabolic and developmental pathways. Of the approximately 1,800 potential TFs on the ATH1 chip, 93 showed marked (>3-fold) changes in transcript abundance (Supplemental Table I; Supplemental Fig. 8) in response to N nutrition. Some interesting examples are depicted in Figure 7.
Figure 7.
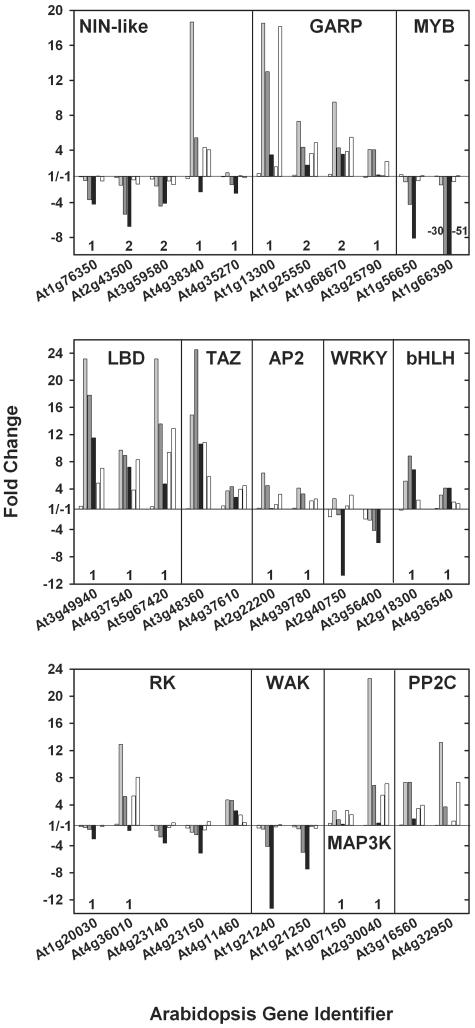
Response of selected regulatory genes to NO3− readdition. The changes in transcript abundance are shown for 22 potential transcriptional regulators from various families (top and middle), nine genes from the receptor, wall-associated, and MAPKK kinases (RK, WAK, and MAP3K, respectively) gene families, as well as two protein phosphatase 2C genes (bottom). The first, i.e. leftmost column (white) in each set of six per gene, represents the change occurring due to the addition of 3 mm KCl to N-depleted seedlings, the second (light gray) and third (dark gray) columns represent the changes after 30 min and 3 h 3 mm KNO3 addition to N-depleted seedlings, the fourth column (black) shows the fold difference in expression between full nutrient conditions and N starvation, whereas the fifth and sixth columns (both in white) are shown for comparison only and depict the changes occurring after 20 min 250 μm NO3− addition in shoots and roots, respectively, of hydroponically grown 10-d-old Arabidopsis plants (Wang et al., 2003). Numbers (1 or 2) mark genes within a given family that arose by segmental gene duplications (http://www.tigr.org/tdb/e2k1/ath1/Arabidopsis_genome_duplication.shtml). Also note the receptor kinase and WAK tandem gene arrays as indicated by the AGI. Further NO3−-responsive genes with potential functions in signaling, transcriptional, and posttranslational regulation are listed in Supplemental Table I.
The plant-specific NIN-like gene family has 16 members in Arabidopsis (Riechmann, 2002). Of the 11 on the ATH1 gene chip, 7 responded quickly and transiently to NO3− readdition (Fig. 7, top; Supplemental Table I), with At4g38340 being the most responsive. NIN-like TFs contain a domain called RWP-RK after a conserved motif at the C terminus of the domain. Interestingly, this domain is found in plant proteins involved in N-controlled development of symbiotic root nodules (Schauser et al., 1999). Many of the Arabidopsis NIN-like genes, including the five shown in Figure 7, also contain an octicosapeptide/Phox/Bem1p domain that is present in many eukaryotic signaling proteins.
Members of the G2-like (MYB-like) GARP family (Riechmann, 2002) are involved in phosphorous metabolism, abaxial cell identity, and photosynthetic development (Eshed et al., 2001; Rubio et al., 2001). Six members are transiently induced by NO3− (four are depicted in Fig. 7). In this group, At1g13300 was the most responsive. It displays root-specific expression, as does At3g25790 (Czechowski et al., 2004; compare with Table I), although expression of the latter is about 10-fold lower (Supplemental Table II). This pair arose by segmental chromosome duplication (Fig. 7). Our data reveal that their expression patterns with respect to NO3− induction and organ specificity have been preserved until the present. Hence, it is tempting to speculate that the two related genes might share considerable overlap in their biological function, as is the case for the MADS box genes SHP1 and SHP2 (Liljegren et al., 2000), SEP1 and SEP2 (Pelaz et al., 2000), and the GRAS genes GAI and RGA (Dill and Sun, 2001; King et al., 2001; see below for further discussion), which are all duplicates of single progenitor genes.
Table I.
NO3−- or N-regulated TF genes identified by real-time RT-PCR
| −N versus +N
|
N 30′ versus −N
|
MAS5 Call
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AGI | Gene | TF Family | Duplication | RT-PCR | ATH1 | RT-PCR | ATH1 | +N | −N | N 30′ |
| At1g01530a | AGL28 | MADS | 1.03 | 1.04 | 32.25 | 1.50 | A | A | A | |
| At1g02230a | NAC | At4g01540 | 0.46 | 0.95 | 10.17 | 1.12 | A | A | A | |
| At1g22130a | MADS | At1g77950 | 0.09 | 0.92 | 0.76 | 0.88 | A | A | A | |
| At1g66380a | MYB | 15.67 | 1.30 | 0.75 | 0.90 | A | A | A | ||
| At1g73870ae | COL7 | CO-like | 0.08 | 0.50 | 1.41 | 1.12 | A | A | A | |
| At1g77950a | MADS | At1g22130 | 18.27 | 1.09 | 0.07 | 1.08 | A | A | A | |
| At2g33720ae | ARP | 18.62 | 0.97 | 1.12 | 1.05 | A | A | A | ||
| At2g38340a | AP2-EREBP | 13.23 | 1.85 | 1.14 | 1.08 | A | A | A | ||
| At3g30260a | AGL8 | MADS | 0.43 | 1.15 | 10.65 | 1.05 | A | A | A | |
| At3g57920a | SPL15 | SBP | At2g42200 | 0.31 | 0.82 | 13.31 | 2.01 | A | A | P |
| At4g01460ad | bHLH | At2g46810 | 0.09 | 0.51 | 0.96 | 0.86 | A | A | A | |
| At4g17490a | AP2-EREBP | At5g47230 | 1.38 | 0.70 | 0.05 | 0.75 | A | A | A | |
| At4g17980a | NAC | At5g46590 | 2.38 | 0.82 | 10.55 | 1.50 | A | A | A | |
| At4g25490ad | CBF1 | AP2-EREBP | 0.81 | 1.09 | 34.83 | 1.51 | A | A | A | |
| At5g38800a | bZIP | 16.79 | 1.61 | 0.63 | 1.12 | A | A | A | ||
| At1g13300be | MYB | At3g25790 | 0.15 | 0.29 | 53.05 | 18.51 | P | A | P | |
| At1g56650b | PAP1 | MYB | 27.89 | 8.10 | 1.18 | 0.61 | A | P | P | |
| At1g66390b | PAP2 | MYB | 155.92 | 51.36 | 0.68 | 0.50 | A | P | P | |
| At1g68190bd | CO-like | 0.09 | 0.29 | 0.75 | 1.23 | A | A | A | ||
| At1g68520be | COL6 | CO-like | At1g25440 | 0.03 | 0.15 | 8.53 | 2.08 | P | P | P |
| At1g68670b | MYB-like | At1g25550 | 0.11 | 0.28 | 19.73 | 9.49 | P | P | P | |
| At1g68880be | bZIP | 0.12 | 0.38 | 14.96 | 4.37 | P | A | P | ||
| At1g77920b | bZIP | At1g22070 | 14.39 | 3.16 | 0.73 | 1.14 | P | P | P | |
| At2g39250bd | AP2 EREBP | At3g54990 | 0.18 | 0.67 | 21.26 | 3.05 | A | A | P | |
| At2g40750b | WRKY | 10.64 | 10.76 | 3.88 | 2.54 | A | P | P | ||
| At2g43500b | NIN-like | At3g59580 | 12.98 | 8.00 | 0.74 | 0.48 | A | P | P | |
| At2g46130b | WRKY | 10.05 | 3.43 | 2.51 | 0.91 | A | P | P | ||
| At3g25790be | MYB-like | 0.04 | 0.84 | 457.35 | 5.34 | A | A | P | ||
| At3g56400b | WRKY | 12.89 | 5.96 | 0.59 | 0.38 | P | P | P | ||
| At4g24020b | NIN-like | At1g64350 | 0.67 | 0.97 | 11.29 | 3.38 | P | P | P | |
| At4g26150b | GATA | At5g56860 | 0.51 | 1.02 | 18.84 | 2.45 | P | P | P | |
| At4g38340b | NIN-like | At4g35590 | 4.07 | 2.78 | 22.80 | 18.65 | A | A | P | |
| At5g22570b | WRKY | 18.13 | 3.90 | 3.08 | 2.25 | P | P | P | ||
| At1g02040c | SPL8 | SBP | 1.48 | 11.69 | NR | |||||
| At1g25440ce | COL16 | CO-like | At1g68520 | 0.10 | 1.61 | NR | ||||
| At2g33550c | MYB-like | 1.10 | 27.87 | NR | ||||||
| At5g10570cd | bHLH | At5g65640 | 0.08 | 0.92 | NR | |||||
| At5g65060c | MAF3 | MADS | 36.32 | 1.20 | NR | |||||
| At1g35560 | TCP | 0.73 | 0.89 | 30.76 | 1.01 | P | P | P | ||
Data for genes exhibiting a more than 10-fold ratio in transcript abundance either between −N and +N conditions or between 30 min 3 mm KNO3 readdition (N 30′) and −N conditions are presented. Gene names, TF family affiliations, and AGIs of segmental duplicated genes are indicated, if available. Data from ATH1 arrays are included for comparison. The absence (A) or presence (P) of transcripts, as determined by Affymetrix MAS5 software, in the conditions compared is indicated, as well as genes not represented (NR).
Induced or repressed transcripts detected by RT-PCR only. Note that these transcripts are usually called absent by MAS5 software.
Transcripts categorized as induced or repressed by RT-PCR and Affymetrix gene chips (≤ or ≥3-fold).
Induced or repressed transcripts detected by RT-PCR; not represented (NR) on the Affymetrix array.
Expression is preferentially in the shoot (shoot/root expression ratio >20) according to Czechowski et al. (2004) or Wang et al. (2003).
Expression is preferentially in the root (shoot/root expression ratio <0.05) according to Czechowski et al. (2004) or Wang et al. (2003).
Eleven MYB genes showed marked changes in transcript abundance following changes in N nutrition (Supplemental Table I). MYB75 (At1g56650; PAP1) and MYB90 (At1g66390; PAP2) were the most affected members of this large family, with transcript levels that plummeted 3 h after NO3− readdition to N-deprived seedlings, being reduced up to 50-fold in N-replete seedlings (Fig. 7, top). Genetic studies suggest PAP1 and PAP2 are involved in anthocyanin and flavonoid biosynthesis (Borevitz et al., 2000), where they are believed to directly act on CHS and genes further downstream in the flavonoid pathway (B. Weisshaar, personal communication). They could contribute to the changes in expression of genes in phenylpropanoid and flavonoid biosynthesis seen in Figure 2, B and C.
Large changes were observed for three members of the plant-specific Lateral Organ Boundary Domain (LBD) gene family (i.e. At5g67420, At3g49940, and At4g36010), as well as for a TAZ finger TF gene At3g48360 (Fig. 7, middle). This family has 43 members, with 37 grouped in class 1 and 6 in class 2. Class 1 includes LOB and LBD6/AS2, which are involved in lateral organ development (Shuai et al., 2002), repression of KNOX expression, and the regulation of adaxial-abaxial patterning (Lin et al., 2003). The three NO3−-responsive genes (LBD37, 38, and 39) belong to class 2. They show a rather broad organ expression pattern by RT-PCR (Shuai et al., 2002) and arose by two independent segmental duplications of chromosome 5, indicating some functional overlap (see above). None of the class 2 members has been functionally characterized to our knowledge.
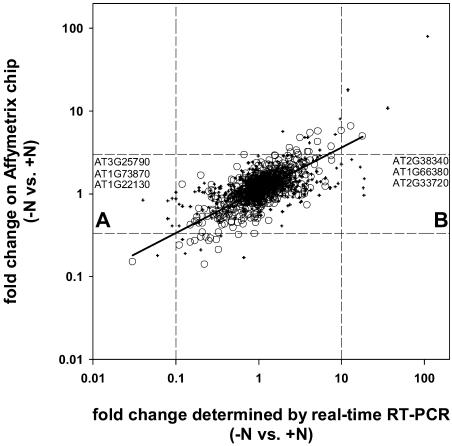
A new real-time RT-PCR platform that measures transcript levels of >1,400 TFs with extremely high sensitivity and precision (Czechowski et al., 2004) was used to confirm the results from the ATH1 arrays (Supplemental Table I; Fig. 7), to search for genes that are expressed at such low levels that they cannot be measured reliably using array technology (see Czechowski et al., 2004; Table I), and to investigate TFs that are absent from the ATH1 array. Figure 8 summarizes the response (−N versus +N) for 1,169 genes that are included in both technology platforms. Hybridization-based technologies typically underestimate changes in transcript abundance, especially for genes expressed at low levels (Holland, 2002; Horak and Snyder, 2002). We chose a real-time RT-PCR expression ratio of 10 as cutoff value for suspect genes, and one of three for ATH1 arrays, which was justified by the linear regression shown in Figure 8. Real-time RT-PCR confirmed that most TF genes did not respond strongly to N availability. Seven genes were identified as suspects with both technologies (Fig. 8) using the chosen cutoff values. Another eleven genes had ratios >10 or <0.1 with real-time RT-PCR but were <3-fold changed on the ATH1 array (Fig. 8, quadrants A and B). All these genes are depicted as cross-hairs, which identify genes called absent by Affymetrix software in at least one condition, due to a low signal. Table I lists TF genes that yielded a >10-fold change of expression as determined by real-time RT-PCR when N-replete seedlings are compared to N-starved seedlings, or when seedlings 30 min after NO3− addition are compared to N-starved seedlings. In total, 15 TF genes were revealed as NO3− responsive by RT-PCR only, 18 genes were revealed by both technologies, and 5 genes were not represented on the ATH1 array. One gene (At1g35560, encoding a TCP-domain TF) gave conflicting results: strong induction was found by real-time RT-PCR after 30 min NO3− resupply but not on ATH1 arrays even though it was called present (and the corresponding probe set appears gene specific). The result from the real-time RT-PCR platform was confirmed by analysis of more biological replicates using a different primer pair (R. Bari and W.-R. Scheible, unpublished data) and by inspection of the Stanford Microarray database (spot history for clone 143C3XP in experiments 3787, 3789, 10849, and 10851). Additional RT-PCR expression ratios for TF genes picked with Affymetrix arrays (>3-fold change; see above) are displayed in Supplemental Table I. It is apparent that data obtained with both technologies are generally consistent.
Figure 8.
Comparison of TF gene expression ratios, as determined by real-time RT-PCR and Affymetrix technology. A total of 1,169 TF genes included in both platforms are shown for a comparison of N-starved versus N-sufficient Arabidopsis seedlings. Circles and cross-hairs denote genes that are called present or absent, respectively, in replicate ATH1 arrays. Dashed lines indicate 10-fold (RT-PCR axis) or 3-fold (Affymetrix axis) changes in expression ratios. A regression line (R2 = 0.59) is shown for the 693 present genes. Quadrants A and B contain genes (see Table I) that are inconspicuous according to Affymetrix analysis but are interesting by RT-PCR.
The additional NO3−/N-responsive TF genes identified by real-time RT-PCR analysis include additional segmental-duplicated gene pairs. Some pairs show conserved expression patterns, such as the two CONSTANS-like TF genes At1g25440 and At1g68520, which both display root-specific expression (Czechowski et al., 2004; Table I) and are strongly repressed in N starvation. Expression of these and other CONSTANS-like genes (e.g. At1g73870; Table I) in roots suggests biological functions beyond promoting flowering (Griffiths et al., 2003). Another example is the two root-specific GARP-like (MYB-like) TF genes At1g13300 and At3g25790, which are induced by >50- and >400-fold in the RT-PCR screen after 30 min NO3− addition (Table I). Conserved expression of gene pairs might represent an evolutionally favored back-up system (see above). However, NO3− regulation of duplicated genes is not always preserved, e.g. the NIN-like genes AT4g38340, At1g76350, and AT4g35270 (Fig. 7) or the GATA-type TF gene At4g26150 (Table I) that was approximately 18-fold induced after 30 min NO3− addition (RT-PCR) and approximately 6-fold after 3 h (ATH1), whereas its duplicated twin At5g56860 was approximately 7-fold and merely approximately 2-fold induced, respectively. Divergence of formerly identical regulatory elements by mutation and subsequent natural selection presumably lead to new biological roles for genes/proteins derived from a single progenitor by duplication, and may be a route to increase fitness and adaptation to a given environment.
Genetic evidence shows that the MADS box gene ANR1 (At2g14210) is required to mediate changes in root architecture in response to NO3− availability. ANR1 was induced in roots within 30 min of adding NO3− to Arabidopsis seedlings (Zhang and Forde, 1998). However, ANR1 was not induced after adding NO3− in our experiments (Supplemental Table II) or those of Wang et al. (2003), and there was only an approximately 2-fold change in full nutrient conditions compared to N starvation. Our studies also failed to confirm two genes reported by Tranbarger et al. (2003; At2g18160 and At3g55770) to be reduced by approximately 50% in 10 mm KNO3-grown plants compared to Gln-supplemented plants. (Supplemental Table II; see also Wang et al., 2003). The reasons for these conflicting results are unclear. NO3− addition leads to an increase of NIA transcript in the presence of protein synthesis inhibitors (see Stitt and Krapp, 1999, for references), indicating that it can also act by modulating preexisting signaling components. It can be speculated that ANR1 might be subject to varying degrees of transcriptional and posttranslational regulation depending on yet unknown external or internal factors.
Signaling and Posttranslational Modification
Expression of several annotated receptor kinases (RK; Shiu and Bleecker, 2001), including a pair of tandemly arrayed RK genes, changed markedly after addition of NO3− to N-deprived seedlings or in full nutrients versus N starvation (Supplemental Table I; Fig. 7, bottom). The most strongly affected RK gene was At4g36010, which was induced transiently by >12-fold 30 min after NO3− addition in our system and almost 8-fold after 20 min in roots from the hydroponic system (Wang et al., 2003), while its duplicated twin At1g20030 showed a different response (Fig. 7, bottom). Interestingly, one CLAVATA3-ESR-like gene (CLE2; At4g18510) was quickly induced by NO3− addition. CLE proteins are believed to act as ligands of RKs (Cock and McCormick, 2001). A tandem pair of wall-associated kinases (WAK; i.e. At1g21250 and At1g21240; Fig. 7, bottom) was strongly repressed in full nutrition conditions. WAKs are covalently bound to pectin in the cell wall, providing evidence that the binding of a structural carbohydrate by a receptor-like kinase may have significance in the control of cell expansion (Anderson et al., 2001).
Five annotated MAP3K genes were induced in at least one of the N transitions (Fig. 7; Supplemental Table I). At2g30040 was by far the most responsive gene in this family, showing a transient >20-fold increase in transcript abundance 30 min after NO3− addition. The segmental-duplicated relative of this gene (i.e. At1g07150) showed a similar but weaker response. There were also marked changes of transcript abundance for a MAP2K gene, four protein phosphatases 2C, three calcineurin B-like interacting protein kinases, and four response regulators (Supplemental Table I). By contrast, genes encoding G proteins and components of the phosphoinositide turnover showed only small responses. Maybe surprisingly, a set of genes assigned on the basis of literature sources to carbon and nutrient signaling, including PII and NIT2 homologs (see introduction), showed little response to N depletion or NO3− addition (Supplemental Table II).
Protein Degradation
More than 1,300 genes are involved in protein degradation via the ubiquitin/26S proteasome pathway, and several mutants in light and hormone signaling have already been mapped to this pathway in Arabidopsis (Vierstra, 2003). In view of the fact that changes in N supply will affect protein turnover, surprisingly few of these genes responded. Only one (i.e. At2g33770) of the approximately 45 Arabidopsis ubiquitin conjugating E2 enzyme genes responded to N availability (5-fold higher transcript level following N depletion). Approximately 1,200 loci encode E3 ubiquitin-protein ligases, including nearly 700 F box and almost 400 RING-finger class proteins (Vierstra, 2003). Of the 350 F-box genes that we traced on ATH1 arrays, one (At1g80440) reacted quickly to NO3− addition (6-fold induced after 30 min), and four showed 4- to 7-fold changes between N-depleted and N-replete seedlings (Supplemental Table I). Of 214 traceable RING-finger genes, one (At1g22500) was rapidly induced after NO3− addition, and two differed between N-starved and N-replete seedlings (Supplemental Table I). Nitrogen also affected the expression of several genes that encode proteases unrelated to the ubiquitin/26S proteasome pathway, including two tandem-arrayed putative Ser carboxypeptidase genes (At2g22980 and At2g22990), two Cys protease genes (At5g45890 and At5g50260), and one subtilisin-like protease gene (At1g32960; Supplemental Table I).
Comparison with Array Data in the Public Domain
Wang et al. (2003) recently published transcriptome data for roots and shoots of Arabidopsis seedlings that were grown hydroponically for 10 d on ammonium and then supplied with 250 μm NO3− for 20 min. A good correlation was found when the global changes in root or shoot transcript levels in response to NO3− addition were compared to our data (Supplemental Fig. 9, A and B). Gene-by-gene comparisons revealed that the vast majority of the NO3−-induced genes (3-fold cutoff) in roots or shoots are also detectable in whole seedlings, and vice versa (Supplemental Fig. 9C). In the few cases where the NO3−-responsive change in transcript abundance did not overlap (Supplemental Fig. 9C), the reasons are probably attributable to the different growth systems or differences in plant morphology (e.g. lateral root mass; Fig. 1). This agreement provides strong support for the reliability of the results published by Wang et al. (2003) and in this study, and suggests that NO3− signaling is largely independent of the general N status of the plant. It also shows that the added work and costs involved in the analysis of individual organs is not required in a first exploratory study of a biological response. Detailed insights into the organ-, stimulus-, or age-dependent expression pattern of a gene can be extracted from large public domain microarray databases (e.g. AtGenExpress at www.arabidopsis.org). It is of course possible that changes of gene expression in small organs or low-abundance cell types may be missed.
To enable more sophisticated comparisons of the data from the two studies, the original data files from Wang et al. (2003) were converted into experimental data files for display by MapMan (Supplemental Figs. 10–12). Comparison with the response in our experiments (see, for example, Fig. 2A) reveals remarkable agreement for most genes in primary metabolism. Identical genes involved in NO3− uptake and assimilation, Suc transport, Suc mobilization, glycolysis, the OPP, and the TCA cycle were found to be NO3− induced. Likewise, a similar response of HB1 and HB2 was seen, and identical genes involved in ammonium and sulfate assimilation and nicotianamine synthesis were identified in both studies. Both experiments show a trend to repression of the shikimate pathway, relative to the other amino acid biosynthesis pathways. There was less agreement for secondary metabolism. There were relatively few differences in the lists of genes found to be differentially expressed in the two studies, including GDH3, which was not found to be induced by NO3− previously (Wang et al., 2003). The close agreement between the two independent datasets for rapid (20–30 min) transcriptional responses to NO3− reflect the general reliability of the methods employed and help to strengthen conclusions drawn from later time points, such as 3 h after NO3− addition. Some of the longer-term (3 h) changes in transcript abundance following NO3− application have already been discussed, including the induction of amino acid and nucleotide synthesis, protein synthesis, widespread changes in secondary metabolism, and the induction of a set of expansins and TIPS. While short-term changes in transcript levels may reflect direct or primary responses to NO3− per se, later changes are more likely to result from secondary responses to NO3− nutrition, which presumably involve products of NO3− assimilation.
Wang et al. (2003) reported the identification of 65 potential regulatory genes in roots and 12 in shoots, which were at least 2-fold induced or repressed after 20 min readdition of NO3−. No statement was made on the overlap between the two groups, and only three genes were named with their AGI code. The two most highly induced regulatory genes were a protein phosphatase and a protein kinase, which were both more than 8-fold induced. As shown in Supplemental Table I, the former is a protein phosphatase 2C (i.e. At4g32950) and the latter is a MAPKKK (i.e. At1g49160), for which we calculated similar induction values, using their data. The strong induction of these genes was confirmed in our experiments in seedlings 30 min after NO3− readdition (Supplemental Table I).
In our ATH1 array analyses, we identified 52 potential regulatory genes (AGI codes included in Supplemental Table I) that were quickly (30 min) and at least 3-fold induced/repressed after NO3− addition to N-starved seedlings. Only 33 of these 52 genes were induced or repressed after 20 min (Wang et al., 2003), using the same 3-fold cutoff value. When we consider the 19 or 9 genes that were at least 5- or 10-fold changed in transcript level after 30 min in our study, 16 and 9 showed an at least 3-fold response in the datasets of Wang et al. (2003), respectively. These differences might be due to different induction times and different experimental systems. Several potential regulatory genes that we identified as highly NO3−/N responsive in our experiments were also induced more than 8-fold in the datasets of Wang et al. (2003) but were not earmarked as of interest: e.g. At1g13300, 18-fold; At5g67420, 13-fold; At3g48360, 11-fold; and At5g09800, 9-fold (compare Fig. 7 with Supplemental Table I). Continuing improvements in genome annotation, including the identification of novel regulatory gene families, could explain why such genes were missed previously. So to summarize: in comparison to Wang et al. (2003), we (1) have independently confirmed a number of the NO3−-responsive regulatory genes by Affymetrix and real-time RT-PCR analysis, (2) identified a substantial number of new NO3−- or N-responsive genes using updated gene annotations, real-time RT-PCR, and additional conditions, and (3) named all potential regulatory genes by their AGI code and provided phylogenetic relationships by analyzing tandem or segmental duplications.
CONCLUSION
Analysis of global gene expression data, using whole-genome ATH1 arrays and real-time RT-PCR of >1,400 transcription factor genes, has yielded an unprecedented overview of the metabolic and regulatory responses of Arabidopsis to changes in N nutrition. This analysis was aided by MapMan visualization software that helps to identify coordinated, system-wide changes in metabolism and other cellular processes. A large number of differentially expressed putative regulator genes were identified, which will provide new impetus for reverse-genetics studies aimed at identifying genes and proteins that play central roles in coordinating the complex responses of plants to changes in N nutrition.
MATERIALS AND METHODS
Plant Growth Media and Conditions
The sterile full nutrition (FN) media contained: 2 mm KNO3, 1 mm NH4NO3, 1 mm Gln, 3 mm KH2PO4/K2HPO4, pH 5.8, 4 mm CaCl2, 1 mm MgSO4, 2 mm K2SO4, 3 mm MES, pH 5.8, 0.5% (w/v) Suc, and microelements (i.e. 40 μm Na2FeEDTA, 60 μm H3BO3, 14 μm MnSO4, 1 μm ZnSO4, 0.6 μm CuSO4, 0.4 μm NiCl2, 0.3 μm HMoO4, 20 nm CoCl2). The low N (−N) media contained 0.1 mm KNO3, 50 μm NH4NO3, no Gln, and 3 mm KCl, and all other components as in FN media.
Wild-type Col-0 Arabidopsis seedlings (100–120) were grown in sterile liquid culture (250-mL Erlenmeyer glass flasks) on orbital shakers with constant, uniform fluorescent light (approximately 50 μE in the flask) and temperature (22°C), in 30 mL of FN media. Shaker speed was low (30 rpm) during the first 3 d and then increased to 80 rpm. Care was taken to prevent the seedlings from significant clumping. After 7 d, the media were changed in all flasks. Some received 30 mL of FN media and others received low N nutrition (−N). FN media was again changed on day 8 in FN cultures to prevent N limitation because by this stage N was rapidly depleted by the growing seedlings. The −N medium included 0.2 mm N to minimize variation between flasks due to different amounts of the original medium being left in the flask and was completely exhausted within hours (measured by HPLC; data not shown), assuring N starvation after 2 d.
Nitrate Addition and Seedling Harvest
On day 9 FN cultures and some of the −N cultures were harvested. At the same time, all the other flasks of N-starved cultures were opened and either reclosed without addition or after adding 180 μL of 500 mm KNO3 (3 mm) or 180 μL of 500 mm KCl (3 mm, control). The added liquid was allowed to disperse without changing the shaking speed. Groups of −N flasks that received no addition, KNO3, or KCl were harvested after 30 min and 3 h. Plant materials from each flask were quickly (<10 s for the entire procedure) blotted on tissue paper, washed twice in an excess of desalted water, blotted on tissue paper, and frozen in liquid N2. Materials were stored in liquid N2 until pulverization using mortar and pestle. Ground materials were kept at −80°C until further use.
Metabolite Measurements
NO3−, Glc, Fru, Suc, starch, oxoglutarate, and amino acids were extracted and measured as described by Scheible et al. (1997a, 1997b), and secondary metabolites as described by Henkes et al. (2001) with slight modifications of the HPLC gradients. Analytes were matched against known secondary compounds based on their spectrotypes (KromaSystem 3000 software; Bio-Tek Instruments, Neufahrn, Germany).
RNA Preparation, Array Hybridization, Data Analysis, and MapMan Display
Two biological replicates were pooled to prepare total RNA. The preparation and quality control of RNA and biotin labeling of the cRNA target were as described by Czechowski et al. (2004). Hybridization, washing, staining, and scanning procedures were performed as in the Affymetrix technical manual. The raw Affymetrix signals (CEL files) were processed using RMA (log-scale Robust Multi-array Analysis) open access software (http://stat-www.berkeley.edu/users/bolstad/RMAExpress/RMAExpress.html). It is based on the Quantile normalization method and has better precision than MicroArray Suite 5.0 (Affymetrix, Santa Clara, CA) and dCHIP (http://www.dchip.org/), especially for low expression values (Irizarry et al., 2003). The averaged signals for a given treatment (2 biological replicates for 30 min 3 mm KNO3 addition, 2 biological replicates for 3 h 3 mm KNO3 addition, 3 biological replicates for full nutrients) were expressed relative to those in N-deficient seedlings (3 biological replicates for no addition), converted to a log2 scale and imported into the MapMan software, which converts the data values to a false color scale and paints them out onto the diagrams. ATH1 datasets for the 30-min and 3-h KCl treatments were used as controls in the analysis of ATH1 TF gene expression but were not used for MapMan displays, since the variation of expression values was within the variation found between biological replicates and not significantly different from those of the no addition controls (Supplemental Fig. 2).
The data were visualized and figures produced using the MapMan software (Thimm et al., 2004). A downloadable version for local application and a servlet version are available at http://gabi.rzpd.de/projects/MapMan/. The Web site contains instructions for the installment and use of the software. The downloadable installers include (1) the Affymetrix experimental datasets presented in the paper, (2) a selection of schematic maps of metabolism and cellular processes, and (3) mapping files that structure the Arabidopsis genes represented on the ATH1 array into BINS and SubBINS for display on the schematic maps of metabolism and cellular processes. There will be periodic releases of improved mapping files. The overview figures in this article are prepared using version 060404. For comparison of our data with those of Wang et al. (2003), we globally normalized our data with MicroArray Suite 5.0 (MAS5), added 10% of the target normalization value of 100 to all signals to dampen responses of lowly expressed genes, and calculated mean expression ratios for the biological replicates. The normalized signal intensities for all 22,750 ATH1 probe sets from our 12 arrays are compiled in Supplemental Table II.
Real-Time RT-PCR Analysis
Sequences of RT-PCR primers for transcription factor genes, real-time PCR conditions, data analysis, and procedures for cDNA synthesis were as described by Czechowski et al. (2004).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
We thank Florian Wagner and his team at RZPD Berlin (German Resource Center for Genome Research, Berlin) for providing expert Affymetrix array service, including all steps from total RNA to data acquisition.
The work was supported by the Max-Planck-Society and the Bundesministerium für Bildung und Forschung-funded project GABI Verbund Arabidopsis III Gauntlets (“Carbon and Nutrient Signaling: Test Systems, and Metabolite and Transcript Profiles”; 0312277A).
The online version of this article contains Web-only data.
References
- Amarasinghe BH, de Bruxelles GL, Braddon M, Onyeocha I, Forde BG, Udvardi MK (1998) Regulation of GmNRT2 expression and nitrate transport activity in roots of soybean (Glycine max). Planta 206: 44–52 [DOI] [PubMed] [Google Scholar]
- Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD (2001) WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol Biol 47: 197–206 [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock J, McCormick S (2001) A large family of genes that share homology with CLAVATA3. Plant Physiol 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131: 1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Dörmann P, Benning C (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M (2001) A small decrease of plastid transketolase activity in antisense tobacco transformants has dramatic effects on photosynthesis and phenylpropanoid metabolism. Plant Cell 13: 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland MJ (2002) Transcript abundance in yeast varies over six orders of magnitude. J Biol Chem 277: 14363–14366 [DOI] [PubMed] [Google Scholar]
- Horak CE, Snyder M (2002) Global analysis of gene expression in yeast. Funct Integr Genomics 2: 171–180 [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Lam HM, van de Loo FJ, Coruzzi G (1998) A PII-like protein in Arabidopsis: putative role in N sensing. Proc Natl Acad Sci USA 95: 13965–13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Froehlich JE, Dörmann P (2003) Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15: 2694–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B, Van Dijck P, Thevelein JM (2001) An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci 6: 510–513 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Lin W, Shuai B, Springer PS (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJ, Filatov V, Herzyk P, Krijger GC, Axelsen KB, Chen S, Green BJ, Li Y, Madagan KL, Sanchez-Fernandez R, et al (2003) Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J 35: 675–692 [DOI] [PubMed] [Google Scholar]
- Marschner M (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press Limited, London
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M (2001. a) The immediate cause of the diurnal changes of N metabolism in leaves of nitrate-replete tobacco: a major imbalance between the rate of nitrate reduction and the rates of nitrate uptake and ammonium metabolism during the first part of the light period. Plant Cell Environ 24: 177–190 [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M (2001. b) Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ 24: 1119–1137 [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Prosser IM, Purves JV, Saker LR, Clarkson DT (2001) Rapid disruption of N metabolism and nitrate transport in spinach plants deprived of sulfate. J Exp Bot 52: 113–121 [PubMed] [Google Scholar]
- Riechmann JL (2002) Transcriptional regulation: a genomic overview. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0085, http://www.aspb.org/publications/Arabidopsis [DOI] [PMC free article] [PubMed]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Suzuki M, Takei M, Deji A, Taniguchi M, Sugiyama T (1998) A response-regulator homolog possibly involved in N signal transduction mediated by cytokinin in maize. Plant J 14: 337–344 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Müller-Röber B, Caboche M, Stitt M (1997. a) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Lauerer M, Schulze ED, Caboche M, Stitt M (1997. b) Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J 11: 671–691 [Google Scholar]
- Scheible WR, Krapp A, Stitt M (2000) Reciprocal diurnal changes of phosphoenolpyruvate carboxylase expression and cytosolic pyruvate kinase, citrate synthase and NADP-isocitrate dehydrogenase expression regulate organic acid metabolism during nitrate assimilation in tobacco leaves. Plant Cell Environ 23: 1155–1168 [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Pena CG, Springer PS (2002) The lateral organ boundaries gene defines a novel plant-specific gene family. Plant Physiol 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 28: 655–662 [DOI] [PubMed] [Google Scholar]
- Smith CS, Weljie AM, Moorhead GBG (2003) Molecular properties of the putative N sensor PII from Arabidopsis thaliana. Plant J 33: 353–360 [DOI] [PubMed] [Google Scholar]
- Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Stitt M, Krapp A (1999) The molecular physiological basis for the interaction between elevated carbon dioxide and nutrients. Plant Cell Environ 22: 583–622 [Google Scholar]
- Stitt M, Müller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A (2002) Steps towards an integrated view of N metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MapMan: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tranbarger TJ, Al-Ghazi Y, Muller B, Teyssendier de la Serve B, Doumas P, Touraine B (2003) Transcription factor genes with expression correlated to nitrate-related root plasticity of Arabidopsis thaliana. Plant Cell Environ 26: 459–469 [Google Scholar]
- Vidmar JJ, Schjoerring JK, Touraine B, Glass ADM (1999) Regulation of the hvst1 gene encoding a high-affinity sulfate transporter from Hordeum vulgare. Plant Mol Biol 40: 883–892 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8: 135–142 [DOI] [PubMed] [Google Scholar]
- Wang Y-H, Garvin DF, Kochian LV (2001) Nitrate-induced genes in tomato roots. Array analysis reveals novel genes that may play a role in N nutrition. Plant Physiol 127: 345–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes that are induced by nitrate. Plant Cell 12: 1491–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1000 rapidly responding genes new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG (1999) Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA 96: 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhou JJ, Fernandez E, Galvan A, Miller AJ (2000) A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett 466: 225–227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.