Abstract
Genetic and physiological data establish that Arabidopsis AtHKT1 facilitates Na+ homeostasis in planta and by this function modulates K+ nutrient status. Mutations that disrupt AtHKT1 function suppress NaCl sensitivity of sos1-1 and sos2-2, as well as of sos3-1 seedlings grown in vitro and plants grown in controlled environmental conditions. hkt1 suppression of sos3-1 NaCl sensitivity is linked to higher Na+ content in the shoot and lower content of the ion in the root, reducing the Na+ imbalance between these organs that is caused by sos3-1. AtHKT1 transgene expression, driven by its innate promoter, increases NaCl but not LiCl or KCl sensitivity of wild-type (Col-0 gl1) or of sos3-1 seedlings. NaCl sensitivity induced by AtHKT1 transgene expression is linked to a lower K+ to Na+ ratio in the root. However, hkt1 mutations increase NaCl sensitivity of both seedlings in vitro and plants grown in controlled environmental conditions, which is correlated with a lower K+ to Na+ ratio in the shoot. These results establish that AtHKT1 is a focal determinant of Na+ homeostasis in planta, as either positive or negative modulation of its function disturbs ion status that is manifested as salt sensitivity. K+-deficient growth of sos1-1, sos2-2, and sos3-1 seedlings is suppressed completely by hkt1-1. AtHKT1 transgene expression exacerbates K+ deficiency of sos3-1 or wild-type seedlings. Together, these results indicate that AtHKT1 controls Na+ homeostasis in planta and through this function regulates K+ nutrient status.
Na+ disequilibrium is a primary consequence of salt stress and leads to catastrophic pathologies affecting cell survival, division, and growth (Blumwald, 2000; Hasegawa et al., 2000; Tester and Davenport, 2003; Zhu, 2003). Some of the critical Na+ homeostasis determinants are now identified because of recent research discoveries (Blumwald, 2000; Hasegawa et al., 2000; Tester and Davenport, 2003; Zhu, 2003). Mutations that render the plasma membrane Na+/H+ antiporter SOS1 dysfunctional cause NaCl hypersensitivity of Arabidopsis plants (Wu et al., 1996; Shi et al., 2002). Overexpression of the vacuolar NHX1 or the plasma membrane SOS1 Na+/H+ antiporter increases NaCl tolerance of plants by controlling Na+ homeostasis under saline stress conditions (Apse et al., 1999; Zhang and Blumwald, 2001; Zhang et al., 2001; Shi et al., 2003). These results confirm that greater capacity to maintain Na+ homeostasis enhances salt adaptation of plants.
Another consequence of Na+ disequilibrium that is caused by salt stress is the detrimental effect of Na+ on K+ acquisition and nutrition. K+ has essential functions in plant metabolism (e.g. charge balance, osmotic adjustment, and enzyme catalysis) and in growth and development (Maathuis and Sanders, 1996; Rigas et al., 2001; Elumalai et al., 2002). Na+ in the soil solution disturbs K+ homeostasis in plants presumably because the cytotoxic ion competes for binding sites in transport systems that mediate K+ uptake (Epstein, 1961; Rains and Epstein, 1965; Niu et al., 1995; Hasegawa et al., 2000). Ca2+ is necessary to control K+/Na+ selective accumulation into plants, which effectively reduces Na+ uptake and increases NaCl tolerance (Epstein, 1961; Cramer et al., 1987; Hasegawa et al., 2000; Demidchik et al., 2002; Zhu, 2003). Data from physiological research of more than 40 years ago identified high- and low-affinity transport systems that mediate K+ uptake into roots and determined that both Ca2+ and Na+ modulate the acquisition of this essential mineral nutrient (Epstein, 1961). K+/Na+ selective accumulation by both systems is Ca2+ dependent with the high-affinity system transporting predominantly K+ over Na+ when Ca2+ is present (Epstein, 1961; Rains and Epstein, 1965). In addition, plants have high- and low-affinity systems for Na+ uptake (Epstein, 1961; Rains and Epstein, 1967). K+ competes effectively with Na+ for uptake by the low-affinity system in the presence of Ca2+, while Na+ uptake by the high-affinity system is predominant even in the presence of K+ and Ca2+ (Epstein, 1961; Rains and Epstein, 1967).
There is now more comprehensive understanding about the transport systems and regulatory mechanisms that mediate K+ and Na+ homeostasis in plants, including some Ca2+-dependent processes that control K+/Na+ selective uptake (Epstein, 1998; Zhu, 2002, 2003). The Ca2+-dependent salt overly sensitive (SOS) ion homeostasis pathway transduces salt stress signals to activate the plasma membrane SOS1 Na+/H+ antiporter, which mediates Na+ efflux and homeostasis necessary for salt adaptation (Qiu et al., 2002; Zhu, 2002, 2003; Shi et al., 2003). sos mutant plants (sos1, sos2, and sos3) are NaCl sensitive and exhibit K+ deficiency at μm external concentrations of the essential nutrient. The later phenotype implicates a positive regulatory effect on K+ acquisition by the SOS signaling pathway, although neither the regulatory components nor the determinants that are regulated have been identified. Pretreatment of seedlings with 50 mm NaCl reduces K+ permeability of plasma membranes from sos1 root cells but not from those of wild-type seedlings (Qi and Spalding, 2001). Reduced K+ uptake into and K+ deficiency of sos1 plants is attributed to elevated cytosolic Na+ levels, which occur because the plasma membrane Na+/H+ antiporter is inoperative (Qi and Spalding, 2001; Qiu et al., 2002; Quintero et al., 2002).
Ca2+ affects K+/Na+ selective uptake also because it inhibits Na+ influx across the plasma membrane, which is attributed to its negative regulation of nonselective cation channels (NSCCs; Roberts and Tester, 1997; Davenport and Tester, 2000; Demidchik and Tester, 2002; Tester and Davenport, 2003). NSCCs are characterized by the lack of monovalent cation selectivity and, for many, by Ca2+ inhibition of ion conductance (Demidchik et al., 2002; Tyerman, 2002; Tester and Davenport, 2003). The similar 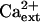 sensitivity of NSCC-mediated Na+ currents and Na+ influx into roots has implicated NSCCs as potential Na+ uptake systems (Roberts and Tester, 1997; Tyerman et al., 1997; Amtmann and Sanders, 1999; Schachtman and Liu, 1999; Davenport and Tester, 2000; Tyerman, 2002). Ca2+-inhibition of NSCC conductance is not absolute so it is possible these ion channels are a substantial leak for Na+ influx, particularly in saline conditions (Schachtman and Liu, 1999; Davenport and Tester, 2000; Demidchik and Tester, 2002). However, the precise contribution of NSCCs to Na+ uptake into plant roots awaits the identification of the molecular determinants and the necessary genetic resources for physiological dissection, although it seems evident that multiple molecular entities contribute to Na+ influx at <mm
sensitivity of NSCC-mediated Na+ currents and Na+ influx into roots has implicated NSCCs as potential Na+ uptake systems (Roberts and Tester, 1997; Tyerman et al., 1997; Amtmann and Sanders, 1999; Schachtman and Liu, 1999; Davenport and Tester, 2000; Tyerman, 2002). Ca2+-inhibition of NSCC conductance is not absolute so it is possible these ion channels are a substantial leak for Na+ influx, particularly in saline conditions (Schachtman and Liu, 1999; Davenport and Tester, 2000; Demidchik and Tester, 2002). However, the precise contribution of NSCCs to Na+ uptake into plant roots awaits the identification of the molecular determinants and the necessary genetic resources for physiological dissection, although it seems evident that multiple molecular entities contribute to Na+ influx at <mm 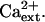
hkt1 alleles are genetic suppressors of sos3-1 NaCl sensitivity (Rus et al., 2001). Physiological characterization of hkt1 genotypes established that AtHKT1 controls plant Na+ homeostasis and is not a K+ transport system. Reduction of TaHKT1 expression by RNAi or cosuppression decreased Na+ uptake into wheat roots and increased plant salt tolerance (Laurie et al., 2002), despite electrophysiological data from experimentation based on expression in heterologous systems that implicated a Na+-dependent K+ cotransport function for TaHKT1 (Rubio et al., 1995). High- and low-affinity Na+ uptake into rice (Oryza sativa) roots is mediated by at least two independent OsHKT transport systems (Garciadeblás et al., 2003). Recently, it was determined that AtHKT1 controls Na+ accumulation in shoots through a proposed mechanism in which the transport system loads Na+ into the shoot phloem and facilitates recirculation of the ion from the shoot to the root (Mäser et al., 2002; Berthomieu et al., 2003). HKT function in the control of Na+ acquisition (Rus et al., 2001; Laurie et al., 2002; Mäser et al., 2002; Berthomieu et al., 2003; Garciadeblás et al., 2003) and hkt1 suppression of K+ deficiency of sos3-1 seedlings establishes that AtHKT1 is a focal determinant of K+/Na+ homeostasis and implicates a function as a downstream effector of the Ca2+-dependent SOS signaling pathway (Rus et al., 2001).
Here, we present genetic (loss- and gain-of-function) and physiological evidence that AtHKT1 controls Na+ homeostasis in planta, a function that apparently is not dependent on Ca2+ or the SOS signal pathway. Furthermore, AtHKT1 regulation of K+ nutrient status is linked to its Na+ homeostasis function and occurs both under conditions of normal or stress Na+ levels.
RESULTS
hkt1 Mutations Are Genetic Suppressors of sos1-1, sos2-2, and sos3-1 NaCl Hypersensitivity
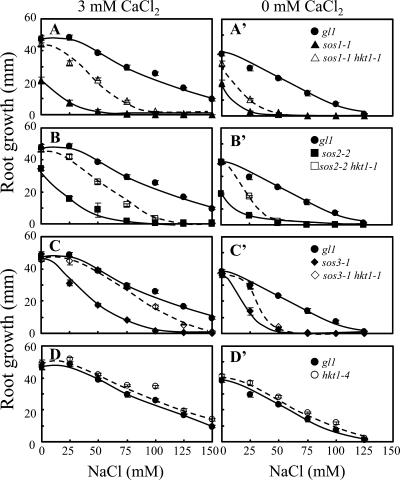
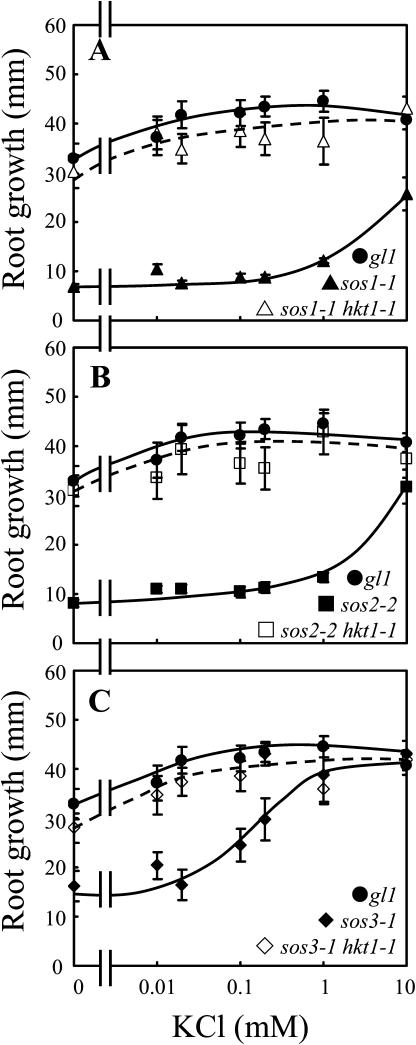
Previously, we determined that AtHKT1 controls Na+ homeostasis in Arabidopsis because hkt1-1 or hkt1-2 can suppress the NaCl sensitivity of sos3-1 seedlings by regulating Na+ accumulation in planta (Rus et al., 2001). hkt1-1 is also a genetic suppressor of sos1-1 and sos2-2 since sos1-1 hkt1-1 and sos2-2 hkt1-1 double mutant seedlings exhibit greater root growth (Fig. 1, A–C) and shoot development with less leaf anthocyanin accumulation (data not shown) when grown in medium supplemented with NaCl. NaCl sensitivity of sos1-1 and sos2-2 seedlings is suppressed to a lesser extent than that of sos3-1 seedlings by hkt1-1, presumably because intracellular Na+ homeostasis is more impaired in sos1-1 and sos2-2 seedlings (Fig. 1, A–C; Qiu et al., 2002). NaCl sensitivity suppression of sos mutations by hkt1-1 is less effective when seedlings are grown in medium without Ca2+ supplement (Fig. 1, A′–C′; Rus et al., 2001), presumably because the Ca2+-sensitive Na+ uptake system(s) is activated due to the reduced 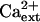 (Tyerman, 2002; Tester and Davenport, 2003). Interestingly, hkt1-1 also suppresses the growth defect of sos1-1 and sos2-2 seedlings that is evident on medium without NaCl supplement (Fig. 1, A and B). AtHKT1 mRNA abundance is not altered in sos1-1, sos2-2, or sos3-1 plants (data not shown), indicating that AtHKT1 is not a transcriptional output of the SOS pathway.
(Tyerman, 2002; Tester and Davenport, 2003). Interestingly, hkt1-1 also suppresses the growth defect of sos1-1 and sos2-2 seedlings that is evident on medium without NaCl supplement (Fig. 1, A and B). AtHKT1 mRNA abundance is not altered in sos1-1, sos2-2, or sos3-1 plants (data not shown), indicating that AtHKT1 is not a transcriptional output of the SOS pathway.
Figure 1.
hkt1-1 is a genetic suppressor of sos mutations. In vitro grown seedlings (4 d on germination medium) were transferred to a basal medium (MS salts without CaCl2, 30 g L−1 Suc, and 12 g L−1 Bacto Agar, DIFCO; Becton-Dickinson, Le Pont de Claix, France) supplemented with various amounts of NaCl, and either 3 mm (A, B, C, and D) or without CaCl2 (A′, B′, C′, and D′). Root length increase was determined 5 d after transfer; values are the mean ± se (n ≥ 8).
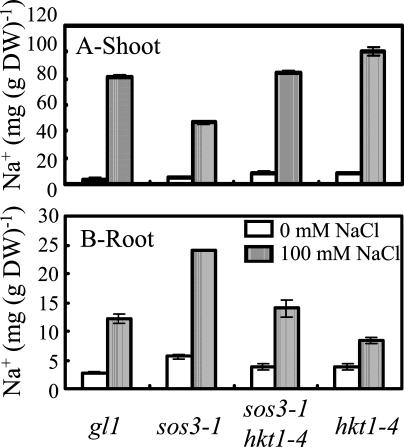
hkt1-1 suppression of sos1-1, sos2-2, and sos3-1 and hkt1-4 suppression of sos3-1 NaCl sensitivity is evident also when plants are grown in soil (Fig. 2) or Turface (Fig. 3) under controlled environmental conditions. The hkt1-4 allele has a 16-bp deletion in the third exon (position 3,496–3,511 of the open reading frame) that is predicted to result in a frame-shift causing a nonsense mutation. hkt1-4 suppresses sos3-1 NaCl sensitivity and K+ deficiency of seedlings grown in vitro to the same extent as hkt1-1 and hkt1-2 (data not shown), indicating that the deletion results in a dysfunctional allele. These results establish that the capacity of hkt1 to attenuate NaCl salt hypersensitivity of sos mutations is operative in plants that are actively transpiring. NaCl-treated sos3-1 hkt1-4 plants exhibit a similar shoot and root Na+ content as wild type (Fig. 4), indicating that hkt1 restores Na+ homeostasis of sos3-1 plants to near that of wild type.
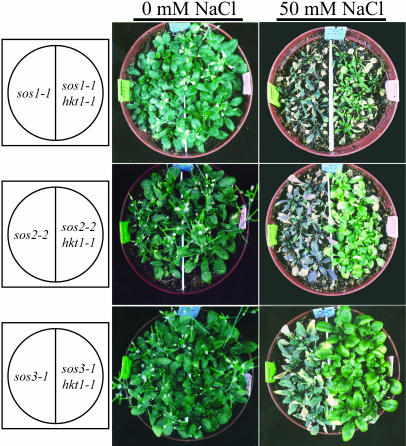
Figure 2.
hkt1-1 suppresses NaCl hypersensitivity of soil-grown sos plants. Three-week-old plants in soil were irrigated by placing the pots in a container filled with deionized water without or with 50 mm NaCl until the soil was saturated by capillarity. Plants were rewatered weekly, and pictures were taken 3 weeks later.
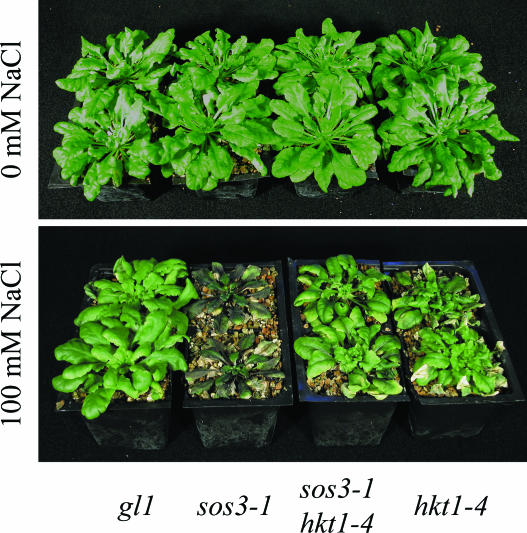
Figure 3.
hkt1-4 suppresses NaCl sensitivity of sos3-1 plants but causes NaCl sensitivity of wild-type (gl1) plants. Stratified seeds in soil were transferred to a growth chamber for germination and plant growth. Plants were irrigated by placing the pots in a container of EXCEL solution until the soil was saturated. EXCEL includes 167 mg L−1 K+, 67 mg L−1 Ca2+, and no detectable Na+, according to the manufacturer's specifications. One-month-old plants were transplanted into Turface. After 1 week of acclimation, plants were watered every 3 or 4 d with EXCEL solution (0 mm NaCl) or supplemented with NaCl (50 mm NaCl for four treatments followed by 100 mm NaCl for 7 treatments). Photographs were taken 5 weeks after beginning of treatment.
Figure 4.
hkt1-4 restores Na+ content of sos3-1 plants to a level similar to that of wild-type plants (gl1). Plants were grown as described in Figure 3. Plants were harvested 5 weeks after the start of NaCl treatment. Shoots and roots (n ≥ 7 for each genotype per treatment) were separated, and materials were rinsed with deionized water and pooled into independent groups (minimum of approximately 50 mg after drying) for ion content analyses. A, Na+ content in the shoot; values are the mean ± se (n = 3). The fresh weight of the plants not treated with NaCl is gl1, 2.95 ± 0.2 g; sos3-1, 2.37 ± 0.25 g; sos3-1 hkt1-4, 2.32 ± 0.21 g; and hkt1-4, 2.63 ± 0.24 g, and of plants treated with NaCl (100 mm final concentration) is gl1, 2.34 ± 0.16 g; sos3-1, 0.44 ± 0.03 g; sos3-1 hkt1-4, 1.23 ± 0.09 g; and hkt1-4, 1.27 ± 0.04 g. B, Na+ content in the root; values are the mean ± se (n ≥ 2), except for sos3-1 treated with 100 mm NaCl. These data are representative of those obtained in replicate experiments.
Salt Responses of hkt1 Plants
hkt1-1 and hkt1-4 alleles were segregated from sos3-1 after backcrossing of plants to wild-type plants (gl1 × sos3-1 hkt1-1 and gl1 × sos3-1 hkt1-4, respectively). There was no difference in root (Fig. 1, D and D′) or shoot growth (data not shown) between hkt1 and wild-type seedlings 6 d after transfer to medium without or supplemented with NaCl. After prolonged growth on medium supplemented with 75 mm NaCl, hkt1 plants exhibited reduced shoot growth and tip senescence of mature leaves (data not shown). hkt1-4 also caused NaCl sensitivity of plants grown in Turface that was manifested as leaf tip senescence and shoot growth inhibition (Fig. 3) and higher and lower Na+ content in the shoot and root, respectively, relative to wild type (Fig. 4). Tip senescence was most pronounced for mature leaves (Fig. 3) 2 weeks after initiation of the salt treatment. The timing of symptom development is indicative of salt rather than hyperosmotic stress pathology (Munns, 2002).
AtHKT1-Mediated Salt Sensitivity Is Na+ Specific
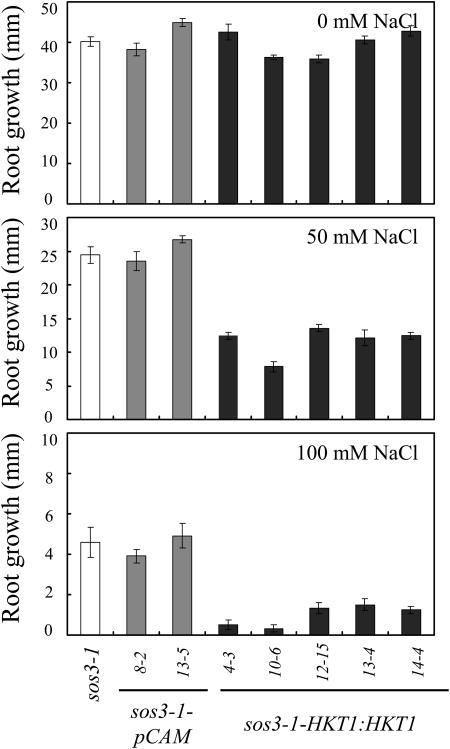
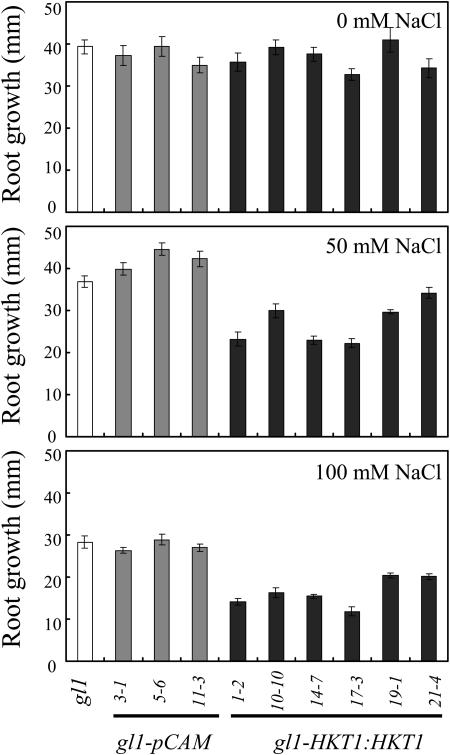
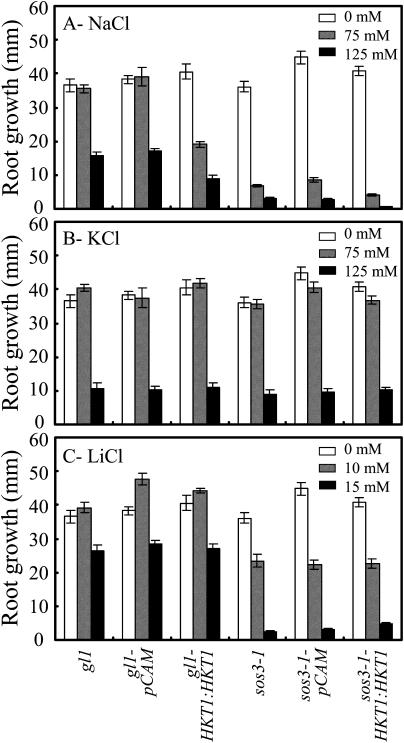
Expression of AtHKT1 (AtHKT1:AtHKT1) in sos3-1 hkt1-1 seedlings results in a phenocopy of sos3-1 Na+ sensitivity (data not shown) based on analysis of progeny from three independent T2 lines that are homozygous for the transgene. This same binary vector was used to express the AtHKT1 transgene in sos3-1 (sos3-1-HKT1:HKT1) and wild-type (gl1-HKT1:HKT1) plants, and five and six independent lines, respectively, were identified, based on segregation for kanamycin resistance of transgenic lines with a single functional insertion, i.e. 3:1 segregation ratio. Plants of all these lines were not preselected for altered NaCl growth response, and all have greater steady-state AtHKT1 transcript abundance (data not shown). NaCl treatment (150 mm NaCl) does not affect AtHKT1 transcript abundance either in plants of wild-type (Uozumi et al., 2000; Rus et al., 2001) or AtHKT1 transgene expressing lines (data not shown). AtHKT1 transgene expression increases NaCl sensitivity of sos3-1 (sos3-1-HKT1:HKT1; Fig. 5) and wild-type gl1 (gl1-HKT1:HKT1; Fig. 6) seedlings. Salt sensitivity is Na+-specific as seedlings expressing the AtHKT1 transgene are not more adversely affected by LiCl or KCl than the nonexpressing lines (Fig. 7).
Figure 5.
AtHKT1:AtHKT1 transgene expression increases NaCl hypersensitivity of sos3-1 seedlings. Four-day-old in vitro-grown seedlings were transferred to a basal medium (MS salts, 30 g L−1 Suc, and 12 g L−1 Bacto Agar) without or supplemented with NaCl; sos3-1, vector control (sos3-1-pCAM, two independent lines) and AtHKT1:AtHKT1 transformed lines (sos3-1-HKT1:HKT1, five independent lines). Root growth was determined 6 d after transfer; values are the mean ± se (n ≥ 11).
Figure 6.
AtHKT1:AtHKT1 transgene expression confers NaCl sensitivity to wild-type seedlings. Four-day-old seedlings were transferred to a basal medium (see Fig. 5 legend) without or supplemented with NaCl; wild-type (gl1), vector control (gl1-pCAM, three independent lines), and AtHKT1:AtHKT1 transformed lines (gl1-HKT1:HKT1, six independent lines). Root growth was determined 6 d after transfer; values are the mean ± se (n ≥ 10).
Figure 7.
AtHKT1:AtHKT1 transgene expression mediates Na+-specific salt sensitivity of wild-type and sos3-1 seedlings. Four-day-old seedlings were transferred to a basal medium (see Fig. 5 legend; white bars) or basal medium supplemented with 75 (hatched bars) or 125 (black bars) mm NaCl (A), 75 (hatched bars) or 125 (black bars) mm KCl (B), and 10 (hatched bars) or 15 (black bars) mm LiCl (C). Root growth of wild-type (gl1), wild-type vector control (gl1-pCAM, line no. 3-1 in Fig. 6), wild-type expressing AtHKT1 transgene (gl1-HKT1:HKT1, line no. 10-10 in Fig. 6), sos3-1, sos3-1 vector control (sos3-1-pCAM, line no. 13-5 in Fig. 5), and sos3-1 expressing AtHKT1 transgene (sos3-1-HKT1:HKT1, line no. 13-4 in Fig. 5) seedlings is illustrated. Root growth was determined 6 d after transfer; values are the mean ± se (n ≥ 10).
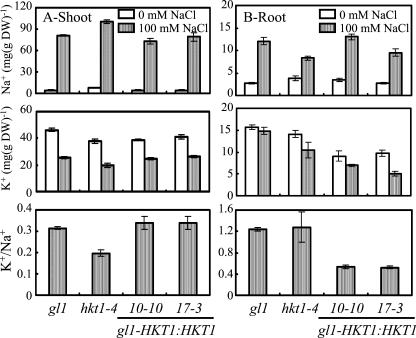
AtHKT1 transgene expression exacerbates NaCl-induced stress symptoms of plants that are grown in soil (data not shown). AtHKT1 transgene expressing plants have a lower K+ to Na+ ratio in the roots due primarily to decreased absolute K+ accumulation (Fig. 8). Together, these results indicate that any modulation of in planta AtHKT1 function causes Na+ disequilibrium and stress symptoms.
Figure 8.
AtHKT1 loss- or gain-of-function affects K+/Na+ homeostasis in planta. Na+ and K+ content in the shoot (A) and root (B) of wild-type (gl1), hkt1-4, or gl1 expressing the AtHKT1 transgene (gl1-HKT1:HKT1 line nos. 10-10 and 17-3 in Fig. 6) plants growing in Turface. Plants were grown as in Figure 3. Shoot content values are the mean ± se (n = 3), root content values are the mean ± se (n ≥ 2), and K+/Na+ was calculated from K+ and Na+ content.
hkt1 Mutations Are Genetic Suppressors of sos1-1, sos2-2, and sos3-1 K+ Deficiency
hkt1-1, hkt1-2, and hkt1-4 mutations suppress the growth defect of sos3-1 seedlings that occurs on medium without or with μm levels of KCl supplement and 0.15 mm CaCl2 (Rus et al., 2001; Fig. 9C; data not shown for hkt1-4). hkt1-1 also suppresses the K+ deficiency of sos1-1 (Fig. 9A) and sos2-2 (Fig. 9B) seedlings. No differences in shoot or root growth of wild-type or hkt1 seedlings occurred on medium without or with KCl supplement (data not shown). Presumably, some growth is supported on medium that is devoid of KCl supplement because there is a trace level of the mineral element in the basal medium (approximately 0.12 mm) and the seedlings accumulate some K+ while on germination medium. These results are genetic evidence that AtHKT1 is not a K+ uptake system in planta but, either directly or indirectly, is a negative regulator of K+ nutrition.
Figure 9.
hkt1-1 suppresses the K+ deficiency of sos seedlings. Four-day-old seedlings were transferred to a basal medium containing 1/20× MS macronutrients [without KNO3, and KH2PO4 replaced with (NH4)2HPO4], MS micronutrients (KI replaced with NaI), 30 g L−1 Suc, and 12 g L−1 Bacto Agar, without or supplemented with various amounts of KCl. The basal medium contains 0.15 mm CaCl2. Illustrated is hk1-1 suppression of sos1-1 (A), sos2-2 (B), and sos3-1 (C). Root growth was determined 9 d after transfer; values are the mean ± se (n ≥ 8).
AtHKT1 Regulation of K+ Nutrition Is Dependent on Na+ Acquisition
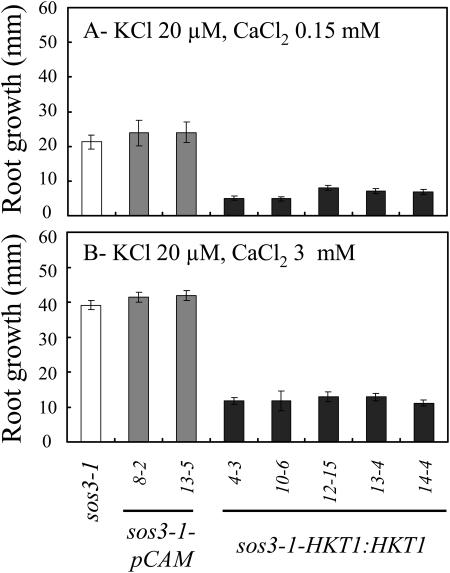
AtHKT1 transgene expression that increases Na+ sensitivity of sos3-1 seedlings (Fig. 5) also causes greater K+ deficiency that is manifested as reduced root growth (Fig. 10) and increased root branching and leaf chlorosis (data not shown) in medium supplemented with 20 μm KCl but without added NaCl. The nutrient medium without added NaCl contains approximately 3 to 4 mm Na+. No difference in growth was observed when sos3-1 or sos3-1-HKT1:HKT1 seedlings were grown on medium supplemented with 10 mm KCl (data not shown), indicating that AtHKT1 negatively affects K+ acquisition by affecting uptake that occurs at μm 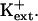 No difference in steady-state transcript abundance of AKT1, TRH1, or SKOR was detected in sos3-1, sos3-1-pCAM (vector control), and sos3-1-HKT1:HKT1 plants (data not shown). These genes encode transport systems that are involved in K+ uptake into roots or loading of the essential element into the xylem (Gaymard et al., 1998; Hirsch et al., 1998; Rigas et al., 2001). Consequently, AtHKT1 modulation of K+ nutrient status in planta does not involve regulating the expression of the genes that encode these K+ transport systems.
No difference in steady-state transcript abundance of AKT1, TRH1, or SKOR was detected in sos3-1, sos3-1-pCAM (vector control), and sos3-1-HKT1:HKT1 plants (data not shown). These genes encode transport systems that are involved in K+ uptake into roots or loading of the essential element into the xylem (Gaymard et al., 1998; Hirsch et al., 1998; Rigas et al., 2001). Consequently, AtHKT1 modulation of K+ nutrient status in planta does not involve regulating the expression of the genes that encode these K+ transport systems.
Figure 10.
AtHKT1:AtHKT1 transgene expression increases the K+ deficiency of sos3-1 seedlings independent of 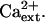 Four-day-old seedlings were transferred to a basal medium (1/20× MS macronutrients [without KNO3, without CaCl2, KH2PO4 replaced with (NH4)2HPO4, with 20 μm KCl], MS micronutrients [KI replaced with NaI], 30 g L−1 Suc, and 12 g L−1 Bacto Agar) and 0.15 (A) or 3 mm CaCl2 (B); genotypes are as in Figure 5. Root growth was determined 6 d after transfer; values are the mean ± se (n ≥ 10).
Four-day-old seedlings were transferred to a basal medium (1/20× MS macronutrients [without KNO3, without CaCl2, KH2PO4 replaced with (NH4)2HPO4, with 20 μm KCl], MS micronutrients [KI replaced with NaI], 30 g L−1 Suc, and 12 g L−1 Bacto Agar) and 0.15 (A) or 3 mm CaCl2 (B); genotypes are as in Figure 5. Root growth was determined 6 d after transfer; values are the mean ± se (n ≥ 10).
Interestingly, K+ deficiency of sos3-1 seedlings that is induced by AtHKT1 transgene expression (65%–75% reduction in root growth) is not affected by increasing the CaCl2 level in the medium from 0.15 to 3 mm (Fig. 10), indicating that AtHKT1 activity is not Ca2+ dependent. An increase in 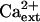 enhances absolute root growth of all seedlings (Fig. 10) and reduces leaf chlorosis (data not shown), which may be attributable to partial Ca2+-activation of the defective sos3-1 protein (Ishitani et al., 2000).
enhances absolute root growth of all seedlings (Fig. 10) and reduces leaf chlorosis (data not shown), which may be attributable to partial Ca2+-activation of the defective sos3-1 protein (Ishitani et al., 2000).
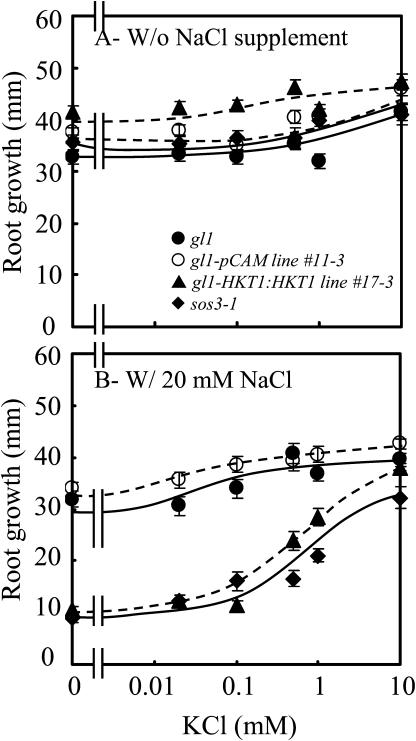
AtHKT1 transgene expression affects K+-dependent growth of wild-type seedlings only if the medium is supplemented with 20 mm NaCl (Fig. 11), unlike the situation of sos3-1 seedlings (Fig. 10). The AtHKT1-induced root growth inhibition (Fig. 11B) apparently is not a Na+ cytotoxicity effect since seedling root growth of all genotypes is similar at this concentration of NaCl if the medium is supplemented with 10 mm KCl (Fig. 11B). The impact of the Ca2+-sensitive Na+ entry system(s) was minimized in this experiment by inclusion of 3 mm CaCl2. Together, these results indicate that AtHKT1 negatively affects K+ acquisition through its function as a Na+ transport system. Presumably, addition of 20 mm NaCl is required to elicit the inhibitory effect of AtHKT1 on K+ nutrition of wild-type seedlings, in contrast to that of sos3-1 seedlings, because the SOS pathway is functional and cells are more capable of intracellular Na+ homeostasis.
Figure 11.
AtHKT1:AtHKT1 transgene expression-mediated K+ deficiency of wild-type (gl1) seedlings requires 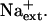 Four-day-old seedlings were transferred to a basal medium (1/20× MS macronutrients [without KNO3, with 3 mm CaCl2, KH2PO4 replaced with (NH4)2HPO4], MS micronutrients [KI replaced with NaI], 30 g L−1 Suc, and 12 g L−1 Bacto Agar) without or supplemented with various amounts of KCl and without NaCl supplement (A) or with 20 mm NaCl (B); plants of wild-type (gl1), vector control (gl1-pCAM, line no. 11-3 in Fig. 6), AtHKT1 transgene expressing line (gl1-HKT1:HKT1, line no. 17-3 in Fig. 6), and sos3-1. Root growth was determined 6 d after transfer; values are the mean ± se (n ≥ 19).
Four-day-old seedlings were transferred to a basal medium (1/20× MS macronutrients [without KNO3, with 3 mm CaCl2, KH2PO4 replaced with (NH4)2HPO4], MS micronutrients [KI replaced with NaI], 30 g L−1 Suc, and 12 g L−1 Bacto Agar) without or supplemented with various amounts of KCl and without NaCl supplement (A) or with 20 mm NaCl (B); plants of wild-type (gl1), vector control (gl1-pCAM, line no. 11-3 in Fig. 6), AtHKT1 transgene expressing line (gl1-HKT1:HKT1, line no. 17-3 in Fig. 6), and sos3-1. Root growth was determined 6 d after transfer; values are the mean ± se (n ≥ 19).
DISCUSSION
AtHKT1 Controls Na+ Homeostasis in Planta
Herein comprehensive evidence from loss- and gain-of-function genetic experimentation establishes that AtHKT1 mediates Na+ homeostasis in Arabidopsis. hkt1 suppresses NaCl sensitivity of sos1-1, sos2-2, and sos3-1 seedlings in vitro and plants that are grown in soil or Turface under controlled environmental conditions. Furthermore, AtHKT1 transgene expression increases NaCl, but not LiCl or KCl, sensitivity of wild-type and sos3-1 seedlings.
In vitro grown hkt1-4 seedlings exhibit NaCl stress symptoms only after a prolonged period. However, actively transpiring hkt1-4 plants (grown in Turface) are acutely more NaCl-sensitive and accumulate more Na+ in the shoot and less in the root compared to wild-type plants (Figs. 3 and 4). Presumably, active transpirational flux is driving greater Na+ transport to the shoot of plants grown in a growth chamber that exacerbates the Na+ stress effects caused by hkt1 mutations. It is hypothesized that AtHKT1 regulates Na+ distribution between the root and the shoot, restricting the accumulation of the toxic cation in the aerial part of the plant (Mäser et al., 2002; Berthomieu et al., 2003). Greater Na+ accumulation in the shoot and less in the root of hkt1 plants indicates that AtHKT1 is not involved in the movement of this cation into the xylem and subsequent movement to the shoot via the transpirational stream (Fig. 4; Mäser et al., 2002; Berthomieu et al., 2003). Since AtHKT1 is expressed predominantly in the phloem, it is hypothesized that, in the shoot, HKT1 loads Na+ into the phloem, which is then translocated to the root, and removal of Na+ from the root phloem occurs by efflux down the electrochemical potential (Berthomieu et al., 2003). Controlling Na+ content of the shoot is a necessary salt adaptation strategy that restricts apoplast accumulation and net influx across the plasma membrane to coordinate effective compartmentalization by the NHX vacuolar Na+/H+ antiporter (Munns, 2002; Munns et al., 2002).
The model of phloem recirculation may not explain all relevant in planta functions of AtHKT1 as revealed by the results of this study that hkt1 mutations suppress Na+ sensitivity of sos mutants and AtHKT1 transgene expression increases NaCl sensitivity of seedlings and plants. The plasma membrane Na+/H+ antiporter SOS1 is proposed to reduce net Na+ flux into cells of the root meristem, which contain vacuoles of insufficient size to compartmentalize Na+ effectively, and to restrict Na+ accumulation in the shoot by controlling xylem loading in the root (Shi et al., 2002). SOS1 overexpression increases NaCl tolerance that is linked to a reduction in Na+ accumulation presumably because the Na+/H+ antiporter is retrieving the toxic cation from the xylem into the surrounding parenchyma cells reducing the salt load transported to the shoot (Shi et al., 2003). It is suggested also that when SOS1 is driven by the 35S promoter, ectopic expression occurs in the epidermis thereby restricting transverse flux of Na+ to the stele (Shi et al., 2003). In planta, Na+ homeostasis apparently is linked tightly to the function of SOS1 and HKT1, and a disruption of either causes ion disequilibrium resulting in salt sensitivity. Combining mutations that reduce the function of these transporters partially restores ion homeostasis that is manifested as suppression of salt sensitivity. Interestingly, suppression of NaCl sensitivity of sos3-1 plants by hkt1 is associated with Na+ profiles in the shoot and root similar to wild-type plants (Fig. 4). Thus hkt1 in the sos background reestablishes Na+ homeostasis of wild-type plants. However, suppression is not equivalent to the salt adaptive capacity of wild-type plants, which have functional SOS1 and HKT1 transport systems.
Plants expressing two additional copies of AtHKT1 under the control of the innate promoter exhibit greater NaCl stress symptoms than parental plants whether wild type or sos3-1. In view of the proposed phloem recirculation mechanism whereby AtHKT1 transports Na+ from the shoot to root (Berthomieu et al., 2003), increased AtHKT1 expression should enhance salt tolerance rather than cause stress symptoms. It is predicted that enhanced Na+ recirculation should reduce accumulation of the toxic cation in the shoot. These results together with hkt1 suppression of Na+ sensitivity of sos mutants implicate other, yet unidentified, function(s) for AtHKT1. It is likely that the Na+ transport function of AtHKT1 is coordinated tightly, both intracellularly and intercellularly, with other transport systems and together these mediate Na+ homeostasis. Either a dysfunctional allele or overexpression of AtHKT1 would modulate Na+ distribution within the plant altering the K+/Na+ homeostasis.
Evidence That AtHKT1 Is Not Regulated by the SOS Pathway
Genetic suppression of sos3-1 and sos2-2 by hkt1 mutations implicate the SOS pathway in the regulation (presumably negative control) of AtHKT1 (Rus et al., 2001; herein). However, AtHKT1 is not a transcriptional output of the SOS signal pathway (data not shown). Furthermore, genetic suppression of sos1-1 by hkt1-1 indicates that AtHKT1 activity is not regulated by the SOS pathway. Both SOS3 and SOS2 are operative in sos1-1 plants, so hkt1-1 should have little, if any, effect on the NaCl phenotype in these plants if AtHKT1 is negatively controlled by the pathway and SOS1 does not have regulatory function. Moreover, there is no indication that Ca2+ modulates the Na+ transport function of AtHKT1 (Fig. 1). Furthermore, AtHKT1 transgene expression increases K+ deficiency of sos3-1 seedlings, and this is 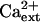 -independent (Fig. 10) even though the K+ deficiency of sos3-1 seedlings is suppressed by
-independent (Fig. 10) even though the K+ deficiency of sos3-1 seedlings is suppressed by 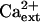 (Liu and Zhu, 1997; Fig. 10). Despite evidence that AtHKT1 may not be regulated by the SOS pathway, genetic data indicate that AtHKT1 and SOS1 are linked in function to the control of Na+ homeostasis in planta.
(Liu and Zhu, 1997; Fig. 10). Despite evidence that AtHKT1 may not be regulated by the SOS pathway, genetic data indicate that AtHKT1 and SOS1 are linked in function to the control of Na+ homeostasis in planta.
K+ Nutrient Status Is Controlled by the Na+ Homeostasis Function of AtHKT1
Herein, evidence establishes that the K+ nutrient status of plants is linked to or is a consequence of the Na+ homeostasis function of AtHKT1. hkt1 mutations suppress the K+ deficient phenotype of sos seedlings, and AtHKT1 transgene expression exacerbates K+ deficiency of wild-type and sos3-1 seedlings. Several lines of evidence indicate that Na+ transport by AtHKT1 controls Na+ homeostasis in planta, which in turn affects K+ acquisition and nutrient status. First, hkt1-induced NaCl sensitivity affects the shoot K+ to Na+ ratio in plants grown under controlled environmental conditions (Figs. 3 and 8). Second, AtHKT1 transgene expression increases Na+ sensitivity of wild-type plants that is linked to a lower K+ to Na+ ratio in the root (Fig. 8). Third, hkt1 mutations suppress K+ deficiency of sos mutants (Fig. 9). Fourth, AtHKT1 transgene expression exacerbates K+ deficiency of sos3-1 seedlings, which are defective in Na+ efflux capacity (Fig. 10; Qiu et al., 2002). Fifth, AtHKT1 transgene expression causes K+ deficiency of wild-type seedlings only if the medium is supplemented with 20 mm NaCl, a level that does not constitute Na+, Cl−, or osmotic stress (Fig. 11). These results are substantial genetic evidence that 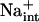 is a controller of K+ acquisition or nutrient status, adding to the knowledge base of more than 40 years that
is a controller of K+ acquisition or nutrient status, adding to the knowledge base of more than 40 years that 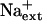 competes with K+ for uptake through high and low-affinity transport systems (Epstein, 1961).
competes with K+ for uptake through high and low-affinity transport systems (Epstein, 1961).
Potential K+ Uptake System Targets of AtHKT1 Regulation
The results herein indicate that AtHKT1 negatively controls K+ acquisition or nutrient status (Figs. 9–11). hkt1 mutations suppress the K+ deficiency of sos seedlings, which exhibit a growth defect that is mitigated by mm 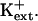 sos1 seedlings exhibit reduced K+ uptake rate in medium supplemented with μm but not mm levels of K+ (Wu et al., 1996). K+ permeability of sos1 root cells is inhibited in the presence of Na+ (Qi and Spalding, 2001). AtHKT1 transgene expression causes K+ deficiency at μm
sos1 seedlings exhibit reduced K+ uptake rate in medium supplemented with μm but not mm levels of K+ (Wu et al., 1996). K+ permeability of sos1 root cells is inhibited in the presence of Na+ (Qi and Spalding, 2001). AtHKT1 transgene expression causes K+ deficiency at μm 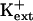 but not at mm
but not at mm 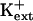 (Fig. 11; data not shown for AtHKT1 transgene expression in sos3-1 seedlings). Together, these results implicate an AtHKT1 function in the control of high-affinity K+ acquisition. However, complete suppression of the growth defect of sos1-1 and sos2-2 seedlings that is evident on medium supplemented with mm K+ may be indicative of a more global function of AtHKT1 to control K+ nutrient status of plants.
(Fig. 11; data not shown for AtHKT1 transgene expression in sos3-1 seedlings). Together, these results implicate an AtHKT1 function in the control of high-affinity K+ acquisition. However, complete suppression of the growth defect of sos1-1 and sos2-2 seedlings that is evident on medium supplemented with mm K+ may be indicative of a more global function of AtHKT1 to control K+ nutrient status of plants.
AKT1 might be a target for negative regulation of AtHKT1 because the 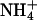 concentration in the medium used in our experimentation (approximately 1.2 mm) should inhibit the activity of the high-affinity K+ transporters (Figs. 9–11; Spalding et al., 1999; Rus et al., 2001).
concentration in the medium used in our experimentation (approximately 1.2 mm) should inhibit the activity of the high-affinity K+ transporters (Figs. 9–11; Spalding et al., 1999; Rus et al., 2001). 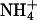 inhibits the activity of HAK1/KUP1 high-affinity K+ transporters in barley (Hordeum vulgare) and rice root cells (Santa-Maria et al., 2000; Bañuelos et al., 2002). Arabidopsis KT/HAK/KUP/TRH transporters are suggested to be components of the
inhibits the activity of HAK1/KUP1 high-affinity K+ transporters in barley (Hordeum vulgare) and rice root cells (Santa-Maria et al., 2000; Bañuelos et al., 2002). Arabidopsis KT/HAK/KUP/TRH transporters are suggested to be components of the 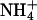 -sensitive pathway for K+ uptake (Spalding et al., 1999). Furthermore, AKT1-mediated K+ currents are inhibited by intracellular Na+ (E. Spalding, personal communication). K+ transport through AKT1 occurs at μm concentrations (Hirsch et al., 1998; Spalding et al., 1999; Dennison et al., 2001), which is consistent with our results that AtHKT1 is a negative regulator of K+ nutrient status in the presence of μm
-sensitive pathway for K+ uptake (Spalding et al., 1999). Furthermore, AKT1-mediated K+ currents are inhibited by intracellular Na+ (E. Spalding, personal communication). K+ transport through AKT1 occurs at μm concentrations (Hirsch et al., 1998; Spalding et al., 1999; Dennison et al., 2001), which is consistent with our results that AtHKT1 is a negative regulator of K+ nutrient status in the presence of μm 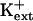 (Figs. 9–11). Our analysis indicates that AKT1, TRH1, or SKOR expression is not controlled by the function of AtHKT1.
(Figs. 9–11). Our analysis indicates that AKT1, TRH1, or SKOR expression is not controlled by the function of AtHKT1.
Evidence is mounting that HKT1 systems are conserved in plant species and that these function in Na+ transport (Rus et al., 2001; Golldack et al., 2002; Laurie et al., 2002; Garciadeblás et al., 2003). It is evident why plants need to control Na+ homeostasis during an episode of salt exposure, as this ion is a necessary osmolyte for vacuolar osmotic adjustment, yet is a cytotoxin (Blumwald, 2000; Hasegawa et al., 2000; Rus et al., 2001; Zhu, 2003). However, the necessity of a specific Na+ transport system in nonsaline environments and the retention of this system in glycophytes are more enigmatic. It has been suggested that Na+ may substitute for K+ physiologically (pH control, volume regulation) under conditions where K+ is limiting (Rodriguez-Navarro, 2000). Na+-mediated pH control may be essential for biotic stress signaling and ontological development of plants, and available data implicate the tonoplast Na+/H+ antiporter as a pH-stat (Fukada-Tanaka et al., 2000; Yamaguchi et al., 2001; Viehweger et al., 2002). Clearly, the existence and function of AtHKT1 together with other transport systems that transport Na+ with high specificity, e.g. SOS1 and perhaps NHX, are indicative that Na+ has a necessary function in nonsaline environments. Also, it is conceivable that 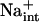 may be a controller of K+ nutrient status, and recent evidence establishes a direct connection between K+ acquisition and regulation of growth and development (Rigas et al., 2001; Elumalai et al., 2002).
may be a controller of K+ nutrient status, and recent evidence establishes a direct connection between K+ acquisition and regulation of growth and development (Rigas et al., 2001; Elumalai et al., 2002).
MATERIALS AND METHODS
Plant Materials
Arabidopsis Col-0 gl1 (wild type) and the other genetic resources reported in this paper are listed in Table I. Plant materials are available upon request.
Table I.
Genotypes and genetic resources used in this study
| Abbreviation Used | Background Genotype | Binary Vector | Reference |
|---|---|---|---|
| Wild type, gl1 | Col-0 gl1 | ||
| sos1-1 | Col-0 gl1 sos1-1 | Shi et al. (2000) | |
| sos2-2 | Col-0 gl1 sos2-2 | Liu et al. (2000) | |
| sos3-1 | Col-0 gl1 sos3-1 | Liu and Zhu (1998) | |
| hkt1-1 | Col-0 gl1 hkt1-1 | This study | |
| hkt1-4 | Col-0 gl1 hkt1-4 | This study | |
| sos1-1 hkt1-1 | Col-0 gl1 sos1-1 hkt1-1 | This study | |
| sos2-2 hkt1-1 | Col-0 gl1 sos2-2 hkt1-1 | This study | |
| sos3-1 hkt1-1 | Col-0 gl1 sos3-1 hkt1-1 | pSKI015 | Rus et al. (2001) |
| gl1-pCAM | Col-0 gl1 | pCAMBIA 2300 | This study |
| gl1-HKT1:HKT1 | Col-0 gl1 | pCAMBIA 2300 [AtHKT1:AtHKT1] | This study |
| sos3-1-pCAM | Col-0 gl1 sos3-1 | pCAMBIA 2300 | This study |
| sos3-1-HKT1:HKT1 | Col-0 gl1 sos3-1 | pCAMBIA 2300 [AtHKT1:AtHKT1] | This study |
sos1-1 hkt1-1 and sos2-2 hkt1-1
sos1-1 hkt1-1 and sos2-2 hkt1-1 were obtained after crossing sos1-1 (Shi et al., 2000) and sos2-2 (Liu et al., 2000), respectively, to sos3-1 hkt1-1 (Rus et al., 2001); sos3-1 hkt1-1 × sos1-1 and sos3-1 hkt1-1 × sos2-2. Genotypic analysis of F2 plants was accomplished by using PCR for detection of mutant and wild-type alleles. Primers used to identify sos3-1 and SOS3 alleles were dub23-3F, 5′-TCTCATGAATTTGCAGTTGC-3′ and dub23-3R, 5′-AAACTGTTTAATCTGGAGGG-3′ (Halfter et al., 2000). The products are 112 (Δ9 bp in sos3-1) and 121 bp, respectively. Specific forward primers were: for SOS2, dub23-2FWT, 5′-AATTTGGATGATATTCGTGCAGTTTTTG-3′ and for sos2-2, dub23-2F, 5′-GGATGATATTCGTGCAGTTTGA-3′ together with the same reverse primer, dub23-2RUH, 5′-TTAACATTTAAATGGAATTGACC-3′. Primers to genotype the SOS1 and sos1-1 alleles were ssr1test-F, 5′-GGCATTGCATCAGTTATTTGG-3′ and ssr1test-R, 5′-GGTCTACAGAAAGACGTACAG-3′; 101 bp for sos1-1 (Δ14 bp) and 115 bp for SOS1. PCR procedures for genotyping the AtHKT1 locus were as described in Rus et al. (2001).
hkt1-1 and hkt1-4
sos3-1 hkt1-1 and sos3-1 hkt1-4 were crossed to Col-0 gl1, and F2 plants were genotyped to identify hkt1-1 and hkt1-4 homozygotes. Primers for analysis of the SOS3 locus were as described above. PCR analysis to genotype hkt1-1 and HKT1 is as described by Rus et al. (2001). hkt1-4 allele was distinguished from HKT1 by using the primer pairs 265F, 5′-TGGTTTCACTACCGGGTACA-3′ and 265R, 5′-GCCAGATTTGGCTGTGAACT-3′ that produced 150- (Δ16 bp in hkt1-4) and 166-bp amplification products, respectively.
AtHKT1 Binary Vector and Generation of Transgenic Plants
An AtHKT1 promoter fragment (−2,000 to −12 bp upstream of the translational initiation site) was amplified by PCR using the bacterial artificial chromosome (BAC) clone F24G24 (GenBank accession no. AL049488) and cloned into the EcoRV-XhoI sites of pBluescript SK(±; Stratagene, La Jolla, CA). The nucleotide sequence of the cloned AtHKT1 promoter fragment was the same as in F24G24. An AtHKT1 open reading frame fragment (spanning from 12 bp upstream of the translational initiation site to 523 bp downstream of the stop codon, including the polyadenylation cis-element) was obtained by digestion of the BAC clone F24G24 (positions 37,557–41,752) with XhoI and HindIII, and this fragment was cloned into pBluescript SK(±). After nucleotide sequence analysis, this fragment containing the AtHKT1 coding sequence was fused, in frame, to the AtHKT1-promoter fragment. The AtHKT1:AtHKT1 genomic fragment was cloned into SacI and HindIII sites of the binary vector pCAMBIA 2300 (CAMBIA, Canberra, Australia). The pCAMBIA 2300 (without insert) and pCAMBIA 2300/AtHKT1:AtHKT1 vectors were introduced into Agrobacterium tumefaciens (strain GV3101) by electroporation.
Wild-type or sos3-1 plants were transformed by spraying flower buds with an Agrobacterium suspension (50 g L−1 Suc and 2 μL mL−1 Silwet L-77 [OSI Specialties, Danbury, CT]; Chung et al., 2000). T1 seeds were collected and then germinated in vitro on medium containing Murashige and Skoog (MS) salts, 30 g L−1 Suc, 8 g L−1 agar, and 50 mg L−1 kanamycin. T2 seeds were collected from individual lines, and only those for which progeny segregated at a 3:1 ratio for kanamycin resistance (i.e. single transgene insertion) were used to obtain T3 progeny. T3 seeds of lines homozygous for kanamycin resistance, and not selected by any other criteria, were used for experimentation. The higher AtHKT1 mRNA abundance in the AtHKT1 transgene expressing lines was confirmed by reverse transcription PCR analysis. The transgenic lines are referred to as gl1-pCAM or gl1-HKT1:HKT1 (expressing AtHKT1 transgene) for wild-type plants transformed with vector without insert or containing AtHKT1:AtHKT1 (as described above), respectively, and sos3-1-pCAM or sos3-1-HKT1:HKT1 for sos3-1 plants transformed with vector without insert or containing AtHKT1:AtHKT1, respectively.
Na+, K+, and Ca2+ Growth Responses of Seedlings in Vitro
After stratification, seeds were germinated in vitro on medium containing MS salts (JRH Biosciences, Lenexa, KS), 30 g L−1 Suc, and 10 g L−1 agar as described by Rus et al. (2001). Four-day-old seedlings were transferred from germination medium to test media and grown in vitro as described (Rus et al., 2001). Test media were prepared as described previously (Rus et al., 2001), and the constituents are listed in the figure legends. The pH of all media was adjusted to 5.7 prior to autoclave sterilization. Data are illustrated as root length increase after transfer to a test medium.
NaCl Growth Response of Plants
Seeds were sown onto moist soil (Scotts Potting Medium; Scotts-Sierra Horticultural Products, Marysville, OH) and stratified at 4°C for 3 d. NaCl growth response of plants in soil or Turface (Profile Products, Buffalo Grove, IL) was evaluated under controlled environmental conditions (8 h light:16 h dark, 150 μmol m−2 s−1 light intensity, 75% relative humidity).
The comparison of the NaCl response of wild-type, sos3-1, sos3-1 hkt1-4, hkt1-4, and two independent transgenic lines expressing AtHKT1 (gl1-HKT1:HKT1 no. 10-10 and no. 17-3) plants was conducted with plants grown on Turface (Figs. 3, 4, and 8). After stratification, pots with seeds were transferred to the growth chamber for germination. One-month-old seedlings were transplanted into Turface and watered with EXCEL solution (Scotts) by capillarity (167 mg L−1 K+, 67 mg L−1 Ca2+, and no detectable Na+ according to specification of the manufacturer; Scotts). NaCl treatment was initiated 1 week after transfer to Turface. Plants were watered every 3 or 4 d with EXCEL solution without or with NaCl using the following procedure: EXCEL solution only; or 50 mm NaCl (four treatments) followed by 7 treatments with 100 mm NaCl.
Na+ and K+ Content Determinations
For determination of shoot and root ion content data, plant material was dried in an oven at 65°C for 4 d. The dried material was ground with a mortar and pestle. Ground dry-matter (around 50 mg/sample) was extracted with HNO3 0.1 N. Na+ or K+ content in the extract was determined by using an atomic absorption spectrophotometer (Varian SpectrAA-10; Victoria, Australia).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AL049488.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the BAC Clone F24G24 (GenBank accession no. AL049488). We also thank Terri Kirk and Brett Lahner for technical assistance with the ion content analyses. This is Purdue University Agricultural Research Program Paper Number 17108.
This work was supported by a Spanish Government Fellowship (to A.M.-M.), by the National Institutes of Health (grant no. R01GM59138 to J.-K.Z.), and by a National Science Foundation Plant Genome Award (DBI–98–13360).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042234.
References
- Amtmann A, Sanders D (1999) Mechanisms of Na+ uptake by plant cells. Adv Bot Res 29: 75–112 [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Bañuelos MA, Garciadeblás B, Cubero B, Rodriguez-Navarro A (2002) Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol 130: 784–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E (2000) Sodium transport and salt tolerance in plants. Curr Opin Cell Biol 12: 431–434 [DOI] [PubMed] [Google Scholar]
- Chung MH, Chen MK, Pan SM (2000) Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res 9: 471–476 [DOI] [PubMed] [Google Scholar]
- Cramer GR, Lynch JL, Laüchli A, Epstein E (1987) Influx of Na+, K+, and Ca2+ into roots of salt-stressed cotton seedlings. Effects of supplemental Ca2+. Plant Physiol 83: 510–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Tester M (2000) A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiol 122: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Davenport RJ, Tester M (2002) Nonselective cation channels in plants. Annu Rev Plant Biol 53: 67–107 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Tester M (2002) Sodium fluxes through nonselective cation in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol 128: 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison KL, Robertson WR, Lewis BD, Hirsch RE, Sussman MR, Spalding EP (2001) Functions of AKT1 and AKT2 potassium channels determined by studies of single and double mutants of Arabidopsis. Plant Physiol 127: 1012–1019 [PMC free article] [PubMed] [Google Scholar]
- Elumalai RP, Nagpal P, Reed JW (2002) A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E (1961) The essential role of calcium in selective cation transport by plant cells. Plant Physiol 36: 437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E (1998) How calcium enhances plant salt tolerance. Science 280: 1906–1907 [DOI] [PubMed] [Google Scholar]
- Fukada-Tanaka S, Inagaki Y, Yamaguchi T, Saito N, Iida S (2000) Colour-enhancing protein in blue petals. Nature 407: 581. [DOI] [PubMed] [Google Scholar]
- Garciadeblás B, Senn ME, Bañuelos MA, Rodriguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34: 788–801 [DOI] [PubMed] [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferriere N, Thibaud J-B, Sentenac H (1998) Identification and disruption of a plant Shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Golldack D, Su H, Quigley F, Kamasani UR, Muñoz-Garay C, Balderas E, Popova OV, Bennett J, Bohnert HJ, Pantoja O (2002) Characterization of a HKT-transporter in rice as a general alkali cation transporter. Plant J 31: 529–542 [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu J-K (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97: 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim C-S, Shi W, Zhu J-K (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJM, Heard PJ, Brown SJ, Leigh RA (2002) A role for HKT1 in sodium uptake by wheat roots. Plant J 32: 139–149 [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K (1997) An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA 94: 14960–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, et al (2002) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Sanders D (1996) Mechanisms of potassium absorption by higher plant roots. Physiol Plant 96: 158–168 [Google Scholar]
- Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239–250 [DOI] [PubMed] [Google Scholar]
- Munns R, Husain S, Rivelli AR, James RA, Condon AG, Lindsay MP, Lagudah ES, Schachtman DP, Hare RA (2002) Avenues for increasing salt tolerance of crops, and the role of physiologically based selection traits. Plant Soil 247: 93–105 [Google Scholar]
- Niu X, Bressan RA, Hasegawa PM, Pardo JM (1995) Ion homeostasis in NaCl stress environments. Plant Physiol 109: 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Spalding EP (2001) Electrophysiological studies of the hypersensitivity of the sos mutants of Arabidopsis (Abstract page 127). In Plant Membrane Biology Meeting. The 12th International Workshop, August 11–16, 2001, University of Wisconsin, Madison, WI
- Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu J-K, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rains DW, Epstein E (1965) Transport of sodium in plant tissue. Science 148: 1611. [DOI] [PubMed] [Google Scholar]
- Rains DW, Epstein E (1967) Sodium absorption by barley roots: role of the dual mechanisms of alkali cation transport. Plant Physiol 42: 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, Dolan L, Hatzopoulos P (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SK, Tester M (1997) A patch clamp study of Na+ transport in maize roots. J Exp Bot 48: 431–440 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro A (2000) Potassium transport in fungi and plants. Biochim Biophys Acta 1469: 1–30 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660–1663 [DOI] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee B-H, Matsumoto TK, Koiwa H, Zhu J-K, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Maria GE, Danna CH, Czibener C (2000) High-affinity potassium transport in barley roots. Ammonium-sensitive and -insensitive pathways. Plant Physiol 123: 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman D, Liu W (1999) Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci 4: 281–287 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu J-K (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Lee B-H, Wu S-J, Zhu J-K (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu J-K (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD (1999) Potassium uptake supporting plant growth in the absence of AKT1 channel activity. Inhibition by ammonium and stimulation by sodium. J Gen Physiol 113: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman SD (2002) Nonselective cation channels. Multiple functions and commonalities. Plant Physiol 128: 327–328 [Google Scholar]
- Tyerman SD, Skerrett M, Garrill A, Findlay G, Leigh R (1997) Pathways for the permeation of Na+ and Cl− into protoplasts derived from the cortex of wheat roots. J Exp Bot 48: 459–480 [DOI] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viehweger K, Dordschbal B, Roos W (2002) Elicitor-activated phospholipase A2 generates lysophosphatidylcholines that mobilize the vacuolar H+ pool for pH signaling via the activation of Na+-dependent proton fluxes. Plant Cell 14: 1509–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-J, Ding L, Zhu J-K (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Fukada-Tanaka S, Inagaki Y, Saito N, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Iida S (2001) Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant Cell Physiol 42: 451–461 [DOI] [PubMed] [Google Scholar]
- Zhang H-X, Blumwald E (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 19: 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang H-X, Hodson JN, Williams JP, Blumwald E (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA 98: 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6: 441–445 [DOI] [PubMed] [Google Scholar]