Abstract
The frd3 mutant of Arabidopsis exhibits constitutive expression of its iron uptake responses and is chlorotic. These phenotypes are consistent with defects either in iron deficiency signaling or in iron translocation and localization. Here we present several experiments demonstrating that a functional FRD3 gene is necessary for correct iron localization in both the root and shoot of Arabidopsis plants. Reciprocal grafting experiments with frd3 and wild-type Arabidopsis plants reveal that the phenotype of a grafted plant is determined by the genotype of the root, not by the genotype of the shoot. This indicates that FRD3 function is root-specific and points to a role for FRD3 in delivering iron to the shoot in a usable form. When grown under certain conditions, frd3 mutant plants overaccumulate iron in their shoot tissues. However, we demonstrate by direct measurement of iron levels in shoot protoplasts that intracellular iron levels in frd3 are only about one-half the levels in wild type. Histochemical staining for iron reveals that frd3 mutants accumulate high levels of ferric iron in their root vascular cylinder, the same tissues in which the FRD3 gene is expressed. Taken together, these results clearly indicate a role for FRD3 in iron localization in Arabidopsis. Specifically, FRD3 is likely to function in root xylem loading of an iron chelator or other factor necessary for efficient iron uptake out of the xylem or apoplastic space and into leaf cells.
Iron is both necessary for plant growth and toxic in excess. It participates as a redox cofactor in a number of metalloenzymes involved in respiration and photosynthesis. These same redox properties allow iron to catalyze the formation of damaging oxygen radicals (Halliwell and Gutteridge, 1992). Although iron is plentiful in the earth's crust, it exists primarily in the insoluble ferric, Fe(III), form. Therefore, plants need specific mechanisms to obtain sufficient amounts of this important nutrient. Dicots rely on acidification of the rhizosphere to solubilize ferric iron, reduction of ferric iron to the more soluble ferrous form, and transport of the ferrous iron into the root epidermal cells. These activities are collectively termed iron uptake responses and are maximally expressed under conditions of iron deficiency. The genes responsible for the root iron-deficiency inducible ferric chelate reductase activity and the major ferrous uptake transporter have been identified as FRO2 and IRT1, respectively, in the model plant Arabidopsis (Eide et al., 1996; Robinson et al., 1999; Vert et al., 2002).
It is well known that iron deficiency causes chlorosis in plants. On a molecular level, this chlorosis is caused by a reduction in the amount of chlorophyll synthesized and an accumulation of both Mg-protoporphyrin IX and Mg-protoporphyrin IX monomethyl ester that are chlorophyll precursors (Spiller et al., 1982). These data imply there is an iron-requiring step between Mg-protoporphyrin IX monomethyl ester and protochlorophyllide (Spiller et al., 1982). Recently, CHL27 has been demonstrated to be this iron-containing protein necessary for protochlorophyllide biosynthesis in Arabidopsis (Tottey et al., 2003). CHL27 is the Arabidopsis homolog to the Chlamydomonas Crd1 protein. Both CHL27 and Crd1 are putative diiron containing enzymes and candidates for the aerobic cyclase enzyme that catalyzes the conversion of Mg-protoporphyrin IX monomethyl ester to divinyl protochlorophyllide that contains the fifth ring characteristic of all chlorophylls (Moseley et al., 2000; Tottey et al., 2003).
Relatively little is known about either the mechanisms that control the expression of iron uptake responses or those involved in iron translocation throughout the plant. Previously, we reported that the frd3 mutant of Arabidopsis constitutively exhibits symptoms of iron deficiency (Rogers and Guerinot, 2002a). It is chlorotic, constitutively expresses its iron uptake responses, and does not accumulate the iron storage protein ferritin in its leaves, which indicates low plastid iron levels. However, when grown under certain conditions, frd3 mutants also have higher total iron concentrations in their leaf tissue. If frd3 is grown under conditions of high iron availability, such as petri plates containing 500 μm Fe(III)EDTA, iron accumulates in the shoots to approximately twice wild-type levels (Rogers and Guerinot, 2002a). However, if frd3 is grown under conditions of much lower iron availability, such as standard potting soil, iron does not overaccumulate and in fact is about 10% lower than in wild-type leaves (Lahner et al., 2003). The FRD3 gene encodes a protein predicted to be a member of the multi-drug and toxin efflux (MATE) family that is expressed only in the root. We proposed two possible roles for FRD3, either in iron deficiency signaling or iron localization within the plant.
Here we report the results of several experiments designed to distinguish between these two models for FRD3 action. First, we show that detached roots of the frd3 mutant are capable of repressing iron uptake responses when cultured under iron-sufficient conditions. Furthermore, grafting experiments show that when frd3 mutant shoots are appropriately supplied with iron, they regreen and the plants are capable of correctly regulating root iron uptake responses. We also show that protoplasts isolated from frd3 mutants have lower iron levels than those from wild-type plants and that frd3 mutant plants accumulate abnormal levels of ferric iron in their root vasculature. FRD3 is expressed in the pericycle and other vascular cylinder cells in the mature portion of the root. All of these data are consistent with a role for FRD3 in the delivery of iron to the shoot in a useable form.
RESULTS
If the wild-type FRD3 protein were involved in transmission or perception of a systemic iron deficiency signal, the loss-of-function frd3 mutants presumably would not be able to down-regulate their iron uptake responses even though there is sufficient iron in the leaf tissues. In this signaling model, shoot chlorosis would be caused by excess iron or manganese. A signaling defect could be shoot-specific, in which case the frd3 mutant shoot would be defective in signaling the shoot iron status to the root. Alternatively, a signaling defect could be root-specific. In this case the frd3 mutant roots would be defective in perceiving a shoot-derived iron status signal or could not regulate their iron uptake responses as directed by the shoot-derived signal. In the other model, the FRD3 protein would be involved in iron localization. In this case, iron would not get to the leaf cells in a usable form in the frd3 mutant, causing the shoot to be functionally iron deficient. This model is consistent with the lack of accumulation of the ferritin protein in frd3 leaf tissue. With this model, the constitutive iron uptake responses exhibited by frd3 mutant roots would be an appropriate response to a shoot iron deficiency signal and shoot chlorosis could be caused by iron deficiency.
Examination of Iron Status Signaling in the frd3 Mutant
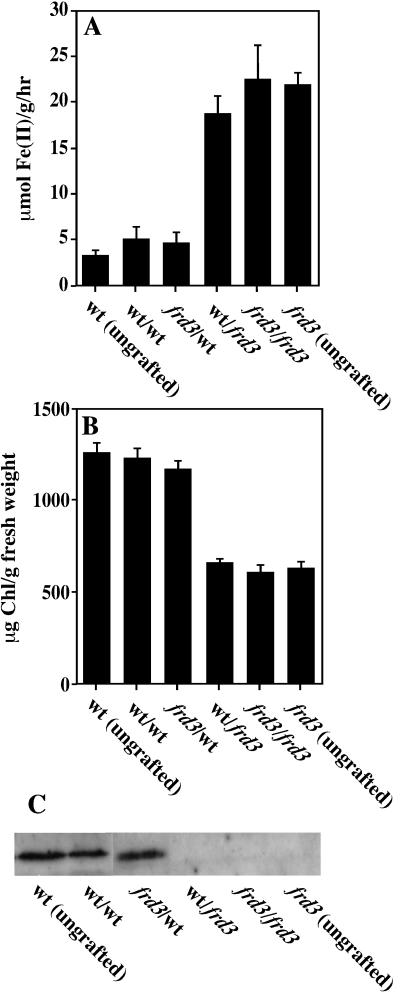
To assess the signaling capabilities of the frd3 mutant, reciprocal grafting experiments were performed with frd3 and wild-type seedlings. Approximately 2 weeks after grafting, plants were transferred to iron-sufficient media for 3 d, after which root ferric chelate reductase activity, and shoot chlorophyll and ferritin levels were measured (Fig. 1). Self-grafted plants behave similarly to the ungrafted controls, indicating that the grafting process itself does not affect iron homeostasis. Examining the reciprocal grafts, it is clear the phenotype of the grafted plant follows the genotype of the root, not the genotype of the shoot. For example, when a frd3 shoot is grafted onto a wild-type root, the frd3 shoot is not chlorotic and accumulates the ferritin protein, and the wild-type root does not express its ferric chelate reductase activity. This indicates that the frd3 shoot is capable of transmitting its iron sufficient status to the root and that when appropriately supplied with iron by the wild-type root, a frd3 shoot does accumulate chlorophyll and ferritin protein. However, when a wild-type shoot is grafted onto a frd3 mutant root, this wild-type shoot becomes chlorotic and no longer accumulates ferritin protein, phenocopying shoots of intact frd3 plants. This result is consistent with a model in which iron signaling is intact in the frd3 mutant, but frd3 roots are not capable of supplying iron to the shoots in a usable form, causing the shoots to become functionally iron deficient.
Figure 1.
The frd3 phenotype is controlled by root genotype in reciprocal grafts. Four-day-old seedlings were grafted, allowed to recover for 2 weeks, and then transferred to iron-sufficient media for 3 d. Root ferric chelate reductase activity (A) and chlorophyll content (B) were measured spectrophotometrically. Ferritin protein was measured by immunoblot using anti-ferritin antibodies (C). Labels are shoot genotype/root genotype.
The expression of root iron uptake responses appears to be regulated by a combination of two signals, a systemic one originating in the shoot and a local one endogenous to the root (Grusak and Pezeshgi, 1996; Schikora and Schmidt, 2001). To examine local, root-endogenous signaling, we determined whether frd3 mutant roots are capable of repressing their iron uptake responses in the absence of a shoot signal. Hypocotyls were cut and the shoot portion removed from 2-week-old seedlings that had been growing on standard media. The root portion was then transferred to either iron-sufficient or deficient media for 5 d before the roots were assayed for ferric chelate reductase activity. Measuring ferric chelate reductase activity is straightforward and was used here as a marker of all three iron deficiency responses. The absolute levels of ferric chelate reductase activity measured in the detached roots from either wild type or frd3 are significantly lower than the reductase activity exhibited by roots of intact plants. This drop in ferric chelate reductase activity occurs immediately after shoot removal and probably reflects the nonphysiological nature of detached roots. Nevertheless, the detached roots continued to grow throughout the experiment and were able to respond to iron levels in the media (see below).
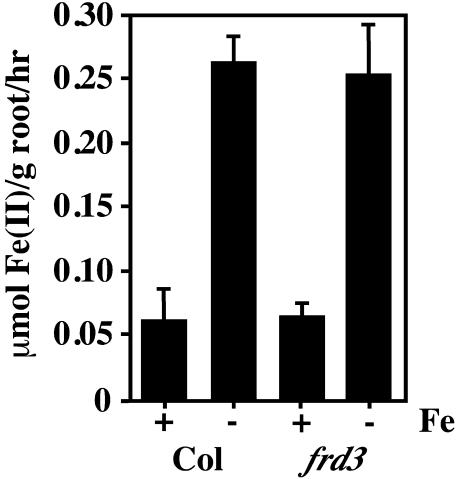
As is shown in Figure 2, wild-type detached roots behaved similarly to roots of intact wild-type plants, with ferric chelate reductase activity repressed under iron-sufficient conditions and elevated under iron deficiency. Interestingly, frd3 mutant roots, in the absence of a shoot signal, were capable of appropriately regulating their ferric chelate reductase activity. By 5 d after transfer, the frd3 mutant roots grown under iron-deficient conditions expressed high levels of reductase activity. More importantly, the detached frd3 mutant roots appropriately repressed their ferric chelate reductase activity under iron-sufficient conditions, a response never seen in intact frd3 mutant seedlings. This result is also consistent with the idea that the root mechanisms that control the iron uptake responses are functional in the frd3 mutant, implying that the defect in the frd3 mutant is not in its ability to down-regulate its root iron uptake responses under conditions of iron sufficiency.
Figure 2.
frd3 mutant roots can appropriately regulate their iron deficiency responses in the absence of a shoot signal. Seedlings had the shoot portion removed prior to transfer to iron-sufficient (+Fe) or iron-deficient (−Fe) media with added Suc. The ferrozine method was used to quantitate root ferric chelate reductase 5 d after transfer. This experiment was repeated three times; a representative experiment is shown.
Causes of Chlorosis in the frd3 Mutant
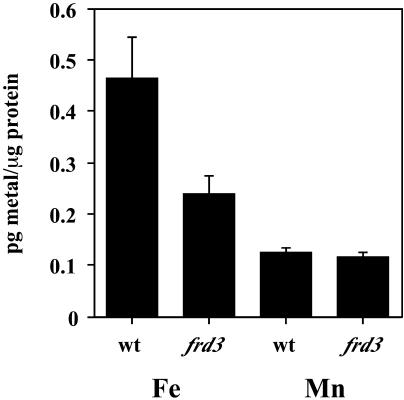
There are two potential causes of the chlorosis observed in frd3 mutant shoots. One is iron deficiency; that in spite of the high iron levels present in whole leaves of the frd3 mutant, the chloroplastic or intracellular iron levels could be low. The other potential cause of chlorosis is an excess of iron or other redox active metals such as manganese (Gonzalez et al., 1998). To distinguish between these two possibilities, we determined what proportion of the iron in a plant leaf is intracellular versus apoplastic. Here, protoplasts were isolated and used as the best available assay of the intracellular fraction. Protoplasts were isolated from shoots of 2-week-old wild-type and frd3 mutant seedlings. Previous elemental analysis on plants grown under these conditions demonstrated that frd3 shoots contain approximately twice as much iron per gram fresh weight as do wild-type shoots (Rogers and Guerinot, 2002a). By isolating protoplasts, we removed the vascular tissue, apoplastic contents, cell walls, and any metals associated with these portions of the shoot. The protoplasts were subjected to elemental analysis and the results are shown in Figure 3. Protoplasts isolated from frd3 mutant seedlings contain only approximately one-half as much iron on a total protein basis as do wild-type protoplasts. On the other hand, manganese levels, which are approximately 3 times higher in frd3 shoots than in wild type, are not significantly different in protoplasts. This indicates that frd3 chlorosis is not caused by manganese excess, since intracellular manganese levels are not increased. We have also grown frd3 plants on manganese deficient medium and do not see a difference in either the chlorosis or constitutive ferric chelate reductase activity phenotypes of the frd3 mutant (data not shown). The protoplast metal data indicate that extracellular or apoplastic iron and manganese must be accumulating to abnormally high levels in the frd3 mutant. The lower intracellular iron levels in frd3 support the hypothesis that iron is mislocalized in the frd3 mutant.
Figure 3.
Protoplasts from frd3 mutant shoots contain less iron than wild-type protoplasts. Protoplasts were isolated from 2-week-old wild-type and frd3 protoplasts and subjected to elemental analysis. This experiment was performed three times; representative results are shown.
Iron Localization in the Root
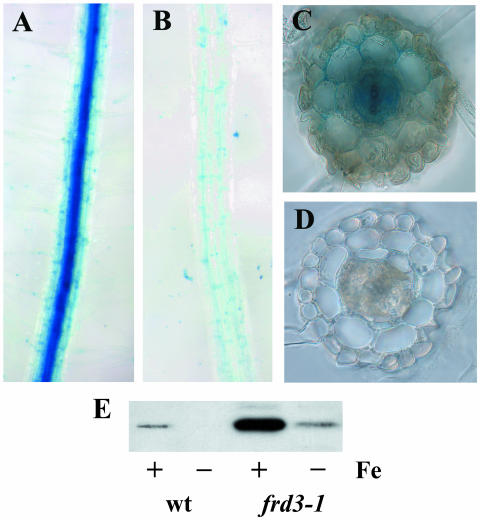
The roots of the frd3 mutant also accumulate excess iron (Delhaize, 1996; Rogers and Guerinot, 2002a). Since it is very difficult to obtain protoplasts from roots, we resorted to a qualitative measure of iron localization in the root tissue. Perls' stain is an acidic solution of potassium ferrocyanide, which reacts with ferric iron to form an insoluble blue precipitate. The insoluble nature of the precipitate formed in this reaction means that the blue color will not diffuse after staining and is an accurate reflection of iron localization in living tissue. As shown in Figure 4, iron accumulates to high levels in the central vascular portion of frd3 roots. This is in contrast to wild-type roots, where iron levels throughout the root are low enough that little blue color forms with Perls' stain. Excess iron accumulation in frd3 mutant roots is in agreement with published root iron levels in frd3-3 (man1; Delhaize, 1996). Perls' staining was also performed on the leaves of wild type and frd3 mutants; however, no staining was observed, indicating that the level of iron in the shoot vasculature of both wild type and frd3 are below the detection limits for Perls' stain. Therefore, no conclusions about iron localization in the shoot can be drawn from this data.
Figure 4.
Iron accumulates in the vascular cylinder of frd3 mutant roots. Five-day-old seedlings were fixed in paraformaldehyde and stained with Perls' stain to visualize ferric iron. Roots were then imbedded and 70-μm sections cut. A, frd3-1 Intact roots; B, wild-type intact roots; C, frd3-1 cross section; D, wild-type cross section. E, Immunoblot showing ferritin protein levels in roots of 2-week-old plants grown under iron-sufficient or deficient conditions for 3 d prior to harvest.
Figure 4, section E shows an immunoblot of total root proteins probed with anti-ferritin antibodies. Root tissue from frd3 mutants grown under iron-sufficient conditions shows significantly higher levels of the ferritin protein than does wild-type root tissue. This result is consistent with the higher iron levels demonstrated by elemental analysis and by Perls' stain. It also indicates that much of the iron found in frd3 roots is intracellular, since it triggers the accumulation of the ferritin protein.
Localization of the FRD3 Protein
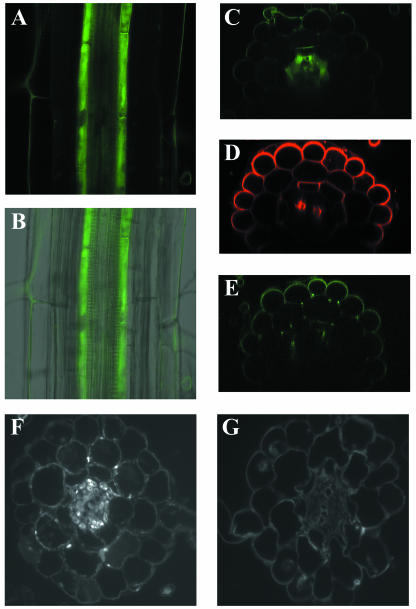
We had previously reported that, by RNA blot and reverse transcription-PCR analysis, the wild-type FRD3 gene was only expressed to detectable levels in Arabidopsis root tissue. To more precisely localize the expression of the FRD3 gene, an FRD3-green fluorescent protein (GFP) fusion construct was used. This fusion protein is capable of complementing the frd3-1 mutant phenotypes. Expression of the fusion protein is driven by the FRD3 promoter to accurately reflect endogenous FRD3 gene expression. Figure 5 shows that the FRD3 gene is expressed in the pericycle and cells internal to the pericycle and surrounding the vascular tissue. The green fluorescence observed in the epidermis is autofluorescence; it is also present in untransformed controls and so is not due to GFP (Fig. 5, section E). The location of FRD3 gene expression is very similar to the site of ferric iron accumulation in the frd3 mutant.
Figure 5.
FRD3 is expressed in the root pericycle and vascular cylinder. A, Confocal fluorescence section of a root expressing FRD3-GFP fusion protein. B, GFP image from A overlaid with a transmitted light image of the same root. C, Optical cross section of a root expressing FRD3-GFP fusion protein. D, Optical cross section of a similar root stained with the cell wall dye FM-143. E, Optical cross section of a wild-type root showing epidermal autofluorescence. F, Immunofluorescence of FRD3-FLAG. G, Immunofluorescence control of a root not expressing the FLAG epitope.
The FRD3 protein is predicted to contain 14 transmembrane domains and to be a member of the MATE family of membrane proteins. Therefore, the FRD3 protein is predicted to localize to a membrane. Close inspection of Figure 5, section A reveals green fluorescence both internal to the pericycle cells and outlining a number of pericycle cells. While this could indicate the presence of FRD3 in the cytoplasm or on intracellular vesicles, we believe this is more likely the result of degradation of some of the FRD3-GFP fusion protein. By immunoblot, there are four protein species that cross-react with anti-GFP antibodies (data not shown). The largest of these is approximately the size of a full-length FRD3 protein fused to GFP; the other three cross-reactive bands are significantly smaller and probably represent degradation products from which some or all of the FRD3 portion of the fusion protein has been removed. For this reason, it is not possible to definitively localize the FRD3 protein on a subcellular level using this FRD3-GFP fusion protein. However, the presence of FRD3-GFP degradation products does not affect the result that the FRD3 gene is expressed in the pericycle and vascular cylinder.
A construct containing FRD3 protein fused to the FLAG epitope tag was also constructed and transformed into wild type and the frd3-1 mutant. By immunoblotting, there is a single anti-FLAG antibody cross-reactive species of the appropriate Mr to be the full-length FRD3-FLAG protein (data not shown). Like the FRD3-GFP fusion, the FRD3-FLAG is able to complement frd3-1. Immunofluorescence experiments were performed on roots expressing the FRD3-FLAG protein and showed that the FRD3-FLAG protein is present in the central vascular cylinder, the pericycle cells and smaller cells surrounding the vascular tissue (Fig. 5, section F), confirming the results obtained for the FRD3-GFP fusion.
DISCUSSION
Previous work suggested two possible models of FRD3 action. The first was that FRD3 is part of an iron signaling pathway. It is likely that plant root iron uptake responses are controlled at least in part by a shoot-derived signal of shoot iron status (Grusak and Pezeshgi, 1996). This signal must both be produced by the shoot and detected in the root. If the frd3 mutant were defective in iron signaling, the defect would likely be specific to either the shoot or the root. The reciprocal grafting experiments presented in Figure 1 indicate that the phenotype of a grafted plant is determined by the genotype of the root, not the genotype of the shoot. The fact that a wild-type root appropriately regulates its iron uptake responses when grafted to a frd3 mutant shoot demonstrates that frd3 mutant shoots are capable of generating a functional signal of iron status.
The competency of frd3 mutant roots to perceive a shoot-generated signal and appropriately regulate iron uptake responses is demonstrated in the detached root experiments presented in Figure 2. Figure 2 clearly demonstrates that in the absence of a shoot and therefore lacking a shoot-derived signal of iron status, frd3 mutant roots appropriately regulate their ferric chelate reductase activity. This includes repressing their reductase activity when grown under iron-sufficient conditions, which is never seen in intact frd3 mutants. These results are inconsistent with the hypothesis that the frd3 mutant is defective in any aspect of iron deficiency signaling.
Our second hypothesis concerning FRD3 action is that FRD3 is involved in shoot iron localization. In this case, the chlorosis observed in the frd3 would be caused by low iron levels in the shoot cells or organelles where it is needed. Here we have presented several lines of evidence that demonstrate that frd3 mutants mislocalize iron in their tissues. Even though whole leaves of frd3 have elevated levels of iron and manganese, frd3 leaf protoplasts have only approximately one-half as much iron as do those from wild type (Fig. 3), and manganese levels in the two are similar. Since frd3 shoots as a whole overaccumulate both iron and manganese, two redox active metals, it was possible that the chlorosis in frd3 shoots was caused by oxidative damage (Gonzalez et al., 1998). However, protoplasts from frd3 contain approximately equivalent manganese levels and significantly lower iron levels, indicating that the excess metals in frd3 mutant are spatially separated from the chloroplasts. Therefore, it is unlikely the frd3 chlorosis is caused by metal overaccumulation. Perls' staining of wild-type and frd3 mutant roots reveals that frd3 roots accumulate high levels of ferric iron in the vascular cylinder (Fig. 4). This is in contrast to wild-type roots, which have much lower iron levels throughout their root tissues. Additionally, wild-type shoots grafted to frd3 roots develop chlorosis and stop accumulating ferritin, indicating problems with iron supply and utilization. Furthermore, frd3 roots grafted to wild-type shoots continue to constitutively express their iron uptake responses, perhaps as a result of a signal from the iron-deficient shoot. This is strong evidence that frd3 mutant roots cannot supply iron to the shoot in a usable form. All of the frd3 mutant phenotypes can be explained if, in the mutant, iron is not delivered to the shoot in a form that is able to be taken up and localized appropriately in the shoot.
It is formally possible that the phenotypes of the frd3 mutant are not caused by changes in iron uptake or localization but rather by the overaccumulation of manganese observed in frd3 (Rogers and Guerinot, 2002a; Lahner et al., 2003). It is possible that the excess manganese in frd3 mutants could compete with iron for uptake into leaf cells and therefore lead to iron deficiency. However, since the intracellular manganese levels in frd3 are no higher than they are in wild type (Fig. 3) it is unlikely that the excess manganese is successfully competing with iron for uptake either into leaf cells or into plastids. Additionally, we have grown frd3 plants under manganese deficient conditions and see no changes in either their chlorosis or constitutive expression of root ferric chelate reductase activity (data not shown); this also argues against manganese excess causing the frd3 mutant phenotypes. Finally, foliar application of iron tended to reduce both the leaf chlorosis and the root ferric chelate reductase activity of frd3 mutant (data not shown). Taken together, this data strongly suggests that the frd3 mutant phenotypes are not caused by manganese overaccumulation. Rather, we would suggest that the manganese overaccumulation is due to the constitutive expression of the IRT1 transporter, which has been shown to transport manganese (Korshunova et al., 1999).
The FRD3 gene is expressed in the central cylinder of the root, in the pericycle cells, and cells surrounding the vascular tissues (Fig. 5). Roots of the frd3 mutant also overaccumulate iron in this central vascular cylinder. The FRD3 gene encodes a protein containing 14 predicted transmembrane domains. Therefore, the FRD3 protein is likely to be associated with a cell membrane. Both the FRD3-GFP fusion fluorescence and the FRD3-FLAG immunofluorescence are brightest at the plasma membranes. However, some of the FRD3-GFP fusion protein is degraded and therefore cannot be used to determine subcellular localization. Additionally, cells in the central vascular cylinder are too small to be able to clearly observe plasma membrane localization. Unfortunately, the FRD3-FLAG protein is expressed at low levels and was not detectable by immunoelectron microscopy (data not shown). Nevertheless, the localization results presented in Figure 5 are consistent with at least some of the FRD3 protein localizing to the plasma membrane. FRD3 belongs to the MATE family of transmembrane proteins (Rogers and Guerinot, 2002a). Other members of the MATE family have been implicated in the efflux of low Mr organic compounds (Morita et al., 1998, 2000).
Together with the data presented, this leads to a new question: how can a putative effluxer of low Mr compounds that is expressed around the root vasculature affect iron localization in the shoot? Iron reaches the shoot tissue in sufficient amounts in the frd3 mutant; even in soil-grown frd3 shoots, shoots grown under conditions of low iron availability, iron levels are only about 10% lower than in wild type (Lahner et al., 2003). This is consistent with iron being loaded into the xylem appropriately in the frd3 mutant since significant amounts of iron do reach the shoot; we do not believe that the FRD3 protein is directly responsible for iron loading into the xylem or for other iron transport steps. The problem in the frd3 mutant appears to be getting iron out of the xylem and into the shoot symplast and then to the chloroplasts. However, since FRD3 is not expressed to detectable levels in the shoots, it is doubtful that FRD3 acts in the shoots, either as an iron transporter or otherwise. This is also consistent with the reciprocal grafting experiments that indicate FRD3 is not needed in the shoots for wild-type iron localization and signaling.
Given FRD3's expression in cells surrounding the root vasculature, we hypothesize that FRD3 effluxes into the xylem a low Mr compound that is necessary for correct iron unloading from the xylem in the shoot. This compound could be an iron chelator. Certainly, given the low solubility of ferric iron at the pH of the xylem, approximately pH 6, ferric iron would need to be chelated to move efficiently. It is widely thought that iron moves in the xylem as ferric citrate (Pich et al., 1994; von Wirén et al., 1999). Although the FRD3 protein could be a citrate transporter, it does not share sequence similarity to other known citrate effluxers. Nicotianamine (NA) is another molecule that has been implicated in long distance iron transport in plants. From gene expression studies of both the chloronerva (chln) gene in tomato and the nicotianamine synthases genes in Arabidopsis, it appears that NA is synthesized throughout the plant (Ling et al., 1999; Suzuki et al., 1999). It is not known where, or via what type of transporter, NA is loaded into the vasculature; this is a possible function for FRD3. However, because the NA-less chln mutant has alterations in copper homeostasis as well as in iron homeostasis (Pich et al., 1994; Herbik et al., 1996) and the frd3 mutant does not have alterations in copper levels (Lahner et al., 2003), it is unlikely that the frd3 mutant has defects in NA metabolism and transport or that the FRD3 protein is involved in NA transport.
The FRD3 protein also could transport one of a variety of compounds necessary for iron uptake in the shoot. Once the ferric iron reaches the shoot apoplast, it is thought to be reduced, probably by a member of the FRO (ferric reductase oxidase) family. FRD3's substrate could be involved in presenting iron to the reductase. After reduction, the ferrous iron could be transported inside the leaf cells either on its own, perhaps by a member of the ZIP (Zrt- Irt- like protein) family (Rogers and Guerinot, 2002b), or as a complex with NA, via one of the YSL (yellow-stripe like) family of transporters (Curie et al., 2001; DiDonato et al., 2003). A number of yellow-stripe like genes are expressed in the shoots and may be responsible for unloading iron from the xylem. It is also possible that FRD3 effluxes a molecule necessary for maintaining pH in the xylem. If the xylem pH was slightly higher in the frd3 mutant than in wild type, that could lower iron solubility in the xylem just enough to cause the observed difficulty in shoot iron uptake. However, the pH of xylem fluid exuded from cut hypocotyls of frd3 mutant plants is approximately 5.5, a little lower than xylem fluid from wild type, which is approximately 5.8 (T.P. Durrett and E.E.Rogers, unpublished data). The xylem pH of iron deficient plants can be lower than that of iron sufficient plants (Lopez-Millan et al., 2000). Therefore, the observed xylem pH difference may simply be caused by frd3's constitutive iron deficiency rather than a defect in xylem loading in the frd3 mutant. The xylem pH difference between wild type and frd3 might affect which ligands preferentially bind iron in the xylem but it would not be expected to reduce iron solubility in the frd3 xylem. Although further work will be required to identify the substrate of FRD3, we can predict that it would have certain characteristics: it will be necessary for iron reduction in or unloading from the xylem or shoot apoplast, and it will have to be loaded into the xylem in the root.
MATERIALS AND METHODS
Arabidopsis Lines and Growth Conditions
The Arabidopsis mutants frd3-1 and frd3-3 and the corresponding wild type Columbia gl-1 have been described previously (Rogers and Guerinot, 2002a). Plants were grown under constant 150 μmol photons/m2/s light at 22°C and under sterile conditions as previously described (Rogers and Guerinot, 2002a), except for the detached root experiments and seedling timecourse where 20 g/L Suc was added to the iron sufficient and iron deficient media (Yi and Guerinot, 1996). Ferric chelate reductase assays also were described previously (Yi and Guerinot, 1996).
Reciprocal Grafting
Micrografting of Arabidopsis seedlings was performed as described (Turnbull et al., 2002). Single-hypocotyl 90° butt grafts were performed on 4- or 5-d-old seedlings; collars were not used. Grafted plants were allowed to recover for 5 d at 26°C and constant 150 μmol photons/m2/s light. Graft integrity was verified by physical strength of the graft site, an absence of adventitious roots on the scion and robust growth of the both the root system and scion.
Detached Root Experiments
Arabidopsis seedlings were grown on Gamborg's B5 medium (Caisson Labs, Sugar City, ID) for approximately 2 weeks. The hypocotyl was cut with a sharp razor blade and shoot was removed. The root was immediately transferred to iron sufficient or iron deficient media prepared as described except for the addition of 20 g/L Suc. After 5 d, ferric chelate reductase activity was assayed as described (Yi and Guerinot, 1996). Under these conditions, the detached roots continued to grow and did not become necrotic in appearance.
Chlorophyll Extraction and Quantitation
Chlorophyll was extracted in methanol and absorbance measured at 652, 665, and 750 nm. Total chlorophyll concentration was calculated as described (Porra et al., 1989).
Immunoblots
Immunoblots were performed as previously described (Connolly et al., 2002). Total protein was prepared from the roots and shoots of plants grown axenically on plates that were either iron-deficient or iron-sufficient. Extracts were prepared by grinding tissue (1–2 mL buffer/1 g wet tissue) on ice in extraction buffer (50 mm Tris, pH 8.0, 5% glycerol, 4% SDS, 1% polyvinyl-polypyrrolidone, 1 mm phenylmethylsulfonyl fluoride), followed by centrifugation at 4°C for 15 min at 14,000g. The supernatant was recovered and total protein was estimated using the BCA protein assay (Pierce, Rockford, IL). Samples for SDS-PAGE were diluted with an equal volume of 2× sample prep buffer (Ausubel et al., 2004) and boiled for 2 min.
Total protein (10 μg) was separated by SDS-PAGE (Laemmli, 1970) and transferred to nitrocellulose membrane by electroblotting (Towbin et al., 1979). Equal loading of blot lanes was verified by Coomassie staining of equivalently loaded gels run in parallel (Ausubel et al., 2004). Membranes were blocked in 1× PBST (0.1% Tween 20 in 1× PBS) with 5% nonfat dry milk for 3 h at 37°C and then washed 2 times in 1× PBST for 5 min each. The membranes were then incubated overnight at 4°C an anti-ferritin antibody (1:1,000-fold dilution in 1× PBST, 1% nonfat dry milk). The ferritin antibody was raised against purified pea seed ferritin (Van Wuytswinkel et al., 1995) and was the kind gift of Dr. Jean-François Briat. It has previously been shown to cross react with Arabidopsis ferritin (Gaymard et al., 1996). Next, the membranes were washed in 1× PBST, four times for 15 min each. Membranes were then incubated for 1 h with goat-anti-rabbit IgG conjugated to Horseradish peroxidase (1:5,000 dilution in 1× PBST, 1% nonfat dry milk; Pierce) followed by 4 washes for 15 min each in 1× PBST. Chemiluminescence was performed using the SuperSignal West Pico chemiluminescent substrate kit (Pierce).
Protoplast Isolation and Elemental Analysis
Arabidopsis protoplasts were isolated as previously described (Fitzpatrick and Keegstra, 2001). Protoplast integrity was verified by microscopic examination. Total protoplast protein levels were measured with a BCA Protein Assay Kit (Pierce). Iron and manganese levels were measured at the MU Research Reactor Center in a VG AXIOM high resolution ion coupled plasma mass spectrometry (ICP-MS).
Perls' Stain for Ferric Iron
Arabidopsis roots were treated with Perls' stain according to established histological methods for mammalian tissues. Briefly, equal amounts of solutions of 4% (v/v) HCl and 4% (w/v) potassium ferrocyanide were mixed immediately prior to use. The stain solution was vacuum infiltrated into 6- or 7-d-old Arabidopsis seedlings for approximately 15 min. Seedlings were rinsed in water and Perls' staining was observed immediately in whole roots. For cross-sections, after staining seedlings were fixed in 4% (w/v) paraformaldehyde and imbedded in 4% (w/v) low-melt agarose. Approximately 70-μm sections were cut with a Lancer series 1000 Vibratome (Vibratome, St. Louis).
FRD3-Fusion Constructs and Localization
The FRD3 genomic sequence cloned into pCAMBIA2300 has been previously described (Rogers and Guerinot, 2002a). A convenient restriction site at the 3′ end of the FRD3 coding sequence in this construct was used to insert DNA coding for the desired fusion. For the FRD3-GFP construct, the GFP gene sequence was obtained by PCR from a version optimized for plant expression (Davis and Vierstra, 1998). For the FRD3-FLAG construct, the FLAG sequence (DYKDDDDK) was encoded in custom synthesized oligonucleotides that were phosphorylated, hybridized to each other, and ligated into the cut FRD3 genomic sequence. All molecular biology techniques were performed using standard protocols (Ausubel et al., 2004). The fusion clones were introduced into wild-type and frd3-1 Arabidopsis by Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). At least six independent transformants of each construct in the frd3-1 mutant were examined to confirm complementation by the FRD3-fusion constructs. With each construct in wild-type Columbia gl-1Arabidopsis, 8 to 10 independent lines were analyzed by segregation to confirm a single insertion event and obtain lines homozygous for the insertion. Preliminary localization was performed on 5 independent FRD3-GFP lines and 2 independent FRD3-FLAG lines. For each, one representative line was chosen for detailed analysis.
GFP fluorescence was visualized in living whole-mount roots using a Bio-Rad (Hercules, CA) Radiance 2000 confocal system coupled to an Olympus IX70 inverted microscope at the University of Missouri Molecular Cytology Core facility. Roots from FRD3-FLAG transgenics were fixed, embeded in methacrylate, sectioned, and stained for immunofluorescence as previously described (Baskin and Wilson, 1997).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
The authors thank Tobias Baskin and Jan Judy-March for help with microscopy and immunofluorescence, Amanda Crawford and Dave Robertson for the ICP-MS analysis, Fritz Bienfait for helpful suggestions, and David Eide, Dirk Charlson, and Mary Lou Guerinot for critical reading of the manuscript.
This work was supported by an MU Research Board grant and a USDA CSREES grant (2002–35100–12331 to E.E.R.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.045633.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2004) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Baskin TI, Wilson JE (1997) Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol 113: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Connolly EC, Fett J, Guerinot ML (2002) Transgenic plants engineered to overexpress the IRT1 metal transporter reveal post-transcriptional regulation by metals. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat J-F, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349 [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36: 521–528 [DOI] [PubMed] [Google Scholar]
- Delhaize E (1996) A metal-accumulator mutant of Arabidopsis thaliana. Plant Physiol 111: 849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato R, Roberts L, Pierson A, Walker E (2003) The Arabidopsis yellow-stripe1-like (YSL) family of metal-nicotianamine transporters. In 1st Pan-American Plant Membrane Biology Workshop, Cuernavaca, Mexico, International Center for Genetic Engineering and Biotechnology
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick L, Keegstra K (2001) A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J 27: 59–65 [DOI] [PubMed] [Google Scholar]
- Gaymard F, Boucherez J, Briat JF (1996) Characterization of ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem J 318: 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Steffen KL, Lynch JP (1998) Light and excess manganese. Implications for oxidative stress in common bean. Plant Physiol 118: 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA, Pezeshgi S (1996) Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol 110: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (1992) Biologically relevant metal ion-dependent hydroxyl radical generation. FEBS Lett 307: 108–112 [DOI] [PubMed] [Google Scholar]
- Herbik A, Giritch A, Horstmann C, Becker R, Balzer H, Bäumlein H, Stephan UW (1996) Iron and copper nutrition-dependent changes in protein expression in a tomato wild type and the nicotianamine-free mutant chloronerva. Plant Physiol 111: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova Y, Eide D, Clark G, Guerinot M, Pakrasi H (1999) The Irt1 protein from Arabidopsis thaliana is a metal transporter with broad specificity. Plant Mol Biol 40: 37–44 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophange T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lahner B, Gong J, Mahmoudian M, Smith E, Abid K, Rogers E, Guerinot M, Harper J, Ward J, McIntyre L, et al (2003) Ionomics: The genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol 21: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Ling H-Q, Koch G, Baumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96: 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Millan AF, Morales F, Abadia A, Abadia J (2000) Effects of iron deficiency on the composition of the leaf apoplastic fluid and xylem sap in sugar beet. Implications for iron and carbon transport. Plant Physiol 124: 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Kataoka A, Shiota S, Mizushima T, Tsuchiya T (2000) NorM of Vibrio parahaemolyticus is a Na+-driven multidrug efflux pump. J Bacteriol 182: 6694–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T (1998) NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 42: 1778–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J, Quinn J, Eriksson M, Merchant S (2000) The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J 19: 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pich A, Scholz G, Stephan UW (1994) Iron-dependent changes of heavy metals, nicotianamine, and citrate in different plant organs and in the xylem exudate of two tomato genotypes. Nicotianamine as possible copper translocator. Plant Soil 165: 189–196 [Google Scholar]
- Porra R, Thompson W, Kreidemann P (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 348–394 [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Guerinot ML (2002. a) FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Guerinot ML (2002. b) Iron Acquisition in Plants. In D Templeton, ed, Molecular and Cellular Iron Transport. Marcel Dekker, New York, pp 359–373
- Schikora A, Schmidt W (2001) Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol 125: 1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller S, Castlefranco A, Castlefranco P (1982) Effects of iron and oxygen on chlorophyll biosynthesis. 1. In vivo observations on iron and oxygen-deficient plants. Plant Physiol 69: 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Higuchi K, Nakanishi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes from Arabidopsis thaliana. Soil Sci Plant Nutr 45: 993–1002 [Google Scholar]
- Tottey S, Block M, Allen M, Westergren T, Albrieux C, Scheller H, Merchant S, Jensen P (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA 100: 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Booker J, Leyser H (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Van Wuytswinkel O, Savino G, Briat J-F (1995) Purification and characterization of recombinant pea-seed ferritins expressed in Escherichia coli: influence of N-terminus deletions on protein solubility and core formation in vitro. Biochem J 305: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot M, Briat J-F, Curie C (2002) IRT1, an Arabidopsis Transporter Essential for Iron Uptake from the Soil and Plant Growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wirén N, Klair S, Bansal S, Briat J-F, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both Fe(III) and Fe(II). Implications for metal transport in plants. Plant Physiol 119: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]