Abstract
There are at least five lipoxygenases (TomloxA, TomloxB, TomloxC, TomloxD, and TomloxE) present in tomato (Lycopersicon esculentum Mill.) fruit, but their role in generation of fruit flavor volatiles has been unclear. To assess the physiological role of TomloxC in the generation of volatile C6 aldehyde and alcohol flavor compounds, we produced transgenic tomato plants with greatly reduced TomloxC using sense and antisense constructs under control of the cauliflower mosaic virus 35S promoter. The expression level of the TomloxC mRNA in some transgenic plants was selectively reduced by gene silencing or antisense inhibition to between 1% and 5% of the wild-type controls, but the expression levels of mRNAs for the four other isoforms were unaffected. The specific depletion of TomloxC in transgenic tomatoes led to a marked reduction in the levels of known flavor volatiles, including hexanal, hexenal, and hexenol, to as little as 1.5% of those of wild-type controls following maceration of ripening fruit. Addition of linoleic or linolenic acid to fruit homogenates significantly increased the levels of flavor volatiles, but the increase with the TomloxC-depleted transgenic fruit extracts was much lower than with the wild-type control. Confocal imaging of tobacco (Nicotiana tabacum) leaf cells expressing a TomloxC-GFP fusion confirmed a chloroplast localization of the protein. Together, these results suggest that TomloxC is a chloroplast-targeted lipoxygenase isoform that can use both linoleic and linolenic acids as substrates to generate volatile C6 flavor compounds. The roles of the other lipoxygenase isoforms are discussed.
Lipoxygenases (LOX; EC1.13.11.12) are nonheme iron-containing dioxygenases that catalyze the incorporation of molecular oxygen into polyunsaturated fatty acids containing a cis, cis-1.4-pentadiene moiety, such as linoleic and linolenic acids, converting them into fatty acid hydroperoxides (HPOs). Multiple isoforms of LOX have been detected in a wide range of plants, animals, and microorganisms (Zimmerman and Vick, 1973; Eskin et al., 1977; Shechter and Grossman, 1983; Hamberg, 1986; Samuelsson et al., 1987; Vick and Zimmerman, 1987). The LOX isoforms are distinguished by differences in reaction pH optimum, pI, substrate and product specificity, tissue-specific or subcellular localization, and synthesis at particular developmental stages (Axelrod, 1974; Bild et al., 1977; Axelrod et al., 1981; Ferrie et al., 1994; Royo et al., 1996; Heitz et al., 1997).
In animals, it is well established that HPOs are the primary metabolites of the pathways that lead to the formation of important regulatory molecules in inflammatory responses such as leukotrienes and lipoxins (Yamamoto, 1991). In higher plants, on the other hand, the physiological role of HPOs generated by individual LOX isoforms is still uncertain. It has been postulated that plant lipoxygenases may be involved in plant growth and development; biosynthesis of regulatory molecules, such as jasmonic acid (JA) and traumatin; biosynthesis of volatile compounds, such as hexanal, hexenal, and hexenol involved in flavor, insect attraction, and defense; and senescence and response to wounding and to stress (Todd et al., 1990; Siedow, 1991; Bell et al., 1995; Heitz et al., 1997; Feussner and Wasternack, 2002).
Tomatoes (Lycopersicon esculentum) are a widely consumed fruit worldwide and there is a particular interest in understanding and improving their flavor. More than 400 volatile compounds have been identified in the ripening tomato fruit (Baldwin et al., 1991), but only a small number of these are important for producing the characteristic fresh tomato aroma (Teranishi et al., 1990). These constituents include fatty acid-derived short-chain volatile aldehydes and alcohols, such as hexanal, (Z)-3-hexenal, (E)-2-hexenal, and (Z)-3-hexenol, which are produced by enzymic action during maceration. Linoleic and linolenic acids are the principal LOX substrates in tomato, and their metabolism by the lipoxygenase pathway is determined by the substrate and product specificities of the lipoxygenases and fatty acid hydroperoxide lyases (HPLs). LOX enzymes can be grouped into two types: 9-LOX, which specifically forms 9-HPOs, and 13-LOX, which specifically forms 13-HPOs. There are also two types of HPLs: 9-HPLs and 13-HPLs, cleaving either 9-HPOs or 13-HPOs and generating either two C9 fragments or a C6 and a C12 fragment, respectively (Hatanaka, 1993). The product of the reaction of linoleic acid with soybean cotyledon lipoxygenase-1 is almost exclusively 13-hydroperoxy linoleic acid (Axelrod, 1974; Axelrod et al., 1981), whereas with lipoxygenases-2 and -3, roughly equal amounts of the 9- and 13-hydroperoxy products are obtained. In potato (Solanum tuberosum), LOX1 is expressed in tubers and roots and uses linoleic acid to produce predominantly 9-hydroperoxides, whereas LOX2 (expressed in leaves) and LOX3 (expressed in leaves and roots) preferentially use linoleic acid to produce 13-hydroperoxides (Royo et al., 1996). In tomato fruit, the majority of the HPOs formed by lipoxygenase activity are the 9-isomers (Galliard and Matthew, 1977; Smith et al., 1997), but there are apparently little or no 9-HPLs to act on 9-HPOs, which therefore accumulate. This property is widely utilized to prepare 9-HPOs in a simple manner (Matthew et al., 1977). The much smaller proportion of the 13-HPOs formed in tomato appear to be metabolized further by the action of the 13-HPLs to give rise to hexanal, hexenal, and the corresponding alcohols, but the subcellular locations and specificities of the lipoxygenase isoforms responsible for the formation of the 13-isomers have not been determined.
Five tomato LOX genes have been shown to be expressed during fruit ripening, TomloxA and TomloxB (Ferrie et al., 1994), TomloxC and TomloxD (Heitz et al., 1997), and TomloxE (National Center for Biotechnology Information [NCBI] accession no. AY008278). TomloxA, TomloxB, and TomloxE are 72% to 77% identical with each other at the amino acid level, while TomloxC and TomloxD show 42% and 47% identity, respectively, to the TomloxA protein, and 46% identity to each other. TomloxC and TomloxD contain chloroplast-targeting signals and are imported into chloroplasts in vitro (Heitz et al., 1997). TomloxD is expressed at a very low level in fruit at the mature green stage and at onset of ripening (Heitz et al., 1997), whereas TomloxC is expressed during ripening. TomloxB and TomloxC expression is enhanced by the ripening hormone ethylene, whereas TomloxA expression declines (Griffiths et al., 1999a). These results indicate that individual LOX isoforms are differentially regulated and may have distinct functions in ripening fruit, such as defense and biosynthesis of flavor compounds.
Potato LOX H1, a 13-lipoxygenase expressed in leaves, is the closest known homolog to TomloxC. Transcripts of LoxH1 are systemically up-regulated in both wounded and nondamaged tissues, indicating a role in signaling or defense (Royo et al., 1996). However, gene silencing of LoxH1 had no effect on the basal or wound-induced levels of jasmonates derived from 13-hydroperoxylinolenic acid (Leon et al., 2002). Potato plants with silenced LOX H3, a TomloxD homolog, showed greatly reduced proteinase inhibitor expression, indicating a role in defense signaling (Royo et al., 1999). We have previously shown that antisense-suppression of TomloxA and TomloxB in tomato fruit causes no significant changes in the production of the known tomato flavor volatiles (Griffiths et al., 1999b). However, in these studies, the expression of TomloxC was unaffected (Griffiths et al., 1999a). We report here that specific depletion of TomloxC in tomato by cosuppression or antisense-inhibition leads to major decreases in the flavor volatiles in both fruit and leaf. The results suggest that TomloxC is specifically involved in the generation of C6 aldehydes and alcohols, which have been shown previously to be important constituents of flavor in tomato.
RESULTS
Cosuppression- and Antisense-Mediated Depletion of TomloxC mRNA
To examine the physiological role of TomloxC, tomato plants were transformed with both sense (pLoxCS) and antisense (pLoxCA) constructs of TomloxC cDNA driven by the constitutive cauliflower mosaic virus (CaMV) 35S promoter, as described in “Materials and Methods” (Fig. 1). Thirteen independent transformants containing construct pLoxCA and 12 containing pLoxCS were selected by their ability to grow on kanamycin and were grown to maturity.
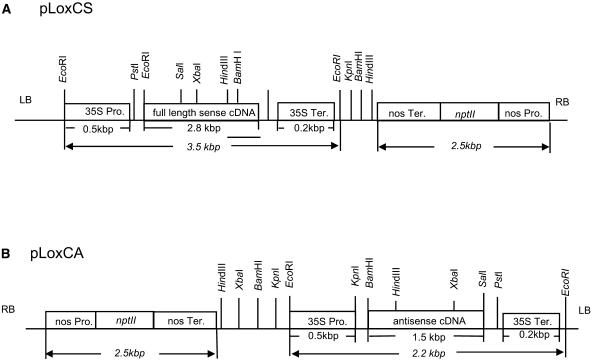
Figure 1.
Structures of the sense and antisense lipoxygenase TomloxC constructs. pLoxCS (A) contained a full-length sequence of TomloxC in the sense orientation and pLoxCA (B) contained a 1.5-kb antisense fragment. Each construct was under the control of the CaMV 35S promoter and included the 35S terminator and the nptII selectable marker gene. The diagrams are not drawn to scale. The design of the constructs is fully described in “Materials and Methods.”
Total RNA was extracted from the fruit and leaves of control and transgenic plants and analyzed by northern blot, using a TomloxC cDNA fragment as a probe. It should be noted that a previous study by Heitz reported the complete absence of TomloxC transcripts in the foliage of wild-type tomato plants (Heitz et al., 1997). However, using a sensitive northern detection method (“Materials and Methods”) we are able to detect TomloxC transcripts at low levels in leaves (Fig. 2).
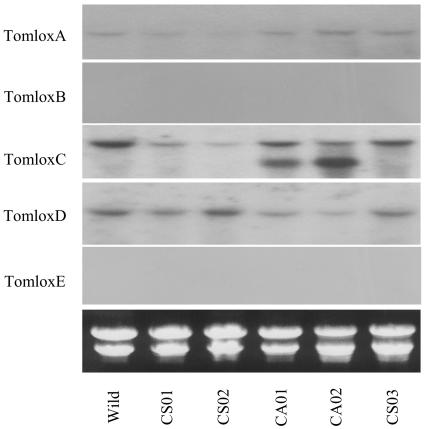
Figure 2.
Expression of TomloxA, TomloxB, TomloxC, TomloxD, and TomloxE mRNAs in leaf in wild-type control and the TomloxC transgenic plants. Leaf material was taken from wild-type control and transgenic plants: CS01, CS02, and CS03 (35S-sense TomloxC construct) and CA01 and CA02 (35S-antisense TomloxC construct). Twenty-microgram samples of total RNA were loaded per lane and hybridized with specific probes for TomloxA, B, C, D, and E. The hybridization signals were detected by autoradiography. Gel RNA loadings are represented below the autoradiograph by inclusion of a UV image of the ethidium bromide-stained rRNAs in each sample lane.
Of the 13 independent primary antisense transformants, 11 lines bearing fruits showed 30% to 90% reduction in the level of TomloxC mRNA compared to wild-type controls at the breaker stage of fruit development and 4 d post-breaker (B+4). Two antisense lines (CA01 and CA02), which showed the greatest reduction in the level of TomloxC mRNA in fruits (Fig. 3), were selected for further analysis.
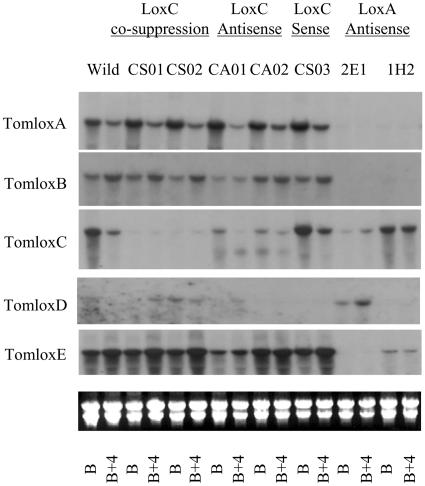
Figure 3.
Expression of TomloxA, TomloxB, TomloxC, TomloxD, and TomloxE mRNAs in fruit at the breaker and B+4 stages in wild-type control and transgenic plants. Total RNA was prepared with fruit pericarp material at the breaker and B+4 stages from wild-type control and transgenic plants: CS01, CS02, and CS03 (35S-sense TomloxC construct), CA01 and CA02 (35S-antisense TomloxC construct), and 2E1 and 1H2 (2A11-antisense TomloxA construct). Twenty-microgram samples of total RNA were loaded per lane and hybridized with specific probes for TomloxA, B, C, D, and E. The hybridization signals were detected by autoradiography. Gel RNA loadings are represented below the autoradiograph by inclusion of a UV image of the ethidium bromide-stained rRNAs in each sample lane.
Of the 12 plants independently transformed with the sense construct, 9 plants bore fruit. Six of these plants showed between 50% to 99% reduction by cosuppression while the other 3 showed no difference in the expression level of TomloxC mRNA compared to wild-type controls at the equivalent stages (breaker and B+4). Two cosuppression lines (CS01 and CS02) showing between 95% and 99% reduction and one sense line (CS03) showing no difference in the level of TomloxC mRNA in either fruit or leaf compared to wild-type controls (Figs. 2 and 3) were selected for further analysis.
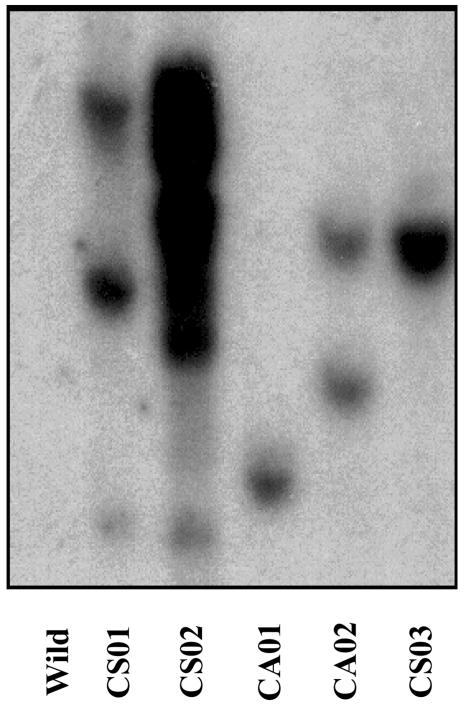
To confirm the presence of the transgene and compare the differences in the transgene copy number in the selected lines (CA01, CA02, CS01, CS02, and CS03), genomic DNA was digested with the restriction enzyme EcoRI, fractionated by agarose gel electrophoresis, Southern blotted, and hybridized with the marker gene nptII sequence. Because nptII is closely adjacent to the target gene in the transformation constructs and not present in the wild-type tomato genome, it should indicate the presence and copy number of the transgene. The result showed that the nptII hybridizing fragments were detected in all five of the selected independent lines, but were not found in wild-type nontransformed plants (Fig. 4). Two cosuppression lines yielded multiple transgene copies (CS01, 3 copies and CS02, more than 6 copies) and another sense line (CS03) yielded only one copy. One antisense line (CA01) yielded one copy, and another antisense line (CA02) yielded two copies.
Figure 4.
Southern blot of wild-type control and the TomloxC transgenic plants. Genomic DNA was prepared from young leaf material from wild-type control and transgenic plants: CS01, CS02, and CS03 (35S-sense TomloxC construct) and CA01 and CA02 (35S-antisense TomloxC construct). The genomic DNA (10 μg/lane) was digested with EcoRI and separated in a 0.8% (w/v) agarose gel. Blotted DNA was hybridized to a probe prepared from the kanamycin resistance (nptII) gene.
The five selected lines were further analyzed by northern blot with TomloxA, TomloxB, TomloxD, or TomloxE cDNA fragments as probes in order to find out whether the cosuppression and antisense-mediated depletion of TomloxC mRNA caused significant changes in the expression levels of any of the other lipoxygenase mRNAs in the transgenic plants. The results showed that the expression levels of TomloxA, TomloxB, and TomloxE mRNA in the fruits at the breaker and B+4 stages in all selected lines were not significantly different from the wild-type control (Fig. 3). Although the endogenous TomloxD message in the tomato fruits was detectable at a very low level (Fig. 3), which was consistent with the previous results (Heitz et al. 1997), the two cosuppression lines (CS01 and CS02) showed significantly higher expression levels of TomloxD mRNA compared to the wild-type control (Fig. 3).
Two fruit-specific (2A11) promoter-driven antisense TomloxA tomato lines (LOX1.2 E1 and LOX1.1 H2), which were previously reported by Griffiths to show significant reduction in the levels of TomloxA and TomloxB mRNAs (Griffiths et al., 1999b), were self-pollinated, and homozygous progenies (2E1 and 1H2) were selected for comparison with the TomloxC transgenic plants. The TomloxA, TomloxB, and TomloxE mRNAs were reduced to very low or undetectable levels in both homozygous lines, whereas TomloxC mRNA was not much affected in the 1H2 line and was partly reduced in the 2E1 line compared to the wild-type control (Fig. 3). Unexpectedly, however, the expression level of TomloxD mRNA was significantly higher than that of the wild-type control in the line 2E1 (Fig. 3).
Volatile Flavor Compounds from Ripening Fruit of Wild-Type and Transgenic Plants
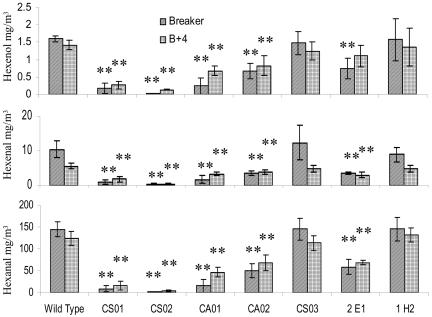
Fatty acid-derived short-chain volatile aldehydes and alcohols, such as n-hexanal, (Z)-3-hexenal, (E)-2-hexenal, and (Z)-3-hexenol, are known to be formed rapidly upon tissue disruption of tomato fruits (Galliard and Matthew, 1977) and play a major role in giving tomato its fresh “top-note” aroma and flavor. To test if the specific depletion of TomloxC in tomato leads to significant changes in the flavor profiles, the contents of n-hexanal, (Z)-3-hexenal, (E)-2-hexenal, and (Z)-3-hexenol were determined for two stages of fruit development, breaker and B+4, from wild-type control plants and the TomloxC transgenic plants from lines CS01, CS02, and CS03 (35S sense-TomloxC), CA01 and CA02 (35S antisense-TomloxC), and the TomloxA antisense transgenic plants from the homozygous lines 2E1 and 1H2 (2A11 antisense-TomloxA).
For fruits of the TomloxC cosuppression lines (CS01 and CS02), in which the endogenous TomloxC mRNA was reduced to about 1% to 5% of that in the wild-type control, (Fig. 3), the levels of hexanal, hexenal, and hexenol were markedly reduced to between 1.5% and 20% of those of wild-type control fruit at both the breaker and B+4 stages of ripening (Fig. 5). For fruits from the TomloxC antisense lines (CA01 and CA02), in which the expression level of TomloxC mRNA was down-regulated to about 10% of that in the wild-type control, the levels of hexanal, hexenal, and hexenol were significantly reduced to between 10% and 50% of those of wild-type control fruit at both the breaker and B+4 stages of ripening (Fig. 5). In the case of the sense line CS03, in which one copy of the sense transgene was detected (Fig. 4) but the expression level of the TomloxC mRNA was similar to that of the wild-type control (Fig. 3), no significant differences in volatile compounds were observed in fruits at either the breaker or B+4 stages of ripening compared to wild-type controls (Fig. 5), thus confirming the relationship between TomloxC mRNA levels and flavor volatile production.
Figure 5.
Changes in the flavor volatile profiles during ripening of wild-type and transgenic fruit. Fruits at the breaker and B+4 stages were taken from wild-type control plants, the 35S sense TomloxC (pLoxCS) transformants (CS01, CS02, and CS03), the 35S antisense TomloxC (pLoxCA) transformants (CA01 and CA02), and the 2A11 antisense TomloxA (p2ALX) transformants (2E1 and 1H2). Data are the means ± sd of five replicates for each sample type. Significant differences are indicated for P < 0.05(*) and P < 0.01(**).
For fruit of the homozygous TomloxA antisense line 2E1, in which the TomloxA, TomloxB, and TomloxE mRNA were reduced to an undetectable level and the TomloxC mRNA level also reduced to between 10% to 20% of that in the wild-type control, but with TomloxD mRNA significantly increased (Fig. 3), hexanal, hexenal, and hexenol were reduced to 30% to 50% of those of wild-type control fruit at the both breaker and B+4 stages (Fig. 5). In fruit of the other homozygous TomloxA antisense line 1H2, the expression of TomloxA and TomloxD was down-regulated to an undetectable level, and the TomloxB and TomloxE mRNAs were just detectable at a very low level in fruit at the stages breaker and B+4, while the expression level of TomloxC was similar to that in the wild-type control (Fig. 3). Fruit of this line showed no significant differences in the contents of the volatile compounds measured at either the breaker or the B+4 stages of ripening compared to wild-type control (Fig. 5).
Levels of Flavor Volatiles Produced in Leaves of Wild-Type and Transgenic Plants
The content of the same volatile compounds was also determined in leaves of wild-type control and transgenic plants. Hexanal, hexenal, and hexenol in leaves of the TomloxC cosuppression lines CS01 and CS02, with markedly reduced TomloxC mRNA (Fig. 2) were also greatly reduced to between 5% and 50% (data not shown). In the case of the antisense lines CA01 and CA02 and the sense line CS03, there was no major reduction in TomloxC mRNA in leaves compared to wild-type control (Fig. 2), and no significant differences were observed in leaf flavor compounds compared to the control (data not shown).
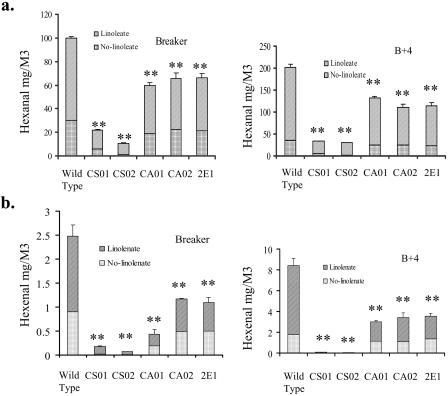
The Effect of Addition of Extra Free Fatty Acids on Hexanal and Hexenal Levels
Hexanal and hexenal are formed from the 13-hydroperoxides of linoleic and linolenic acids, respectively (Galliard and Matthew, 1977), and addition of extra linoleic or linolenic acids increases respectively the level of hexanal and hexenal produced by tomato fruit homogenates (Boukobza et al., 2001). When extra linoleic acid was added to the homogenates prepared from fruit of wild-type control and incubated for 15 min, the hexanal levels increased by approximately 131% and 355% at the breaker and B+4 stages, respectively. Similarly, the hexenal levels were increased by 72% and 278%, respectively, by addition of linolenic acid (Fig. 6, A and B). However, with homogenates prepared from the fruits of transgenic plants showing much lower TomloxC mRNA level compared to wild-type controls (Fig. 3), the increases in the hexanal and hexenal levels were markedly lower than in wild-type controls (Fig. 6, A and B). For the TomloxC co-suppression lines CS01 and CS02, the hexanal level was about 15% to 25% and the hexenal level was about 1% to 10% of those of wild-type control at the breaker and B+4 stage fruits following addition of linoleic and linolenic acids, respectively (Fig. 6, A and B). In the case of the TomloxC antisense lines CA01 and CA02, after addition of linoleic or linolenic acids, the hexanal and hexenal levels in the breaker and B+4 stage fruits were from 15% to 60% of those of the wild-type control. After addition of linoleic or linolenic acids to fruit extracts of the TomloxA antisense line 2E1, the hexanal and hexenal levels were about 35% to 65% of those of wild-type control (Fig. 6, A and B). These results suggest that TomloxC recognizes both linoleic and linolenic acids as substrates.
Figure 6.
Effect of extra linoleate or linolenate on the hexanal and hexenal contents in fruit homogenates prepared from wild-type and transgenic fruits at different developmental stages or leaves. Breaker (B) and B+4 fruit pericarp was ground to a fine powder in liquid nitrogen and 5 g of the powder was mixed well with 5 mL emulsion, which was prepared by mixing 2 g of linoleic acid (a) or linolenic acid (b) with Tween 20 (2 g) in sodium phosphate buffer (120 mL) at pH 7.0 by stirring for 15 min. After incubation for 15 min at room temperature with gentle shaking in a food blender, the monitoring of the headspace was carried out for a further 3 min. sds of the means of three replicates for each sample type are indicated by the error bars. Significantly different results are marked at P < 0.05(*) and P < 0.01(**).
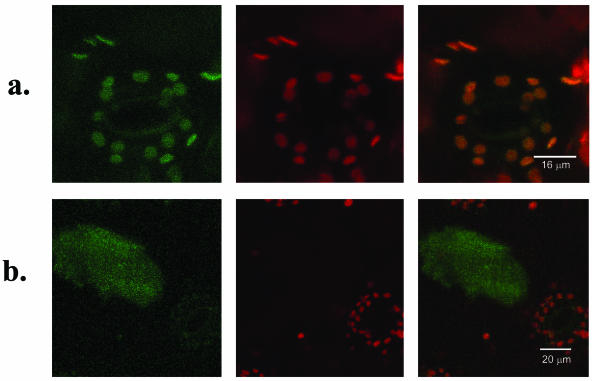
Expression of GFP-Constructs in Tobacco
Constructs encoding TomloxC-GFP fusions, one with and one without the presumed 42-amino acid N-terminal chloroplast targeting sequence, were generated as described in “Materials and Methods” and verified by DNA sequencing. The recombinant genes were introduced into tobacco (Nicotiana tabacum) leaf cells by particle gun transformation. Confocal imaging of targeted cells revealed that the localization of fluorescence was dependent on the presence of the chloroplast transit peptide. With the full-length TomloxC-GFP fusion, green fluorescence was colocalized with red fluorescence from chlorophyll-containing plastids in the guard cells imaged in Figure 7A. Fusions lacking the N-terminal TomloxC transit peptide sequence produced green fluorescence throughout the cytosol that was not colocalized with red chlorophyll fluorescence (Fig. 7B).
Figure 7.
Cellular localization of TomloxC-GFP fusions. Tobacco leaf cells were transiently transformed with vector encoding either full length TomloxC-GFP fusion (a) or TomloxC-GFP fusion lacking the N-terminal chloroplast transit peptide (b). Images were obtained by confocal microscopy of leaf tissue excited with a 488-nm Ar laser line. GFP fluorescence emissions are shown in green (left) and chlorophyll fluorescence emissions are shown in red (center), with combined signals on the right.
DISCUSSION
We have shown that the specific depletion of TomloxC mRNA by cosuppression or antisense-inhibition in transgenic tomato plants leads to significant reductions in the contents of fatty acid-derived short-chain volatile aldehydes and alcohols, such as n-hexanal, (Z)-3-hexenal, (E)-2-hexenal, and (Z)-3-hexenol, in both fruit and leaf tissues. Major reductions have been observed in five transgenic lines obtained independently and, moreover, the extent of reduction in the hexanal and hexenal levels correlated well with the residual TomloxC mRNA present in the transgenic lines (Figs. 3 and 5). These findings relate to plants in which the levels of other LOX isoforms were not significantly different from wild-type controls. Furthermore, no significant differences in the hexanal and hexenal levels were observed in transgenic plants with different levels of other LOX isoforms, providing the TomloxC mRNA levels were not significantly reduced. These results demonstrate that the specific tomato lipoxygenase isozyme TomloxC appears to play a key role in generation of fatty acid-derived short-chain volatile aldehydes and alcohols in tomato. Lipoxygenases are also involved in JA synthesis, in generation of volatiles that attract some insects or insect predators, and in defense responses against insect pests. For example, depletion of potato LOX H3 reduced wound induction of proteinase inhibitors and increased weight gain of insect pests (Leon et al., 2002). This suggests that our range of transgenic plants with altered levels of different LOX isoforms (Fig. 3) could provide a useful resource for studying the role of specific LOX isoforms in JA synthesis, insect defense, and flavor generation.
Multiple isoforms of LOX have been detected in a wide range of organisms (Eskin et al. 1977; Samuelsson et al. 1987; Vick and Zimmerman 1987). The lipoxygenase isoforms are distinguished by differences in substrate and product specificity (9-lipoxygenase and 13-lipoxygenase according to the position of addition of the −OOH moiety in the primary hydroperoxide product using linoleic or linolenic acids as substrates). Fatty acid-derived C6 short-chain volatile aldehydes and alcohols, such as n-hexanal, (Z)-3-hexenal, (E)-2-hexenal, and (Z)-3-hexenol, are important flavor compounds in fruit (Buttery et al., 1989; Hildebrand et al., 1989) and are derived only from 13-hydroperoxy products, but not from the more abundant 9-hydroperoxides. Therefore, only lipoxygenase isozymes that produce 13-hydroperoxy products from linoleic and linolenic acids would be expected to play an important role in generation of fatty acid-derived volatile aldehydes and alcohols in tomato.
Although it is known that there are at least five lipoxygenase isozymes present in tomato (Fig. 3; Ferrie et al., 1994; Heitz et al., 1997), their substrate and product specificity and role during ripening is not clear. We previously reported that antisense-suppression of TomloxA and TomloxB to an undetectable level in tomato fruit caused no significant changes in the content of C6 flavor volatile aldehydes and alcohols in tomato fruit compared to wild-type control (Griffiths et al., 1999b). We have shown here that the levels of TomloxE are also significantly reduced in fruit of the progeny of these plants. The sequences of TomloxA, TomloxE, and TomloxB have strong homology to that of potato Lox1 and show 92%, 75%, and 69% identity, respectively, at the amino acid level. Taking into account that the majority of the HPOs formed by lipoxygenase activity in tomato are the 9-isomers (Galliard and Matthew, 1977; Smith et al., 1997) and that TomloxB, TomloxE, and TomloxA have high expression levels in fruit, it is most likely that TomloxA, TomloxB, and TomloxE may produce 9-hydroperoxides, which are not directly involved in generation of the C6 flavor volatile aldehydes and alcohols.
It has been suggested that key components of the volatile production pathway may be compartmentalized in the intact cells of the tomato fruit and that maceration breaks down these compartments allowing the components to mix and generate new volatiles (Baldwin et al., 2000). The results from the TomloxC-GFP fusion experiments confirm the earlier in vitro work of Heitz et al. (1997) suggesting a chloroplast localization of the enzyme (Fig. 7). The targeting of TomloxC to the chloroplasts may therefore act to isolate this key enzyme activity from other components of the volatile production pathway so that volatile production does not occur until cells are disrupted. In contrast, it has also been suggested that lipoxygenases may play a role in the disruption of chloroplast thylakoid membranes that occurs during the chloroplast-to-chromoplast transition during fruit ripening (Thelander et al., 1986; Ferrie et al., 1994). It is generally assumed that the substrate for LOX is unesterified polyunsaturated fatty acids released from complex lipids by the action of lipases. It has been proposed that chloroplasts are the major site for fatty acid hydroperoxide metabolism. Linoleic and linolenic acids may come from disruption of these thylakoid membranes. As TomloxC is a chloroplast targeted lipoxygenase expressed at high levels during fruit ripening, then it is clearly a prime candidate to perform such a role. The product of TomloxD, which also has a chloroplast target signal, is expressed principally in leaves (Fig. 2) and may be involved in defense signaling in response to herbivore and pathogen attack, forming a component of the octadecanoid signaling pathway (Heitz et al., 1997). In transgenic Arabidopsis plants, suppression of the chloroplast-targeted LOX2 gene resulted in the absence of wound-inducible JA accumulation and reduced expression of the wound- and JA-inducible vsp gene (Bell et al., 1995). These present results indicate a dual role for LOX in fruit development, including a defense component and a contribution to aroma and flavor generation, which ultimately aids seed dispersal. Addition of extra linoleic and linolenic acids to tomato homogenates significantly increased the level of hexanal and hexenal, respectively, in wild-type control and transgenic plants; however, the increase in the transgenic plants that showed a very low level of the TomloxC mRNA was significantly lower than that in wild-type control (Fig. 6). Taken together, all data suggest that the TomloxC lipoxygenase isoform can use both linoleic and linolenic acids as substrates and produce 13-hydroperoxy products, which are specifically used as the precursors of C6 short-chain flavor compounds, such as hexanal, hexenal, and hexenol.
Our results (Fig. 3; data not shown) are consistent with the previous work of Heitz et al. (1997), which concluded that TomloxD is mainly expressed in tomato leaf and is up-regulated in leaves in response to wounding and systemin or methyl jasmonate treatment and may play a role as a component of the octadecanoid defense-signaling pathway (Heitz et al., 1997). Interestingly, compared to wild-type control, there was a higher expression level of the TomloxD transcript in fruit of the TomloxC cosuppression lines CS01 and CS02, which have a very low level of TomloxC mRNA, and homozygous TomloxA antisense line 2E1, which not only showed undetectable expression levels of TomloxA, TomloxB, and TomloxE but a very low level of TomloxC mRNA. This suggests that deficiency of other LOX isoforms may induce enhanced production of TomloxD (Fig. 3). Sequence comparison showed that the TomloxD protein sequence has 92% identity at the amino acid level with that of potato Lox3, which is mostly expressed in potato leaves and roots and produces almost exclusively 13-hydroperoxides (Royo et al., 1996). In leaves of the cosuppression lines CS01 and CS02, which showed a very low level of the TomloxC transcript but no significant difference in the TomloxD level compared to wild-type control, the contents of hexanal, hexenal, and hexenol were greatly reduced. This result suggests that TomloxD does not play a key role in the generation of flavor volatiles in fruit even though it may be capable of generating 13-hydroperoxides. 13-Hydroperoxides, which are the products of 13-lipoxygenases, can be used as substrates by several different enzymes (Schaller, 2001), but only 13-HPLs can convert them to hexanal and hexenal. Thus, the metabolic fate of 13-hydroperoxides may depend on their colocalization or association with specific downstream enzymes. In conclusion, in tomato, TomloxC is a key lipoxygenase specifically involved in the generation of fatty acid-derived C6 short-chain flavor compounds and is important in generation of fruit flavor. It will be of interest to see whether fruits with high activity of TomloxC or its equivalent have better flavor.
MATERIALS AND METHODS
Plant Materials
All experiments were performed using a near-isogenic line of diploid tomato (Lycopersicon esculentum Mill. cv Ailsa Craig) plants that has been grown at Sutton Bonington, Leicestershire, UK, for over 20 years. Unless otherwise stated, plants were grown in 24-cm diameter pots in M2 compost (Levington Horticulture, Ipswich, Suffolk, UK) under identical glasshouse conditions used for nontransformed control plants. Plants were watered daily and fed with high-nitrogen liquid fertilizer at regular intervals. Flowers were tagged at anthesis and fruit development recorded as DPA. Mature green fruit were defined as 35 DPA and were characterized as being green and shiny with no obvious ripening-associated color change. Breaker fruit were defined as those fruit showing the first signs of ripening-associated color change from green to yellow. Fruit of subsequent ripening stages were defined in days post-breaker. B+4 fruit were orange-red in color. All plant samples for the preparation of total RNA were taken at the same time each day, frozen in liquid nitrogen, and stored at −70°C until required.
Construction of Transgenes and Plant Transformation
Two transgene constructs were produced. The first (pLoxCA) was designed to constitutively express an antisense TomloxC RNA under control of the CaMV 35S promoter. The second (pLoxCS) was designed to cosuppress specifically the endogenous TomloxC mRNA or constitutively overexpress a functional TomloxC transgene. Throughout, all basic methods were as described by Sambrook et al. (1989).
To construct pLoxCA, a 1.5-kb SalI/BamHI cDNA fragment from the clone pTomloxC (Heitz et al., 1997) was first ligated into pDH51 (Pietrzak et al., 1986), which was previously digested with the same enzymes, in the antisense orientation between the CaMV 35S promoter and terminator to yield pDHLoxCA. The CaMV 35S-driven antisense gene and CaMV 35S terminator cassette were excised from pDHLoxCA with EcoRI and ligated into similarly digested pBIN19 (Bevan, 1984) to yield pLoxCA.
To construct pLoxCS, a 2.8-kb SmaI/XhoI cDNA fragment, including the full coding sequence and poly(A+) tail from pTomloxC, was first ligated in the sense orientation between the CaMV 35S promoter and terminator of SmaI/SalI-digested pDH51 to yield pDHLoxCS. The sense gene was then excised from pDHLoxCS by partial digestion with EcoRI and ligated into similarly digested pBIN19 to yield pLoxCS.
The correct orientations of ligated fragments used to produce the antisense and sense constructs were verified by restriction digest analysis and by sequencing. After transfer to Agrobacterium tumefaciens strain LBA4404 (Bevan, 1984) by the freeze-thaw method of An et al. (1988), the constructs were used to transform tomato cotyledon explants as described by Bird et al. (1988). Transgenic plants that rooted on kanamycin were transferred to compost and grown as described above.
Extraction and Analysis of RNA
RNA was extracted from tomato fruit pericarp and leaf as previously described (Smith et al., 1986) except that contaminating carbohydrates and DNA were removed by differential precipitation of the RNA from 4 m LiCl at −20°C for 1 h. RNA was quantified by spectrophotometry and, following formamide denaturation, 20-μg samples and RNA size markers (GIBCO BRL Life Technologies, Inchinnan, Paisley, UK) were fractionated in 1% (w/v) agarose gels containing 3% (v/v) formaldehyde. RNA was capillary-blotted onto GeneScreen Plus (NEN Life Science Products, Hounslow, UK) membranes, which were then prehybridized at 65°C in 5× SSPE (1× SSPE = 150 mm NaCl, 10 mm NaH2PO4, 1 mm Na2EDTA, pH 7.4), 1% (w/v) SDS, 0.1 m phosphate buffer, pH 6.8, 10% dextran sulfate, 50% formamide, 0.01% sodium pyrophosphate, and 150 μg mL−1 sheared, denatured salmon sperm DNA for 4 h. The RNA was hybridized at 42°C in the same buffer to 32P-labeled specific probes generated from cDNA sequences (TomloxA, 147–1,414; TomloxB, 585–1,435; TomloxC, 841–2,345; TomloxD, 44–604; and TomloxE, 1–790) using the Rediprime labeling system from Amersham International, Little Chalfont, Buckinghamshire, UK. After hybridization, membranes were washed in 0.2× SSPE and 0.1% (w/v) SDS at 42°C and were autoradiographed.
Extraction and Analysis of Genomic DNA
Genomic DNA was extracted by grinding 5 g of young leaf tissue in 25 mL of ice-cold homogenization buffer (25 mm Tris-HCl, pH 7.6, 20% [v/v] glycerol, 2.5% [w/v] ficoll 400, 0.44 m Suc, 10 mm β-mercaptoethanol, and 0.1% [v/v] Triton X-100). The homogenate was filtered through muslin and nuclei pelleted by centrifugation (1,000g, 4°C, 15 min). The nuclei in the pellet were lysed at 70°C in urea buffer (42% [w/v] urea, 25 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 50 mm EDTA, 1% [w/v] N-lauryl sarcosine) and the DNA allowed to dissolve. The solution was extracted twice with phenol/chloroform (1:1, v/v) and the DNA precipitated from the aqueous phase by the addition of an equal volume of ethanol. The DNA was washed successively with 50 mm potassium acetate in 70% (v/v) ethanol, 70% (v/v) ethanol, and 95% (v/v) ethanol, allowed to partially air dry, and was dissolved in sterile distilled water containing 10 μg mL−1 DNase-free calf pancreatic RNase A (Boehringer Mannheim UK, Lewes, East Sussex, UK) and stored at 4°C until required. Individual genomic DNA (30 μg) samples were completely digested with EcoRI, separated in 0.8% (w/v) agarose gels, and capillary blotted to GeneScreen Plus membranes. Membranes were prehybridized as for northern analysis and the DNA hybridized to probes generated from either the cDNA sequence of pTomloxC or from the DNA sequence of the neomycin phosphotransferase gene (nptII; Pridmore, 1987) located within the T-DNA borders of pBIN19. Membranes were washed at 42°C in 0.2× SSPE and 0.1% (w/v) SDS and were autoradiographed.
Volatile Measurement
Fruits and leaves from wild-type and transgenic plants were analyzed for their volatile profiles using the method described by Boukobza et al. (2001). The individual intact tomato fruit (35–50 g) or 15 g of leaves was placed inside the food blender (volume 355 mL; Phillips, Eindhoven, The Netherlands; type HR2810/A), which was then sealed and macerated (2–5 s). The headspace within the device was continually flushed with air (170 mL/min) and, on the outlet side, a portion of the airflow was continuously sampled into the Atmospheric Pressure Chemical Ionization-Mass Spectrometer (Micromass, Altrincham, UK) at 11.5 mL/min through a heated transfer line (0.53-mm i.d. fused silica tube held at 100°C). The excess of airflow was vented to atmosphere. Monitoring of the headspace above the macerate was carried out for a further 3 min to follow the release of the selected volatile compounds as described by Boukobza et al. (2001). Because resolution was by mass alone, the system was unable to differentiate between positional isomers or stereoisomers. Therefore, the signal for hexenals represented the sum of the isomers (E)-2-hexenal and (Z)-3-hexenal. For each fruit stage at least five fruits from each of the plant types were analyzed separately.
Addition of Free Fatty Acids
Emulsions were prepared by mixing linoleic or linolenic acid (2 g, Sigma, St. Louis) with Tween 20 (2 g) in sodium phosphate buffer (120 mL) at pH 7 by stirring for 15 min. Tomato fruit pericarp or leaf were frozen in liquid nitrogen and ground to a fine powder. The powder (5 g) was mixed with 5 mL of the emulsion and incubated for 15 min at room temperature with a gentle shake in the food blender. After incubation, the volatile compounds were analyzed as above. For each fruit stage at least three fruits from each of the plant types were analyzed.
Construction of TomLoxC-GFP Fusions
Two TomloxC-GFP fusion constructs were produced. The first (pGWB5-C1) was designed to express full-size TomloxC with a C-terminal green fluorescent protein (GFP) fusion. The second (pGWB5-C2) was designed to express TomloxC lacking the N-terminal 42-amino acid transit peptide with a C-terminal GFP fusion. pGWB5-C1 was constructed using the primers DCW21 (CACCATGTTGAAGCCTCAATTTCAACAATC) and DCW22 (AATTGAAATACTATAAGGTACACCTTTTCC) to amplify the full-size TomloxC coding sequence using the proof reading DNA polymerase Pwo (Roche Diagnostics, Basal). The primers included sequences for directional TOPO cloning into the pENTR/D-TOPO entry vector (Invitrogen, Carlsbad, CA). The TomloxC sequence could then be inserted in-frame with the GFP sequence from the GATEWAY vector pGWB5 (Tsuyoshi Nakagawa, Shimane University, Japan) by performing an attL × attR → attB + attP reaction (LR reaction) between the entry clone and pGWB5. C-terminal GFP fusions in pGWB5 are under the control of the 35S promoter. pGWB5-C2 was constructed as above but substituting primer DCW29 (CACCATGAATTTTAGGGTTCATCATAATTA) for primer DCW21 to give a truncated TomloxC-GFP fusion.
Transient Expression of GFP Fusions in Tobacco
Tobacco (Nicotiana tabacum) leaves and tobacco seedlings were transformed with pGWB5-C1 and pGWB5-C2 using a PDS-1000/He Biolistic particle delivery system (Bio-Rad, Hercules, CA) and supplied protocol. Briefly, a suspension of gold microprojectiles (0.4–1.2 μm) was vortexed vigorously for 30 min before dispensing 50-μL aliquots into sterile Eppendorf tubes. Under continuous vortexing 5 μg of plasmid DNA followed by 50 μL of 2.5 m CaCl2 and 20 μL of fresh 0.1 m spermidine were added and vortexing continued for a further 30 min. The DNA coated microprojectiles were then pelleted by centrifuging for 10 s and the supernatant discarded. The gold particles were washed with 250 μL of absolute ethanol before pelleting and resuspending in 75 μL of absolute ethanol. Ten milliliters of freshly coated microprojectiles were transferred to the center of sterile macrocarrier and left to dry. The loaded macrocarriers were then installed into the macrocarrier launch assembly of the Biolistic apparatus prior to bombardment.
Confocal Imaging of Transformed Tobacco Cells
Transformed tobacco cells were imaged with a Leica TCS-SP2 confocal microscope equipped with an Argon ion laser (Leica, Heidelberg). GFP fluorescence was excited with the 488-nm Ar laser line and confocal sections were collected using 498 to 525 nm (GFP) and 640 to 720 nm (chlorophyll) emission settings.
Novel Materials
Upon request, all novel materials described in this publication will be made available in reasonable quantities in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the materials. Obtaining any permission will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number U37839.
Acknowledgments
Our special thanks go to Dr. Allen Griffiths for supplying seeds of 2A11 antisense TomloxA transgenic plants and Mr. Suthat Surawang and Dr. Fabienne Boukobza for the technical assistance with the measurement of flavor compounds.
This work was supported by the Biotechnology and Biological Science Research Council.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.041608.
References
- An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors. In S Gelvin, RA Schilperoot, DPS Verma, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A3/1–19
- Axelrod B (1974) Lipoxygenases. ACS Adv Chem Ser 136: 324–348 [Google Scholar]
- Axelrod B, Cheesbrough TM, Laasko S (1981) Lipoxygenase from soybeans. Methods Enzymol 71: 441–451 [Google Scholar]
- Baldwin EA, Nisperos-Carriedo MO, Moshonas MG (1991) Quantitative analysis of flavor and other volatiles and for certain constituents of two tomato cultivars during ripening. J Am Soc Hortic Sci 116: 265–269 [Google Scholar]
- Baldwin EA, Scott JW, Shewmaker CK, Schuch W (2000) Flavour trivia and tomato aroma: biochemistry and possible mechanisms for control of important aroma components. HortScience 35: 1013–1021 [Google Scholar]
- Bell E, Creelman RA, Mullet JE (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92: 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 24: 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild GS, Ramadoss CS, Axelrod B (1977) Effect of substrate polarity on the activity of soybean lipoxygenase isoforms. Lipids 12: 732–735 [Google Scholar]
- Bird CR, Smith CJS, Ray JA, Moureau P, Bevan MW, Bird AS, Hughes S, Morris PC, Grierson D, Schuch W (1988) The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Mol Biol 11: 651–662 [DOI] [PubMed] [Google Scholar]
- Boukobza F, Dunphy PJ, Taylor AJ (2001) Measurement of lipid oxidation-derived volatiles in fresh tomatoes. Postharvest Biol Technol 23: 117–131 [Google Scholar]
- Buttery RG, Teranishi R, Flath RA, Ling LC (1989) Fresh tomato volatiles-composition and sensory studies. In R Teranishi, RG Buttery, F Shahidi, eds, Flavour Chemistry: Trends and Developments. ACS Symposium Series 338. American Chemical Society, Washington, DC, pp 213–222
- Eskin NAM, Grossman S, Pinsky A (1977) Biochemistry of lipoxygenase in relation to food quality. Crit Rev Food Sci Nutr 9: 1–41 [DOI] [PubMed] [Google Scholar]
- Ferrie BJ, Beaudoin N, Burkhart W, Bowsher CG, Rothstein SJ (1994) The cloning of two tomato lipoxygenase genes and their differential expression during tomato fruit ripening. Plant Physiol 106: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53: 275–297 [DOI] [PubMed] [Google Scholar]
- Galliard T, Matthew JA (1977) Lipoxygenase-mediated cleavage of the fatty acids to carbonyl fragments in tomato fruits. Phytochemistry 16: 339–343 [Google Scholar]
- Griffiths A, Barry C, Alpuche-Soli AG, Grierson D (1999. a) Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J Exp Bot 50: 793–798 [Google Scholar]
- Griffiths A, Prestage S, Linforth R, Zhang J, Taylor A, Grierson D (1999. b) Fruit-specific lipoxygenase suppression in antisense-transgenic tomatoes. Postharvest Biol Technol 17: 163–173 [Google Scholar]
- Hatanaka A (1993) The biogeneration of green odour by green leaves. Phytochemistry 34: 1201–1218 [Google Scholar]
- Hamberg M (1986) Isolation and structure of lipoxygenase from Saprolegnia parasitica. Biochim Biophys Acta 876: 688–692 [Google Scholar]
- Heitz T, Bergey DR, Ryan CA (1997) A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol 114: 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand DF, Rodriguez JG, Legg CS, Brown GC, Bookjans G (1989) The effect of wounding and mite infestation on soybean leaf lipoxygenase levels. Z Naturforsch 44c: 655–659 [Google Scholar]
- Leon J, Royo J, Vancanneyt G, Sanz C, Sikowski H, Griffiths G, Sanchez-Serrano JJ (2002) Lipoxygenase H1 gene silencing reveals a specific role in supplying fatty and hydroperoxides for aliphatic aldehyde production. J Biol Chem 277: 416–423 [DOI] [PubMed] [Google Scholar]
- Matthew JA, Chan HW-S, Galliard T (1977) A simple method for the preparation of pure 9-D-hydroperoxide of linoleic acid and methyl linoleate based on the positional specificity of lipoxygenase in tomato fruit. Lipids 12: 324–326 [DOI] [PubMed] [Google Scholar]
- Pietrzak M, Shillito RD, Hohn T, Potrykus I (1986) Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res 14: 5857–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridmore RD (1987) New and versatile cloning vectors with kanamycin-resistance marker. Gene 56: 309–312 [DOI] [PubMed] [Google Scholar]
- Royo J, Leon J, Vancanneyt G, Albar JP, Rosahl S, Ortego F, Castanera P, Sanchez-Serrano JJ (1999) Antisense-mediated depletion of a potato lipoxygenase reduces wound induction of proteinase inhibitors and increases weight gain of insect pests. Proc Natl Acad Sci USA 96: 1146–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo J, Vancanneyt G, Pérez AG, Sanz C, Störmann K, Rosahl S, Sánchez-Serrano JJ (1996) Characterization of three potato lipoxygenases with distinct enzymatic activities and different organ-specific and wound-regulated expression patterns. J Biol Chem 271: 21012–21019 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Samuelsson B, Dahlen S-E, Lindgren JA, Rouzer CA, Serhan CN (1987) Leucotrienes and lipoxins: structures, biosynthesis and biological effects. Science 237: 1171–1176 [DOI] [PubMed] [Google Scholar]
- Schaller F (2001) Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot 52: 11–23 [PubMed] [Google Scholar]
- Shechter G, Grossman S (1983) Lipoxygenase from baker's yeast: purification and properties. Int J Biochem 15: 1295–1304 [DOI] [PubMed] [Google Scholar]
- Siedow JN (1991) Plant lipoxygenases: structure and function. Annu Rev Plant Physiol Plant Mol Biol 42: 145–188 [Google Scholar]
- Smith JJ, Linforth R, Tucker GA (1997) Soluble lipoxygenase isoforms from tomato fruit. Phytochemistry 45: 453–458 [Google Scholar]
- Smith CJS, Slater A, Grierson D (1986) Rapid appearance of an mRNA correlated with ethylene synthesis encoding a protein of molecular weight 35,000. Planta 168: 94–100 [DOI] [PubMed] [Google Scholar]
- Teranishi R, Buttery RG, Stern DJ, Takeoka G (1990) Computer assisted correlation of gas chromatographic data with aroma. In Y Bessiere, AF Thomas, eds, Flavour Science and Technology. Wiley, Chichester, UK, pp 267–270
- Todd JF, Paliyath G, Thompson JE (1990) Characteristics of membrane-associated lipoxygenase in tomato fruit. Plant Physiol 94: 1225–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander M, Narita JO, Gruissem W (1986) Plastid differentiation and pigment biosynthesis during tomato fruit ripening. Curr Top Plant Biochem Physiol 5: 128–141 [Google Scholar]
- Vick BA, Zimmerman DC (1987) Oxidative systems for modification of fatty acids: the lipoxygenase pathway. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants: A Comprehensive Treatise, Vol 9. Academic Press, New York, pp 53–91
- Yamamoto S (1991) Enzymatic lipid peroxidation: Reactions of mammalian lipoxygenases. Free Radic Biol Med 10: 149–159 [DOI] [PubMed] [Google Scholar]
- Zimmerman DC, Vick BA (1973) Lipoxygenase in Chlorella pyrenoidosa. Lipids 8: 264–266 [DOI] [PubMed] [Google Scholar]