Summary
Early human embryonic stem cell (hESC)-derived neural populations consist of various embryonic neural progenitors (ENPs) with broad neural developmental propensity. Here, we sought to directly convert human somatic cells into ENP-like phenotypes using hESC-ENP-enriched neural transcription factors (TFs). We demonstrated that induced ENP could be efficiently converted from human fibroblasts using two TF combinations. The iENPs exhibit cellular and molecular characteristics resembling hESC-ENPs and can give rise to astrocytes, oligodendrocytes, and functional neuronal subtypes of the central and peripheral nervous system. Nevertheless, our analyses further revealed that these two iENP populations differ in terms of their proliferation ability and neuronal propensity. Finally, we demonstrated that the iENPs can be induced from fibroblasts from patients with Huntington's disease and Alzheimer’s disease, and the diseased iENPs and their neuronal derivatives recapitulated the hallmark pathological features of the diseases. Collectively, our results point toward a promising strategy for generating iENPs from somatic cells for disease modeling and future clinical intervention.
Keywords: cell fate conversion, induced neural progenitors, transcription factors, disease modeling
Graphical Abstract
Highlights
-
•
iENPs can be converted from fibroblasts using hESC-ENP enriched factors
-
•
iENPs exhibit cellular and molecular characteristics resembling hESC-ENPs
-
•
iENPs induced by different TF combinations exhibit different neural propensity
-
•
iENP and their neuronal derivatives recapitulated HD and AD phenotypes
In this paper, Kuo and colleagues show that induced embryonic neural progenitors (iENPs) can be directly converted from human fibroblasts using defined factors. The iENPs exhibit functional characteristics resembling hESC-derived ENPs, and their neuronal derivatives recapitulate the hallmark pathological features of neurodegenerative diseases. These results point toward a promising strategy for generating NPs from somatic cells for future clinical intervention.
Introduction
Certain progressive, degenerative, and ultimately fatal, neurological disorders, such as Huntington's disease (HD) and Alzheimer's disease (AD), cannot be effectively treated; therefore, there remains a need to elucidate the pathological progress behind these disorders, and further effective clinical interventions (Bredesen et al., 2006). By taking advantage of pluripotency reprogramming technology, researchers can readily reprogram disease-specific induced pluripotent stem cells (iPSCs) from patients' somatic cells, and subject them to in vitro differentiation for generation of various disease-relevant cell types for disease modeling and drug development (HD iPSC Consortium, 2012). However, tumorigenic and spontaneous differentiation of iPSCs remains a concern. In addition to iPSCs, induced neurons (iNs), which can be directly converted from fibroblasts (FBs) by defined transcription factors (TFs) (Vierbuchen et al., 2010), provide another source of neuronal cells for in vitro disease modeling and drug testing. The advantages of iN technology are that it can provide a fast and simple method for the generation of specific neuronal subtypes, and its use may avoid certain problems, such as uncontrolled cell differentiation and tumor formation, which are associated with hiPSCs. However, the induction of each neuronal subtype requires different combinations of defined factors (Ang and Wernig, 2014), and the yield of such iNs is still too low for meaningful clinical applications. Therefore, developing strategies that allow direct conversion of somatic cells into expandable neural stem cell/progenitor (NSC/NP) populations that possess multiple neural differentiation potentials is an important step toward the generation of patient-specific neural cell types on a scalable level.
Previously, it was demonstrated that induced NP (iNPs) can be directly converted from mouse somatic cells by overexpressing various TF combinations (Han et al., 2012, Kim et al., 2011, Lujan et al., 2012). Kim et al. (2011) first demonstrated that expandable iNPs could be generated from FBs via a modified pluripotency reprogramming procedure, and the resulting iNPs were able to differentiate into neurons and glial cells. Subsequently, several studies reported the generation of iNPs through the introduction of neural-enriched factors with/without iPSC factors (Han et al., 2012, Lu et al., 2013, Lujan et al., 2012), and the resulting iNPs were able to differentiate into all three major neural cell types of the CNS. Meanwhile, reports show that human iNPs can also be converted from somatic cells via the introduction of TFs (Lu et al., 2013, Ring et al., 2012, Wang et al., 2013). In these studies, several TF combinations, including at least one of the iPS factors, were used for hiNP generation (Ring et al., 2012), and the differentiation propensity of the iNPs described in the aforementioned studies was mainly restricted to CNS neurons.
hESCs can be used as an in vitro differentiation model to generate neural phenotypes of various developmental stages, including embryonic NPs (ENPs) populations, and the critical neural genetic factors that contribute to the neural fate acquisition have begun to be uncovered (Hou et al., 2013, Rosa and Brivanlou, 2011, Zhang et al., 2010). Given that hESC-ENP populations possess broad differentiation potential to give rise to both CNS and PNS neural cell types, it may be possible to directly convert FBs into iNPs resembling hESC-ENPs through the use of TFs highly expressed in the hESC-ENP population.
Here, we identified a panel of neural TFs (nTFs) highly enriched in hESC-ENPs compared with FBs, through comparative gene expression profiling. We defined two TF combinations, the overexpression of which can efficiently convert human FBs into multipotent iENPs. The iENP populations generated in this manner resemble hESC-ENPs in many respects, including their pattern of proliferation, gene expression profile, and in vitro and in vivo differentiation propensity. Importantly, we found that different combinations of TFs can induce iENP populations with varying proliferative features and regional differentiation preferences. We also demonstrated that neurons derived from AD- and HD-iENPs, recapitulated the major disease pathological features in vitro. Taken together, our results point toward a promising and reproducible strategy for generating iENPs from somatic cells for disease modeling and future clinical intervention.
Results
Conversion of iENPs from Human FBs
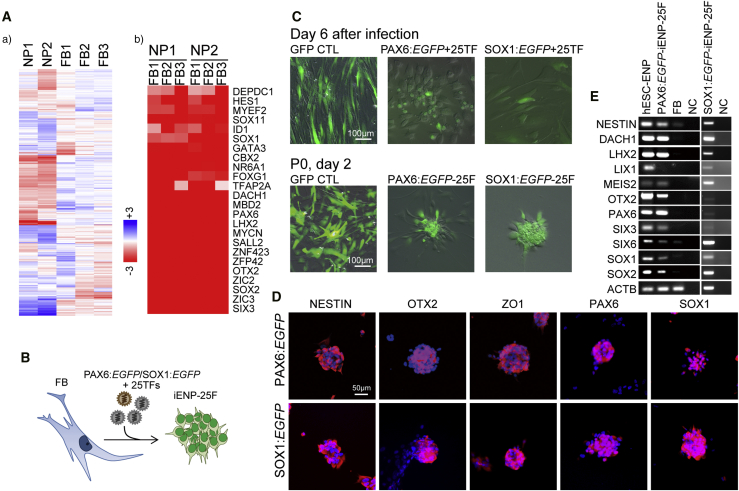
To screen potential TFs for iENP generation, we compared the global gene expression profiling of multiple hESC-ENP and FB populations by microarray analysis (Figure 1Aa). Twenty-four TFs were selected based on their greater levels of expression in hESC-ENPs than in FBs (Figure 1Ab). NR2F2 was also selected because it was previously reported to be crucial for neural differentiation (Rosa and Brivanlou, 2011). As the hESC-ENP-TFs were highly expressed in heterogeneous ENP populations derived from hESCs, there is a possibility that certain combinations of our 25 hESC-ENP-TFs may be able to induce different types of ENPs from FBs. To this end, two reporter systems, PAX6:EGFP and SOX1:EGFP, were created to monitor the progression of neural fate conversion and to evaluate ENP induction efficiency, as both PAX6 and SOX1 have been reported to be expressed in hESC-ENPs (Zhang et al., 2010). We confirmed that these reporters are expressed in hESC-ENPs (Figure S1).
Figure 1.
Induction of Human FBs into iENPs by 25 nTFs Highly Expressed in hESC-ENPs
(A) Identification of hESC-ENP-enriched neural TFs by comparative gene expression profiling of FBs and hESC-ENPs. (a) Heatmap analysis of the global gene expression profiles of hESC-ENPs (NP1, E-MEXP-2668, ArrayExpress database; Zhang et al., 2010; NP2 from H9-SOX1:EGFP sorted ND day18-NP) and FBs (FB1, -2, and -3). (b) Selected TFs with higher expression in hESC-ENPs than in FBs.
(B) Schematic depiction of the experimental strategy for directly converting FBs into iENPs.
(C) Infection of FBs with lentivirus encoding hESC-ENP TFs (25TF) and neural reporter, and the growth of FACS-sorted cells. Cells infected with UbC:EGFP were used as controls.
(D) ICC analysis of iENP-25F clusters resembling NP colonies/spheres using antibodies against the indicated antigen.
(E) RT-PCR analysis of the indicated genes in iENP-25F.
FB, fibroblast; NC, negative control (H2O). See also Figure S1.
To generate iENPs, we simultaneously infected FBs with lentiviruses encoding each of the aforementioned 25 TFs as well as the neural reporter, PAX6:EGFP or SOX1:EGFP (Figure 1B). At around 6 days post lentiviral infection, PAX6:EGFP+ cells with a rounded shape started to emerge, whereas no morphological change was noted in the control FBs infected with UbC:EGFP (Figure 1C). Similar results were also observed in FBs transfected with the 25 TFs and SOX1:EGFP (Figure 1C). The proportion of PAX6:EGFP+ and SOX1:EGFP+ cells was 5.31% ± 0.38% and 6.31% ± 0.45%, respectively (Figures 2D and 3D). After purification of the PAX6:EGFP+ or SOX1:EGFP+ cells by fluorescence-activated cell sorting (FACS) and subsequent culture (Figure 1B), the purified cells started to spontaneously form neural sphere-like structures (Figure 1C) at 2 days after re-plating, whereas no neural sphere-like structures were observed in the control cells (Figure 1C). We then characterized the putative iENPs generated using the 25 TFs (iENP-25F) with various assays. Through immunocytochemical (ICC) and RT-PCR analyses, we showed that PAX6:EGFP- and SOX1:EGFP-iENP-25F expressed common neural markers, such as NESTIN, OTX2, and ZO1 (Figure 1D), and neural genes (Figure 1E).
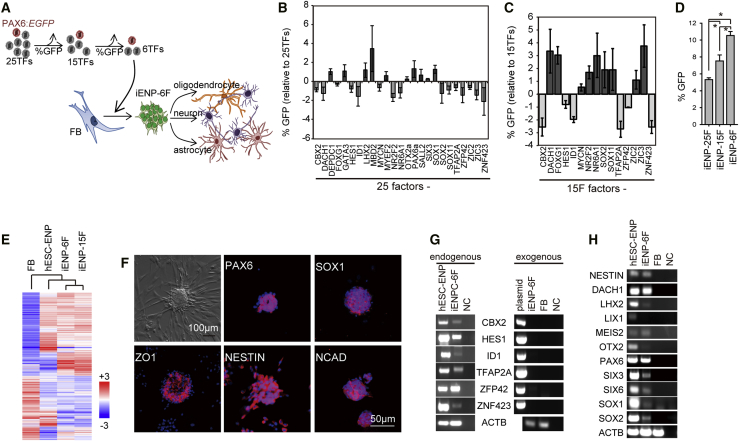
Figure 2.
Induction of FBs into iENPs by Six TFs Selected Using the PAX6:EGFP Neural Reporter
(A) Schematic depiction of the experimental strategy for reducing the number of TFs (from 25 to 6) for iENP induction using the neural reporter PAX6:EGFP.
(B and C) Stepwise selection of potent iENP factors for iENP-6F generation by single TF dropouts from the original 25-TF set (B) and the 15-TF set (C). The results are expressed as the relative percentage of PAX6:EGFP+ cells after each TF was removed from the 25-TF or 15-TF combinations.
(D) Comparison of the efficiency of induction of PAX6:EGFP+ cells from FBs by 25-, 15-, and 6-TF combinations.
(E) Global gene expression heatmap of FB, hESC-ENP, iENP-6F, and iENP-15F as determined by microarray analysis.
(F) ICC staining of iENPs-6F using antibodies against the indicated NP markers.
(G) RT-PCR analysis of endogenous and exogenous expression of the 6 TFs using mRNA isolated from iENP-6F.
(H) RT-PCR analysis of the indicated neural genes using mRNA isolated from iENP-6F.
FB, fibroblast; NC, negative control (H2O); plasmid, expression plasmids for the indicated genes. All quantitative data were obtained from three independent experiments and are presented as means ± SD. ∗p < 0.05. See also Figures S2–S4.
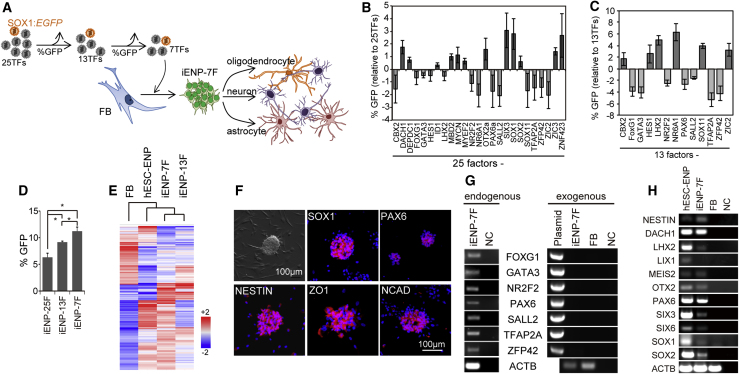
Figure 3.
Induction of FBs into iENPs by the Seven TFs Selected Using the SOX1:EGFP Neural Reporter
(A) Schematic depiction of the experimental strategy for reducing the number of TFs (from 25 to 7) for iENP induction using the neural reporter SOX1:EGFP.
(B and C) Stepwise selection of potent iENP factors for iENP-7F generation by single TF dropouts from the original 25-TF set (B) and 13-TF set (C). The results are expressed as the relative percentage of SOX1:EGFP+ cells after each TF was removed from the TF combination.
(D) Comparison of the efficiency of induction of SOX1:EGFP+ cells from FBs by the 25-, 13-, and 7-TF combinations.
(E) Global gene expression heatmap of FBs, hESC-ENP, iENP-7F, and −13F, as determined by microarray analysis.
(F) ICC staining of iENPs-7F using antibodies against the indicated NP markers.
(G) RT-PCR analysis of endogenous and exogenous expression of the seven TFs using mRNA isolated from iENP-7F.
(H) RT-PCR analysis of the indicated neural genes using mRNA isolated from iENP-7F.
FB, fibroblasts; NC, negative control (H2O); plasmid: expression plasmids for the indicated genes. All quantitative data were obtained from three independent experiments and are presented as means ±SD. ∗p < 0.05. See also Figures S2–S4.
To reduce the number of TFs for iENP generation (Figures 2A and 3A), we performed a two-step selection of TFs for iENP induction by removing one TF from the 25-TF pool each time, and infecting the FBs with the remaining 24 TFs with PAX6:EGFP or SOX1:EGFP reporters. The impact of the missing TF on the induction of PAX6:EGFP+ or SOX1:EGFP+ cells was then evaluated by flow cytometric analysis (Figures 2B and 3B). Accordingly, we identified 15 factors (CBX2, DACH1, FOXG1, HES1, ID1, MYCN, NR2F2, NR6A1, SOX2, SOX11, TFAP2A, ZFP42, ZIC2, ZIC3, ZNF423) and 13 (CBX2, FOXG1, GATA3, HES1, LHX2, NR2F2, NR6A1, PAX6, SALL2, SOX11, TFAP2A, ZFP42, ZIC2) whose removal significantly decreased the generation of the PAX6:EGFP or SOX1:EGFP+ cells, respectively, compared with their counterparts generated with 25 TFs (Figures 2B and 3B). To determine whether iENPs can be induced from FBs using the deduced TF combinations, we infected FBs with lentiviruses encoding the selected 15 or 13 TFs under a doxycycline-inducible overexpression system. After purification of PAX6:EGFP+ or SOX1:EGFP+ cells by FACS, our subsequent analysis demonstrated that iENP-15F and iENP-13F, like iENP-25F, can spontaneously form neural sphere-like structures, and expressed neural markers and genes as revealed by ICC and RT-PCR analyses (Figures S2A and S2B). Through PCR and RT-PCR analyses, we also confirmed integration of the exogenous transgenes into genomic DNA and activation of endogenous ENP gene expression after doxycycline withdrawal, respectively (Figures S2C and S2D). Further microarray analysis revealed that the global gene expression profiles of iENP-15F and iENP-13F were overtly different from those of their respective parental FBs, and more similar to those of hESC-ENPs (Figures 2E and 3E). Importantly, in vitro differentiation of iENP-15F and -13F demonstrated that they are able to spontaneously give rise to TUJ1+ neurons, GFAP+ astrocytes, and GALC+ oligodendrocytes (Figure S2E). These results suggested that iENP-15F and iENP-13F possess the common characteristics of NPs and are able to give rise to the major components of the human nervous system.
To define the minimal number of TFs required for iENP generation, we carried out an extra run of TF selection (through a procedure similar to that described above) to select the most potent TFs for iENP induction (Figures 2C and 3C). After the second TF selection, we found that removal of CBX2, HES1, ID1, TFAP2A, ZFP42, or ZNF423 (6F) and FOXG1, GATA3, NR2A2, PAX6, SALL2, TFAP2A, or ZFP42 (7F) from the 15-TF and 13-TF combinations caused a significant reduction of the generation of PAX6:EGFP+ or SOX1:EGFP+ cells, respectively (Figures 2C and 3C). After infection with the identified 6 TFs or 7 TFs, 10.54% ± 0.47% of PAX6:EGFP+ cells and 11.22% ± 0.44% of SOX1:EGFP+ cells were purified by FACS (Figures 2D and 3D). Similar to our observations for iENP-25F, -15F, and -13F (Figures 1C and S2A), FACS-isolated iENP-6F and iENP-7F also spontaneously formed neural sphere-like structures (Figures 2F and 3F). Notably, removal of any individual factor from the 6- or 7-TF combination significantly compromised the generation of PAX6:EGFP+ or SOX1:EGFP+ cells, respectively (Figures S4A–S4B), as well as neural sphere-like structure formation (Figure S4C). Collectively, these results suggested that each TF in the 6- and 7-TF combinations is essential for iENP generation. Further, PCR analysis confirmed the integration of exogenous transgenes in genomic DNA of iENP-6F and iENP-7F (Figure S2C), and RT-PCR analysis revealed that the expression of the exogenous transgenes was completely silenced, while the expression of the corresponding endogenous genes was activated in iENP-6F and -7F after doxycycline withdrawal (Figures 2G and 3G). Clustering global gene expression analysis by GeneSpring software showed that the gene expression profiles of iENP-6F and -7F were closer to those of hESC-ENPs than those of their parental FBs (Figures 2E and 3E) and expressed ENP markers and genes, as revealed by ICC and RT-PCR analyses (Figures 2F, 2H, 3F, and 3H). Moreover, iENP-6F and -7F could be sub-cultivated for more than 20 passages while maintaining a normal karyotype (Figure S2A), and subsequently cryopreserved and thawed for further expansion without losing their NP characteristics. Together, these results demonstrated that the morphological, biochemical, and molecular traits of both iENP-6F and -7F resemble those of hESC-ENPs.
Multipotency of iENP In Vitro
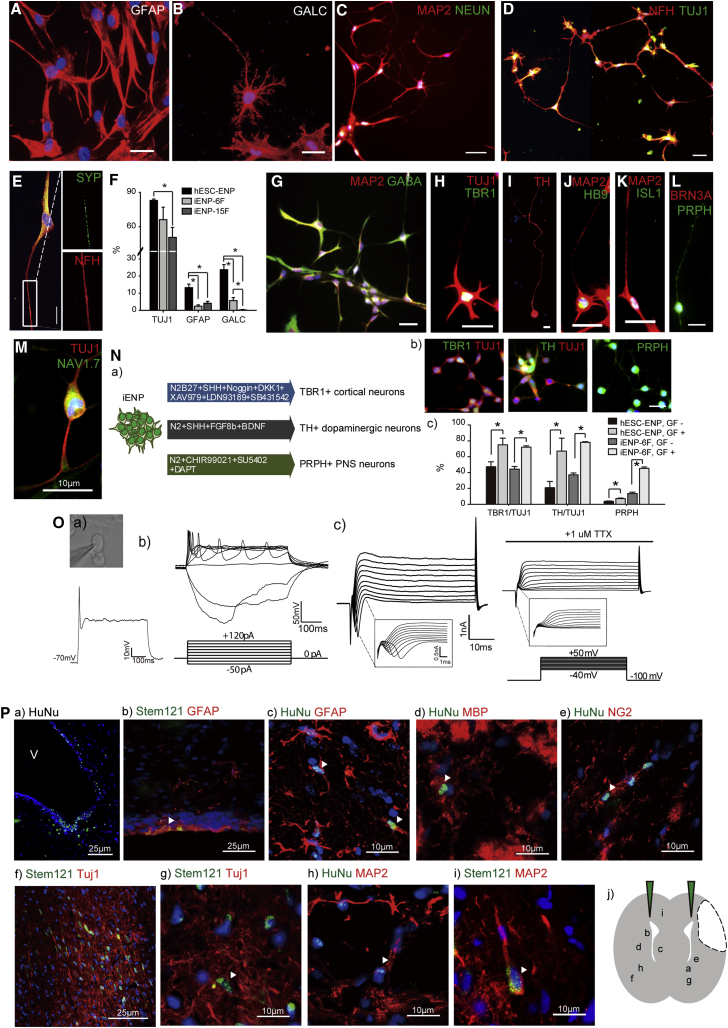
As functional ENPs can differentiate into astrocytes, oligodendrocytes, and neurons, we examined the ability of our iENPs to differentiate in vitro (Figures 4 and 5). After 2–3 weeks of differentiation, GFAP+ and GALC+ cells and abundant neuron-like cells exhibiting neuronal process and expressing neuronal marker MAP2, NEUN, or TUJ1 were readily observed in both differentiating iENP-6F (Figures 4A–4D) and -7F cells (Figures 5A–5C) under neural differentiation conditions (Supplemental Experimental Procedures). Notably, the synapse marker synaptophysin (SYP) was also found co-expressed with mature neuronal marker, NFH (Figure 4E). Further, quantification of the proportion of cell expressing TUJ1 GFAP, and GALC in the differentiating iENPs by ICC analysis revealed that the neuronal differentiation ability of iENP-6F was similar to that of hESC-ENPs, whereas iENP-15F was less able to generate neurons compared with hESC-ENPs and iENP-6F, suggesting that removal of the 9 TFs from the 15-TF combination further enhanced the neuronal propensity of iENP-6F (Figure 4F). The astrocyte and oligodendrocyte differentiation abilities of iENP-6F and -15F were significantly lower than those of hESC-ENPs (Figure 4F), and iENP-15F exhibited poorer ability to generate oligodendrocytes compared with iENP-6F (Figure 4F). On the other hand, the neuronal differentiation abilities of iENP-7F and -13F were similar to but lower than that of hESC-ENPs (Figure 5D). Both iENP-7F and -13F exhibited significantly lower ability to generate astrocytes and oligodendrocytes (Figure 5D) compared with hESC-ENPs.
Figure 4.
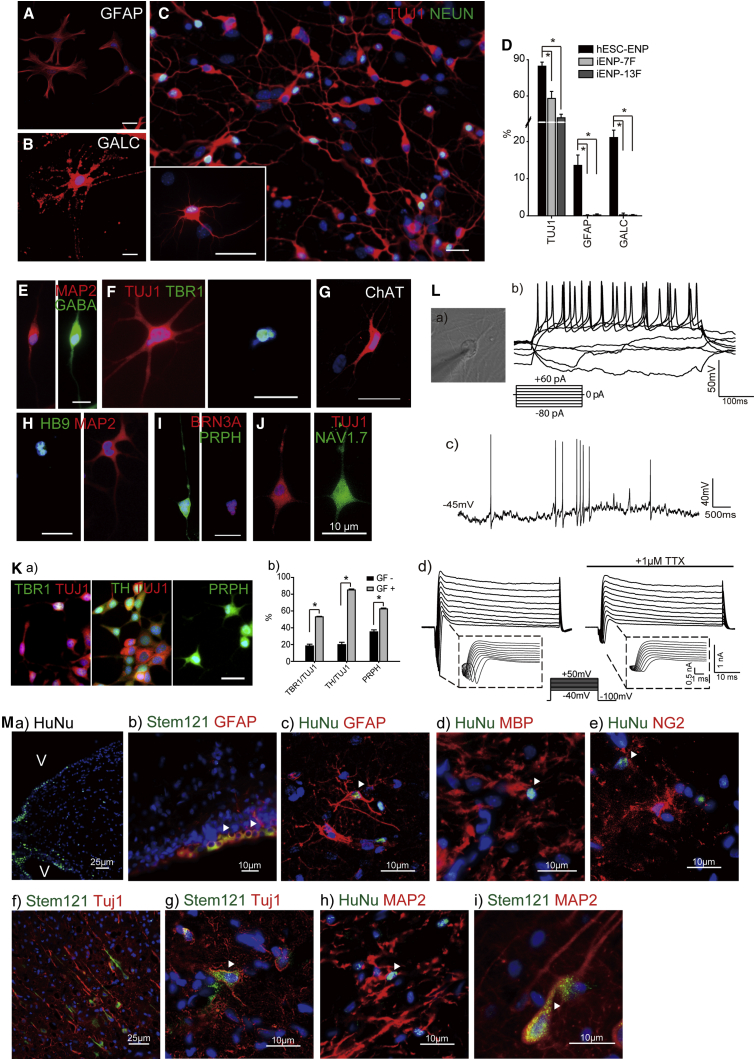
Multipotency of iENP-6F In Vitro and In Vivo
(A–E) ICC staining of differentiated iENP-6F using antibodies against the glial marker GFAP (A), oligodendrocyte marker GALC (B), and neuronal markers, as indicated (C, D), and synapse marker SYN (E).
(F) Quantification and comparison of TUJ1+, GFAP+, and GALC+ cells in differentiated hESC-NPs, iENP-6F, and iENP-15F.
(G–M) ICC staining of differentiated iENP-6F using antibodies against CNS and PNS neuronal antigens, as indicated.
(N) Lineage-specific cues promote the generation of specific neuronal subtypes from iENP-6F. (a) Schematic depiction of the Experimental Procedures used to induce specific neuronal subtypes from iENP-6F. (b) ICC characterization of differentiated iENP-6F under neuronal subtype-specific differentiation conditions by ICC using antibodies against CNS and PNS neuronal antigens, as indicated. (c) Quantification of the indicated neuronal subtypes induced by the conditions described in (Na). GF−, without inducers; GF+, with inducers.
(O) Whole-cell patch-clamp recording of iENP-6F-derived neurons. (a) Current recording of a neuron at 4–6 weeks post differentiation. (b) Action potentials were induced by current steps from −50 to +120 pA. (c) Inward Na+ currents and outward Ca2+ currents were induced by voltage steps from −40 to +50 mV. The inward Na+ currents could be blocked with tetrodotoxin (TTX).
(P) In vivo transplantation of iENP-6F. (a) IHC staining of the corpus callosum containing iENP-6F transplants using an antibody against human nuclear antigen (HuNu). (b–i) IHC analysis of brain cryosections at 12 weeks post-transplantation using antibodies against HuNu or Stem121 and the indicated neural antigens. (j) Scheme showing the relative position of the indicated cells after transplantation. Arrowheads indicate the cells expressing human-specific markers and neural markers.
All quantitative data were obtained from three independent experiments and are presented as means ± SD. ∗p < 0.05. (A–E, G–L, and Nb) Scale bar, 10 μm. See also Figure S3.
Figure 5.
Multipotency of iENP-7F In Vitro and In Vivo
(A–C) ICC staining of differentiated iENP-7F using antibodies against the glial marker GFAP (A), oligodendrocyte marker GALC (B), and neuronal markers, as indicated (C).
(D) Quantification and comparison of TUJ1+, GFAP+, and GALC+ cells in differentiated hESC-ENPs, iENP-7F, and iENP-13F.
(E–J) ICC staining of differentiated iENP-7F with antibodies against CNS and PNS neuronal antigens, as indicated.
(K) Lineage-specific cues promote the generation of specific neuronal subtypes from iENP-7F. (a) ICC characterization of differentiated iENP-7F under neuronal subtype-specific differentiation conditions using antibodies against CNS and PNS neuronal antigens, as indicated. (b) Quantification of iENP-7F-derived neuronal subtypes induced by the conditions described in Figure 4Na. GF−, without inducers; GF+, with inducers.
(L) Whole-cell patch-clamp recordings of iENP-7F-derived neurons. (a) Current recording from a neuron at 4–6 weeks. (b) Action potentials were induced by current steps from −80 to +60 pA. (c) Spontaneously firing action potentials were recorded at a subthreshold oscillatory potential of −40 mV. (d) Inward Na+ currents and outward Ca2+ currents were induced by voltage steps from −40 to +50 mV. The inward Na+ currents could be blocked by tetrodotoxin (TTX).
(M) In vivo transplantation of iENP-7F. (a) IHC staining of the corpus callosum containing iENP-7F transplants using an antibody against human nuclear antigen (HuNu), revealing migration of iENPs into ventricular zones. (b–i) IHC analysis of brain cryosections at 12 weeks post-transplantation using antibodies against HuNu or Stem121 and the indicated neural antigens. Arrowheads indicate the cells expressing human-specific markers and neural markers.
All quantitative data were obtained from three independent experiments and are presented as means ± SD. ∗p < 0.05. (A–I and K) Scale bar, 10 μm. See also Figure S3.
To determine whether iENPs can differentiate into various neuronal subtypes, we interrogated the iENP-6F- and -7F-derived neuronal population with a panel of neuronal markers (Figures 4G–4M and 5E–5J). ICC analysis revealed that both iENP-6F and -7F can give rise to various neuronal subtypes, including GABA+ (Figures 4G and 5E), TBR1+ cortical (Figures 4H and 5F), TH+ dopaminergic (Figures 4I and 5Ka), HB9+/ISL1+ motor (Figures 4J, 4K, and 5H), and BRN3A+, PRPH+, or NAV1.7+ peripheral neurons (Figures 4L, 4M, 5I, and 5J). As hESC-ENPs can be coaxed by extrinsic stimuli to differentiate into specific neuronal subtypes, we examined whether our iENPs respond in a similar manner. To this end, iENPs were exposed to differentiation conditions for cortical (Maroof et al., 2013), dopaminergic (Nguyen et al., 2011), and peripheral neuron (Chambers et al., 2012) generation (Figures 4N and 5K). ICC analysis with antibodies against TBR1, TH, or PRPH revealed that exposure to specific neuronal differentiation conditions significantly improved the generation of the representative neuronal subtypes (cortical, dopaminergic, or peripheral neurons, respectively) from the iENPs (Figures 4N and 5K). These findings suggest that iENPs are multipotent and able to respond to specific differentiation cues in a manner similar to hESC-ENPs.
Next, we explored whether the iENP-derived neurons possess functional electrophysiological properties similar to those of neurons. We cultured iENP-derived neurons in neuronal maturation medium for 2 weeks, and then subjected them to whole-cell patch-clamp recoding, revealing that the resting membrane potential was −35.25 ± 0.64 mV in iENP-6F-derived neurons (Figure 4Oa) and −64.3 ± 17.96 mV in iENP-7F-derived neurons (Figure 5La). Action potentials could be elicited by membrane depolarization in current clamp mode (Figures 4Ob and 5Lb), and spontaneous action potentials were recorded in iENP-derived neurons (Figure 5Lc). Sodium-channel-mediated inward currents were blocked by tetrodotoxin (TTX), a Na+ ion-channel-specific inhibitor (Figures 4Oc and 5Ld). These results suggested that the iENP-derived neurons possess functional electrophysiological properties similar to those of neurons.
Transplanted iENPs Integrate and Differentiate in Rat Brains
To assess the in vivo differentiation potency of iENP, we transplanted the iENPs into the corpus callosum of rat brains, and analyzed the brains 12 weeks after transplantation (Figures 4P and 5M). We first examined whether iENP transplantation caused tumor formation in the brain. H&E staining of brain sections and further RT-PCR and immunohistochemical (IHC) analyses revealed no expression of tumor-associated markers or tumor formation in iENP-transplanted brains at 12 weeks post-transplantation (Figure S3B). Interestingly, we found some of the transplanted cells migrated to the ventricular zones, a brain region where neurogenesis takes place, and GFAP, a radial glia progenitor marker, is expressed (Figures 4Pa,4Pb, 5Ma, and 5Mb). Consistent with the in vitro differentiation results, the transplanted iENPs were found to have differentiated into GFAP+ astrocytes (Figures 4Pc and 5Mc), NG2+ oligodendrocytes (Figures 4Pd, 4Pe, 5Md, and 5Me), and TUJ1+ or MAP2+ neurons (Figures 4Pf–4Pi) in the brains. Together, our results indicate that iENPs integrate into adult brain tissue and differentiate into major neural cell types in vivo.
The iENP-6F and -7F Populations Exert Differential Developmental Propensity
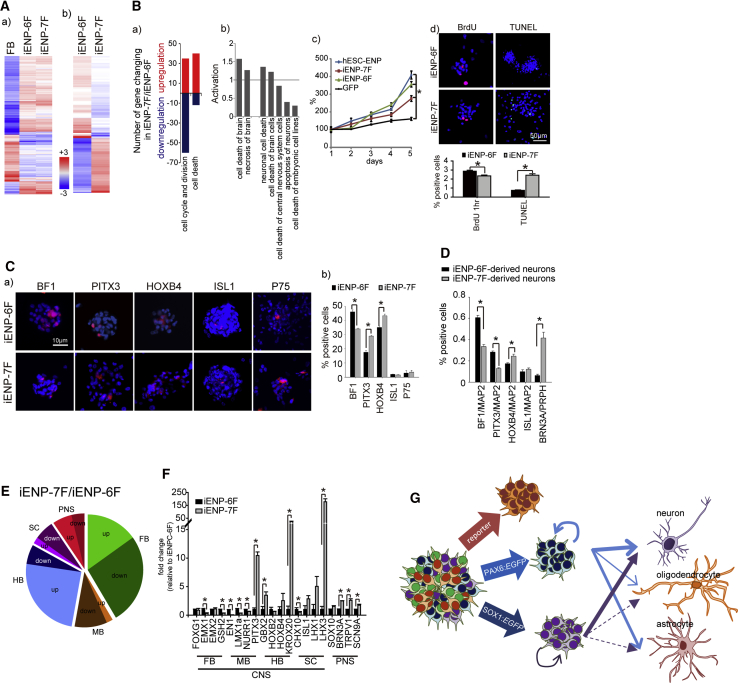
As described above, our results indicated that iENP-6F and -7F possess different neural differentiation propensity. This observation prompted us to further explore the differences between these two populations. To this end, we first examined the gene expression profiles of these two iENP populations by microarray analysis. Heatmap analysis revealed that the global gene expression profiles of iENP-6F and -7F are similar (Figure 6Aa). Through Ingenuity Pathway Analysis (IPA) and examination of combined fold change and gene ontology, the expression levels of 170 genes were found to be significantly different (≥2-fold) between iENP-6F and -7F (Figure 6Ab). Of these, a panel of genes related to cell cycle and division exhibited lower expression in iENP-7F than in iENP-6F (Figure 6Ba), and IPA analysis showed that cell-death-associated pathways were activated in iENP-7F compared with iENP-6F (Figure 6Bb). Consistently, the growth curve of iENP-6F was found to be similar to that of hESC-ENPs, whereas iENP-7F exhibited a slower proliferation rate (Figure 6Bc). Further analysis showed greater BrdU+ and reduced TUNEL+ proportions in iENP-6F compared with iENP-7F (Figure 6Bd).
Figure 6.
The Differential Properties of iENP-6F and iENP-7F
(A) Heatmap analysis of global gene expression profiles of undifferentiated iENP-6F, iENP-7F, and FBs.
(B) (a) Dynamic changes in the expression of genes characterized by the indicated GO terms. Red, upregulated; blue, downregulated. (b) IPA of the activated pathways associated with cell death. (c) Growth curve analysis of the indicated cell populations. (d) ICC staining and quantification of iENPs by BrdU incorporation and TUNEL assays. Nuclei were counterstained with DAPI (blue).
(C) Preferential expression of brain regional markers in iENPs. (a) ICC staining of iENPs with antibodies against brain regional antigens, as indicated. (b) Quantification of the percentage of cells expressing brain regional markers, as indicated, in iENPs.
(D) Quantification of the percentage of cells expressing the indicated brain regional markers in iENP-derived neurons.
(E) Pie chart depicting the proportion of brain regional subtype-associated genes up- and downregulated between iENP-7F and -6F.
(F) Relative expression of brain regional-associated genes in iENP-7F and -6F, as measured by RT-qPCR analysis.
(G) Model illustrating the strategy for direct conversion of iENPs with differential differentiation propensities through the use of different combinations of hESC-ENP-TFs and neural reporters.
FB, forebrain; MB, midbrain; HB, hindbrain; SC, spinal cord. The weight of the solid/dashed arrow indicates the neural differentiation potential of the iENP populations.
All quantitative data were obtained from three independent experiments and are presented as means ± SD. ∗p < 0.05.
To further explore the developmental propensity of iENP-6F and -7F, we interrogated these populations with a panel of regional markers (Figure 6Ca). ICC analysis of the undifferentiated iENP populations and their derived neurons (iENP-Ns) revealed that the proportion of iENPs/iENP-Ns expressing BF1 (forebrain marker) was significantly higher in iENP-6F/-Ns than in iENP-7F/-Ns, whereas the proportion of cells expressing PITX3 (midbrain marker), HOXB4 (hindbrain marker), and p75 or BRN3A (PNS marker), were lower in iENP-6F/-Ns than in iENP-7F/-Ns (Figures 6Cb and 6D). Consistent with our ICC analysis, comparative global gene expression profiling and RT-qPCR analysis between iENP-6F and 7F revealed that iENP-6F preferentially expressed more forebrain, midbrain, and spinal cord-related genes compared with iENP-7F, whereas iENP-7F preferentially expressed more hindbrain and PNS-related genes compared with iENP-6F (Figures 6E and 6F). Together, these results suggest that iENP-6F and -7F are different NP subpopulations with different neural gene expression, growth rates, and developmental propensities.
Recapitulation of Pathological Features in Diseased iENPs
To explore the potential of iENPs for disease modeling, we generated iENPs from FBs derived from an AD patient with an APOE4/E4 mutation (AD1), two familial AD (fAD) patients with the PSEN1 mutation (fAD, AD2, and AD3), and two HD patients (male and female, 41 CAG repeats in the HTT gene). Similar to wild-type FBs, AD- and HD-FBs could be converted into PAX6:EGFP− and SOX1:EGFP+ cells with the 6-TF or 7-TF combination, respectively, and these populations formed neural sphere-like structures and expressed NP markers/genes (Figures 7A and S5). Further, we showed that the putative AD- and HD-iENPs were able to give rise to TUJ1+ neurons, GFAP+ astrocytes, and GALC+ oligodendrocytes (Figure 7B).
Figure 7.
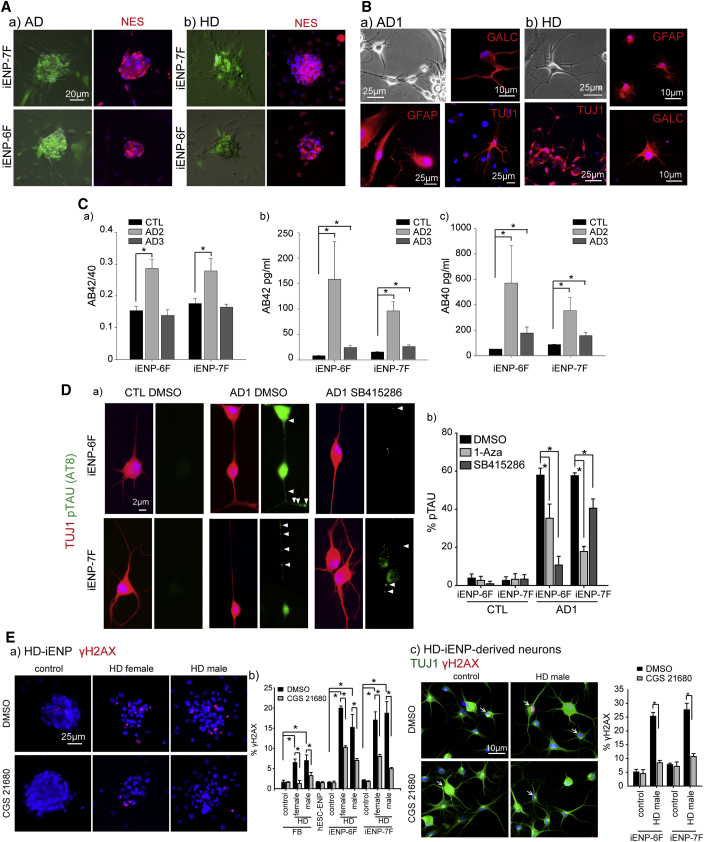
Recapitulation of Disease Phenotypes in the Diseased iENPs and Their Neuronal Derivatives
(A) Representative images of the morphology and ICC staining for Nestin in (a) AD-iENPs and (b) HD-iENPs.
(B) Phase-contrast image of AD-iENP-derived (a) and HD-iENP-derived (b) neurons and ICC staining of AD-iENP (a) and HD-iENP (b) derivatives using antibodies against GFAP, GALC, and TUJ1.
(C) Secreted Aβ42/40 ratio; Aβ42 and Aβ40 from AD-iENP-derived neurons. AD2 and AD3, patients carrying PSEN1 mutations.
(D) (a) ICC staining analysis of pTAU expression in AD-iENP-derived neurons using antibodies against TUJ1 and pTAU (AT8). (b) Quantification of the effect of 1-Aza and SB415286 on the reduction in pTAU expression in AD-iENP-derived neurons. AD1, patient carrying the APOE4/E4 mutation. Controls were treated with DMSO. Arrowheads indicated pTAU accumulation.
(E) ICC staining (a) and quantification (b) of γH2AX+ cells in vehicle (DMSO)- and CGS 21680-treated control and HD-iENPs. (c) ICC staining and quantification of γH2AX+ cells in vehicle (DMSO)- and CGS 21680-treated controls and HD-iENP-derived neurons. Arrows indicated γH2AX+ cells.
All quantitative data were obtained from three independent experiments and are expressed as means ± SD. ∗p < 0.05. See also Figure S5.
Next, we examined whether AD- and HD-iENPs and their neuronal derivatives exhibit the pathological features of the relevant diseases. As an increase of amyloid β (Aβ) and accumulation of phosphorylated TAU (pTAU) are the major pathological features present in the neurons of AD patients (Choi et al., 2014), we first measured the level of extracellular Aβ40 and Aβ42 in conditioned media of neurons differentiated from AD- or control-iENPs. The results of ELISA analysis revealed that the level of both Aβ isoforms was significantly elevated in the neurons of AD-iENP derived from two fAD-FB populations with the PSEN1 mutation (AD2 and AD3, Figure 7C) compared with the control-iENP-derived neurons. The Aβ42/Aβ40 ratio was also increased in the fAD-iENP-derived neurons induced from fAD-FBs with the PSEN1 E184D mutation (AD2), although no significant increase in the Aβ42/Aβ40 ratio was detected in the fAD-iENP-derived neurons induced from another fAD-FB population with the PSEN1 P264L mutation (AD3), which was previously reported to be associated with a slight increase in the Aβ42/Aβ40 ratio in PSEN1 P264L-overexpressing cells (Dumanchin et al., 2006). To investigate pTAU pathologies in the AD-iENP-derived neurons, we first subjected AD1- and control-iENP-derived neurons to ICC analysis with an antibody recognizing pTAU (AT8); in this way, we readily detected pTAU in the processes of certain TUJ1+ neurons and observed patched pTAU aggregates in the cell body of AD-iENP-derived neurons, as previously reported in the cortex of AD patients and AD-iPS-derived neurons (Figure 7Da) (Choi et al., 2014, Goedert et al., 1993). In addition, treatment of AD-iENP-derived neurons with GSK3β inhibitors (SB415286 and 1-Aza) significantly reduced pTAU aggregation compared with DMSO-treated and control-iENP-derived neurons (Figure 7Db).
We previously reported that HD-iPSC-derived neurons are vulnerable to DNA damage, and that stimulation of A2AR using selective agonists reduced DNA damage in HD-iPSC-derived neurons (Chiu et al., 2015). To identify whether the HD-iENPs and their neuronal derivatives recapitulate the above features of HD, we treated HD-iENPs and control-iENPs with a selective A2AR agonist, CGS21680. ICC analysis of the expression of phosphorylated γH2AX, a hallmark feature of DNA damage (Rogakou et al., 1998), revealed that the number of γH2AX+ nuclei is significantly higher in HD-iENPs (Figure 7Ea and b) and their neuronal derivatives (Figure 7Ec) compared with their counterparts derived from control cells. Furthermore, CGS21680 stimulation significantly decreased γH2AX expression in the HD-iENPs and their neuronal derivatives, suggesting that activation of A2AR might reduce DNA damage in these cells (Figures 7Eb and 7Ec). Taken together, these results reveal that the diseased iENPs and their neuronal derivatives recapitulate the pathological features of AD and HD.
Discussion
Previously, various TF combinations have been used to directly convert FBs into iNPs (Lu et al., 2013, Ring et al., 2012, Wang et al., 2013). These iNPs possessed the general properties of neural progenitors, such as neural marker/gene expression, proliferation, and differentiation propensity. Unlike the hESC-ENPs, which were demonstrated to differentiate into both CNS and PNS lineages (Elkabetz et al., 2008, Lee et al., 2007), previous reports have shown that iNPs exhibit developmental potentials primarily toward CNS subtypes (Lu et al., 2013, Wang et al., 2013). However, studies have rarely addressed whether these iNPs possess the ability to give rise to PNS neuron subtypes. In this study, we showed that iENP-6F and iENP-7F are able to differentiate into not only CNS lineages but also PNS lineages. Furthermore, they responded to the same extracellular stimuli as hESC-ENPs and gave rise to specific neuronal subtypes. In line with these observations, genome-wide transcriptome profiling also confirmed a high similarity between FB-induced ENPs and their hESC-derived counterparts. Thus, our results suggest that the iENP population reprogramed by hESC-ENP-TFs may be more similar to embryonic NPCs than adult brain-derived NPCs.
Although the two iENP populations generated in this study shared similar NP characteristics, further investigation revealed that they exhibit different functional features. First, our analysis demonstrated that iENP-6F exhibited higher proliferation and reduced apoptosis compared with iENP-7F. Second, iENP-7F showed stronger differentiation potency toward neuronal lineages than glial lineages. Third, dissection of the neuronal differentiation potential of the iENP populations revealed that iENPs-7F have a regional preference toward caudal identity, whereas iENPs-6F have a regional preference toward rostral identity. The above differences between iENP-6F and 7F may be explained by the neural reporters used for TF and iENP selection. We used two neural reporters, PAX6 and SOX1, to monitor and evaluate the efficiency of neural fate conversion by hESC-ENP-nTFs, through which we identified a 6- and 7-TF combinations for iENP-6F and -7F induction, respectively. On the other hand, PAX6 and SOX1 were also used to select the iENP-6F and 7F populations, respectively. Thus, it is tempting to suggest that neural reporter selection may decide the functional characteristics of the resulting iENP populations. It is well known that hESC-derived neural rosettes and neural epithelia consist of various ENSCs/ENPs, which are responsible for the subsequent neural development of the CNS and PNS (Pankratz et al., 2007). Therefore, the originally selected 25 nTFs highly expressed in hESC-ENPs are likely essential for the formation of heterogeneous NP populations. Accordingly, induction of FBs with specific nTF combinations selected from the 25-TF pool should result in the formation of an iENP population with specific neural characteristics. Together, these results suggest that the scheme described in this study may provide an excellent way for generating desirable iENP populations through the selection of specific TF combinations from the original 25-TF pool and iENP populations using different neural reporters. Future studies will be required to determine whether specific combinations of hESC-ENP-nTFs can define the functional aspects of the resulting iENPs, and elucidate the mechanisms by which the TF combinations reprogram FBs into iENPs.
Previously, it has been reported that iNPs can be directly converted from human or mouse FBs by TF combinations including all iPSC factor(s) (Lu et al., 2013), only certain factors (Wang et al., 2013, Yu et al., 2015), or single factors (Ring et al., 2012). Starting from a panel of 25 TFs, we identified two TF combinations, six TFs and seven TFs, that can induce FBs into iENPs. Overall, the functional aspects of the TFs used for iENP induction are associated with neural development or neural identity maintenance. In the 6-TF combination, the majority of the component TFs have been reported to be involved in neuronal differentiation (Morey et al., 2012, Tsai and Reed, 1997) and maintenance of NSC fates (Bai et al., 2007, Nakamura et al., 2000). In the 7-TF combination, most TFs were reported to be associated with the early CNS (Reinchisi et al., 2012, Rosa and Brivanlou, 2011, Zhang et al., 2010), PNS development (Reinchisi et al., 2012, Rosa and Brivanlou, 2011), and early neural regional specification (Pata et al., 1999). Unlike most of the reported TF sets used for iNP generation, none of the TFs identified by our strategy are functionally associated with human iPSC generation, suggesting the induction of iENPs from FBs does not require iPSC factors. Thus, this precludes the possibility that the iENPs were generated through a transient pluripotent state, thereby circumventing the increased tumorigenic risk associated with iPSC factors. Of note, two TFs, TFAP2A and ZFP42/REX1, were shared between both TF combinations. TFAP2A is well documented to participate in the development of many tissues during embryogenesis, especially in neural development. ZFP42/REX1 is expressed in ESCs and NPs, but is dispensable for mouse pluripotency (Masui et al., 2008). However, infection of cells with lentivirus encoding TFAP2A and ZFP42 did not result in the generation of iENPs (data not shown), suggesting they may be essential, but insufficient, to induce iENPs.
One of the advantages of iNP generation is that it provides a cell-based platform for neurodegenerative disease modeling and drug discovery. As proof of principle, we induced iENPs from the FBs of AD and HD patients and demonstrated that the diseased iENPs and their neuronal derivatives exhibited pathological features of HD and AD (Choi et al., 2014, Jackson and Bartek, 2009). For example, our data showed a dramatic increase of Aβ variants and Aβ42/Aβ40 ratio and increased pTAU expression in the AD-iENPs-derived neurons; expression of pTAU could be reduced by GSK3β inhibitors, suggesting the AD-iENP-derived neurons recapitulate some, if not all, of the AD pathological features (Choi et al., 2014, Dumanchin et al., 2006). Several lines of evidence have indicated that stress factors can cause DNA damage and increase profound neuronal death in cells derived from HD patients (Jackson and Bartek, 2009). Also, it has been reported that A2AR agonists are beneficial in HD transgenic animal models and an HD-iPSC-derived neuronal population (Chiu et al., 2015). In line with these observations, our results demonstrated that HD-iENPs and their neuronal derivatives were more susceptible to DNA damage than their counterparts derived from normal FBs. Moreover, CGS21680 treatment decreased DNA damage in the HD-iENPs and their neuronal derivatives. Together, these findings suggest that, to some extent, the iENP model can recapitulate neurodegenerative disease-relevant pathogenesis, and thus may be suitable for characterization of the disease mechanism and for screening therapeutic agents.
Through in vivo transplantation of iENPs into rat brains, we demonstrated that iENPs can survive and differentiate into various neural subtypes in the adult brain environment. This observation confirms that iENPs possess an in vivo differentiation propensity similar to that of hESC-ENPs (Ben-Hur et al., 2004), suggesting that the iENPs established in this study could serve as an autologous cell source to treat neurodegenerative diseases, such as AD and HD. Nevertheless, further efforts are required to explore the tumorigenic potential of the transplanted iENPs in brains, although our results showed that brains are free from tumor formation at 12 weeks post-transplantation.
Collectively, our studies have demonstrated a paradigm for direct conversion of multipotent iENPs from human somatic cells through overexpression of hESC-NP-enriched TFs. This system will allow generation of expandable iNP populations with desirable neural differentiation propensities (Figure 6G) and also facilitate the discovery of disease mechanisms and drugs for treatment of neurodegenerative diseases and use in regenerative medicine.
Experimental Procedures
Human Materials
Human samples were obtained with written informed consent from tissue donors in accordance with the protocol approved by the Internal Research Board of Academia Sinica.
Construct Generation
Constructs carrying candidate neural TFs were generated from the coding sequences of the 25 TFs (see Supplemental Experimental Procedures). The coding sequences were cloned into FUW or FUW-tetO vectors for further experiments. Reporter constructs were generated by cloning a 1.3-kb PAX6 P1 promoter (Plaza et al., 1995) and a 1-kb SOX1 promoter into the FUW vector to generate PAX6:EGFP and SOX1:EGFP, respectively.
Generation of iENPs
Lentiviral particles carrying candidate TFs were generated in 293FT cells using standard procedures. FBs (see Supplemental Experimental Procedures) were infected with lentiviruses and then cultured in FB media (DMEM, 10% fetal bovine serum). One day after infection, media were replaced with neural induction media (see Supplemental Experimental Procedures). After 1 week of induction, GFP+ cells were purified using a BD FASCAriaII sorter and plated on Matrigel-coated dishes containing iENP media (see Supplemental Experimental Procedures). Cells spontaneously formed neural sphere-like structures after 2 or 3 days. These neural sphere-like structures were collected and trypsinized into single cells and then plated on ornithine-laminin-coated dishes in iENP media containing 2 μg/mL doxycycline. The efficiency of iENP generation was measured by combining two parameters: the percentage of GFP+ cells driven by either PAX6:EGFP or SOX1:EGFP on day 6 post lentiviral infection and the percentage of neurosphere formation on day 2 post purification. After 2 to 3 passages, doxycycline was removed from the culture media and the cells were maintained and split every 7 days.
Differentiation and Drug Testing
General neural differentiation was examined using differentiation media (see Supplemental Experimental Procedures), while specific neuronal differentiation was examined using cortical neuronal differentiation media, dopaminergic neuronal differentiation media, and PNS neuronal differentiation media. For AD drug testing, AD- and control-iENPs were subjected to cortical differentiation. At 7 days after differentiation, cells were treated with SB415286, 1-azakenpaullone (Selleckchem), or DMSO (Sigma) for 2 days. For HD studies, cells were induced to differentiate and then treated with CGS21680 as previously described (Chiu et al., 2015).
Cell Transplantation and Ethics Statement
Long-Evans rats (7–8 weeks old) were subjected to ischemia by right middle cerebral artery occlusion and common carotid arteries for 30 min, and then 50,000 undifferentiated iENP-6F and iENP-7F were injected into the cerebrum (anterior/posterior, 0.3 cm; medial/lateral, −2.0 cm; dorsal/ventral, −2.8 cm; top/bottom, −3.0 cm). After 12 weeks, rats were killed and perfused with 4% paraformaldehyde in 0.1 M phosphate buffer, and then the brains were isolated. All the animal experiments were approved by the Animal Care and Use Committee of Academia Sinica and performed in accordance with the Institutional Animal Care and Use Committee of Academia Sinica.
Microarray Analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). Two biological duplicates per cell type were examined. Chips were scanned with an Affymetrix GeneChip Scanner 7G, and data were analyzed by GeneSpring X software (Agilent). Raw data were normalized independently for each experiment using the Robust Multichip Average. Gene expression patterns were analyzed by Genespring Software and Ingenuity Pathway Analysis Software. The accession numbers for the microarray data reported in this article are GEO: GSE81554 (for NP2, FB1, hESC-ENP, and iENPs); GEO: GSE27280 (for FB2 and 3); ArrayExpress: E-MEXP-2668 (for NP1).
Immunocytochemical and Immunohistochemical Analysis
The ICC procedure was performed as previously described (Hou et al., 2013). For IHC analysis, transplanted rat brains were dehydrated using 20% sucrose in PBS and embedded in OCT compound (Tissue-Tek). Consecutive coronal sections (12 μm) were performed using a Leica CM3050S sliding microtome. Tissue slices were post-fixed with 4% paraformaldehyde for 30 min at room temperature and cold methanol for 30 min. The primary and secondary antibodies are listed in Table S2. Nuclei were counterstained with DAPI. Signals were recorded using a Zeiss microscope and Spot software.
Statistical Analyses
All quantitative data were obtained from three independent experiments and determined as means ± SDs (error bars) (∗p < 0.05; two-tailed two-sample t test) or otherwise indicated.
Author Contributions
Conceptualization, P.S.H, C.Y.C., and H.C.K.; Methodology, P.S.H., C.Y.C., C.H.Y., and W.C., T.N.L. and H.J.L.; Investigation, P.S.H., C.Y.C., C.H.Y., W.C., H.J.L., and H.C.K.; Writing – Original Draft, P.S.H., C.Y.C., and H.C.K.; Writing – Review & Editing, P.S.H., C.Y.C., C.H.Y., and H.C.K.; Visualization, P.S.H., C.Y.C., W.C., and C.H.Y.; Supervision and Funding acquisition, H.C.K.
Acknowledgments
This research was supported by Academia Sinica (Thematic project, AS-104-TP-B09 and AS-103-TP-B10) and the Ministry of Science and Technology (MOST 104-0210-01-09-02, MOST 105-0210-01-13-01).
Published: December 8, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.11.006.
Accession Numbers
The accession number for NP2, FB1, hESC-ENP, and iENPs reported in this article is GEO: GSE81554.
Supplemental Information
References
- Ang C.E., Wernig M. Induced neuronal reprogramming. J. Comp. Neurol. 2014;522:2877–2886. doi: 10.1002/cne.23620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G., Sheng N., Xie Z., Bian W., Yokota Y., Benezra R., Kageyama R., Guillemot F., Jing N. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell. 2007;13:283–297. doi: 10.1016/j.devcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Ben-Hur T., Idelson M., Khaner H., Pera M., Reinhartz E., Itzik A., Reubinoff B.E. Transplantation of human embryonic stem cell-derived neural progenitors improves behavioral deficit in Parkinsonian rats. Stem Cells. 2004;22:1246–1255. doi: 10.1634/stemcells.2004-0094. [DOI] [PubMed] [Google Scholar]
- Bredesen D.E., Rao R.V., Mehlen P. Cell death in the nervous system. Nature. 2006;443:796–802. doi: 10.1038/nature05293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Qi Y., Mica Y., Lee G., Zhang X.J., Niu L., Bilsland J., Cao L., Stevens E., Whiting P. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu F.L., Lin J.T., Chuang C.Y., Chien T., Chen C.M., Chen K.H., Hsiao H.Y., Lin Y.S., Chern Y., Kuo H.C. Elucidating the role of the A2A adenosine receptor in neurodegeneration using neurons derived from Huntington's disease iPSCs. Hum. Mol. Genet. 2015;24:6066–6079. doi: 10.1093/hmg/ddv318. [DOI] [PubMed] [Google Scholar]
- Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D'Avanzo C., Chen H., Hooli B., Asselin C., Muffat J. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanchin C., Tournier I., Martin C., Didic M., Belliard S., Carlander B., Rouhart F., Duyckaerts C., Pellissier J.F., Latouche J.B. Biological effects of four PSEN1 gene mutations causing Alzheimer disease with spastic paraparesis and cotton wool plaques. Hum. Mutat. 2006;27:1063. doi: 10.1002/humu.9458. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y., Panagiotakos G., Al Shamy G., Socci N.D., Tabar V., Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M., Jakes R., Crowther R.A., Six J., Lubke U., Vandermeeren M., Cras P., Trojanowski J.Q., Lee V.M. The abnormal phosphorylation of tau protein at Ser-202 in Alzheimer disease recapitulates phosphorylation during development. Proc. Natl. Acad. Sci. USA. 1993;90:5066–5070. doi: 10.1073/pnas.90.11.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Hermann A., Hemmer K., Hoing S., Arauzo-Bravo M.J., Zaehres H., Wu G., Frank S., Moritz S. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- HD iPSC Consortium Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P.S., Chuang C.Y., Kao C.F., Chou S.J., Stone L., Ho H.N., Chien C.L., Kuo H.C. LHX2 regulates the neural differentiation of human embryonic stem cells via transcriptional modulation of PAX6 and CER1. Nucleic Acids Res. 2013;41:7753–7770. doi: 10.1093/nar/gkt567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Efe J.A., Zhu S., Talantova M., Yuan X., Wang S., Lipton S.A., Zhang K., Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. USA. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Kim H., Elkabetz Y., Al Shamy G., Panagiotakos G., Barberi T., Tabar V., Studer L. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat. Biotechnol. 2007;25:1468–1475. doi: 10.1038/nbt1365. [DOI] [PubMed] [Google Scholar]
- Lu J., Liu H., Huang C.T., Chen H., Du Z., Liu Y., Sherafat M.A., Zhang S.C. Generation of integration-free and region-specific neural progenitors from primate fibroblasts. Cell Rep. 2013;3:1580–1591. doi: 10.1016/j.celrep.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E., Chanda S., Ahlenius H., Sudhof T.C., Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. USA. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof A.M., Keros S., Tyson J.A., Ying S.W., Ganat Y.M., Merkle F.T., Liu B., Goulburn A., Stanley E.G., Elefanty A.G. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S., Ohtsuka S., Yagi R., Takahashi K., Ko M.S., Niwa H. Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells. BMC Dev. Biol. 2008;8:45. doi: 10.1186/1471-213X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L., Pascual G., Cozzuto L., Roma G., Wutz A., Benitah S.A., Di Croce L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Sakakibara S., Miyata T., Ogawa M., Shimazaki T., Weiss S., Kageyama R., Okano H. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J. Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.N., Byers B., Cord B., Shcheglovitov A., Byrne J., Gujar P., Kee K., Schule B., Dolmetsch R.E., Langston W. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz M.T., Li X.J., Lavaute T.M., Lyons E.A., Chen X., Zhang S.C. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pata I., Studer M., van Doorninck J.H., Briscoe J., Kuuse S., Engel J.D., Grosveld F., Karis A. The transcription factor GATA3 is a downstream effector of Hoxb1 specification in rhombomere 4. Development. 1999;126:5523–5531. doi: 10.1242/dev.126.23.5523. [DOI] [PubMed] [Google Scholar]
- Plaza S., Dozier C., Turque N., Saule S. Quail Pax-6 (Pax-QNR) mRNAs are expressed from two promoters used differentially during retina development and neuronal differentiation. Mol. Cell. Biol. 1995;15:3344–3353. doi: 10.1128/mcb.15.6.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinchisi G., Ijichi K., Glidden N., Jakovcevski I., Zecevic N. COUP-TFII expressing interneurons in human fetal forebrain. Cereb. Cortex. 2012;22:2820–2830. doi: 10.1093/cercor/bhr359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring K.L., Tong L.M., Balestra M.E., Javier R., Andrews-Zwilling Y., Li G., Walker D., Zhang W.R., Kreitzer A.C., Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rosa A., Brivanlou A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y., Reed R.R. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J. Neurosci. 1997;17:4159–4169. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang L., Huang W., Su H., Xue Y., Su Z., Liao B., Wang H., Bao X., Qin D. Generation of integration-free neural progenitor cells from cells in human urine. Nat. Methods. 2013;10:84–89. doi: 10.1038/nmeth.2283. [DOI] [PubMed] [Google Scholar]
- Yu K.R., Shin J.H., Kim J.J., Koog M.G., Lee J.Y., Choi S.W., Kim H.S., Seo Y., Lee S., Shin T.H. Rapid and efficient direct conversion of human adult somatic cells into neural stem cells by HMGA2/let-7b. Cell Rep. 2015;10:441–452. doi: 10.1016/j.celrep.2014.12.038. [DOI] [PubMed] [Google Scholar]
- Zhang X., Huang C.T., Chen J., Pankratz M.T., Xi J., Li J., Yang Y., Lavaute T.M., Li X.J., Ayala M. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.