Abstract
The spontaneous and deleterious conversion of l-asparaginyl and l-aspartyl protein residues to l-iso-Asp or d-Asp occurs as proteins age and is accelerated under stressful conditions. Arabidopsis (Arabidopsis L. Heynh.) contains two genes (At3g48330 and At5g50240) encoding protein-l-isoaspartate methyltransferase (EC 2.1.1.77; PIMT), an enzyme capable of correcting this damage. The gene located on chromosome 5 (PIMT2) produces two proteins differing by three amino acids through alternative 3′ splice site selection in the first intron. Recombinant protein from both splicing variants has PIMT activity. Subcellular localization using cell fractionation followed by immunoblot detection, as well as confocal visualization of PIMT:GFP fusions, demonstrated that PIMT1 is cytosolic while a canonical nuclear localization signal, present in PIMT2ψ and the shorter PIMT2ω, is functional. Multiplex reverse transcription-PCR was used to establish PIMT1 and PIMT2 transcript presence and abundance, relative to β-TUBULIN, in various tissues and under a variety of stresses imposed on seeds and seedlings. PIMT1 transcript is constitutively present but can increase, along with PIMT2, in developing seeds presumably in response to increasing endogenous abscisic acid (ABA). Transcript from PIMT2 also increases in establishing seedlings due to exogenous ABA and applied stress presumably through an ABA-dependent pathway. Furthermore, cleaved amplified polymorphic sequences from PIMT2 amplicons determined that ABA preferentially enhances the production of PIMT2ω transcript in leaves and possibly in tissues other than germinating seeds.
The proteome is subject to deleterious alteration over time through aging or due to stressful conditions. Aging is a conflict between the unrelenting production of undesirable side products of metabolism or aberrant molecules through spontaneous modification versus an organism's ability to eliminate, repair, or tolerate the change (Clarke, 2003). Protein damage can be of two general types: (1) conformational damage to three-dimensional structure or (2) covalent damage to primary structure (Galletti et al., 1995; Visick and Clarke, 1995). These detrimental conversions can lead to the recognition, tagging, and destruction of the altered peptides, requiring that they be resynthesized de novo. It is, therefore, orders of magnitude more efficient to rectify damaging protein modifications if possible (Reissner and Aswad, 2003). Hence, organisms encode and deploy a plethora of damage repair systems that are capable of recognizing and repairing specific changes to polypeptides at a fraction of the “cost” associated with protein degradation and synthesis (Schumacher et al., 1996; Zou et al., 1998; Pliyev and Gurvits, 1999; Hoshi and Heinemann, 2001). Protein methyltransferase is one such system, capable of repairing abnormal isoaspartyl (isoAsp) residues, a predominate form of protein damage accrued under normal physiological conditions (Lowenson and Clarke, 1992).
Protein-l-isoaspartate methyltransferase (EC 2.1.1.77; PIMT) is a repair enzyme that catalyzes the S-adenosylmethionine (AdoMet)-dependent methyl esterification of the α-carboxyl group of l-isoAsp residues (as well as, in some instances, the β-carboxyl group of d-aspartyl residues). l-IsoAsp is an unencoded amino acid arising from the spontaneous degradation of Asn and Asp in proteins through deamidation and isomerization in the former, and dehydration and isomerization in the latter, instance (Geiger and Clarke, 1987; Bhatt et al., 1990; Aswad et al., 2000). Formation of isoAsp in the protein introduces an extra carbon into the peptide backbone, which now proceeds through the R-group carboxy terminus and produces a kink in the secondary structure of the protein. These alterations are frequently detrimental to enzymatic function (Szymanska et al., 1998; Mamula et al., 1999; Esposito et al., 2000; Tarcsa et al., 2000). Repair begins with an enzyme-mediated methylation reaction followed by nonenzymatic steps that, through reiteration of the methylation/demethylation process, eventually result in the net conversion of l-isoAsp residues to l-Asp (Aswad et al., 2000) and, usually, a reconstitution of normal protein function (Brennan et al., 1994; Clarke, 1999).
Found in nearly all eukaryotic cells as well as most archaebacteria and gram-negative eubacteria (Johnson et al., 1991; Kagan et al., 1997a), PIMT activity is vitally important in those organisms employing it (Kagan et al., 1997b; Kim et al., 1997; Visick et al., 1998; Yamamoto et al., 1998; David et al., 1999). Nonetheless, although several isoforms of PIMT have been identified in a variety of organisms, only a single gene encoding PIMT has been reported per eukaryotic organism studied to date (Kagan et al., 1997a). PIMT isoforms in human erythrocytes, bovine brain, mouse testis, and wheat germ are proposed to be alternative splicing variants (Potter et al., 1992; Romanik et al., 1992; Mudgett and Clarke, 1993). In plants, protein-l-iso-Asp methyltransferase, but not protein-d-Asp methyltransferase, activity has been identified in monocots, dicots, and green algae (Mudgett and Clarke, 1993). Organisms outside of Plantae encode enzymes with activity against both substrates (Lowenson and Clarke, 1992; Thapar et al., 2002). Although some PIMT enzymatic activity is detectable in a few nonseed tissues (Thapar et al., 2001), the greatest PIMT specific activity is found in seeds where nonenzymatic protein damage is hypothesized to accrue during dehydration and quiescence (Mudgett and Clarke, 1994). Such damage should be repaired for normal, vigorous germination and subsequent radicle protrusion (Mudgett and Clarke, 1993; Kester et al., 1997). In Arabidopsis (Arabidopsis L. Heynh.), a PIMT gene located on chromosome 3 (PIMT1) has been characterized previously (Mudgett and Clarke, 1996; Thapar and Clarke, 2000). In this work, we identify a second gene for PIMT on Arabidopsis chromosome 5 (PIMT2), the first instance of a eukaryote encoding two PIMTs. Similar to PIMTs present in other organisms, this gene produces two transcripts through alternative splicing. We have analyzed the enzymatic properties of the PIMT proteins and characterized relative transcript amounts and protein subcellular localization.
RESULTS
Identification and Isolation of Full-Length cDNAs from a Second PIMT Gene in Arabidopsis
We identified a gene on chromosome 5 (At5g50240) that is highly similar to the previously identified PIMT gene on chromosome 3 encoding a protein repair methyltransferase. Two full-length mRNA sequences of the chromosome 5 PIMT2 gene, differing by only a 9-nucleotide (nt) alternatively spliced segment, were assembled from reverse transcription (RT)-PCR amplicons and 5′- and 3′-RACE products (GenBank accession no. AY496702; Fig. 1). These transcripts possess a 23-bp 5′ untranslated region (UTR), a 930- (PIMT2ψ) or 921- (PIMT2ω) bp coding region, and a 111-bp 3′ UTR (Fig. 1). These results reveal that the splicing pattern predicted for At5g50240 (GenBank accession no. 124403) is incorrect. Specifically, the 3′ splice site in the first intron was predicted incorrectly (nts 412–413, rather than nts 440–441 or 449–450; Fig. 1), a large portion of exon 2 was excluded (nts 501–712; Fig. 1), and a fragment of intron 2 was included (nts 775–787; Fig. 1). The two transcripts recovered in a 5′-RACE specific for capped mRNA terminated at the same 5′ site, supporting transcriptional initiation at this site. The splicing scheme presented (Fig. 1) is consistent with previously identified expressed sequence tags, which lack some residues at the 5′ and 3′ ends (GenBank accession no. AK118104). The 9-bp (5′-CAATTCCAG-3′) difference in the coding region is a result of alternative 3′splice site selection in the first intron (Fig. 1).
Figure 1.
Nucleotide sequence (GenBank accession no. AY496702) of the PIMT2 gene and its deduced amino acid sequence. Nucleotide and amino acid numbers are indicated on the left and right, respectively. The putative ABRE motif in the promoter is identified as such and is in bold type. The transcription initiation site (TIS) is demarcated with an arrow. Italicized, lowercase sequence denotes the 5′- and 3′-UTRs, with a polyA signal in the latter underlined. Introns are designated by lowercase letters. The 9-nt alternative 3′ splice site is boxed and the Hpy188 III recognition site used in CAPS is in bold text. The predicted NLS in the amino acid sequence is boxed. Amino acids in common among Arabidopsis PIMT1, PIMT2, and PIMT from wheat (GenBank accession no. Q43209) are in black boxes. The three regions conserved among different families of methyltransferase utilizing S-AdoMet as a cosubstrate are labeled and highlighted in black. The two regions that appear to be unique to protein-l-iso-Asp (d-Asp) methyltransferase are also highlighted in black and labeled pre-region I and post-region III.
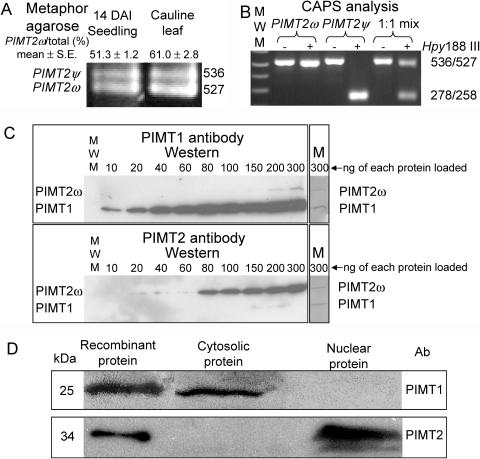
Alternative splicing was verified by the production of two amplicons (Fig. 2A) from RT-PCR reactions using primers CH5F-0 and CH5R-0 (Supplemental Table I, available at www.plantphysiol.org). Additionally, the longer splicing variant encodes an extra endonuclease restriction site (Hpy188 III, TCNNGA) that enabled distinction between PIMT2ψ and PIMT2ω following RT-PCR using cleaved amplified polymorphic sequences (CAPS; Konieczny and Ausubel, 1993; Fig. 2B). A putative polyadenylation signal (AATAAA) was detected in the 3′ UTR of PIMT2 transcripts. A plant cis-acting regulatory DNA elements (PLACE) signal scan search (Higo et al., 1999) identified a putative ABA response element (ABRE; 25–36 bp) in the PIMT2 promoter ending −2 nts from the transcriptional start site (Fig. 1). Comparison of PIMT2 with PIMT1 nucleotide sequences revealed that both contained three introns. The introns are situated between the same nucleotides in both genes (contingent on adopting PIMT2ψ splicing junctions in intron 1).
Figure 2.
A, Amplicons from RT-PCR reactions from DNA-free, seedling, and leaf RNA produced using primers CH5F-0 and CH5R-0 (Supplemental Table I) were separated in a 2% MetaPhor gel, stained using SYBR-Gold, and photographed. Two bands were discernable of a molecular mass consistent with an alternative splicing event that retained (PIMT2ψ) or eliminated (PIMT2ω) the 9 3′-most nt of the first intron. The average ± se of multiple estimates of PIMT2ω abundance relative to total PIMT2 transcript based on CAPS, metaphor agarose, or polyacrylamide separation are provided. B, Inclusion of an extra 9 nt in the larger of the two transcripts through alternative 3′ splice site selection results in a CAPS using Hpy188 III. The 536-bp PIMT2ψ amplicon is cleaved into two smaller, comigrating products of 278 and 258 bp that are discernable from the larger 527-bp PIMT2ω amplicon on a 1% w/v agarose gel. −, No restriction endonuclease was added. +, Hpy188 III was added to the amplicon. PIMT2ω, Amplicon from plasmid containing PIMT2ω insert; PIMT2ψ, amplicon from plasmid containing PIMT2ψ insert; 1:1, amplicon generated from an equal mixture of plasmid containing PIMT2ψ or PIMT2ω. C, Polyclonal antibodies to PIMT1 and PIMT2 were produced and affinity purified. Recombinant PIMT1 and PIMT2 proteins produced from pET23d with and without a carboxy-terminal hexahistidyl tag (HIS-tag), respectively, were mixed in equal proportions and loaded in increasing amounts (from 10–300 ng isozyme−1 well−1) in two 12% SDS-polyacrylamide gels. Following electrophoresis and transfer to membrane, representative blots were challenged with either PIMT1 or PIMT2 antibody. After chemiluminescent detection, the membranes (M) were stained with Coomassie Blue to exhibit recombinant protein amounts. D, Western blots of the cytosolic and nuclear fractions from 24-h imbibed seeds challenged with affinity-purified, polyclonal antibodies to PIMT1 or PIMT2 protein. Recombinant protein, produced from pET23d with (PIMT1) or without (PIMT2) a carboxy-terminal HIS-tag, were run as controls.
When the coding regions of PIMT2 and PIMT1 are compared, it is clear that the 5′-most 237 bp of PIMT2 are unique (Fig. 1). Excluding this region, nucleotide identity over the 693-bp coding region shared with PIMT1 is 68%. The unique 5′ 237-bp portion of PIMT2 encodes a 79-amino acid N-terminal sequence in which both iPSORT (Bannai et al., 2002) and PredictProtein algorithms (Rost, 1996) identified a nuclear localization signal (NLS). Excluding the 79-amino-terminal extension, PIMT2 is 68% identical, 93% similar to PIMT1, and 65% identical, 90% similar to wheat isoaspartyl methyltransferase (GenBank accession no. Q43209). All three of these methyltransferase sequences are most identical in the PIMT regions I, II, and III (Fig. 1) proposed to be involved in binding S-AdoMet (Kagan and Clarke, 1994). Two additional regions, pre-I and post-III, proposed to be unique to PIMTs and possibly important in determining substrate specificity (Kagan et al., 1997a), are also present in PIMT2.
Subcellular Localization of PIMT1, PIMT2ψ, and PIMT2ω
Protein extracts of flowers, seeds, or vegetative organs that had or had not undergone abiotic stress were assayed for PIMT activity and the presence of PIMT protein using affinity-purified antibodies (Fig. 2C). Enzyme activity was detectable from protein extracts from mature seeds, the sole plant tissue tested where western blots also detected immunoreactive bands (data not shown; Fig. 2D). Cell fractionation of seed tissue followed by SDS-PAGE and western blot localized PIMT1 to the cytoplasm and PIMT2 to the nuclear fraction (Fig. 2D). The largest immunoreactive band detected using PIMT2-specific antibody is comparable in size to that of untagged, recombinant PIMT2 protein produced from the first in-frame translational start site in the PIMT2 cDNA (Figs. 1 and 2C), adding credence to the transcriptional start site identified using 5′-RACE, though the smaller band discernable in the PIMT2 western was too small to be PIMT2ω.
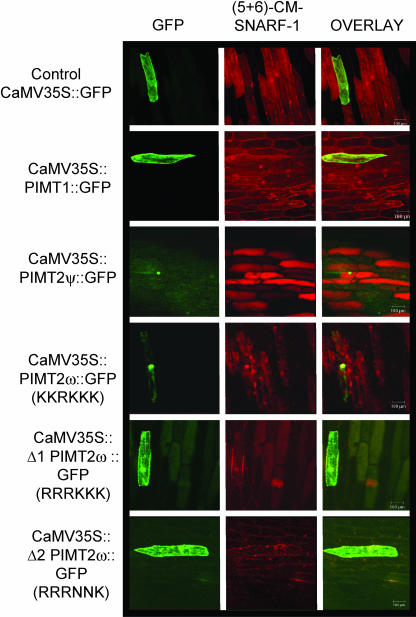
To directly test the subcellular localization of the different PIMTs, the coding regions of PIMT1, PIMT2ψ, or PIMT2ω were cloned behind the cauliflower mosaic virus (CaMV) 35S promoter, contiguous at the 3′ end with the coding region for the green fluorescent protein (GFP). These constructs were then biolistically introduced into onion (Allium cepa) epidermal cells, a model plant cell amenable for live imaging of the GFP (Scott et al., 1999). The GFP, when expressed without an amino-terminal fusion, was distributed throughout the cytoplasm but was also able to traverse the nuclear pore complex (Talcott and Moore, 1999; Fig. 3). Fluorescence was again detectable in both the cytoplasm and the nucleus for carboxy-terminal fusions of GFP with PIMT1, and with altered PIMT2ω proteins with either two or four of the five Lys, comprising the presumptive NLS, substituted (Fig. 3). With the NLS intact, PIMT2ψ:GFP and PIMT2ω:GFP fusions were efficiently sequestered in the nucleus (Fig. 3).
Figure 3.
Introduction of constructs with the CaMV 35S driving expression of chimeric genes ultimately producing PIMT1 or PIMT2 isoforms fused at their carboxy terminus with the GFP indicated a nuclear localization for PIMT2 isoforms in onion epidermal cells. GFP fluorescence was found throughout the cytoplasm and the nucleus when GFP was expressed in native form (CaMV 35S:GFP) or in a carboxy-terminal fusion with PIMT1 (CaMV 35S:PIMT1:GFP), or versions of PIMT2ω in which two (CaMV 35S:Δ1 PIMT2ω:GFP) or four (CaMV 35S:Δ2 PIMT2ω:GFP) of the five Lys comprising the NLS were substituted. When the NLS was intact, and regardless of the presence (CaMV 35S:PIMT2ψ:GFP) or absence (CaMV 35S: PIMT2ω:GFP) of the differentially spliced fragment from intron 1, GFP was sequestered in the nucleus. (5 + 6)−CM−SNARF, (5 and 6) Chloromethyl-seminaphthorhodafluors.
Purification and Activity Assay of Recombinant PIMT1, PIMT2 Proteins
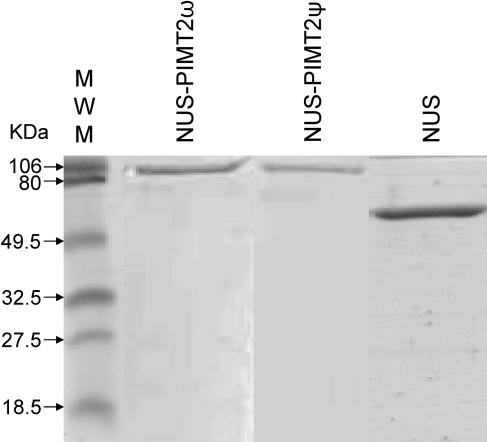
All recombinant proteins were produced in Escherichia coli, originally from pET23d with or without a carboxy-terminal, hexahistidyl-tag, and three linking amino acids (Trp, Asp, Pro; PIMT1; and Trp, Val, Glu; PIMT2). Because the PIMT2 enzymes were insoluble, PIMT2 splicing variants were also expressed from pET43.1 as N utilization substance (NUS)-fusion (491 amino acid) proteins including an amino-terminal 38-amino acid linker (His-tag, S-tag, thrombin site). The exact cloning sites used in each instance are provided in “Materials and Methods.” PIMT1 fortuitously retained the initiating Met when expressed in E. coli (Flinta et al., 1986; Hu et al., 1999; data not shown). Proteins were purified by Ni-affinity chromatography and the affinity-purified enzyme preparations (Fig. 4) assessed for an l-isoaspartyl methyltransferase activity using Val-Tyr-Pro-l-isoAsp-His-Ala as a methyl-accepting peptide. PIMT1 and both NUS-PIMT2 proteins (ψ and ω), with or without thrombin-mediated removal of the NUS fusion, had an l-isoaspartyl methyltransferase activity, confirming the initial identification based on nucleotide and deduced amino acid homology. The NUS-PIMT2ψ and NUS-PIMT2ω fusion proteins (Fig. 4) exhibited specific activities of 83.6 ± 3.1 and 87.1 ± 3.7 pmol CH3OH min−1 mg protein−1, respectively, while the NUS protein alone (Fig. 4) had no activity. Additionally, PIMT2 isoforms purified as inclusion bodies exhibited an isoaspartyl methyltransferase activity when solubilized in 6 m urea and used in the assay. This relatively rare ability among enzymes to retain activity even in the presence of urea has been documented previously as being a characteristic of PIMTs (Thapar and Clarke, 2000).
Figure 4.
The purity of the recombinant, affinity-purified isoforms of protein isoaspartyl methyltransferase2, synthesized as a NUS fusion, was analyzed using SDS-PAGE.
Relative PIMT1, PIMT2ω, and PIMT2ψ RNA Abundance in Different Plant Tissues
Using multiplex RT-PCR it was possible to detect and distinguish PIMT1 from PIMT2, relate both PIMT transcript amounts to that of β-TUBULIN, and, using CAPS, discriminate between PIMT2ω and PIMT2ψ and determine their relative abundance. Results obtained using multiplex RT-PCR were verified by performing all amplifications two or more times. When the relative abundance of PIMT2ψ and PIMT2ω varied due to treatment or tissue, results were assessed two or more times using CAPS, or by separating amplicons using Metaphor agarose, or PAGE. In these instances, PIMT2ω transcript abundance is reported as an average of two or more assays with the se of the mean. RNA was extracted twice from stressed or unstressed seedlings grown on two separate occasions and used to assess transcript abundance and splicing variation independently. Excluding results for the stem (see below), in no instance was the se greater than 15% of the mean PIMT2ω percent, relative to the total PIMT2 transcript amount.
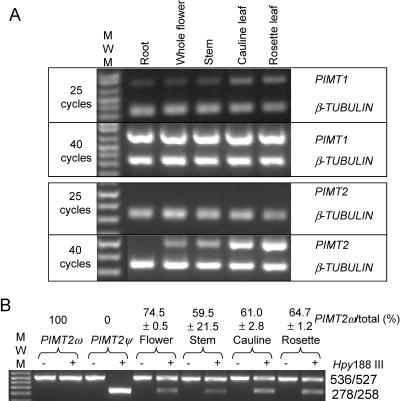
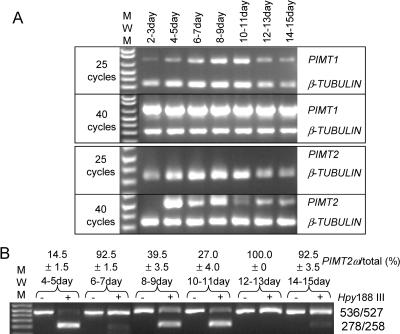
Both PIMT1 and PIMT2 transcripts were less abundant than β-TUBULIN transcripts (compare with 25 cycles, Fig. 5A). PIMT1 appeared constitutively expressed in all tissues tested (Fig. 5A). PIMT2 transcript, if detectable, was in less abundance than PIMT1, as evident by the appearance of PIMT1 but not PIMT2 product after 25 cycles. PIMT2 was usually undetectable in roots although faint product was apparent after 40 cycles in one experiment (data not shown). PIMT2 was present in stems and flowers, and was in somewhat greater abundance in leaves (Fig. 5A). The production of PIMT2ω was consistently favored over that of PIMT2ψ in these tissues except for the stem (Fig. 5B). While the amount of total PIMT2 transcript detected in the stem by multiplex RT-PCR was consistent, analysis of splicing variant amounts in stem tissue was highly variable. The most extreme percentages of PIMT2ω relative to total PIMT2 transcript measured are provided (Fig. 5B).
Figure 5.
Qualitative multiplex RT-PCR analysis of RNA transcripts extracted from various tissues and CAPS of PIMT2 splicing variants. A, The RT-PCR products at 25 and 40 reaction cycles for target cDNAs (PIMT1 and PIMT2) and the β-TUBULIN cDNA were separated on 1.5% (w/v) agarose gels. B, Amplicons of PIMT2 were not (−) or were (+) digested with Hpy188 III and PIMT2ψ and PIMT2ω splicing variant amounts visualized by ethidium bromide staining after electrophoresis through a 1.5% (w/v) agarose gel. The specificity of the CAPS analysis was demonstrated in the first two lanes using respective plasmid DNA templates under identical PCR and digestion conditions. The average ± se of multiple estimates of PIMT2ω abundance relative to total PIMT2 transcripts is indicated on the top of each lane.
PIMT1 was present in siliques and seeds, increased in abundance at least from 4 to 8 d after pollination (DAP) and remained high up to at least 11 DAP (Fig. 6A). Amplicon intensity following ethidium bromide staining quantified PIMT1 in siliques and seeds at 15 DAP as 52% and, in mature, dehydrated seeds, as 48% of β-TUBULIN amplicon intensity (data not shown). Amounts of PIMT1 transcript in mature, dehydrated seeds was similar to that in whole siliques (including seeds) at 15 DAP (Figs. 6A and 7A). PIMT2 transcript was undetectable in flowers 2 to 3 DAP. Its abundance increased and then declined by 10 DAP (Fig. 6A). The increase in abundance of PIMT2 4 to 5 DAP was almost entirely due to the production of PIMT2ψ (approximately 90%) but the situation was reversed by 6 to 7 DAP, and from 12 onward to 15 DAP with PIMT2ω comprising the bulk of the transcripts (Fig. 6B). Between 8 and 11 DAP PIMTψ was in greater abundance (Fig. 6B).
Figure 6.
Qualitative multiplex RT-PCR analysis of RNA transcripts extracted from developing seeds and the siliques enclosing them at various developmental time points (DAP). A, The RT-PCR products were obtained as described in Figure 4A. B, Amplicons of PIMT2 were analyzed as in Figure 4B.
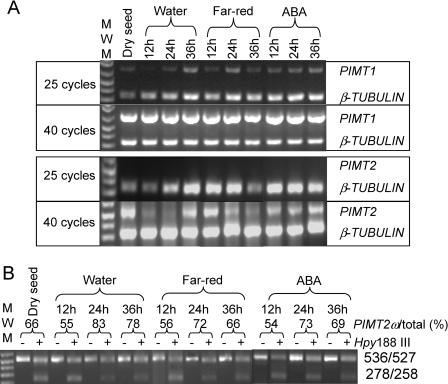
Figure 7.
Qualitative multiplex RT-PCR analysis of RNA transcripts extracted from mature seeds during a germination time course on water, under far-red light, and on 100 μm ABA. A, The RT-PCR products were obtained as described in Figure 4A. B, Amplicons of PIMT2 were analyzed as in Figure 4B.
PIMT activity and immunoreactive protein were detected only in extracts from mature seeds prompting an analysis of PIMT transcript abundance during seed germination on water, on 100 μm abscisic acid (ABA) and under continuous far-red illumination 0, 12, 24, and 36 h after imbibition (HAI). The general trend, normalizing for β-TUBULIN among the different lanes at 25 cycles, was for transcripts from PIMT1 to remain more or less in constant abundance (Fig. 7A). On water under fluorescent lights, PIMT2 mRNA declined following imbibition until 36 HAI, at which time PIMT2 transcript abundance increased (Fig. 7A). The decline of PIMT2 transcript during the first 24 HAI observed for seeds on water under fluorescent light was delayed by exposure to constant far-red light and mitigated by ABA (Fig. 7A). Additionally, the accumulation 36 HAI observed when seeds were imbibed on water or ABA under fluorescent lights was absent when illuminated by far-red light (Fig. 7A). The PIMT2ω-splicing variant tended to be in greater abundance 24 and 36 HAI regardless of treatment (Fig. 7B). However, within an imbibition time, there were no striking differences in PIMTω abundance among treatments (Fig. 7B). In fact, when the splicing variant estimates among treatments were used as replications to obtain an average PIMT2ω percentage for each HAI examined the greatest se of the estimate encountered was only 5% of the average (12 HAI, 55.0 ± 0.6%; 24 HAI, 76.0 ± 3.5%; and 36 HAI, 71.0 ± 3.6%).
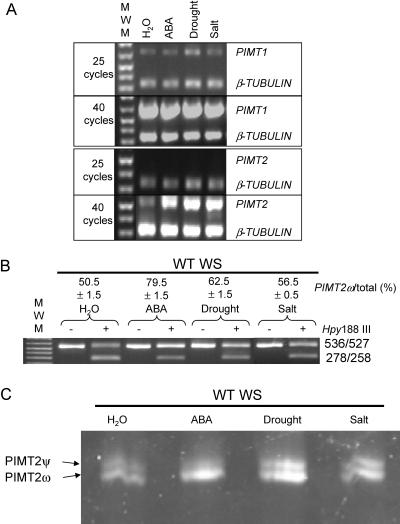
Expression of PIMT1, PIMT2ψ, and PIMT2ω in Establishing Seedlings during Stress
Promoter analysis of both PIMT1 (Mudgett and Clarke, 1996) and PIMT2 (Fig. 1) identified an ABRE. To determine if the Arabidopsis PIMT2 gene was responsive to ABA, as well as to salt- and drought-stress, seedlings were exposed, 12 d after imbibition, to different stress treatments and transcript abundance analyzed. PIMT1 expression was more or less similar regardless of stress. In contrast, PIMT2 transcript abundance increased due to ABA, drought-, or salt-stress (Fig. 8A). Methyltransferase activity assays and western blot of control and stressed seedlings detected neither PIMT activity nor immunoreactive protein (data not shown). The increase in PIMT2 transcript abundance by ABA is accompanied by the predominant production of PIMT2ω relative to PIMT2ψ (Fig. 8, B and C).
Figure 8.
Qualitative multiplex RT-PCR analysis of RNA transcripts extracted from stressed or unstressed, 14-d-old seedlings. A, RNA was extracted from seedlings on water, after incubation on ABA, or following exposure to drought or salt stress. The RT-PCR products were obtained as described in Figure 4A. B, Amplicons of PIMT2 were analyzed as in Figure 4B. C, A separate experiment on a different set of seedlings was conducted and amplicons were separated on 3.5% acrylamide nondenaturing gels at 25mA constant current. Gels were stained for 20 min in SYBER-Gold (Molecular Probes), visualized, and quantified (data not shown) for comparison with values obtained from the first set of stressed seedlings using CAPS.
DISCUSSION
Arabidopsis is the first eukaryotic organism in which two PROTEIN L-ISOASPARTYL METHYLTRANSFERASE genes have been identified. If there are two PIMT genes present in other plants, it may be possible to explain, to some extent, reported differences in gene expression patterns between Plantae and other kingdoms or among plant species (Mudgett and Clarke, 1993, 1994; Mudgett et al., 1997; Thapar et al., 2001). For example, PIMT1 transcription is akin to that of constitutive bacterial and animal PIMT gene expression, while transcript dynamics of PIMT in wheat during seed development, seed germination on water, and stress or ABA treatment applied to seedlings (Mudgett and Clarke, 1994) are more similar to that of PIMT2 transcripts in Arabidopsis at the same developmental period or experiencing the same stress.
Regardless of the presence of detectable transcript amounts, western blots using antibodies specific to PIMT1 or PIMT2 and a sensitive chemiluminescent detection system did not detect immunoreactive protein in any tissue other than that of seeds. As was alluded to in Mudgett and Clarke (1996), the previously reported lack of substantial PIMT activity in Arabidopsis tissues other than seeds is probably due to a posttranscriptional control of protein production from PIMT mRNA (Mudgett et al., 1997).
The amino acid composition of the N-terminal region of PIMT2 isoforms influenced their subcellular residence (predominately nuclear) relative to PIMT1 (predominately cytosolic). PIMT activity and protein have been localized to the nucleus in other organisms (O'Connor, 1987; O'Connor and Germain, 1987). Histone H2B is a major in vivo target for methytransferase repair in the nucleus of rat PC12 cells (Young et al., 2001). It is thought that PIMTs (smaller in size than the postulated 50-kD exclusion limit for passive entry into the nucleus; Talcott and Moore, 1999) are sufficiently compact to arrive in the nucleus by crossing the nuclear pore complex without assistance from an NLS (Clarke, 2003). Why Arabidopsis should produce proteins that depart from this paradigm is not apparent. There are neither published instances of differences in the exclusion limit for, or selectivity of, nuclear pore complexes in Plantae versus other kingdoms, nor any obvious requirement for the retention of a PIMT activity in the plant nucleus relative to that of other kingdoms. Searches using the unique, N-terminal portion of the nuclear-targeted PIMT2 proteins for domains of relevance to a nuclear localization (DNA binding, etc.) were unfruitful. Nevertheless, PIMT2ψ and PIMT2ω proteins are efficiently sequestered in the nucleus of onion epidermal cells and Arabidopsis seeds (Figs. 2D and 3). Perhaps the NLS is not to assist PIMT2 into the nucleus but to retain it once there, although no reasons for such a scenario are apparent.
Other than seeds, routinely detectable PIMT activity in plants is limited to vegetative parts of a few species (Thapar et al., 2001). The use of a sensitive, PCR-based detection strategy has determined that, contrary to a previous report using northern blot which would detect all PIMT transcripts regardless of the gene producing them or posttranscriptional modifications (Mudgett and Clarke, 1996), PIMT1 transcript was present in more or less similar amounts in all tissues. This transcript ubiquity makes PIMT1 expression more similar to the constitutive PIMT expression pattern present in bacteria and animals (Clarke, 2003). PIMT2 transcripts were consistently in less abundance than those of PIMT1, occasionally below the level of even PCR-based detection (roots and flowers 2–3 DAP), but PIMT2 was more influenced by developmental stage, ABA, and abiotic stress than PIMT1. In particular, PIMT2 accumulated following the application of ABA to seedlings. The increase in PIMT2 abundance in response to ABA might also explain the accumulation of PIMT2 8 to 9 DAP in silique/seed development, approximately the period of greatest concentration of maternally synthesized ABA in the seed (Karssen et al., 1983). That PIMT1 expression was not noticeably up-regulated by ABA when applied to seedlings in this study (Fig. 8A) may be a reflection on the nondiscriminatory nature of the probe used in the northern blots and/or the different developmental stages the seedlings were at upon treatment in the two studies. Additionally, Mudgett and Clarke (1996) used the ecotype Columbia while Wassilewskija (Ws) was the ecotype used throughout this study. In agreement with the reported up-regulation of PIMT1 by ABA (Mudgett and Clarke, 1996), transcript abundance increased in developing seeds/siliques from 4 to 11 DAP (Fig. 6A), presumably also in response to increasing maternal ABA concentrations in the seeds which reaches a peak at approximately 10 DAP (Karssen et al., 1983). The increase in PIMT2 transcript abundance in drought- or salt-stressed seedlings may also be through an ABA-dependent stress response pathway. That PIMT transcript abundance should increase in response to ABA lends some credence to the postulated role of PIMT during plant stress: the repair of spontaneously formed isoaspartyl residues known to accumulate under stressful conditions (Clarke, 2003). Both genes possess distinguishable ABREs in their promoters (Mudgett and Clarke, 1996; this report).
The dynamics of PIMT2ψ/PIMT2ω relative abundance during silique/seed development and upon ABA application to seedlings is intriguing. In seedlings placed on ABA not only did PIMT2 abundance increase but also transcript splicing consistently altered to favor accumulation of the shorter PIMT2ω form. This was not the case when seedlings were stressed with drought or salt, which, although they enhanced the overall accumulation of PIMT2 transcripts, did not favor PIMT2ω production as dramatically as ABA (Fig. 8). Examining the silique/seed development time course, PIMT2ω was favored 6 to 7, and 12 to 15 DAP, the latter period corresponding to the stage of development when embryo-derived ABA is maximal in developing Arabidopsis seeds (Karssen et al., 1983).
The relevance of the conditional inclusion of the tri-peptide Gln-Phe-Gln (QFQ) through alternative 3′-splice site selection of the nucleotides encoding it is not currently understood. It did not affect the nuclear localization of the PIMT2 proteins. PIMT1 and the deduced amino acid sequence for the cloned wheat PIMT both have Gln-Phe-Trp (QFW) in this site and PIMT1 has no sequence amenable to alternatively splicing the 9 nt encoding the QFW tri-peptide (Mudgett and Clarke 1993; 1996). The low amount of PIMT activity in Arabidopsis tissues other than seeds, despite detectable amounts of PIMT1 and, usually, PIMT2 transcript, prompted speculation that perhaps QFW/Q was destabilizing the PIMT proteins in tissues other than seeds. To bolster this surmise, far fewer onion cells were observed to exhibit considerable GFP signal when the reporter was linked in frame with PIMT2ψ than when linked with PIMT2ω. However, despite the necessary inclusion of the codons encoding QFW and access to the nucleus, onion cells expressing PIMT1:GFP fusions (Fig. 3) were abundant following bombardment. Neither was recombinant PIMT2ψ significantly more prone to degradation than PIMT2ω in lysates during a time course at 25°C (data not shown).
The only other documented instance of alternative splice site selection in a PIMT is in the substitution of nucleotides encoding the usual carboxy terminus of the Type-I PIMT isoform in bovine brain for those encoding an endoplasmic reticulum retention signal on the Type-II PIMT isoform (Maclaren et al., 1992; Potter et al., 1992). A SET-domain-containing methyltransferase (Trievel et al., 2002), with the N-terminal Met of the processed form of the small subunit of Rubisco as its target, is also alternatively spliced to provide protein isoforms differing toward the carboxy terminus by WVQQ (Ying et al., 1999), the relevance of which also is not understood.
It is acknowledged that there is an increase in functional complexity during the transition from the genome to the proteome and this has stimulated interest in differential splicing of mRNA as a powerful means of generating diversity (Stamm, 2002). Particularly relevant is the observation that plant and animal genes whose products are associated with stress responses have a greater propensity to be differentially spliced (for review, see Kazan, 2003). A realization of ABA effects in altering posttranscriptional modification of transcripts, including alternative splicing events, in the plant cell is at its genesis (Himmelbach et al., 2003; Kuhn and Schroeder, 2003). PIMT2 appears to be subject to precisely this form of control although the physiological relevance of such control remains to be discovered.
MATERIALS AND METHODS
Using the amino acid sequence of PIMT1 (At3g48330) as query in a TBLASTN search, a second gene was identified in the Arabidopsis (Arabidopsis L. Heynh.) genome (PIMT2; At5g50240) putatively encoding a protein l-isoapartyl methyltransferase. RT-PCR of mature leaf total RNA using gene-specific primers to the predicted coding region was used to amplify a PIMT2 cDNA. The full-length cDNA of PIMT2 was delineated using 3′-RACE and a 5′-RACE strategy designed to amplify capped mRNA specifically (GeneRacer; Invitrogen, Carlsbad, CA). Amplicons from the latter experiment determined that two transcripts were produced from the PIMT2 gene through alternative 3′-splice site selection. These were designated as PIMT2ψ (longer) and PIMT2ω (shorter).
Recombinant Protein Production, Purification, and Enzyme Assay
All coding regions of PIMT1 and PIMT2, with or without the stop codon, were cloned into the NcoI, XhoI sites of pET23d (Novagen, La Jolla, CA). For PIMT1 a 5′-NcoI and 3′-XhoI site was introduced into the coding region by PCR. The second codon of PIMT1 commenced with a guanine, the same nucleotide terminating the NcoI site, which maintained the amino-terminal PIMT1 amino acid sequence. Both PIMT2 coding regions were altered by PCR to introduce a 5′-BspHI site and 3′-SalI site. These enzymes cleave to leave NcoI or XhoI ligation compatible overhangs, respectively. The 5′ modification retained the adenine in position one of the second PIMT2 codon (Fig. 1) and the 3′ modification avoided an internal XhoI site in the PIMT2 coding region. No PIMT2ψ or PIMT2ω protein produced from pET23d was soluble. This problem was overcome by cloning the coding regions, including stop codons, into the XmaI, XhoI sites of pET43.1 (Novagen) by introducing BspEI, SalI sites into the PIMT2 5′ and 3′ termini, respectively.
For all constructs, recombinant protein production was induced in the Escherichia coli strain BL21(DE3)RIL (Stratagene, La Jolla, CA) growing at 37°C in 500 mL of Luria-Bertani media containing appropriate selection once the OD600 reached 0.4. Upon induction with 0.1 mm isopropylthio-β-galactoside, the culture was shifted to 22°C and grown for an additional 8 h. Bacterial cells were pelleted (4,000g for 5 min at 4°C), resuspended in 1/10 volume of lysis buffer (0.2 m sodium phosphate and 5 m NaCl, pH 7.4), and lysed with two passages through a French press at 1,200 pounds square inch−1. The lysate was centrifuged at 16,000g for 20 min at 4°C. The supernatant was collected, the presence of soluble recombinant protein confirmed on 10-μL aliquots using SDS-PAGE, and recombinant proteins loaded onto and eluted from nickel charged, affinity columns (Hi-Trap; Amersham-Pharmacia Biotech, Piscataway, NJ) using an FPLC (Amersham-Pharmacia) to wash the column and elute the protein with an imidazol gradient in lysis buffer. Sixteen-microliter aliquots from the 1-mL fractions were run on SDS-PAGE to determine those fractions containing recombinant protein and the purest fractions pooled and dialysed overnight against HEPES buffer (0.05 m HEPES-KOH, pH 7.5) to remove the imidazol. These fractions were then used in PIMT assays before and following cleavage of the NUS fusion from the native PIMT using thrombin (Sigma-Aldrich, St. Louis) 2 h at 22°C. Additionally, insoluble inclusion bodies of PIMT2 protein generated from the pET23 plasmid in E. coli were purified by washing according to the manufacturers instructions (Novagen) and resolubilized in freshly prepared 6 m urea. An aliquot of the resolubilized protein was used in a PIMT assay. The PIMT assay was a vapor diffusion assay performed on recombinant proteins as described in Mudgett and Clarke (1993).
Antibody Production
pET23d-expressed, recombinant PIMT1 (purified with a carboxy-terminal HIS-tag) and PIMT2ω (lacking a HIS-tag and purified as an inclusion body according to specifications supplied by the vector manufacturer [Novagen]) were subjected to SDS-PAGE and transferred to Immobilon-PSQ (Millipore, Billerica, MA) membranes. Blotted protein was subjected to automated Edman degradative sequencing (Procise 494; PE-Applied Biosystems, Foster City, CA) using the facilities at the University of Kentucky Macromolecular Structure Analysis Facility. In both cases the first five amino acids corresponded to those predicted for PIMT1 (MKQFW) and PIMT2ω (MNTNT).
The purified proteins were used as antigen for production of polyclonal antibodies (Strategic BioSolutions, Ramona, CA). Antibodies were affinity purified and tested against both recombinant PIMT substrates for specificity and titred using known amounts of purified recombinant protein to determine sensitivity. In all instances, protein concentration of enzyme assays, extracts, and lysates was determined using Pierce Coomassie Blue Plus reagent (Pierce Biotechnology, Rockford, IL).
Plant Material
Arabidopsis (Wassilewskija) seeds were cold treated on moist germination blotter (Grade 628; Stults Scientific Engineering, Springfield, IL) in a petri dish (Fisher Scientific, Springfield, NJ) at 4°C for 3 d prior to being transferred to 22°C in a germinator with continuous fluorescent light (135 μmol m−2 s−1). Seven days after transfer to 22°C, seedlings were transferred to soil and grown to maturity in a growth chamber (Conviron, Winnipeg, Canada) at 22°C with 16 h fluorescent light (200 μmol m−2 s−1).
Three weeks after sowing on soil, wild-type Ws plants were harvested, the soil removed from the roots, and the plants divided into roots, stem, flowers, cauline, and rosette leaves. Tissue was bulked and flash frozen in liquid nitrogen prior to being stored at −80°C until extracted for RNA.
Opening flowers of wild-type Ws plants grown in the growth chamber were tagged every 2nd d and, upon the siliques from the earliest tagged flowers turning brown, all tagged siliques in the following developmental intervals were harvested: 2 to 3 d, 4 to 5 d, 6 to 7 d, 8 to 9 d, 10 to 11 d, 12 to 13 d, 14 to 15 d, and mature dehydrated seeds. The siliques were stored in microcentrifuge tubes on dry ice while harvesting followed by flash freezing in liquid nitrogen, and storage at −80°C until RNA extraction. Dehydrated seeds were stored at −20°C.
Wild-type Ws seeds were surface sterilized (70% ethanol, 2 min; 30% bleach, 0.1% Triton X-100, 15 min), the seeds washed at least five times with sterile water, and resuspended in sterile 0.1% agarose. Seed suspensions were plated on sterile germination paper saturated with Murashige and Skoog medium and situated on solid media (1× Murashige and Skoog salts, pH 5.7, 1% Suc, and 0.8% agar). The seeds were germinated at 4°C for 3 d and then under continuous light (135 μmol m−2 s−1) at 22°C. After 12 d of growth at 22°C, seedling populations were subjected to stress treatments. The filter paper supporting the seedlings was transferred to: (1) a petri dish with two layers of dry, sterilized germination blotter on the bottom to impose drought stress for 48 h; (2) 12 mL of 250 mm NaCl solution (salt stress) for 48 h; (3) 12 trans-ABA in deionized water for 48 h; and (4) as a control, seedlings were transferred to dishes containing 12 mL of deionized water for 48 h. Following stress, the seedlings were harvested immediately and flash frozen in liquid nitrogen before being stored at −80°C until RNA extraction. This experiment was performed on two separate occasions.
Approximately 100 mg wild-type Ws seeds were imbibed on two layers of germination blotter moistened with 17 mL water under continuous white (135 μmol m−2 s−1) or far-red (0.3 ± 0.03 μmol m−2 s−1) light or imbibed on 17 mL of 100 μm cis, trans-ABA in deionized water under continuous white light at 22°C for 0, 12, 24, and 36 h. At each time point, a petri dish containing imbibed seeds under each of the three conditions (white, far-red light, ABA) was removed from treatment and the seeds within harvested immediately and flash frozen in liquid nitrogen before being stored at −80°C until RNA extraction.
Total RNA Isolation from Plant Material
Total RNA was isolated from roots, stems, flowers, cauline, and rosette leaves using a kit (RNeasy Plant kit; Qiagen USA, Valencia, CA) and from unstressed and stressed seedlings using Trizol Reagent (Invitrogen) according to the respective manufacturer's instructions. Total RNA was isolated from developing siliques using a method modified from the RNeasy Plant kit (Qiagen; Gehrig et al., 2000) and from dry or germinating seeds using a hot borate technique (Wan and Wilkins, 1994).
5′- and 3′-Rapid Amplification of cDNA Ends
Total RNA isolated from the aboveground parts of Arabidopsis from plants grown under normal conditions was pretreated with DNase I using a kit (DNA-free, Ambion, Austin, TX) according to the manufacturer. For 3′-RACE, the first strand cDNA was synthesized using an oligo(dT18) anchor primer with a 5′-nonhomologous extension (Supplemental Table I) and SUPERSCRIPT III (Invitrogen) according to the manufacturer's directions. 5′-RACE procedures designed to amplify only capped mRNA were performed on 2.5 μg of DNase I-treated leaf total RNA using gene specific primers and a kit (GeneRacer; Invitrogen) according to the instructions of the manufacturer (Supplemental Table I).
PCR was performed on 3 μL of first strand cDNA, using gene-specific primers in conjunction with primers designed to the nonhomologous extension on the 3′-RACE anchor primer or the reverse transcribed RNA-adaptor ligated to the 5′ end of full-length RNA in 5′-RACE (Invitrogen; Supplemental Table I). Amplicons were isolated from 1% (w/v) agarose gels following electrophoresis, cloned into a T/A vector, and sequenced.
Sequence Analysis
Sequencing was performed at the Advanced Genetics Technologies Center (University of Kentucky, Lexington, KY) using a Beckman Coulter CEQ 8000, 8 capillary electrophoresis Genetic Analysis system to read cycle sequencing reactions employing a combination of universal and gene-specific primers (Integrated DNA Technologies, Coralville, IA). Sequences were viewed and assembled using Sequencer software (version 4.0; Gene Codes Corporation, Ann Arbor, MI). Gene and cDNA sequences were analyzed using PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/; Higo et al., 1999), iPSORT (http://www.hypothesiscreator.net/iPSORT/index.html; Bannai et al., 2002), and PredictProtein (http://www.embl-heidelberg.de/predictprotein/ predictprotein.html; Rost, 1996) algorithms.
Subcellular Localization of PIMT1, PIMT2ψ, and PIMT2ω
Isolation of Nuclei and Extraction of Nuclear Proteins
Arabidopsis nuclei were isolated according to Sarnowski et al. (2002). Nuclear proteins for immunoblot analysis were isolated according to procedures from Martinez-Garcia et al. (1999). Up to 100 μg of cytosolic or nuclear protein were suspended in Laemmli's buffer (Laemmli, 1970), boiled, and electrophoresed on 12% SDS-PAGE gels. Protein was transferred to PVDF membrane (Immobilon P; Millipore) using semi-dry transfer (Milliblot SDE System; Millipore). Blots were incubated (1 h, 25°C) in Tris-buffered saline plus Tween 20 (TBST; 20 mm Tris-HCl, 500 mm NaCl, and 0.05% [v/v] Tween 20, pH 7.6), containing 3% (w/v) bovine serum albumin (fraction V), and washed in 50 mL of TBST (1× 15 min, 2× 5 min). Blots were challenged (1.5 h, 25°C) with a 1,000-fold dilution of affinity-purified primary antibody in 20 mL of TBST, 3% (w/v) bovine serum albumin. The membrane was washed (TBST), incubated (1.5 h, 25°C) in 20 mL of deionizied water, 0.5% (w/v) reconstituted milk powder, and 30,000-fold diluted goat anti-rabbit peroxidase conjugate followed by 3× 5 min washes with TBST. The membrane was incubated with chemiluminescent substrates (Kirkegaard & Perry Laboratories, Gaithersburg, MD) for 1 min and bands visualized using x-ray film (Eastman Kodak, Rochester, NY).
Transient Expression of PIMT:GFP Fusions
The PIMT1, PIMT2ψ, and PIMT2ω coding sequences were cloned down stream from the CaMV 35S promoter in frame with a 3′-situated GFP reporter in the binary vector, pKYLX80 (Schardl et al., 1987; Dinkins et al., 2003). As a negative control the presumptive NLS of the PIMT2ω:GFP fusion was altered through successive rounds of site-directed mutagenesis (QuikChange; Stratagene).
Transient GFP assays were conducted by introducing 100 ng plasmid DNA coated onto 1-μm diameter, gold microcarriers (Bio-Rad Laboratories, Hercules, CA) into onion epidermal cells using a PDS1000 DuPont/Bio-Rad Microprojectile delivery system (Bio-Rad Laboratories). Epidermal peels were cultured for 36 h on media (Scott et al., 1999), and 200 μL of 10 μm 5-(and 6)-chloromethyl SNARF-1 acetate (Molecular Probes, Eugene, OR), used as a counter-stain, spread on the bombarded area 15 min prior to observation. The peels were mounted on slides, and viewed using a Leica TCS laser confocal imaging microscope (Leica Microsystems Heidelberg GmbH, Heidelberg) with a numerical aperture of 1.2 and a working distance of 100 to 300 μm.
Multiplex RT-PCR and PIMT2 CAPS
All RNA was pretreated with DNase I (DNA-free, Ambion). The first strand cDNA for RT-PCR analysis was synthesized with oligo(dT)18 primers using 1 μg of total RNA and SUPERSCRIPT III (Invitrogen) at 50°C for 1 h. The reaction was terminated (75°C, 15 min) and treated with RNase cocktail (Ambion; 37°C, 20 min).
The alternative 3′ splice site selection identified during 5′-RACE was verified by size fractionating amplicons produced using primers CH5F-0 and CH5R-0 in 2% MetaPhor agarose (Cambrix Bio Science, Rockland, ME) or 3.5% nondenaturing poly-acrylamide gels (Ausubel et al., 1994). In both instances, resolved gels were stained with SYBR-Gold, visualized, and the image captured (Alpha Innotech, San Leandro, CA).
Due to the size similarity of PIMT2ψ and PIMT2ω transcripts, and their low abundance, qualitative RT-PCR (semiquantitative RT-PCR) was used to determine the presence of PIMT2 and compare it to transcript abundance from PIMT1. Forward and reverse primers amplifying (1) a 699-bp fragment of PIMT1 or (2) a 536-bp fragment of PIMT2 were tested with or without primers amplifying a 333-bp fragment of β-TUBULIN (GenBank accession no. Q43209; locus tag At5g62690). Following ethidium bromide staining, the amplicons from multiplex PCR reactions at 25, 30, 35, and 40 cycles were analyzed for so called interference, i.e. any decrease in intensity relative to reactions amplifying a single template at the same number of cycles (Joyce, 2002). There was no interference of PIMT1 and β-TUBULIN primers or templates, and both amplicons increased in intensity in successive rounds of PCR beyond those at which aliquots were taken and used to quantify amplicon intensity (25 PCR cycles). However, no primer combination or input cDNA amount avoided interference between β-TUBULIN and PIMT2 in a multiplex PCR. Decreasing β-TUBULIN primer input avoided interference but β-TUBULIN amplicon accumulation plateaued prior to PIMT2 amplicon becoming quantifiable. PIMT2 and β-TUBULIN were, therefore, amplified in separate tubes at primer concentrations sufficient to maintain exponential accumulation of both amplicons beyond 30 cycles. However, even at the greater β-TUBULIN primer concentration, PIMT2 at 30 cycles was not in sufficient abundance to be quantifiable prior to β-TUBULIN amplification plateauing at 35 cycles. Amplicon amounts were, therefore, not quantified and a multiplex PCR analysis was conducted with decreased β-TUBULIN primer concentrations for both PIMT1 and PIMT2. β-TUBULIN amounts in the multiplex were visualized at 25 cycles, while amplicon accumulation was still in the logarithmic phase and PIMT2 was visualized at 40 cycles along with β-TUBULIN, which had plateaued before this point.
Following PIMT2 amplification, bands were removed from the gel using a kit (QIAquick; Qiagen), reamplified, and cleaved at 37°C for at least 8 h with Hpy188 III (New England Biolabs, Beverly, MA). The CAPS (Konieczny and Ausubel, 1993) resulted in two visible bands when electrophoresed in a 1.5% (w/v) agarose gel, a larger amplicon (527 bp; uncleaved PIMT2ω) and two colocalized fragments (278 and 258 bp; cleaved PIMT2ψ. Controls of PIMT2ψ and PIMT2ω amplicons uncut and cut with Hpy188 III (depicted in Figs. 2B and 5B but not shown in subsequent figures) were run with each experiment.
Densitometric Analysis and Relative Quantitation of PCR Products
TIF files of photographs of amplicons or CAPS were quantified using the 1D electrophoresis gel analysis module of ImageQuant TL (Amersham Pharmacia). The ratio of the band intensity of total, uncleaved PIMT2 amplicon (PIMT2ψ + PIMT2ω) was compared to that of comigrating bands (278/258; PIMT2ψ) from identical aliquots exhaustively digested with Hpy188 III.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining such permission will be the responsibility of the applicant.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY46702, 124403, AK118104, Q43209, and Q43209.
Supplementary Material
Acknowledgments
We thank Dr. Sharyn Perry for advice on performing the chemilumnescent detection with the western blots and Ms. Kristine Hill and Dr. Huai Wang for some of the RNA samples. Prof. Glenn Collins allowed the use of his gene gun and the University of Kentucky Department of Plant Pathology generously allowed the use of their gel documentation system and phosphorimager. Prof. Robert L. Houtz provided advice on antibody production and immunopurification while Dr. Lynnette Dirk, Dr. Sharyn Perry, and an anonymous reviewer critically read an earlier version of the manuscript and made many suggestions for its improvement.
This work was supported by the University of Kentucky Department of Horticulture (stipend to Q.X.) and by the National Institutes of Health (grant nos. GM26020 and AG18000 to S.G.C.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046094.
References
- Aswad DW, Paranandi MV, Schurter BT (2000) Isoaspartate in peptides and proteins: formation, significance, and analysis. J Pharm Biomed Anal 21: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K[r] (1994) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 [DOI] [PubMed] [Google Scholar]
- Bhatt NP, Patel K, Borchardt RT (1990) Chemical pathways of peptide degradation. I. Deamidation of adrenocorticotropic hormone. Pharm Res 7: 593–599 [DOI] [PubMed] [Google Scholar]
- Brennan TV, Anderson JW, Jia Z, Waygood EB, Clarke S (1994) Repair of spontaneously deamidated HPr phosphocarrier protein catalyzed by the L-isoaspartate-(D-aspartate) O-methyltransferase. J Biol Chem 269: 24586–24595 [PubMed] [Google Scholar]
- Clarke S (1999) A protein carboxyl methyltransferase that recognizes age-damaged peptides and proteins and participates in their repair. In X Cheng, RM Blumenthal, eds, S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, pp 123–148
- Clarke S (2003) Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res Rev 2: 263–285 [DOI] [PubMed] [Google Scholar]
- David CL, Keener J, Aswad DW (1999) Isoaspartate in ribosomal protein S11 of Escherichia coli. J Bacteriol 181: 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins RD, Conn HM, Dirk LMA, Williams MA, Houtz RL (2003) The Arabidopsis thaliana PEPTIDE DEFORMYLASE 1 protein is localized to both mitochondria and chloroplasts. Plant Sci 165: 751–758 [Google Scholar]
- Esposito L, Vitagliano L, Sica F, Sorrentino G, Zagari A, Mazzarella L (2000) The ultrahigh resolution crystal structure of ribonuclease A containing an isoaspartyl residue: hydration and sterochemical analysis. J Mol Biol 297: 713–732 [DOI] [PubMed] [Google Scholar]
- Flinta C, Persson B, Jornvall H, von Heihne G (1986) Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem 154: 193–196 [DOI] [PubMed] [Google Scholar]
- Galletti P, Ingrosso D, Manna C, Clemente G, Zappia V (1995) Protein damage and methylation-mediated repair in the erythrocyte. Biochem J 306: 313–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig HH, Winter K, Cushman J, Borland A, Taybi T (2000) An improved RNA isolation method for succulent plant species rich in polyphenols and polysaccharides. Plant Mol Biol Report 18: 369–376 [Google Scholar]
- Geiger T, Clarke S (1987) Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides: succinimide-linked reactions that contribute to protein degradation. J Biol Chem 262: 785–794 [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6: 470–479 [DOI] [PubMed] [Google Scholar]
- Hoshi T, Heinemann S (2001) Regulation of cell function by methionine oxidation and reduction. J Physiol 531: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YJ, Wei Y, Zhou Y, Rajagopalan PT, Pei D (1999) Determination of substrate specificity for peptide deformylase through the screening of a combinatorial peptide library. Biochemistry 38: 643–650 [DOI] [PubMed] [Google Scholar]
- Johnson BA, Nigo SQ, Aswad DW (1991) Widespread phylogenetic distribution of a protein methyltransferase that modifies L isoaspartyl residues. Biochem Int 24: 841–848 [PubMed] [Google Scholar]
- Joyce C (2002) Quantitative RT-PCR. In J O'Connell, ed, RT-PCR Protocols, Vol 193. Humana Press, Totowa, NJ, pp 83–102
- Kagan RM, Clarke S (1994) Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys 310: 417–427 [DOI] [PubMed] [Google Scholar]
- Kagan RM, McFadden HJ, McFadden PN, O'Connor C, Clarke S (1997. a) Molecular phylogenetics of a protein repair methyltransferase. Comp Biochem Physiol 117B: 379–385 [DOI] [PubMed] [Google Scholar]
- Kagan RM, Niewmierzycka A, Clarke S (1997. b) Targeted gene disruption of the Caenorhabditis elegans L-isoaspartyl protein repair methyltransferase impairs survival of dauer stage nematodes. Arch Biochem Biophys 348: 320–328 [DOI] [PubMed] [Google Scholar]
- Karssen CM, Brinkhorst-van der Swan DLC, Breekland AE, Koornneef M (1983) Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157: 158–165 [DOI] [PubMed] [Google Scholar]
- Kazan K (2003) Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Pharmacol Sci 8: 468–471 [DOI] [PubMed] [Google Scholar]
- Kester ST, Geneve RL, Houtz RL (1997) Priming and accelerated aging affect L-isoaspartyl methyltransferase activity in tomato (Lycopersicon esculentum Mill.) seed. J Exp Bot 309: 943–949 [Google Scholar]
- Kim E, Lowenson JD, MacLaren DC, Clarke S, Young SG (1997) Deficiency of a protein-repair enzyme results in the accumulation of altered proteins, retardation of growth, and fatal seizures in mice. Proc Natl Acad Sci USA 94: 6132–6137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kuhn JM, Schroeder JI (2003) Impacts of altered RNA metabolism on abscisic acid signaling. Curr Opin Plant Biol 6: 463–469 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lowenson JD, Clarke S (1992) Recognition of D-aspartyl residues in polypeptides by the erythrocyte L-isoaspartyl/D-aspartyl protein methyltransferase: implications for the repair process. J Biol Chem 267: 5985–5995 [PubMed] [Google Scholar]
- Maclaren DC, Kagan RM, Clarke S (1992) Alternative splicing of the human isoaspartyl protein carboxyl methyltransferase RNA leads to the generation of a C-terminal-RDEL sequence in isozyme II. Biochem Biophys Res Commun 185: 277–283 [DOI] [PubMed] [Google Scholar]
- Mamula MJ, Gee RJ, Elliott JI, Sette A, Southwood S, Jones PJ, Blier PR (1999) Isoaspartyl post-translational modification triggers autoimmune responses to self-proteins. J Biol Chem 274: 22321–22327 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Monte E, Quail PH (1999) A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Mudgett MB, Clarke S (1993) Characterization of plant L-isoaspartyl mehtyltransferases that may be involved in seed survival: purification, cloning and sequence analysis of the wheat germ enzyme. Biochemistry 32: 11100–11111 [DOI] [PubMed] [Google Scholar]
- Mudgett MB, Clarke S (1994) Hormonal and environmental responsiveness of a developmentally regulated protein repair L-isoaspartyl methyltransferase in wheat. J Biol Chem 41: 25606–25612 [PubMed] [Google Scholar]
- Mudgett MB, Clarke S (1996) A distinctly regulated protein repair L-isoaspartyl methyltransferase from Arabidopsis thalinana. Plant Mol Biol 30: 723–737 [DOI] [PubMed] [Google Scholar]
- Mudgett MB, Lowenson JD, Clarke S (1997) Protein repair L-isoaspartyl methyltransferase in plants: phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds. Plant Physiol 115: 1481–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor CM (1987) Regulation and subcellular distribution of a protein methyltransferase and its damaged aspartyl substrate sties in developing Xenopus oocytes. J Biol Chem 262: 10398–10403 [PubMed] [Google Scholar]
- O'Connor CM, Germain BJ (1987) Kinetic and electrophoretic analysis of transmethylation reactions in intact Xenopus oocytes. J Biol Chem 262: 10404–10411 [PubMed] [Google Scholar]
- Pliyev BK, Gurvits BY (1999) Peptidyl-prolyl cis-trans isomerases: structure and functions. Biochemistry 64: 738–751 [PubMed] [Google Scholar]
- Potter SM, Johnson BA, Henschen A, Aswad DW (1992) The type II isoform of bovine brain protein L-isoaspartyl methyltransferase has an endoplasmic reticulum retention signal (…RDEL) at its C-terminus. Biochemistry 31: 6337–6347 [DOI] [PubMed] [Google Scholar]
- Reissner KJ, Aswad DW (2003) Deamidation and isoaspartate formation in proteins: unwanted alterations or surreptitious signals? Cell Mol Life Sci 60: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanik EA, Ladino CA, Killoy LC, D'Adrenne SC, O'Conner CM (1992) Genomic organization and tissue expression of the murine gene encoding the protein β-aspartate methyltransferase. Gene 118: 217–222 [DOI] [PubMed] [Google Scholar]
- Rost B (1996) PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol 266: 525–539 [DOI] [PubMed] [Google Scholar]
- Sarnowski TJ, Swiezewski S, Pawlikowska K, Kaczanowski S, Jerzmanowski A (2002) AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res 30: 3412–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DF, Hunt AG (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61: 1–12 [DOI] [PubMed] [Google Scholar]
- Schumacher RJ, Hansen WJ, Freeman BC, Alnemri E, Litwack G, Toft DO (1996) Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry 35: 14889–14898 [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt S, Tsou PL, Roberston D, Strömgren Allen N (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125–1132 [DOI] [PubMed] [Google Scholar]
- Stamm S (2002) Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum Mol Genet 11: 2409–2416 [DOI] [PubMed] [Google Scholar]
- Szymanska G, Leszyk JD, O'Connor CM (1998) Carboxyl methylation of deamidated calmodulin increases its stability in Xenopus oocyte cytoplasm: implications for protein repair. J Biol Chem 273: 28516–28523 [DOI] [PubMed] [Google Scholar]
- Talcott B, Moore MS (1999) Getting across the nuclear pore complex. Trends Cell Biol 9: 312–318 [DOI] [PubMed] [Google Scholar]
- Tarcsa E, Szymanska G, Lecker S, O'Connor CM, Goldberg AL (2000) Ca2+ free calmodulin and calmodulin damaged by in vitro aging are selectively degraded by 26 S proteasomes without ubiquitination. J Biol Chem 275: 20295–20301 [DOI] [PubMed] [Google Scholar]
- Thapar N, Clarke S (2000) Expression, purification, and characterization of the protein repair L-isoaspartyl methyltransferase from Arabidopsis thaliana. Protein Expr Purif 20: 237–251 [DOI] [PubMed] [Google Scholar]
- Thapar N, Griffith SC, Yeates TO, Clarke S (2002) Protein repair methyltransferase from the hyperthermophilic archaeon Pyrococcus furiosus: unusual methyl-accepting affinity for D-aspartyl and N-succinyl-containing peptides. J Biol Chem 277: 1058–1065 [DOI] [PubMed] [Google Scholar]
- Thapar N, Kim A-K, Clarke S (2001) Distinct patterns of expression but similar biochemical properties of protein L-isoaspartyl methyltransferase in higher plants. Plant Physiol 125: 1023–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Beach BM, Dirk LMA, Houtz RL, Hurley JH (2002) Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111: 91–103 [DOI] [PubMed] [Google Scholar]
- Visick JE, Cai H, Clarke S (1998) The L-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol 180: 2623–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick JE, Clarke S (1995) Repair, refold, recycle: how bacteria can deal with spontaneous and environmental damage to proteins. Mol Microbiol 16: 835–845 [DOI] [PubMed] [Google Scholar]
- Wan C-Y, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Takagi H, Kitamura D, Tatsuoka H, Nakano H, Kawano H, Kuroyanagi H, Yahagi Y, Kobayashi S, Koizumi K, et al (1998) Deficiency in protein L-isoaspartyl methyltransferase results in a fatal progressive epilepsy. J Neurosci 18: 2063–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z, Mulligan RM, Janney N, Houtz RL (1999) Rubisco small and large subunit N-methyltransferases: Bi- and mono-functional methyltransferases that methylate the small and large subunits of RUBISCO. J Biol Chem 274: 36750–36756 [DOI] [PubMed] [Google Scholar]
- Young AL, Carter WG, Doyle HA, Mamula MJ, Aswad DW (2001) Structural integrity of histone H2B in vivo requires the activity of protein L-isoaspartate O-methyltransferase, a putative repair enzyme. J Biol Chem 276: 37161–37165 [DOI] [PubMed] [Google Scholar]
- Zou Y, Crowley DJ, Van Houten B (1998) Involvement of molecular chaperonins in nucleotide excision repair: DnaK leads to increased thermal stability of UvrA, catalytic UvrB loading, enhanced repair, and increased UV resistance. J Biol Chem 273: 12887–12892 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.