Abstract
The cellular and subcellular localization of endogenous nitric oxide (NO˙) in leaves from young and senescent pea (Pisum sativum) plants was studied. Confocal laser scanning microscopy analysis of pea leaf sections with the fluorescent probe 4,5-diaminofluorescein diacetate revealed that endogenous NO˙ was mainly present in vascular tissues (xylem and phloem). Green fluorescence spots were also detected in the epidermal cells, palisade and spongy mesophyll cells, and guard cells. In senescent leaves, NO˙ generation was clearly reduced in the vascular tissues. At the subcellular level, by electron paramagnetic resonance spectroscopy with the spin trap Fe(MGD)2 and fluorometric analysis with 4,5-diaminofluorescein diacetate, NO˙ was found to be an endogenous metabolite of peroxisomes. The characteristic three-line electron paramagnetic resonance spectrum of NO˙, with g = 2.05 and aN = 12.8 G, was detected in peroxisomes. By fluorometry, NO˙ was also found in these organelles, and the level measured of NO˙ was linearly dependent on the amount of peroxisomal protein. The enzymatic production of NO˙ from l-Arg (nitric oxide synthase [NOS]-like activity) was measured by ozone chemiluminiscence. The specific activity of peroxisomal NOS was 4.9 nmol NO˙ mg−1 protein min−1; was strictly dependent on NADPH, calmodulin, and BH4; and required calcium. In senescent pea leaves, the NOS-like activity of peroxisomes was down-regulated by 72%. It is proposed that peroxisomal NO˙ could be involved in the process of senescence of pea leaves.
The gaseous free radical nitric oxide (NO˙) is a widespread intracellular and intercellular messenger with a broad spectrum of regulatory functions in many physiological processes (Moncada et al., 1991; Ignarro, 2002; Wendehenne et al., 2001; Lamattina et al., 2003; Neill et al., 2003; del Río et al., 2004). In recent years, NO˙ was reported to be involved in many key physiological processes of plants, such as ethylene emission (Leshem and Haramaty, 1996), response to drought (Leshem, 1996), disease resistance (Delledonne et al., 1998, 2001; Durner et al., 1998; Clarke et al., 2000), growth and cell proliferation (Ribeiro et al., 1999), maturation and senescence (Leshem et al., 1998), apoptosis/programmed cell death (Magalhaes et al., 1999; Clarke et al., 2000; Pedroso and Durzan, 2000; Pedroso et al., 2000a; Zhang et al., 2003), and stomatal closure (García-Mata and Lamattina, 2001, 2002; Neill et al., 2002a; García-Mata et al., 2003).
The application of exogenous NO˙ to plants has been used as a tool to study how this molecule affects some physiological processes, such as inhibition of certain enzyme activities (Clark et al., 2000; Navarre et al., 2000), cell wall lignification (Ferrer and Ros Barceló, 1999), the alternative oxidase pathway (X. Huang et al., 2002), cell death (Pedroso et al., 2000b; Beligni et al., 2002; Saviani et al., 2002), accumulation of ferritin (Murgia et al., 2002), wound signaling (Orozco-Cárdenas and Ryan, 2002), and root organogenesis (Pagnussat et al., 2002).
In animal systems, a considerable attention is being dedicated to this molecule and the enzyme responsible for its production from l-Arg, nitric oxide synthase (NOS; EC 1.14.13.39; Hemmens and Mayer, 1998; Alderton et al., 2001). On the contrary, in plants comparatively much less is known on the source of NO˙ production (Neill et al., 2003; Wendehenne et al., 2003; del Río et al., 2004). There are several enzymes that have been shown to produce NO˙ in plants, such as nitrate reductase (Yamasaki et al., 1999; Yamasaki and Sakihama, 2000; Rockel et al., 2002), a variant of the P protein of the Gly decarboxylase complex (GDC; Chandok et al., 2003), horseradish (Armoracia lapathifolia) peroxidase (Boucher et al., 1992; J. Huang et al., 2002), a plasma membrane-bound enzyme (Stöhr et al., 2001), and an Arabidopsis protein (AtNOS1; Guo et al., 2003). Cueto et al. (1996) and Ninnemann and Maier (1996) were the first to show the presence of NOS activity in higher plants by using the Arg-citrulline method. Since then, a significant number of reports have shown the presence in different plant species of NOS-like activity and immunorelated proteins with animal NOS (Wendehenne et al., 2001; Modolo et al., 2002; Neill et al., 2003; del Río et al., 2004).
Peroxisomes are single membrane-bounded subcellular organelles with an essentially oxidative type of metabolism and a simple morphology that does not reflect the complexity of their enzymatic composition (Tabak et al., 1999; Baker and Graham, 2002). The main functions described for peroxisomes in plant cells are the photorespiration cycle, fatty acid β-oxidation, the glyoxylate cycle, and the metabolism of ureides (Huang et al., 1983; Baker and Graham, 2002). These roles indicate that peroxisomes are involved in distinct metabolic networks, mainly by establishing interconnections between different cell compartments (Corpas et al., 2001; Igamberdiev and Lea, 2002; Minorsky, 2002). During the last decade, different evidence was obtained showing that peroxisomes are a source of reactive oxygen species (ROS), which could function as signal molecules in the communication between peroxisomes and other cell compartments (Corpas et al., 2001; del Río et al., 2002).
In this work, evidence is provided that NO˙ is present in several cell types of pea leaves and is an endogenous metabolite of peroxisomes. On the basis of the results obtained with young and senescent pea plants, it is proposed that peroxisomes, as a source of NO˙, could participate in the senescence process of leaves.
RESULTS
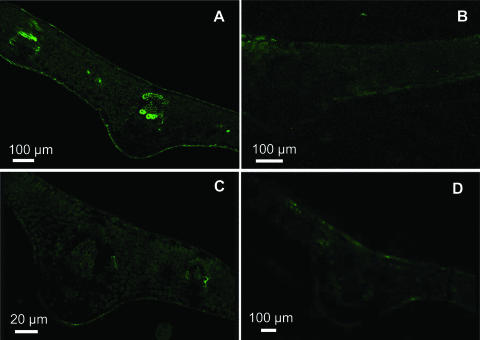
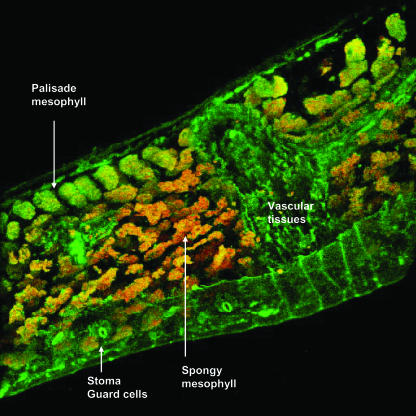
The production of NO˙ in higher plants is well established, although very little is known about where this free radical is produced in healthy plant tissues. The cellular localization of endogenous NO˙ in pea leaf sections by confocal laser scanning microscopy (CLSM) with the fluorescent probe 4,5-diaminofluorescein diacetate (DAF-2 DA) is shown in Figure 1. An intense green fluorescence was found in vascular tissues (xylem and phloem) and a lower brightness in the upper and lower epidermal cells (Fig. 1A). However, fluorescence was not observed when the pea sections were incubated under the same conditions but without the fluorescent probe (Fig. 1B). Likewise, NO˙-derived fluorescence was not detected either in pea leaf sections preincubated with 5 mm NG-nitro-l-Arg methyl ester (l-NAME), a well-known inhibitor of NOS activity in animals (Fig. 1C), or in leaves from senescent plants (Fig. 1D). Using the capability of CLSM, a digital three-dimensional reconstruction of a pea leaf section is showed in Figure 2, where, like in Figure 1A, the brightness of green fluorescence was observed in vascular tissues. However, green fluorescence spots were also distributed throughout the epidermal tissue, palisade and spongy mesophyll, and guard cells. The orange-yellow color corresponds to the chlorophyll autofluorescence distributed in all cells.
Figure 1.
Representative images illustrating the confocal laser immunofluorescent detection of endogenous NO˙ in pea leaves. Pea leaf sections 60 to 100 μm thick were used, and NO˙ was detected by its bright green fluorescence. A, Leaf from young pea plants. B, Leaf from young pea plants incubated with 10 mm Tris-HCl, pH 7.4, and without DAF-2 DA. C, Leaf from young pea plants incubated with 5 mm l-NAME (NOS inhibitor). D, Leaf from senescent pea plants.
Figure 2.
Three-dimensional composition showing the cellular localization of NO˙ in pea leaves. It was prepared from 153 pea leaf sections analyzed by CLSM. Strong and bright green fluorescence is observed in vascular tissues and green fluorescence spots are observed in epidermal, palisade and spongy mesophyll, and guard cells. The orange-yellow color corresponds to the chlorophyll autofluorescence.
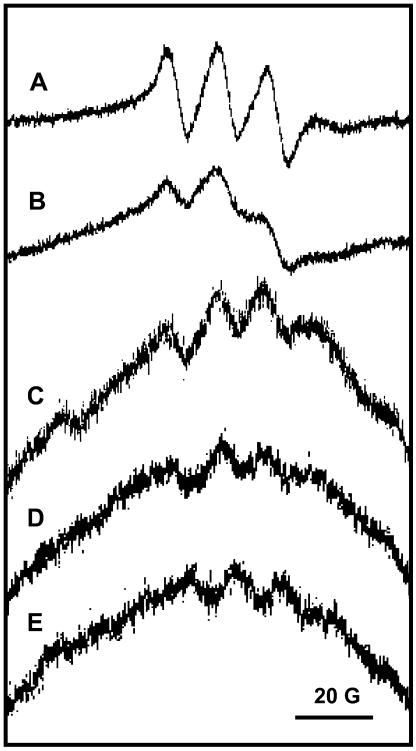
In isolated leaf peroxisomes, NOS-like activity from l-Arg was previously detected by measuring the production of l-citrulline (Barroso et al., 1999). Although the presence of NO˙ in peroxisomes appears to be logical, the production of NO˙ in these organelles has not been demonstrated so far, and some authors have cast doubts on the occurrence of NOS activity in peroxisomes. Therefore, in peroxisomal fractions isolated from pea leaves, the presence of NO˙ was studied by two complementary approaches: electron paramagnetic resonance (EPR) spectroscopy and fluorometric analysis. EPR measurements were performed by using the spin trap Fe(MGD)2 that reacts with NO˙ forming a stable NO˙-Fe(MGD)2 complex with a characteristic three-line EPR spectrum. As positive control of NO˙ production, pure nNOS (Merck Biosciences, Darmstadt, Germany) was used under the standard assay conditions. In Figure 3A, a representative EPR spectrum of the NO˙-Fe(MGD)2 spin adduct produced by nNOS in its enzymatic reaction is shown, with the typical triplet signal of this spin adduct (g = 2.05 and aN = 12.8 G). When crude extracts and isolated peroxisomes from pea leaves were assayed for NO˙, similar three-line signals with the same values for g and aN were found (Fig. 3, B and C, respectively). The preincubation of peroxisomal samples with a known NOS inhibitor (1 mm l-NAME; Fig. 3D) or without NADPH (Fig. 3E) produced spectra with lower signal intensities than control peroxisomes (Fig. 3C).
Figure 3.
EPR spectra of the NO˙-spin adduct of the Fe(MGD)2 complex. For the detection of NO˙, samples were added to a reaction mixture containing the substrate and all the cofactors of the NOS reaction, plus the NO˙-spin trap Fe(MGD)2, and were incubated for 1 h at 37°C. Then, samples were analyzed by EPR as described in “Materials and Methods.” A, Pure nNOS (15 μg) from Merck Biosciences. B, Crude extracts of pea leaves (2.5 mg protein). C, Peroxisomes purified from pea leaves (270 μg protein). D, Purified pea leaf peroxisomes (270 μg protein) preincubated with 1 mm l-NAME. E, Purified pea leaf peroxisomes (270 μg protein) without NADPH in the reaction mixture. The EPR parameters of the spectra were g = 2.05 and aN = 12.8 G. Representative spectra from three to five independent measurements are shown. Spectra B to E were obtained by the accumulation of five recordings.
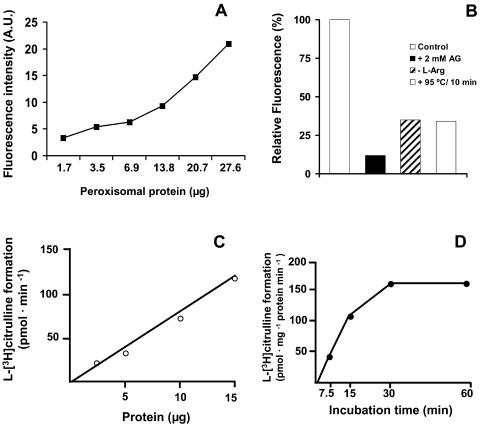
The second approach used to detect the presence of NO˙ in pea leaf peroxisomes was spectrofluorometric analysis with DAF-2 DA. An increasing protein concentration-dependent fluorescence was observed, which indicated the existence of a linear relationship between the levels of NO˙ generated and the amount of peroxisomal proteins (Fig. 4A). To corroborate these results several additional assays were carried out: (1) the peroxisomal samples were preincubated with 2 mm aminoguanidine, a known inhibitor of NOS; (2) l-Arg, the substrate of NOS, was not added to the reaction mixtures; and (3) prior to the addition of DAF-2 DA, the peroxisomal samples were denatured by heating at 95°C for 10 min. In these cases, reductions of 65% to 88% in the relative fluorescence compared to the control reaction were obtained (Fig. 4B). Therefore, it can be concluded that at least 65% of the NO˙ detected by spectrofluorometric analysis has an enzymatic origin. When the NOS-like activity was measured as production of l-[3H]citrulline from l-[3H]Arg, a linear correlation with the amount of peroxisomal protein was observed (Fig. 4C), which is in agreement with the fluorometric results of Figure 4A. This NOS-like activity increased during the first 30 min of incubation, and then reached a plateau (Fig. 4D).
Figure 4.
Spectrofluorometric detection of NO˙ in pea leaf peroxisomes with DAF-2 DA. Peroxisomes freshly isolated from pea leaves were added to the reaction mixtures and the fluorescence measured. Values shown are means of three independent experiments. A, Different volumes of peroxisomal fractions were added to the reaction mixture and the fluorescence produced was expressed as arbitrary units (A.U.). B, Effect on the NO˙ generation of: (1) preincubating peroxisomes with 2 mm aminoguanidine (AG); (2) removing l-Arg from the reaction mixture (l-Arg); and (3) denaturing the samples by heating (95°C/10 min). The amount of peroxisomal proteins in the reaction mixtures was 12 μg, and fluorescence was expressed as percent of the control (relative fluorescence). C, NOS activity of peroxisomal fractions determined as protein-dependent formation of l-[3H]citrulline from l-[3H]Arg. D, Influence of the incubation time on the NOS activity of peroxisomal fractions determined by measuring the formation of l-[3H]citrulline from l-[3H]Arg.
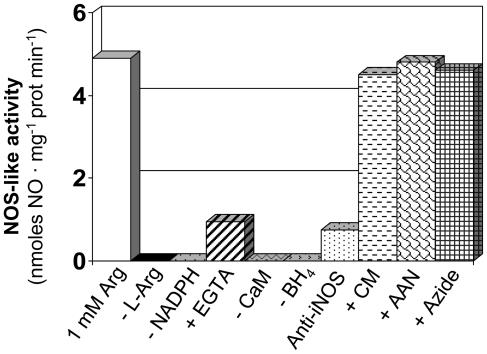
The enzymatic production of NO˙ in isolated leaf peroxisomes, measured by ozone chemiluminescence, is shown in Figure 5. In this case, peroxisomes generated NO˙ from l-Arg (NOS-like activity), and this production was strictly dependent on l-Arg, NADPH, BH4, and calmodulin. In the presence of EGTA, which complexes calcium, the NO˙ generation was reduced about 80%, and when the peroxisomal fractions were preincubated with an antibody against murine iNOS, the production of NO˙ was inhibited by 85%. These data are in agreement with the protein-dependent production of NO˙ determined by spectrofluorometric analysis (Fig. 4A). Additionally, when the peroxisomal samples were preincubated either with 200 μm carboxymethoxylamine (CM) and 0.01% aminoacetonitrile (AAN), two inhibitors of the P protein of the GDC, or 1 mm azide (an inhibitor of nitrate reductase), the peroxisomal NOS activity was not affected.
Figure 5.
Determination in pea leaf peroxisomes of NO˙ production from l-Arg (NOS activity) by ozone chemiluminiscence. Reaction mixtures containing peroxisomal fractions were incubated in the absence and presence of l-Arg (1 mm), NADPH (1 mm), EGTA (0.5 mm), calmodulin (10 μg/mL), BH4 (10 μm), antibody against iNOS, 200 μm CM, 0.01% AAN, and 1 mm azide. Then the NO˙ production was assayed using a 1 mm l-Arg concentration and an incubation time of 30 min. The NO˙ produced was quantified by ozone chemiluminiscence using a nitric oxide analyzer. Results are means of samples from at least three different Suc-density gradients.
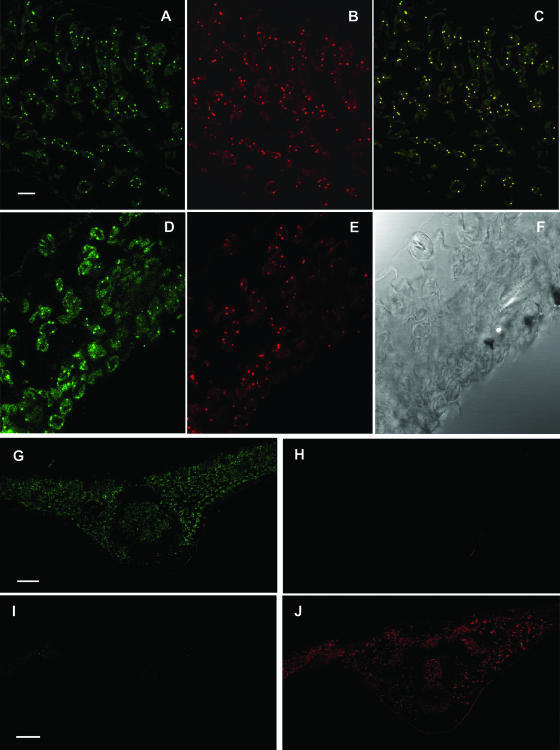
To verify the subcellular localization of the NOS-like protein in peroxisomes, colocalization studies with the peroxisomal marker enzyme catalase were carried out in pea leaves by CLSM. With the immunocytochemical procedure used for iNOS, a green punctuate fluorescence pattern of Cy2-streptavidin was detected in the spongy mesophyll and guard cells of pea leaf sections (Fig. 6, A and D). On the other hand, with the immunocytochemical procedure to detect catalase, a red fluorescence pattern of Cy3 was observed in the same spots where iNOS was detected (Fig. 6, B and E). Peroxisomes appeared as small peripheral fluorescent spots within the spongy mesophyll cells. The colocalization of Cy2-streptavidin and Cy3 immunofluorescence patterns is shown in Figure 6C, where the nearly complete overlapping of the two punctuate patterns indicated that the protein immunorelated to mouse iNOS was localized in peroxisomes. Additionally, a similar pattern was detected in guard cells (Fig. 6, D and E). These data are in agreement with the localization of NO˙ in pea leaves with the fluorescence probe DAF-2 DA (Fig. 2), which showed a green fluorescence punctuate pattern throughout different cells of the leaf. As control for background staining, primary antibodies to either catalase (Fig. 6, G and H) or iNOS (Fig. 6, I and J) were omitted in the colocalization experiments. Under these conditions, immunofluorescence background was not observed, indicating the absence of interferences and the specificity of the procedure used.
Figure 6.
Colocalization of NOS and catalase in pea leaves. Representative images illustrating the CLSM detection of NOS and catalase in spongy mesophyll and guard cells of pea leaf sections (60 μm thick). A and D, Cy2-streptavidin immunofluorescence punctuates (green) attributable to anti-iNOS. B and E, Cy3 immunofluorescence punctuates (red) attributable to anti-catalase. C, Colocalized immunofluorescence punctuates (yellow). F, Bright-field image of the same pea leaf area. G and H, Images of the same pea leaf section where the catalase antiserum was omitted but not the other components of the colocalization mixture; therefore, only the anti-iNOS fluorescence (green) is observed. I and J, Images of the same pea leaf section where the iNOS antiserum was omitted but not the other components of the colocalization mixture; therefore, only the anti-catalase fluorescence (red) is observed. Scale bar represents 10 μm (A–F) and 20 μm (G–J).
The effect of senescence of pea plants on the subcellular production of NO˙ was studied in peroxisomes isolated from pea leaves. As shown in Figure 1D, the green fluorescence due to NO˙ was undetectable in leaf sections from senescent plants. To evaluate the physiological significance of peroxisomes in the senescence process, the enzymatic production of NO˙ from l-Arg (NOS-like activity) was analyzed in peroxisomes isolated from pea leaves. The activity of the peroxisomal enzyme malate synthase was also studied. This is a key enzyme of the glyoxylate cycle, which is induced in peroxisomes by leaf senescence when it takes place the metabolic conversion of leaf peroxisomes into glyoxysomes (De Bellis et al., 1990; del Río et al., 1998). In peroxisomes from pea leaves, malate synthase was up-regulated by senescence and catalase activity underwent a significant reduction (Table I). Under these conditions, the NO˙ production in peroxisomes was reduced by 72%, and this is in agreement with the data of NO˙ detection in senescent pea leaves by CLSM (Fig. 1D).
Table I.
NO˙ production from l-Arg (NOS-like activity), and catalase and malate synthase activity in leaf peroxisomes from young and senescent pea plants
| Enzymes | Young Plants | Senescent Plants |
|---|---|---|
| NOS-like activity (nmol NO˙ mg−1 protein min−1) | 4.97 ± 0.38 | 1.40 ± 0.07 |
| Malate synthase (nmol mg−1 protein min−1) | 4.58 ± 2.05 | 40.00 ± 9.89 |
| Catalase (nmol mg−1 protein min−1) | 460.0 ± 24.4 | 339.0 ± 66.8 |
Peroxisomes were purified from leaves of young plants (15-d-old) and senescent plants (50-d-old). Each value is the mean of at least three different experiments ± SEM.
DISCUSSION
In higher plants, results obtained in recent years have shown that NO˙ has an important role as messenger in many important physiological processes. However, little is known about the cellular and subcellular production sites of NO˙ (Neill et al., 2003). In this work, a subcellular localization of NO˙ in pea leaves has been identified. It has been demonstrated that NO˙ is an endogenous metabolite of peroxisomes, where it is produced from l-Arg by a protein immunorelated to mouse iNOS, and the NO˙ production is down regulated by leaf senescence.
NO˙ Is Localized in the Vascular Tissue and Other Different Cell Types
The plant vascular tissues have evolved to keep a specialized role mediating the exchange of essential nutrients between different organs. Phloem has gone through a tremendous evolutionary transformation from a simple pathway for photoassimilate transport into a highly complex system for translocation, messaging, and plant integration (Thompson and Schulz, 1999; Ruiz-Medrano et al., 2001). Recently, it has been shown that phloem sap of cucumber (Cucumis sativus) and pumpkin (Cucurbita maxima) plants include antioxidant defense enzymes, such as copper, zinc-containing superoxide dismutase, and monodehydroascorbate reductase, and these activities were increased by drought stress (Walz et al., 2002). The data obtained by CLSM revealed a strong accumulation of NO˙ in vascular tissue, suggesting that it could be involved in some aspects of long-distance communications inside the plant, perhaps as a signaling effector or receptor. These results support the mode of action of NO˙ in cell expansion proposed by Leshem (2000). As NO˙ is highly diffusible, it can probably reach the apoplast and get in contact with the cell wall and perhaps weaken its intermicellar links. This would result in a loosening and growth-promoting effect, allowing cell turgor to cause cell expansion. From a plant physiologic viewpoint, this effect could be equivalent to the vasodilatation induced in mammals by NO˙.
The localization of NO˙ in guard cells has been reported by several authors (García-Mata and Lamattina, 2002; Neill et al., 2002a), but its production has been attributed to the enzyme nitrate reductase (Desikan et al., 2002). In this work, a strong accumulation of NO˙ in different cell types of leaves (Fig. 2) and the presence of a protein immunorelated to mouse iNOS in guard cells was observed (Fig. 6D), indicating that this could be an alternative source of NO˙ in this type of cell. This result is in agreement with the data reported by Neill et al. (2002a) in pea plants where guard cells in response to abscisic acid generate NO˙, via a NOS-like activity, to produce the stomatal closure. Additionally, similar results have been shown in Arabidopsis where the AtNOS1 protein is required for abscisic acid-induced NO˙ generation and stomatal closure (Guo et al., 2003).
NO˙ Is an Endogenous Metabolite of Peroxisomes
Considering the green fluorescent spots due to NO˙ detected in different cell types (parenchymal, epidermical, and guard cells) and the previous report on the presence of NOS-like activity in peroxisomes (Barroso et al., 1999), a study was carried out to get conclusive information on the production of NO˙ in these organelles.
EPR spectroscopy using as spin trap the complex Fe(MGD)2 has been demonstrated to be a specific method to determine the direct formation of NO˙ in biological systems and unequivocal evidence for its presence (Kotake et al., 1996; Xia et al., 2000; Nagano and Yoshimura, 2002). In our experimental conditions, the biological source of NO˙ used as control (commercial nNOS) gave an identical NO˙-Fe(MGD)2 spectrum to that obtained by Xia and Zweier (1997) with NOS from animal origin. This EPR method has also been used in plant systems to detect NO˙ in homogenates of soybean (Glycine max) embryonic axes (Caro and Puntarulo, 1999), sorghum (Sorghum bicolor) seeds (Simontacchi et al., 2004) and cucumber explants (Pagnussat et al., 2002), and in cell suspension cultures of maize (Zea mays; Dordas et al., 2004) and Arabidopsis (Huang et al., 2004). The relatively weak three-line signal of the NO˙-spin adduct detected in isolated peroxisomes could be due to a low enzymatic generation of the signal molecule NO˙, as has been reported in animal systems for the constitutive NOSs (nNOS and eNOS; Joshi et al., 1999). However, another possibility for the low intensity of the EPR signal derives from the well-known affinity of NO˙ for heme proteins (Sharma et al., 1987; Kanner et al., 1992). The heme-enzyme catalase is very abundant in peroxisomes and can bind NO˙ and even catalyze its breakdown in the presence of H2O2 (Brown, 1995). Catalase may, therefore, compete with the spin trap for the NO˙ generated in the peroxisomal-NOS reaction, thus leading to a lower NO˙-Fe(MGD)2 EPR signal. On the other hand, peroxisomal membranes contain NAD(P)H-dependent O2˙−-generating systems (del Río et al., 1989; López-Huertas et al., 1999), and the spin adduct NO˙-Fe(MGD)2 can react with O2˙− radicals yielding EPR-silent species (Nagano and Yoshimura, 2002). This could equally lead to an underestimation of the endogenous NO˙ present in peroxisomes.
The use of DAF-2 DA as fluorescent probe has become a common and very sensitive technique to detect NO˙ in animal and plant systems (Nakatsubo et al., 1998; Nagano, 1999; Nagano and Yoshimura, 2002). This probe has been used in plant cells to obtain real-time bioimaging of NO˙ with fine temporal and spatial resolution (Foissner et al., 2000; Pedroso et al., 2000a; García-Mata and Lamattina, 2002; Neill et al., 2002a). In the spectrofluorometric method used in this work, the detection limit of DAF-2 DA is less than 2 to 5 nm and shows a linear correlation up to around 1,000 nm NO˙ (Nakatsubo et al., 1998). The positive correlation found between the fluorescence intensity and the amount of peroxisomal protein seems to indicate a protein origin for this NO˙. Results obtained with DAF-2 DA, both spectrofluorometrically and by CLSM, support those obtained by EPR based on the paramagnetism of the NO˙ molecule and clearly demonstrate the presence of NO˙ in pea leaf peroxisomes.
The occurrence of NO˙ in peroxisomes suggests different interactions of this molecule with other components of the ROS metabolism. Besides catalase, several antioxidative enzyme systems have been demonstrated in plant peroxisomes, including different superoxide dismutases, the four enzymes of the ascorbate-glutathione cycle plus ascorbate and glutathione, and three NADP-dependent dehydrogenases (del Río et al., 2002). The NADP-dehydrogenases found in the peroxisomal matrix (Corpas et al., 1998, 1999) could provide the necessary NADPH for the NO˙-producing NOS reaction. But inside peroxisomes NO˙ could react with O2˙− radicals generated in the peroxisomal matrix by xanthine oxidase (del Río et al., 1989, 2002) to form peroxynitrite (ONOO−), a powerful oxidizing reactive nitrogen species. On the other hand, NO˙ can react with reduced glutathione, present in peroxisomes (Jiménez et al., 1998), to generate S-nitrosoglutathione, which is a powerful inducer of defense genes (Durner et al., 1998) and can also function as a long-distance signal molecule, transporting glutathione-bound NO˙ (Durner and Klessig, 1999).
NO˙ could also have a role in the activity regulation of different peroxisomal heme-flavin enzymes involved in the metabolism of ROS, such as catalase and ascorbate peroxidase (Clark et al., 2000; Brunelli et al., 2001) or xanthine oxidase (Sakuma et al., 1997). Recently, the presence of a calcium-dependent protein kinase in peroxisomes of Arabidopsis has been described (Dammann et al., 2003), which could be another target of NO˙ in these organelles and participate in the signaling process. Considering the high diffusion coefficient of NO˙ (Denicola et al., 1996) it could function as a signal molecule in other organelles of the same cell or even in other cells. In plant cells, NO˙ produced in peroxisomes could act on plant mitochondria, where apparently NOS is not present (Barroso et al., 1999), and modulate the activity of the alternative oxidase (Huang et al., 2002). Additionally, peroxisomal NO˙ could also regulate the activity of cytosolic aconitase (Navarre et al., 2000).
Peroxisomal NO˙ Is Generated by a Protein Immunorelated to Animal iNOS
In higher plants, the production of NO˙ by different enzymes has been described, mainly including nitrate reductase (Yamasaki et al., 1999), a variant of the P protein of the mitochondrial GDC (Chandok et al., 2003), and the protein AtNOS1 codified by an Arabidopsis NOS gene (Guo et al., 2003), although different evidence available indicates that there are other potential enzymatic sources of NO˙ in plants (Corpas et al. 2004; del Río et al., 2004). Taking into account that none of these proteins are present in peroxisomes and that the existence of NOS-like activity in these organelles has been questioned by some authors, the measurement of this activity as l-Arg-dependent NO˙ production, reported in this work, provides additional evidence of the presence of NOS-like activity in peroxisomes. The NO˙ determination by ozone chemiluminiscence has a minimum detection limit of 1 pg NO˙, and it is widely used to quantify NO˙ production in many biological systems. The NOS-specific activity determined in peroxisomes in this work (about 5 nmol NO˙ mg−1 protein min−1) was of the same order as that obtained previously in pea leaf peroxisomes by monitoring the conversion of l-[3H]Arg to l-[3H]citrulline (Barroso et al., 1999). The specific activity was also very similar to that reported for the AtNOS1 protein, which was determined with the Greiss reagent (Guo et al., 2003). In addition, in this work it was proved that the NO˙ production from l-Arg required NADPH, BH4, calmodulin, and calcium, and its production was inhibited by an antibody against mouse iNOS. Moreover, CM and AAN, two characteristic inhibitors of the P protein of the mitochondrial GDC (Douce et al., 2001) and the nitrate reductase inhibitor azide, did not have any effect on the peroxisomal NOS activity. This indicates that the enzymatic-generating system of NO˙ in peroxisomes is different from that reported for the variant P protein of the GDC (Chandok et al., 2003). Likewise, the peroxisomal NOS activity is distinct from the NOS activity of the AtNOS1 protein of Arabidopsis, which did not depend on BH4, FAD, and FMN as cofactors (Guo et al., 2003).
On the other hand, by CLSM, a complementary approach to immunogold electron microscopy, the coincident immunofluorescence punctuate patterns obtained in pea leaves with antibodies against iNOS and catalase, confirmed that peroxisomes have a NOS-like protein. This peroxisomal location of NOS is not unique for pea plants since this protein has also been immunolocalized in peroxisomes of olive (Olea europaea) leaves and sunflower (Helianthus annuus) cotyledons (R. Valderrama, unpublished data). Recently, the localization of iNOS in animal cell peroxisomes has been reported for the first time, in organelles of cultured rat hepatocytes (Stolz et al., 2002). This finding supports the idea that the enzymatic-NOS activity of leaf peroxisomes could be characteristic of peroxisomes from different origins.
NO˙ Generation Is Down-Regulated during the Natural Senescence of Leaves
Leaf senescence is a developmentally programmed degeneration process that constitutes the final step of leaf development and is controlled by multiple developmental and environmental signals (Lim et al., 2003). This process is mainly characterized by a cessation of photosynthesis, disintegration of organelle structures, intensive losses of chlorophyll and proteins, and dramatic increases in lipid peroxidation and membrane leakiness (Buchanan-Wollaston, 1997). These latter oxidative changes are mainly originated by the strong enhancement in the generation of ROS and the deterioration of the antioxidative defense systems that takes place during the senescence process (del Río et al., 1998; Thompson et al., 1999; Navabpour et al., 2003).
Senescence brings about important alterations in the oxidative metabolism of peroxisomes. The down-regulation of catalase and the photorespiration enzymes is a characteristic feature of leaf senescence (Strother, 1988; Pastori and del Río, 1997), whereas the enzymes of the glyoxylate cycle, malate synthase and isocitrate lyase, are up-regulated (De Bellis et al. 1990; Pastori and del Río, 1997). Moreover, the endogenous endoprotease activity is increased up to six times in peroxisomes from senescent plants and four additional isoenzymes are detected (Distefano et al., 1999). Another case of senescence-induced changes in peroxisomes is NADP-isocitrate dehydrogenase, whose specific activity does not change while its Km is 11 times decreased by senescence (Corpas et al., 1999).
The implication of the reactive oxygen metabolism of leaf peroxisomes in the oxidative mechanism of leaf senescence has been demonstrated. The NADH-dependent generation of O2˙− radicals by the peroxisomal membranes and the H2O2 concentration in intact peroxisomes, as well as the rate of lipid peroxidation, increased significantly in these organelles during senescence. Enhanced activities of xanthine oxidase and urate oxidase in senescent peroxisomes led to an increase of O2˙− and H2O2 in these organelles (Pastori and del Río, 1997; del Río et al., 1998). On the other hand, the peroxisomal enzymes of the ascorbate-glutathione cycle were notably affected by senescence (Jiménez et al., 1998). All these data indicate that, during senescence, peroxisomes undergo very significant changes in its metabolism and, particularly, in the activity of those enzymes involved in the regulation of the ROS level. The down-regulation of NO˙ production during senescence, reported in this work, suggests that reactive nitrogen species and ROS present in peroxisomes could function as sensor molecules and, therefore, these organelles could participate in the intra- and inter-cellular communication networks.
To our knowledge, there is little information on the relationship between NO˙ and natural senescence in leaves, and most of the data available were obtained from the application of exogenous NO˙ to plants. Leshem and Haramaty (1996) showed that application of an NO˙ donor to senescent pea leaves reduced the generation of ethylene, which is involved in the regulation of this process. It has also been described that exogenous NO˙ can retard the flower and vegetable senescence and fruit ripening (Leshem, 2000). Exogenous NO˙ counteracts the senescence of rice (Oryza sativa) leaves induced by abscisic acid (Hung and Kao, 2003), and it has been shown that NO˙ can delay programmed cell death in barley (Hordeum vulgare) aleurone layers (Beligni et al., 2002). All these data are in agreement with the results obtained in this work in leaf sections with DAF-2 DA and in isolated peroxisomes on the down-regulation of the NO˙ generation from l-Arg.
An important feature of eukaryotic cells is the compartmentation of proteins within membranous organelles, which is essential to coordinate, regulate, and integrate the different metabolic pathways (Igamberdiev and Lea, 2002). Considering that a characteristic property of peroxisomes is the participation in metabolic pathways shared with other cell compartments (Huang et al., 1983; Baker and Graham, 2002; Igamberdiev and Lea, 2002), the presence of NOS activity and NO˙ in peroxisomes can have an impact on the metabolic regulation of cell organelles. Peroxisomes can be considered as cellular compartments with the capacity to generate and release into the cytosol important signal molecules such as NO˙ and probably S-nitrosoglutathione, apart from O2˙− and H2O2 (Corpas et al., 2001). Moreover, in plants the cellular population of peroxisomes can proliferate during senescence and under different stress conditions (del Río et al., 1998, 2002; Romero-Puertas et al., 1999; López-Huertas et al., 2000). This has prompted us to propose that plant peroxisomes could act as subcellular indicators or sensors of plant stress and senescence by releasing the signaling molecules NO˙, O2˙−, and H2O2 to the cytosol and triggering a specific gene expression (Corpas et al., 2001). These messenger molecules could be used in different signaling pathways (Bolwell, 1999; Neill et al., 2002b) and contribute to a more integrated communication among cell compartments particularly in response to biotic and abiotic stresses.
Further research is necessary to achieve the purification and characterization of the peroxisomal protein responsible for the NOS activity and the cloning of the corresponding gene.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Pea (Pisum sativum L. cvs Lincoln and Phoenix) seeds were obtained from Royal Sluis (Enkhuizen, Holland) and Südwestdeutsche Saatzucht (Rastatt, Germany), respectively. Seeds were surface sterilized with 3% (v/v) commercial bleaching solution for 3 min, then were washed with distilled water and germinated in vermiculite for 14 d under greenhouse conditions (28–18°C, day–night temperature; 80% relative humidity). Healthy and vigorous seedlings were selected and grown in aerated optimum-nutrient solutions for 15 d (young plants) or 50 d (senescent plants) under the greenhouse conditions indicated above. The nutrient solutions had a pH of 5.5 and the following composition in mM: NO3−, 12.1; H2PO4−, 4.0; SO42−, 1.5; K+, 5.0; Ca2+, 4.5; Mg2+, 1.5; and in μm: Fe, 91.3; B, 46; Cu, 1.1; Zn, 2.3; Mn, 9.3.
Preparation of Leaf Extracts
All operations were carried out at 0 to 4°C. Leaves were ground in liquid N2 with 40 mm HEPES buffer, pH 7.2 (1:4, w/v), with mortar and pestle. Homogenates were filtered through two layers of Miracloth and centrifuged at 27,000g for 20 min. Aliquots of supernatants were immediately used for the assays.
Purification of Peroxisomes
All operations were performed at 0°C to 4°C. Peroxisomes were purified from pea leaves by differential and Suc density-gradient centrifugation (35%–60%, w/w), as described by López-Huertas et al. (1995).
EPR Spectroscopy
NO˙ was detected by EPR spectroscopy using the spin trap Fe(MGD)2 by the method of Kotake et al. (1996), as modified by Caro and Puntarulo (1999). The spin trap was prepared by mixing N-methyl-d-glucamine dithiocarbamate and FeSO4 stock solutions to give a final concentration in the reaction mixture of 10 mm and 1 mm, respectively. For the NO˙ detection, samples (freshly isolated peroxisomes, crude extracts, or pure nNOS) were added to reaction media containing the substrate l-Arg and all the cofactors of the NOS reaction, plus the NO˙-spin trap Fe(MGD)2. The composition of the reaction mixtures (1 mL) was: 40 mm HEPES buffer, pH 7.2, containing 1 mm l-Arg, 1 mm β-NADPH, 1.25 mm CaCl2, 10 μm FAD, 10 μm FMN, 10 μm BH4, 10 μg/mL calmodulin, 10 mm N-methyl-d-glucamine dithiocarbamate, and 1 mm FeSO4. Reaction mixtures were incubated for 1 h at 37°C and then were placed in a 2-mm-thick quartz flat cell and introduced in the EPR resonator cavity. The EPR spectra were recorded at room temperature using a Bruker ESP 300 E EPR spectrometer (X-band = 9.5–10.0 GHz; Bruker Instruments, Billerica, MA). EPR conditions were as follows: microwave frequency, 9.77 GHz; microwave power, 100 mW; field modulation frequency, 50 kHz; modulation amplitude, 5.2 G; center field, 3,410 G; sweep range, 3,350 to 3,470 G; and time constant, 81.92 ms.
Spectrofluorometric Assays
To freshly isolated peroxisomes l-Arg and DAF-2 DA (Merck Biosciences) were added at 1 mm and 10 μm final concentrations, respectively. Then, reaction mixtures were incubated at 37°C in the dark for 2 h, and the fluorescence was measured in a spectrofluorophotometer Shimadzu RF-540 (Shimadzu, Columbia, MD) at excitation and emission wavelengths of 485 and 515 nm, respectively (Nakatsubo et al., 1998). Three control reaction mixtures of peroxisomes were prepared: (1) without l-Arg; (2) preincubated with 2 mm aminoguanidine; and (3) preheated at 95°C for 10 min.
Quantification of NO˙ Production from l-Arg (NOS Activity)
NOS activity was determined by monitoring the conversion of l-[3H]Arg to l-[3H]citrulline, as previously described by Barroso et al. (1999). The NOS activity was also estimated by measuring the production of NO˙ by ozone chemiluminescence using an NO˙ analyzer (NOA 280i; Sievers Instruments, Boulder, CO). NOS activity was performed in duplicate for each sample in a reaction medium containing 40 mm HEPES buffer, pH 7.2, 0.2 mm CHAPS, 10 μm/mL calmodulin, 1.25 mm CaCl2, 1 mm β-NADPH, 10 μm FAD, 10 μm FMN, 10 μm BH4, and 1 mm l-Arg. The reaction mixture was incubated at 37°C for 30 min. In this method the NO˙ produced is quickly converted to its stable end products, nitrite and nitrate. At 0 time and at 30 min, 200 μL of the reaction medium were deproteinated by the addition of 100 μL 0.8 n NaOH and 100 μL 16% ZnSO4, and the mixture was shaken vigorously for 30 s and centrifuged for 10 min at 13,000 rpm. A 40-μL aliquot of supernatant was injected into a reaction chamber online with the analyzer, where nitrite-nitrate content was reduced back to NO˙ with a vanadium reducing mixture at 90°C. NO˙ was carried on a constant stream of nitrogen gas to a reaction chamber, and the light emitted from the chemiluminescence reaction between NO˙ and ozone was detected by a photomultiplier tube. Calibration of the magnitude of NO˙ production was determined from the integral of the signal over time compared with that from nitrite concentration standards. The production of NO˙ in samples was calculated by subtracting the blank value (zero time), which represented the nonenzymatic NO˙ production, and the activity was expressed as nmole of NO˙ mg−1 protein min−1. In some experiments, the reaction mixtures containing peroxisomal fractions were preincubated in the absence of l-Arg, NADPH, CaCl2, calmodulin, BH4, and in the presence of 200 μm CM and 0.01% AAN (inhibitors of the P protein of the GDC), 1 mm azide (inhibitor of nitrate reductase), EGTA, and an antibody against murine iNOS (Barroso et al., 1999).
NO˙ Detection by CLSM
Pea leaf segments of approximately 25 mm2 were incubated for 1 h at 25°C, in darkness, with 10 μm DAF-2 DA (Calbiochem-Novabiochem) prepared in 10 mm Tris-HCl, pH 7.4, this probe being highly specific for NO˙ (Kojima et al., 1998; Nakatsubo et al., 1998). Then, they were washed twice in the same buffer for 15 min each. After washing, leaf sections were embedded in a mixture of 15% acrylamide-bisacrylamide stock solution as described elsewhere (Peinado et al., 2000), and 100 mm-thick sections, as indicated by the vibratome scale, were cut under 10 mm phosphate-buffered saline (PBS). Sections were then soaked in glycerol:PBS (containing azide; 1:1 v/v) and mounted in the same medium for examination with a confocal laser scanning microscope system (Leica TCS SL; Leica Microsystems, Wetzlar, Germany), using standard filters and collection modalities for DAF-2 green fluorescence (excitation 495 nm; emission 515 nm) and chlorophyll autofluorescence (chlorophyll a and b, excitation 429 and 450 nm, respectively; emission 650 nm and 670 nm, respectively) as orange. Background staining, routinely negligible, was controlled with leaf sections unstained or preincubated with 5 mm l-NAME (1 h, 25°C), a competitive NOS inhibitor.
Colocalization of Catalase and NOS-Like Protein by CLSM
Pea leaves were cut into 4 to 5 mm pieces and fixed in 4% (w/v) p-formaldehyde in 0.1 m phosphate buffer (PB), pH 7.4, for 3 h at room temperature. Then they were cryoprotected by immersion in 30% (w/v) Suc in PB overnight at 4°C. Serial sections, 60 μm thick, were obtained by means of a cryostat (2800 Frigocut E; Reichert-Jung, Vienna). Colocalization studies were carried out by confocal analysis of double immunofluorescence-stained sections as described by Esteban et al. (2001). Free floating sections were incubated overnight at room temperature with an antibody to murine iNOS (Uttenthal et al., 1998) diluted 1:2,500 in 5 mm Tris buffer, pH 7.2, 0.9% (w/v) NaCl, containing 0.05% (w/v) sodium azide, 0.1% (w/v) bovine serum albumin, and 0.1% (v/v) Triton X-100 (TBSA-BSAT). After several washes with TBSA-BSAT, sections were incubated with biotinylated goat anti-rabbit IgG (Pierce Chemical, Rockford, IL), diluted 1:1,000 in TBSA-BSAT for 1 h at room temperature. Then, sections were washed again and incubated with Cy2-streptavidin (Amersham, Buckinghamshire, UK), diluted 1:1,000 in TBSA-BSAT for 1.5 h at room temperature, this and the following steps being carried out in the dark. After three additional washes, the sections were soaked in goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA) diluted 1:200, for 30 min at room temperature. The sections were then washed 20 times, 15 min each, and after an additional overnight wash, they were incubated with an antibody against pumpkin (Cucurbita maxima) catalase (Yamaguchi and Nishimura, 1984) diluted 1:500 in TBSA-BSAT, for 3 d at 4°C. After several washes, sections were incubated with Cy3-labeled anti-rabbit IgG (Amersham) diluted 1:1,000 in TBS-BSAT, for 1 h at room temperature, and then mounted in PBS:glycerol 1:1. Controls for background staining, which was usually negligible, were performed by replacing the corresponding primary antiserum by preimmune serum. Leaf sections were examined with a confocal laser scanning microscope (Leica TCS SL; Leica).
Other Assays
Catalase activity was determined according to Aebi (1984) and malate syntase was determined by measuring the formation of the complex CoA-DTNB at 412 nm (Hock and Beevers, 1966). Proteins were assayed according to Bradford (1976) using bovine serum albumin as standard.
Acknowledgments
A.M.L. and M.C.R.-P. acknowledge a Ph.D. fellowship (F.P.I.) from the Ministry of Education and Science and Junta de Andalucía, respectively. We are grateful to Prof. José Rodrigo, Instituto Cajal, Consejo Superior de Investigaciones Científicas, Madrid, and Prof. Mikio Nishimura, National Institute for Basic Biology, Okazaki, Japan, for their generous donation of antibodies against murine iNOS and pumpkin catalase, respectively. The valuable advice of Dr. Susana Puntarulo, School of Pharmacy and Biochemistry, University of Buenos Aires, in the spin-trapping EPR method used is appreciated. The EPR and CLSM analyses were carried out at the Centre of Scientific Instrumentation of the University of Granada and the Technical Services of the University of Jaén, respectively.
This work was supported by the Dirección General de Investigación, Ministry of Education and Science (grant no. PB98–0493–01), the European Union (contract no. HPRN–CT–2000–00094), and Junta de Andalucía (groups CVI 0192 and CVI 0286).
This article is dedicated to the loved and esteemed memory of Prof. Dr. Julio López-Gorgé, Estación Experimental del Zaidín, Consejo Superior de Investigaciones Científicas, Granada, who died of a stroke on June 7, 2004, at the age of 69.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042812.
References
- Aebi H (1984) Catalase in vitro. Methods Enzymol 105: 121–126 [DOI] [PubMed] [Google Scholar]
- Alderton WK, Cooper CE, Knowles RG (2001) Nitric oxide synthases: structure, function and inhibition. Biochem J 357: 593–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A, Graham I (2002) Plant Peroxisomes: Biochemistry, Cell Biology and Biotechnological Applications. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupiáñez JA, del Río LA (1999) Localization of nitric oxide synthase in plant peroxisomes. J Biol Chem 274: 36729–36733 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol 129: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP (1999) Role of active oxygen species and NO in plant defense responses. Curr Opin Plant Biol 2: 287–294 [DOI] [PubMed] [Google Scholar]
- Boucher JL, Genet A, Vadon S, Delaforge M, Mansuy D (1992) Formation of nitrogen oxides and citrulline upon oxidation of NW-hydroxyl-l-arginine by hemeproteins. Biochem Biophys Res Commun 184: 1158–1164 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quatities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brown GC (1995) Reversible binding and inhibition of catalase by nitric oxide. Eur J Biochem 232: 188–191 [DOI] [PubMed] [Google Scholar]
- Brunelli L, Yermilov V, Beckman JS (2001) Modulation of catalase peroxidatic and catalatic activity by nitric oxide. Free Radic Biol Med 30: 709–714 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V (1997) The molecular biology of leaf senescence. J Exp Bot 48: 181–199 [Google Scholar]
- Caro A, Puntarulo S (1999) Nitric oxide generation by soybean embryonic axes: possible effect on mitochondrial function. Free Radic Res 31: S205–S212 [DOI] [PubMed] [Google Scholar]
- Chandok MR, Ytterberg AJ, van Wijk KJ, Klessig DF (2003) The pathogen-inducible nitric oxide synthase (iNOS) in plants is a variant of the P protein of the glycine decarboxylase complex. Cell 113: 469–482 [DOI] [PubMed] [Google Scholar]
- Clark D, Durner J, Navarre DA, Klessig DF (2000) Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol Plant Microbe Interact 13: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Clarke A, Desikan R, Hurst RD, Hancock JT, Neill SJ (2000) NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J 24: 667–677 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Río LA (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6: 145–150 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Río LA (2004) Enzymatic sources of nitric oxide in plant cells: beyond one protein-one function. New Phytol 162: 246–248 [Google Scholar]
- Corpas FJ, Barroso JB, Sandalio LM, Distefano S, Palma JM, Lupiáñez JA, del Río LA (1998) A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem J 330: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Sandalio LM, Palma JM, Lupiáñez JA, del Río LA (1999) Peroxisomal NADP-dependent isocitrate dehydrogenase: characterization and activity regulation during natural senescence. Plant Physiol 121: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueto M, Hernández-Perea O, Martín R, Ventura ML, Rodrigo J, Lamas S, Golvano MP (1996) Presence of nitric oxide synthase activity in roots and nodules of Lupinus albus. FEBS Lett 398: 159–164 [DOI] [PubMed] [Google Scholar]
- Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, Harper JF (2003) Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol 132: 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bellis L, Picciarelli P, Pistelli L, Alpi A (1990) Localization of glyoxylate-cycle enzymes in peroxisomes of senescent leaves and green cotyledons. Planta 180: 435–439 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denicola A, Souza JM, Radi R, Lissi E (1996) Nitric oxide diffusion in membranes determined by fluorescence quenching. Arch Biochem Biophys 328: 208–212 [DOI] [PubMed] [Google Scholar]
- Desikan R, Griffiths R, Hancok J, Neill S (2002) A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 16314–16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distefano S, Palma JM, McCarthy I, del Río LA (1999) Proteolytic cleavage of plant proteins by peroxisomal endoproteases from senescent pea leaves. Planta 209: 308–313 [DOI] [PubMed] [Google Scholar]
- Dordas C, Hasinoff BB, Rivoal J, Hill RD (2004) Class 1 hemoglobins, nitrate and NO levels in anoxic maize cell suspension cultures. Planta 219: 66–72 [DOI] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuberger M, Rébeillé F (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Durner J, Klessig DF (1999) Nitric oxide as a signal in plants. Curr Opin Plant Biol 2: 369–374 [DOI] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban FJ, Jiménez A, Fernández AP, Delmoral ML, Sánchez-López AM, Hernández R, Garrosa M, Pedrosa JA, Rodrigo J, Peinado MA (2001) Neuronal nitric oxide synthase immunoreactivity in the guinea-pig liver: distribution and colocalization with neuropeptide Y and calcitonin gene-related peptide. Liver 21: 374–379 [DOI] [PubMed] [Google Scholar]
- Ferrer MA, Ros Barceló A (1999) Differential effects of nitric oxide on peroxidase and H2O2 production by the xylem of Zinnia elegans. Plant Cell Environ 22: 891–897 [Google Scholar]
- Foissner I, Wendehenne D, Langebartels C, Durner J (2000) In vivo imaging of an elicitor-induced nitric oxide burst in tobacco. Plant J 23: 817–824 [DOI] [PubMed] [Google Scholar]
- García-Mata C, Gay R, Sokolovski S, Hills A, Lamattina L, Blatt MR (2003) Nitric oxide regulates K+ and Cl- channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc Natl Acad Sci USA 100: 11116–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2001) Nitric oxide induce stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol 126: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2002) Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol 128: 790–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F-Q, Okamoto M, Crawford NM (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- Hemmens B, Mayer B (1998) Enzymology of nitric oxide synthases. Methods Mol Biol 100: 1–32 [DOI] [PubMed] [Google Scholar]
- Hock B, Beevers H (1966) Development and decline of the glyoxylate-cycle enzymes in watermelon seedlings (Citrulus vulgaris Schrad): effect of actinomycin and cycloheximide. Z Pflanzenphysiol 55: 405–414 [Google Scholar]
- Huang AHC, Trelease RN, Moore TS Jr (1983) Plant Peroxisomes. Academic Press, New York
- Huang J, Sommers EM, Kim-Shapiro DB, King SB (2002) Horseradish peroxidase catalyzed nitric oxide formation from hydroxyurea. J Am Chem Soc 124: 3473–3480 [DOI] [PubMed] [Google Scholar]
- Huang X, Rad U, Durner J (2002) Nitric oxide induces transcriptional activation of the nitric-oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215: 914–923 [DOI] [PubMed] [Google Scholar]
- Huang X, Stettmaier K, Michel C, Hutzler P, Mueller MJ, Durner J (2004) Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218: 938–946 [DOI] [PubMed] [Google Scholar]
- Hung KT, Kao CH (2003) Nitric oxide counteracts the senescence of rice leaves induced by abscisic acid. J Plant Physiol 160: 871–879 [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Lea PJ (2002) The role of peroxisomes in the integration of metabolism and evolutionary diversity of photosynthetic organisms. Phytochemistry 60: 651–674 [DOI] [PubMed] [Google Scholar]
- Ignarro LI (2002) Nitric oxide as a unique signaling molecule in the vascular system: a historical overview. J Physiol Pharmacol 53: 503–514 [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, Pastori GM, del Río LA, Sevilla F (1998) Role of the ascorbate-glutathione cycle of mitochondria and peroxisomes in the senescence of pea leaves. Plant Physiol 118: 1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi MS, Ponthier JL, Lancaster JR Jr (1999) Cellular antioxidant and pro-oxidant actions of nitric oxide. Free Radic Biol Med 27: 1357–1366 [DOI] [PubMed] [Google Scholar]
- Kanner J, Harel S, Granit R (1992) Nitric oxide, an inhibitor of lipid oxidation by lipoxygenase, cyclooxygenase and hemoglobin. Lipids 27: 46–49 [DOI] [PubMed] [Google Scholar]
- Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T (1998) Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins. Anal Chem 70: 2446–2453 [DOI] [PubMed] [Google Scholar]
- Kotake Y, Tanigawa T, Tanigawa M, Ueno Y, Allen DR, Lai C-S (1996) Continuous monitoring of cellular nitric oxide generation by spin trapping with an iron-dithiocarbamate complex. Biochim Biophys Acta 1289: 362–368 [DOI] [PubMed] [Google Scholar]
- Lamattina L, García-Mata C, Graziano M, Pagnussat G (2003) Nitric oxide: the versatility of an extensive signal molecule. Annu Rev Plant Biol 54: 109–136 [DOI] [PubMed] [Google Scholar]
- Leshem YY (1996) Nitric oxide in biological systems. Plant Growth Regul 18: 155–159 [Google Scholar]
- Leshem YY (2000) Nitric Oxide in Plants: Occurrence, Function and Use. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Leshem YY, Haramaty E (1996) The characterization and contrasting effects of the nitric oxide free radical in vegetative stress and senescence of Pisum sativum Linn. foliage. J Plant Physiol 148: 258–263 [Google Scholar]
- Leshem YY, Wills RBH, Veng-Va Ku V (1998) Evidence for the function of the free radical gas-nitric oxide (NO) as an endogenous maturation and senescence regulating factor in higher plants. Plant Physiol Biochem 36: 825–833 [Google Scholar]
- Lim PO, Woo HR, Nam HG (2003) Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci 8: 272–278 [DOI] [PubMed] [Google Scholar]
- López-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A (2000) Stress induces peroxisome biogenesis genes. EMBO J 19: 6770–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Huertas E, Corpas FJ, Sandalio LM, del Río LA (1999) Characterization of membrane polypeptides from pea leaf peroxisomes involved in superoxide radical generation. Biochem J 337: 531–536 [PMC free article] [PubMed] [Google Scholar]
- López-Huertas E, Sandalio LM, del Río LA (1995) Integral membrane polypeptides of pea leaf peroxisomes: characterization and response to plant stress. Plant Physiol Biochem 33: 295–302 [Google Scholar]
- Magalhaes JR, Pedroso MC, Durzan D (1999) Nitric oxide, apoptosis and plant stresses. Physiol Mol Biol Plants 5: 115–125 [Google Scholar]
- Minorsky PV (2002) Peroxisomes: organelles of diverse function. Plant Physiol 130: 517–518 [Google Scholar]
- Modolo LV, Cunha FQ, Braga MR, Salgado I (2002) Nitric oxide synthase-mediated phytoalexin accumulation in soybean cotyledons in response to the Diaporthe phaseolorum f. sp. meridionalis elicitor. Plant Physiol 130: 1288–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada S, Palmer RMJ, Higgs EA (1991) Nitric oxide: physiology, pathophysiology and phamarcology. Pharmacol Rev 43: 109–142 [PubMed] [Google Scholar]
- Murgia I, Delledonne M, Soave C (2002) Nitric oxide mediates iron-induced ferritin accumulation in Arabidopsis. Plant J 30: 521–528 [DOI] [PubMed] [Google Scholar]
- Nagano T (1999) Practical methods for detection of nitric oxide. Luminescence 14: 283–290 [DOI] [PubMed] [Google Scholar]
- Nagano T, Yoshimura T (2002) Bioimaging of nitric oxide. Chem Rev 102: 1235–1269 [DOI] [PubMed] [Google Scholar]
- Nakatsubo N, Kojima H, Kikuchi K, Nagoshi H, Hirata Y, Maeda D, Imai Y, Irimura T, Nagano T (1998) Direct evidence of nitric oxide production from bovine aortic endothelial cells using new fluorescence indicators: diaminofluoresceins. FEBS Lett 427: 263–266 [DOI] [PubMed] [Google Scholar]
- Navabpour S, Morris K, Allen R, Harrison E, A-H-Mackerness S, Buchanan-Wollaston V (2003) Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot 54: 2285–2292 [DOI] [PubMed] [Google Scholar]
- Navarre DA, Wendehenne D, Durner J, Noad R, Klessig DF (2000) Nitric oxide modulates the activity of tobacco aconitase. Plant Physiol 122: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT (2002. a) Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol 128: 13–16 [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT (2003) Nitric oxide signaling in plants. New Phytol 159: 11–35 [DOI] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT (2002. b) Hydrogen peroxide and nitric oxide as signaling molecules in plants. J Exp Bot 53: 1237–1247 [PubMed] [Google Scholar]
- Ninnemann H, Maier J (1996) Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem Photobiol 64: 393–398 [DOI] [PubMed] [Google Scholar]
- Orozco-Cárdenas ML, Ryan CA (2002) Nitric oxide negatively modulates wound signaling in tomato plants. Plant Physiol 130: 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Simontacchi M, Puntarulo S, Lamattina L (2002) Nitric oxide is required for root organogenesis. Plant Physiol 129: 954–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori GM, del Río LA (1997) Natural senescence of pea leaves: an activated oxygen-mediated function for peroxisomes. Plant Physiol 113: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso MC, Durzan DJ (2000) Effect of different gravity environments of DNA fragmentation and cell death in Kalanchoë leaves. Ann Bot (Lond) 86: 983–994 [DOI] [PubMed] [Google Scholar]
- Pedroso MC, Magalhaes JR, Durzan DJ (2000. a) A nitric oxide burst precedes apoptosis in angiosperm and gymnosperm callus cells and foliar tissues. J Exp Bot 51: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Pedroso MC, Magalhaes JR, Durzan DJ (2000. b) Nitric oxide induces cell death in Taxus cells. Plant Sci 157: 173–180 [DOI] [PubMed] [Google Scholar]
- Peinado MA, Torres MI, Thompson RP, Esteban FJ (2000) Immunolocalization of the HNK-1 epitope in the autonomic innervation to the liver and upper digestive tract of the developing rat embryo. Histochem J 32: 439–446 [DOI] [PubMed] [Google Scholar]
- Ribeiro EA, Cunha FQ, Tamashiro WMSC, Martins IS (1999) Growth phase-dependent subcellular localization of nitric oxide synthase in maize cells. FEBS Lett 445: 283–286 [DOI] [PubMed] [Google Scholar]
- del Río LA, Corpas FJ, Barroso JB (2004) Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry 65: 783–792 [DOI] [PubMed] [Google Scholar]
- del Río LA, Corpas FJ, Sandalio LM, Palma JM, Gómez M, Barroso JB (2002) Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J Exp Bot 53: 1255–1272 [PubMed] [Google Scholar]
- del Río LA, Fernández VM, Rupérez FL, Sandalio LM, Palma JM (1989) NADH induces the generation of superoxide radicals in leaf peroxisomes. Plant Physiol 89: 728–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río LA, Pastori GM, Palma JM, Sandalio LM, Sevilla F, Corpas FJ, Jiménez A, López-Huertas E, Hernández JA (1998) The activated oxygen role of peroxisomes in senescence. Plant Physiol 116: 1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53: 103–110 [PubMed] [Google Scholar]
- Romero-Puertas MC, McCarthy I, Sandalio LM, Palma JM, Corpas FJ, Gómez M, del Río LA (1999) Cadmium toxicity and oxidative metabolism of pea leaf peroxisomes. Free Radic Res 31: S25–S31 [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ (2001) The phloem as a conduit for inter-organ communication. Curr Opin Plant Biol 4: 202–209 [DOI] [PubMed] [Google Scholar]
- Sakuma S, Fujimoto Y, Sakamoto Y, Uchiyama T, Yoshioka K, Nishida H, Fujita T (1997) Peroxynitrite induces the conversion of xanthine dehydrogenase to oxidase in rabbit liver. Biochem Biophys Res Commun 230: 476–479 [DOI] [PubMed] [Google Scholar]
- Saviani EE, Orsi CH, Oliveira JFP, Pinto-Maglio CAF, Salgado I (2002) Participation of the mitochondrial permeability transition pore in nitric oxide induced plant cell death. FEBS Lett 510: 136–140 [DOI] [PubMed] [Google Scholar]
- Sharma VS, Traylor TG, Gardiner R, Mizukami H (1987) Reaction of nitric oxide with heme proteins and model compounds of hemoglobin. Biochemistry 26: 3837–3842 [DOI] [PubMed] [Google Scholar]
- Simontacchi M, Jasid S, Puntarulo S (2004) Nitric oxide generation during early germination of Sorghum seeds. Plant Sci 167: 839–847 [Google Scholar]
- Stöhr C, Strube F, Marx C, Ullrich WR, Rockel P (2001) A plasma membrane-bound enzyme of tobacco roots catalyses the formation of nitric oxide from nitrite. Planta 212: 835–841 [DOI] [PubMed] [Google Scholar]
- Stolz DB, Zamora R, Vodovotz Y, Loughran PA, Billiar TR, Kim YM, Simmons RL, Watkins SC (2002) Peroxisomal localization of inducible nitric oxide synthase in hepatocytes. Hepatology 36: 81–93 [DOI] [PubMed] [Google Scholar]
- Strother S (1988) The role of free radicals in leaf senescence. Gerontology 34: 151–156 [DOI] [PubMed] [Google Scholar]
- Tabak HF, Braakman I, Distel B (1999) Peroxisomes: simple in function but complex in maintenance. Trends Cell Biol 9: 447–453 [DOI] [PubMed] [Google Scholar]
- Thompson GA, Schulz A (1999) Macromolecular trafficking in the phloem. Trends Plant Sci 4: 354–360 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Froese CD, Madey E, Smith MD, Hong Y (1999) Lipid metabolism during plant senescence. Prog Lipid Res 37: 119–141 [DOI] [PubMed] [Google Scholar]
- Uttenthal LO, Alonso D, Fernández AP, Campbell RO, Moro MA, Leza JC, Lizasoain I, Esteban FJ, Barroso JB, Valderrama R, et al (1998) Neuronal and inducible nitric oxide synthase and nitrotyrosine immunoreactivities in the cerebral cortex of the aging rat. Microsc Res Tech 43: 75–88 [DOI] [PubMed] [Google Scholar]
- Walz C, Juenger M, Schad M, Kelr J (2002) Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J 31: 189–197 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Lamotte O, Pugin A (2003) Plant iNOS: conquest of the holy grail. Trends Plant Sci 8: 465–468 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J (2001) Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci 6: 177–183 [DOI] [PubMed] [Google Scholar]
- Xia Y, Cardounel AJ, Vanin AF, Zweier JL (2000) Electron paramagnetic resonance spectroscopy with N-methyl-D-glucamine dithiocarbamate iron complexes distinguishes nitric oxide and nitroxyl anion in a redox-dependent manner: applications in identifying nitrogen monoxide products from nitric oxide synthase. Free Radic Biol Med 29: 793–797 [DOI] [PubMed] [Google Scholar]
- Xia Y, Zweier JL (1997) Direct measurement of nitric oxide generation from nitric oxide synthase. Proc Natl Acad Sci USA 94: 12705–12710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi J, Nishimura M (1984) Purification of glyoxysomal catalase and immunochemical comparison of glyoxysomal and leaf peroxisomal catalase in germinating pumpkin cotyledons. Plant Physiol 262: 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y (2000) Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett 468: 89–92 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y, Takahashi S (1999) An alternative pathway for nitric oxide production in plants: new features of an old enzyme. Trends Plant Sci 4: 128–129 [DOI] [PubMed] [Google Scholar]
- Zhang C, Czymmek KJ, Shapiro AD (2003) Nitric oxide does not trigger early programmed cell death events but may contribute to cell-to-cell signaling governing progression of the Arabidopsis hypersensitive response. Mol Plant Microbe Interact 16: 962–972 [DOI] [PubMed] [Google Scholar]