Abstract
ZPT2-related proteins that have two canonical Cys-2/His-2-type zinc-finger motifs in their molecules are members of a family of plant transcription factors. To characterize the role of this type of protein, we analyzed the function of Arabidopsis L. Heynh. genes encoding four different ZPT2-related proteins (AZF1, AZF2, AZF3, and STZ). Gel-shift analysis showed that the AZFs and STZ bind to A(G/C)T repeats within an EP2 sequence, known as a target sequence of some petunia (Petunia hybrida) ZPT2 proteins. Transient expression analysis using synthetic green fluorescent protein fusion genes indicated that the AZFs and STZ are preferentially localized to the nucleus. These four ZPT2-related proteins were shown to act as transcriptional repressors that down-regulate the transactivation activity of other transcription factors. RNA gel-blot analysis showed that expression of AZF2 and STZ was strongly induced by dehydration, high-salt and cold stresses, and abscisic acid treatment. Histochemical analysis of β-glucuronidase activities driven by the AZF2 or STZ promoters revealed that both genes are induced in leaves rather than roots of rosette plants by the stresses. Transgenic Arabidopsis overexpressing STZ showed growth retardation and tolerance to drought stress. These results suggest that AZF2 and STZ function as transcriptional repressors to increase stress tolerance following growth retardation.
Drought, high salinity, and low temperature are adverse environmental conditions that limit the growth of plants. Plants respond and adapt to these stresses in order to survive. These stresses induce various biochemical and physiological changes, including growth inhibition, to acquire stress tolerance. A number of genes have been described that respond to stresses at the transcriptional level (Ingram and Bartels, 1996; Bray, 1997; Hasegawa et al., 2000; Zhu, 2002; Shinozaki et al., 2003). Although various genes are induced by these stresses, many stress-down-regulated genes are also reported (Seki et al., 2002). Analysis of stress-down-regulated as well as stress-up-regulated genes is important for understanding molecular responses to abiotic stresses. The plant hormone abscisic acid (ABA) is a key regulator mediating abiotic-stress-signaling pathways. ABA treatment also induces not only stress tolerance but also growth inhibition of plants.
Recently, cis- and trans-acting factors involved in stress-inducible gene expression have been analyzed extensively in Arabidopsis L. Heynh. (Zhu, 2002; Shinozaki et al., 2003). As positive cis-elements, dehydration-responsive element (DRE)/C-repeat, ABA-responsive element (ABRE), MYB recognition sequence (MYBRS), and MYC recognition sequence (MYCRS) were reported previously and are essential for the induction of some stress-responsive genes. Trans-acting factors, which specifically bind to these cis-elements and act as transcriptional activators, have been studied. DREB1/CBF and DREB2 are characterized as DRE-binding proteins, which contain an ethylene response factor (ERF)/AP2 DNA-binding domain (Stockinger et al., 1997; Liu et al., 1998). AREB/ABRE-binding factor and ABI5 are characterized as ABRE-binding proteins, which contain basic Leu zipper motifs (Choi et al., 2000; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Uno et al., 2000). AtMYB2 and AtMYC2 (rd22BP1) are MYB- and bHLH-related transcription factors that bind to MYBRS and MYCRS, respectively (Abe et al., 1997, 2003). All these transcription factors function as transcriptional activators in the expression of stress-inducible genes. Although understanding of the down-regulation of gene expression under stress conditions is also important for understanding molecular responses to abiotic stresses, little is known about cis- and trans-acting factors involved in repression of stress-down-regulated genes.
We have previously reported a gene family of Cys-2/His-2-type zinc-finger proteins in Arabidopsis (Sakamoto et al., 2000). Some members of this family were up-regulated by abiotic stress in Arabidopsis. The Cys-2/His-2-type zinc finger, also called the classical or TFIIIA-type finger, is one of the best-characterized DNA-binding motifs found in eukaryotic transcription factors. This motif is represented by the signature CX2-4CX3FX5LX2HX3-5H, consisting of about 30 amino acids and 2 pairs of conserved Cys and His bound tetrahedrally to a zinc ion (Pabo et al., 2001). In plants, many Cys-2/His-2-type zinc-finger proteins have the same structural features as each other, which are unique to plant zinc-finger proteins (Takatsuji, 1999). Most fingers have a six-amino acid stretch, QALGGH (underlined are the conserved Leu and His), at a position corresponding to the N-terminal part of the recognition helix. In multiple-fingered proteins, the adjacent fingers are separated by a long spacer that is highly variable in length and sequence from one protein to another. This is in striking contrast with animal and yeast proteins, which in general contain a cluster of multiple fingers separated by a short spacer (6–8 amino acids) known as an HC-link (Klug and Schwabe, 1995). Such structural features suggest that the mechanism for target recognition and consequently regulation of downstream genes may be different between plant and animal proteins. Because petunia (Petunia hybrida) ZPTs were the first reported among this type of protein, we call this zinc-finger motif the ZPT type.
Plant ZPT-type proteins have one to four fingers each and can be classified according to the number of fingers. Two-fingered protein genes, here called ZPT2-related genes, constitute the major class of the ZPT-type family genes and include 14 ZPT2 genes in petunia (Takatsuji et al., 1992, 1994; Kubo et al., 1998); WZF1 in wheat (Triticum aestivum; Sakamoto et al., 1993); STZ (Lippuner et al., 1996), five ZAT genes (Meissner and Michael, 1997), and RDH41 (Iida et al., 2000) in Arabidopsis; Pszf1 in pea (Pisum sativum; Michael et al., 1996); Mszpt2-1 in alfalfa (Medicago sativa; Frugier et al., 2000); and SCOF-1 in soybean (Glycine max; Kim et al., 2001).
To elucidate the functions of plant ZPT2-related proteins, we analyzed four Arabidopsis ZPT2-related genes, AZF1, AZF2, AZF3, and STZ, which had been cloned previously using PCR techniques (Sakamoto et al., 2000). STZ had already been cloned by complementation of the salt-sensitive phenotype of a yeast calcineurin mutant (Lippuner et al., 1996). In this study, we analyzed the role of these AZF and STZ proteins in transcriptional regulation in plants. All four proteins were localized in nuclei and bound to DNA in a sequence-specific manner. The binding site of these proteins functions as a negative cis-acting element, and all four proteins function as transcriptional repressors in plant cells. Among the four genes, AZF2 and STZ showed strong induction by various stresses and ABA. We analyzed transgenic plants overexpressing STZ and found growth inhibition. These transgenic plants also showed tolerance to drought stress. We discuss the function of AZF2 and STZ as transcriptional repressors under various stress conditions.
RESULTS
Targeting of the AZF and STZ Proteins to the Nucleus
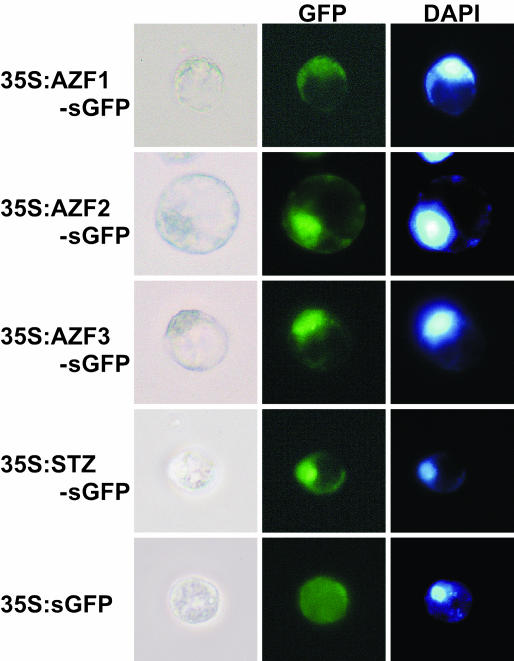
All of the putative AZF1, AZF2, AZF3, and STZ proteins contained a short stretch of basic amino acids near the N terminus. This region is conserved among most ZPT2-related proteins and might function as a potential nuclear localization signal. To investigate the nuclear localization of individual proteins, we performed an in vivo targeting experiment using the synthetic green fluorescent protein (sGFP) gene of the jellyfish Aequorea victoria (Chiu et al., 1996). The coding regions of the AZF and STZ cDNAs were fused in-frame to the sGFP gene, and the resulting constructs were introduced into protoplasts prepared from Arabidopsis T87 cultured cells by polyethylene glycol-mediated transformation. Localization of each fusion protein expressed transiently was determined by visualization with fluorescence microscopy. Each fusion protein was localized to the nuclei of the T87 cells (Fig. 1). The nuclei in the cells, made visible by 4′,6-diamino-phenylindole (DAPI) staining, corresponded to the region of sGFP fluorescence. By contrast, the control sGFP without AZF or STZ was distributed throughout the cells, thus indicating that AZFs and STZ are nuclear-localized proteins.
Figure 1.
Nuclear localization of the AZF and STZ proteins. Constructs carrying 35S:AZF-sGFP, 35S:STZ-sGFP, or 35S:sGFP were introduced into protoplasts prepared from Arabidopsis T87 cells by polyethylene glycol-mediated transformation. Transfected protoplasts were observed after 3 d by optical (left sections) or fluorescence microscopy (middle and right sections). Nuclei are shown with DAPI staining.
Specific Binding of AZFs and STZ to the A(G/C)T Repeat within the EP2 Sequence
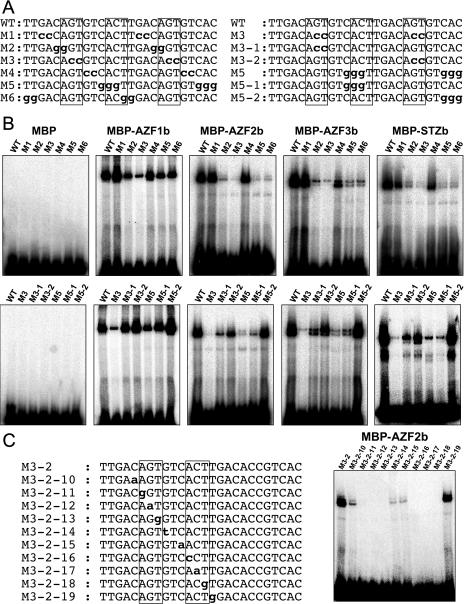
Since the AZFs and STZ contained the Cys-2/His-2-type zinc-finger motif that has been identified in many transcription factors as a DNA-binding motif, we observed whether they could have DNA-binding ability in vitro by gel-shift assay. We tested the EP2 sequence as a probe because it had already been reported that some of the petunia ZPT2 proteins bind to this probe in a sequence-specific manner (Takatsuji et al., 1992). EP2 is a 26-bp sequence composed of two 13-bp EP1S sequences repeated tandemly. EP1S was originally identified as a cis-element within the PESPS gene promoter in petunia (Takatsuji et al., 1992). Truncated AZF and STZ proteins including two canonical zinc-finger motifs were expressed as in-frame fusion proteins with maltose-binding protein (MBP) in Escherichia coli. All of the recombinant fusion proteins bound to the EP2 (wild-type) sequence (Fig. 2). The binding activity of each fusion protein was abolished by incubation with a high concentration (10 mm) of EDTA (data not shown), suggesting that all four recombinant proteins might bind to EP2 directly through their zinc-finger domains.
Figure 2.
Characterization of the DNA-binding affinity of the recombinant AZF and STZ proteins to the EP2 (26-bp) fragment. A, Upper-strand sequences of EP2 (wild-type) and its mutated fragments used as probes are shown with mutated bases indicated by bold letters. Boxes indicate A(G/C)T sequences. B, Gel-shift assays of sequence-specific binding to the EP2 of the recombinant AZF and STZ proteins. C, Effect of single-base substitutions within M3-2 on DNA-binding activity of AZF2. Upper-strand sequences of M3-2 and its mutated fragments used as probes are shown at left, with mutated bases indicated by bold letters. The A(G/C)T sequences necessary for binding are boxed.
To identify the target sequences within EP2 of the AZF and STZ proteins, we prepared a series of probes (M1–M6) by substitution of two or three bases at the same position in the tandemly repeated sequences (Fig. 2A). The binding ability of each fusion protein was examined by using these base-substituted probes (Fig. 2B). Each protein bound to M1 and M4 strongly but weakly to M2, M3, M5, and M6. Three petunia ZPT2-related proteins, ZPT2-1, ZPT2-2, and ZPT2-4, bind to two copies of the AGT core sequence separated by 10 bp in the EP2 sequence (Takatsuji et al., 1994). By the use of Southwestern assay, the wheat WZF1 protein has been shown to interact with a DNA fragment containing tandem copies of a CACTC sequence (ACT box; Sakamoto et al., 1993). As the ACT box has the AGT core sequence in the reverse orientation, we speculated on the importance of A(G/C)T in EP2 for the binding of the AZF and STZ proteins. The EP2 sequence contains three A(G/C)T sequences, two in the 13-bp EP1S sequence and the other at the junction of the tandem repeats. The M2, M3, M5, and M6 sequences lacked one or two of the three A(G/C)T sequences (Fig. 2A).
We prepared some more probes by base substitution to identify which A(G/C)T is necessary in order for the AZFs and STZ to bind to EP2. These probes, M3-1, M3-2, M5-1, and M5-2, had a substitution in one of the two parts of M3 or M5. All four proteins showed similar binding specificities as each other to the base-substituted fragments (Fig. 2B). All bound to the M3-1 and M3-2 probes, in which the first and third AGT, respectively, were disrupted, though the shifted bands were weaker than that of the wild-type EP2 fragment. On the other hand, the proteins did not bind to the M5-1 probe except MBP-AZF1b, in which the second ACT was disrupted, just as they did not bind to M5. The substitution in M5-2, which did not disrupt any A(G/C)T, had little effect on the binding specificities of the proteins. These results indicate that the second ACT is most important for the binding and that the existence of either the first or the third AGT is necessary for the binding of the proteins to EP2.
To confirm the importance of the A(G/C)T repeat sequences within EP2 for the binding of the AZFs and STZ, we used the AZF2 fusion protein and analyzed how it interacts with the M3-2 fragment. M3-2 retained the first AGT and second ACT but not the third ACT. M3-2-10 to M3-2-19 are the 1-base-substituted fragments of M3-2. The AGT and ACT repeats within M3-2 are important for AZF2 binding because AZF2 did not bind to M3-2-11, -12, -13, -16, -17, or -18 (Fig. 2C). Flanking sequences around the AGT and ACT are also important to AZF2 binding because the binding to M3-2-10, M3-2-14, and M3-2-15 was reduced in comparison with that to M3-2 (Fig. 2C). We expect that AZF2 interacts with the second ACT and third AGT of M3-1 directly in a similar way as it does with the first AGT and second ACT of M3-2. These results indicate that the binding of the AZF and STZ fusion proteins to the EP2 sequence is specific. Two pairs of the A(G/C)T repeats within the EP2 sequence seem to be the core target site for the AZF and STZ proteins, and sequences around the A(G/C)T repeats in this fragment might have an influence on binding.
Function of AZFs and STZ as Transcriptional Repressors in Arabidopsis
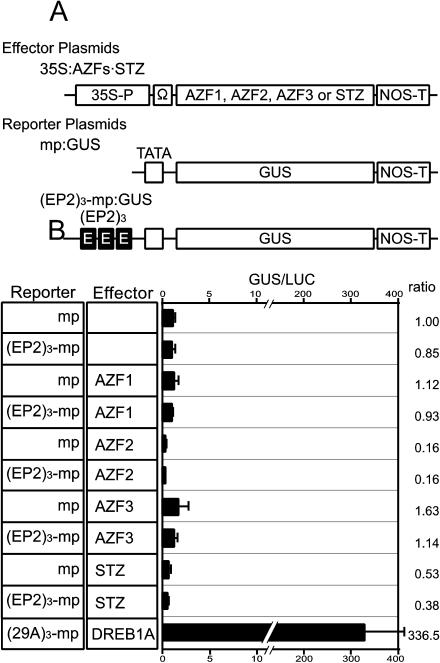
To determine whether the AZF and STZ proteins are capable of transactivating EP2-dependent transcription in plant cells, we performed transient analysis using protoplasts prepared from Arabidopsis leaves (Fig. 3). Protoplasts were cotransfected with a β-glucuronidase (GUS) reporter gene with or without the trimeric EP2 fragments and effector plasmids. The effector plasmids consisted of the cauliflower mosaic virus (CaMV) 35S promoter fused to the AZF or STZ cDNAs. Coexpression of the AZF and STZ proteins in protoplasts did not show any induction in the expression of the GUS reporter gene, with or without the EP2 sequences. The result suggests that the AZF and STZ proteins cannot function as transcription activators in the EP2-dependent gene expression.
Figure 3.
Lack of transactivation potential of AZFs and STZ. A, Schematic diagram of the effector and reporter constructs used in the transient assay. All effectors were driven by the CaMV 35S promoter. 35S-P, Ω, and NOS-T represent the CaMV 35S promoter, the translational enhancer of tobacco mosaic virus, and the polyadenylation signal of the nopaline synthase gene, respectively. The reporter gene (GUS) was driven by the rd29A minimal promoter (mp) with or without three repeats of EP2 [(EP2)3]. B, Transient assay using (EP2)3-mp:GUS, 35S:AZFs, and 35S:STZ. The reporter gene was transfected with each effector plasmid and the vector as control treatments. Each transfection contained 30 μg each of the reporter and effector plasmids. Transactivation of (29A)3-mp:GUS by DREB1A was used as a positive control (Liu et al., 1998). The error bar indicates the se of each set of replicates. Ratios indicate the relative amount of expression compared with the value obtained with the mp:GUS reporter only.
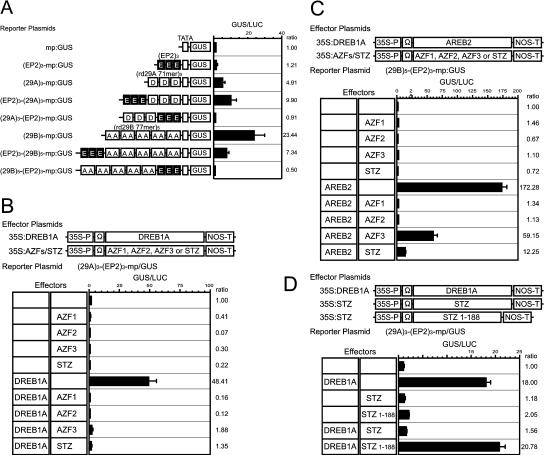
Then we examined whether EP2 can function as a cis-acting element in chimeric promoters by using transient analysis (Fig. 4A). To construct these promoters, we used the 71-bp and 77-bp fragments of the rd29A and rd29B promoters, respectively, in addition to EP2. Both fragments are characterized as Arabidopsis promoters containing positive cis-elements—DRE and ABRE—for dehydration-responsive gene expression (Yamaguchi-Shinozaki and Shinozaki, 1994; Uno et al., 2000). When three copies of the 71-bp fragment of the rd29A promoter or five copies of the 77-bp fragment of the rd29B promoter were directly fused to the rd29A minimum promoter, reporter activities were 5 and 23 times higher than that of the minimum promoter. When three copies of EP2 were inserted between these positive fragments and the minimum promoter in both the rd29A and rd29B fragments, the reporter activities were negated. When three copies of EP2 were fused upstream of these positive fragments, reporter activity was induced in the case of rd29A but reduced in the case of rd29B, in comparison with the positive fragments only. These results suggest that the EP2 sequence functions as a negative cis-element in Arabidopsis cells. However, the effect of EP2 as a negative cis-element in the promoter is influenced by the position relations between other cis-elements, and it may be stronger when EP2 is downstream of the other positive cis-element.
Figure 4.
Repression activity of AZFs and STZ. A, Function of the EP2 fragment as a negative cis-element in the chimeric promoter. The schematic diagrams of the reporter constructs used in the transient assay are shown at the left. To make the chimeric promoters, we used the 71-bp fragment of the rd29A promoter and the 77-bp fragment of the rd29B promoter in addition to the EP2 fragment. White boxes indicate the promoter fragments. Black boxes show EP2. D and A indicate DRE in the 71-bp fragment of the rd29A promoter and ABRE in the 77-bp fragment of the rd29B promoter, respectively, which were identified as positive cis-elements. The error bar indicates the se of each set of replicates (n = 5). Ratios indicate the relative amount of expression compared with the value obtained with the mp:GUS reporter. B, Negative effect of AZFs and STZ on DREB1A-mediated transactivation. The reporter plasmid was transfected with different sets of effector plasmids. To normalize for transfection efficiency, the CaMV 35S promoter-luciferase plasmid was cotransfected in each experiment. The error bar indicates the se of each set of replicates. Ratios indicate the relative amount of expression compared with the value obtained with the reporter only. C, Negative effect of AZFs and STZ on AREB2-mediated transactivation. D, Repressive effect of the DLN region of STZ. STZ or truncated STZ without the DLN region (STZ 1-188) was used as an effector plasmid.
Because EP2 was shown to be a negative cis-element, we speculated that the AZFs and STZ might act as transcriptional repressors mediated by binding to this element. Therefore, we analyzed the influences of coexpression of the AZF or STZ proteins on transactivation of transcriptional activators (Fig. 4, B and C). We used two different activators, DREB1A and AREB2. DREB1A and AREB2 transactivate via interaction with DRE and ABRE, respectively. The 71-bp fragment of the rd29A promoter contains one DRE, and the 77-bp fragment of the rd29B promoter contains two ABREs (Yamaguchi-Shinozaki and Shinozaki, 1994). Expression of DREB1A or AREB2 in protoplasts transactivated the expression of the GUS reporter gene driven by DRE and ABRE about 50 and 170 times, respectively. However, reduction of transactivation activities was observed in the cells in which the AZFs or STZ were coexpressed with these activators. In particular, the proteins effectively repressed the expression of the reporter gene transactivated by DREB1A. Thus, these results indicate that the AZFs and STZ function as transcriptional repressors via interaction with the EP2 sequence.
Previously, a fusion protein of STZ and the GAL4 DNA-binding domain was shown to function as an active repressor of transcription, and the DLN box in the C-terminal region was identified as a repression domain (Ohta et al., 2001). Therefore, we carried out a transient analysis using STZ without the DLN region (STZ 1-188) as an effector plasmid to show the repressive effect of the DLN region of STZ. The transactivation activity was reduced when STZ was coexpressed with DREB1A but not when STZ 1-188 was coexpressed with DREB1A (Fig. 4D).
Expression of the AZF and STZ Genes under Various Stress Conditions or Hormone Treatments
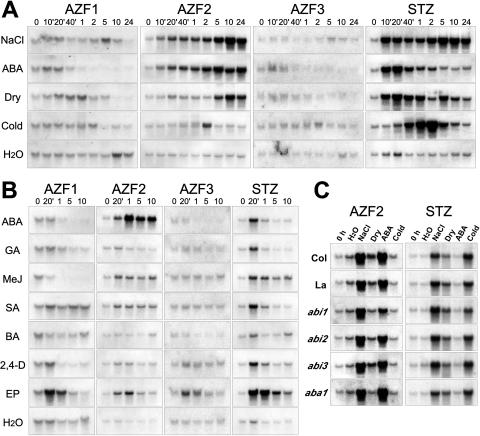
Expression patterns of the AZF and STZ genes were analyzed under various stress conditions by using RNA gel-blot analysis (Fig. 5A). Among the four genes, AZF2 and STZ exhibited strong induction by various stresses. Expression of both was induced by high salt and dehydration, and expression of STZ was induced also by cold. Expression of both was induced by ABA treatment. The level of AZF2 mRNA increased slowly and peaked within 24 h with high-salt, dehydration, and ABA treatments. On the other hand, the level of STZ mRNA increased rapidly and peaked within 10 min with high-salt stress. It decreased by 1 h and thereafter increased again by 24 h. With dehydration stress and ABA treatment, the level of STZ mRNA increased rapidly and peaked within 10 min, similar to that with high-salt stress, then it decreased slowly. The level of STZ mRNA by cold treatment increased from 20 min, peaked by 2 h, and thereafter decreased. By contrast, little expression of AZF1 and AZF3 was induced by the stresses. The expression levels of these genes were weaker than those of AZF2 and STZ. We analyzed the expression of AZFs and STZ by plant hormones other than ABA (Fig. 5B). AZF2 showed the strongest induction by ABA and a little induction by methyl jasmonate and salicylic acid. STZ showed similar induction by GA, methyl jasmonate, salicylic acid, and 2,4-dichlorophenoxyacetic acid to that by ABA. Expression of all four AZF and STZ genes was induced by ethephon treatment.
Figure 5.
RNA gel-blot analysis of expression of the AZF and STZ genes in different treatments. A, Expression of the AZF and STZ genes in response to high salt, ABA, dehydration, or cold. Each lane was loaded with 20 μg of total RNA from 3-week-old, unbolted Arabidopsis plants that had been transferred to hydroponic growth in 250 mm NaCl, transferred to hydroponic growth in 100 μm ABA, dehydrated (Dry), transferred to 4°C (Cold), or transferred to water. B, Expression of the AZF and STZ genes in response to plant hormones. Each lane was loaded with 20 μg of total RNA from 3-week-old, unbolted Arabidopsis plants that had been transferred to hydroponic growth in 100 μm ABA, 100 μm GA, 100 μm methyl jasmonate (MeJ), 100 μm salicylic acid (SA), 100 μm benzyladenine (BA), 100 μm 2,4-dichlorophenoxyacetic acid (2,4-D), or 1 mm ethephon (EP), or transferred to water. The number above each lane indicates the number of minutes or hours after the initiation of treatment before isolation of RNA. C, Effects of various stress treatments on the expression of AZF2 and STZ in the abi1, abi2, abi3, and aba1 mutants. Each lane was loaded with 20 μg of total RNA. Plants were treated for 5 h. Col, Columbia ecotype; La, Landsberg erecta ecotype.
Expression of both AZF2 and STZ was strongly induced by various stresses and ABA treatment. To analyze the role of ABA in the expression of these two genes, we used the abi1, abi2, abi3, and aba1 mutants of Arabidopsis (Fig. 5C). The aba mutant is deficient in ABA biosynthesis, and the three abi mutants have an impaired response to ABA. ABI1 and ABI2 encode PP2C-type protein phosphatases and are thought to be essential for negative regulation of ABA signaling in both vegetative tissues and seeds (Leung et al., 1994, 1997; Meyer et al., 1994). ABI3 encodes a homolog of the maize (Zea mays) VP-1 protein and a seed-specific transcription factor (Giraudat et al., 1992). Compared with the wild type, AZF2 induction in the aba1 and abi1 mutants was reduced by dehydration but not by NaCl stress. No changes of STZ expression between the wild-type and mutant plants were observed under any stress conditions. These results suggest that a part of the AZF2 expression induced by dehydration is induced via the ABA-dependent pathway and that this ABA-dependent expression is mediated by the ABI1 protein. However, the high-salt-responsive expression of AZF2 and the expression of STZ were mainly induced via an ABA-independent pathway.
Tissue-Specific Expression of the AZF and STZ Genes
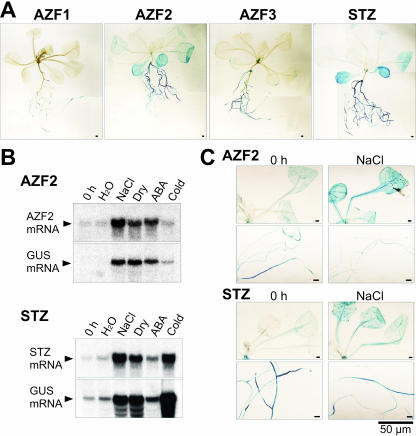
To study the tissue specificity of AZF and STZ expression, we constructed chimeric genes consisting of the AZF or STZ promoter regions fused to the GUS reporter gene and introduced them into Arabidopsis. We used a 1.6-kb DNA fragment for AZF1, 2.2-kb for AZF2, 1.3-kb for AZF3, and 2.6-kb for STZ upstream sequences to make these chimeric gene constructs. We examined the localization of the GUS reporter gene expression under the control of the AZF and STZ promoters in transgenic plants. Histochemical analysis of GUS activity in rosette plants revealed that all AZF and STZ genes were commonly expressed in roots but at different levels in each root tissue under normal growth conditions (Fig. 6A). Expression of AZF1 and AZF3 was restricted to roots at a low level, whereas expression of AZF2 and STZ was detected not only in roots but also in aerial parts. This result is in agreement with the pattern of RNA gel-blot analysis previously reported for STZ (Lippuner et al., 1996; Meissner and Michael, 1997; Sakamoto et al., 2000) or AZFs (Sakamoto et al., 2000).
Figure 6.
Histochemical localization of GUS activity and RNA gel-blot analysis in transgenic Arabidopsis plants. A, Histochemical analysis of GUS activity in transgenic Arabidopsis plants containing the AZF1, AZF2, AZF3, or STZ promoters fused to the GUS reporter gene. Three-week-old, unbolted, whole plants were stained. B, RNA gel-blot analysis of the AZF2 and STZ genes in transgenic Arabidopsis plants. Each lane was loaded with 20 μg of total RNA. Plants were untreated or treated with the stresses indicated for 5 h. C, Histochemical analysis of GUS activity in transgenic Arabidopsis plants containing the AZF2 or STZ promoters fused to the GUS reporter gene. Three-week-old unbolted plants were untreated or treated with 250 mm NaCl for 5 h.
To determine where AZF2 and STZ expression is induced under stress conditions, we analyzed the expression of these genes in transgenic Arabidopsis containing the promoter::GUS fusion genes. First, we checked the level of induction of the GUS gene driven by the AZF2 and STZ promoters by using RNA gel-blot analysis (Fig. 6B). The expression patterns of the GUS gene under stress conditions were almost the same as those of the endogenous AZF2 and STZ genes. This result confirms that both the AZF2 and STZ promoters used here contain cis-acting elements involved in stress-responsive gene expression. Then we determined the spatial pattern of AZF2 and STZ expression under stress conditions by using GUS staining (Fig. 6C). Both AZF2 and STZ were up-regulated mainly in rosette leaves of salt-treated plants.
Effect of Overexpression of the STZ Gene in Transgenic Plants
To examine the role of STZ and AZF2 in plants under stress conditions, we tried to generate transgenic Arabidopsis plants that constitutively overexpress AZF2 or STZ under the control of the CaMV 35S promoter. The transformation efficiency of these plants was very low, and we repeated the transformation again and again. Finally, we generated 49 lines of AZF2 transformants and 26 lines of STZ. Transgenic plant lines overexpressing AZF2 or STZ were selected by RNA gel-blot analysis. However, no AZF2 overexpressors were obtained, and STZ was overexpressed in only two independent transgenic lines.
The growth of the STZ transgenic plants on germination medium (GM) agar plates or on soil was compared with that of the wild-type plants at 2 or 5 weeks, respectively, after sowing. Both transgenic lines on both GM agar plates and soil showed growth retardation compared with the wild-type plants (Fig. 7, A and B), and the level of the growth retardation was correlated with that of STZ expression in the transgenic plants (Fig. 7C).
Figure 7.
Phenotypes and drought-stress tolerance of the 35S::STZ and wild-type plants. A, Transgenic and wild-type plants grown on agar plates for 21 d. We used plants transformed with the vector pBI121 as controls. B, Expression of the STZ gene in the transgenic and wild-type plants. Each lane was loaded with 20 μg of total RNA from 3-week-old Arabidopsis plants. C, Plants grown in soil pots for 36 d. D, Drought-stress tolerance of the transgenic and control plants. Both 35S::STZa and 35S::STZb plants were highly tolerant to drought stress (P < 0.001; χ2 test). Number codes = number of surviving plants out of total number. E, Difference in recovery after rehydration among the wild-type, 35S::STZa, and 35S::STZb plants. Photographs show the plants dehydrated on dry plastic plates in air for 4 h and then rehydrated overnight. F, Electrolyte leakage was evaluated after dehydration treatment. A 17-d-old plant was used in each experiment. Plants were removed from the agar plates and dehydrated on dry plastic plates for 4 h. The values are means of 15 independent samples. Statistical significance compared with the value of the control plants was determined by Welch's test (P < 0.005).
To examine whether overexpression of STZ affects the tolerance to drought stress, we grew the wild-type and transgenic plants in pots for 3 weeks and then left them unwatered for 2 weeks. Both STZ transgenic lines showed growth retardation, but these 3-week-old soil plants did not show big difference between the wild-type (pBI121) and STZ transgenic plants. We measured the soil water contents for all three lines during the drought stress experiment and found that they are not so different among three plans (data not shown). Almost all the wild-type plants died within this 2-week period, whereas nearly all the transgenic plants of both STZ lines survived this level of drought stress and continued to grow when rewatered (Fig. 7D). We also tried to explore the differences in recovery after desiccation using plants grown on the agar plates. Seventeen-day-old wild-type and transgenic plants were removed from the agar plates and were kept on plastic plates for 4 h (20% ± 10% relative humidity) followed by rehydration overnight. The wild-type plants had wilted and crinkled leaves, whereas the transgenic plants had green leaves that were spread out and appeared healthier as shown in Figure 7E. Then, we calculated the survival rates of the wild-type and transgenic plants. Only 5.5% of the wild-type plants (pBI121) survived, whereas 88.9% and 63.9% of the 35S:STZa and 35S:STZb survived, respectively (Fig. 7E).
The leakage of electrolytes is a sensitive measure of loss-of-membrane integrity and it is commonly used to assay osmolality injury (Gilmour et al., 1998; Vannini et al., 2004). Therefore, we also analyzed the stress tolerance of these transgenic plants using the ion leakage test. When plants were dehydrated for 4 h, the ion leakage of the wild-type plants was 88.1%, whereas for the 35S:STZa and 35S:STZb, the ion leakage was about 48.0% and 63.4%, respectively (Fig. 7F). These results indicate that both STZ transgenic lines clearly showed higher tolerance to drought stress than control plants.
DISCUSSION
We characterized four Arabidopsis genes, AZF1, AZF2, AZF3, and STZ, encoding plant-specific transcription factors with two Cys-2/His-2-type zinc-finger motifs (ZPT2-related proteins). The ZPT2-related proteins form a relatively large family of transcription factors in plants (Takatsuji, 1999). We searched the completed Arabidopsis genome sequence and found 18 genes encoding ZPT2-related proteins. These 18 genes were divided into two groups on the basis of similarities in their DNA-binding domains. The first group contains 6 genes, including AZF1, AZF2, AZF3, and STZ, and the second group contains the other 12 genes. The six genes of the first group have high homology not only in their DNA-binding domains but also in their N-terminal basic regions (B box); have regions consisting of three acidic residues followed by hydrophobic residues rich in Leu (L box); and have C-terminal short hydrophobic regions containing DLNL sequences (DLN box). The B box is a potential nuclear localization signal, and the L box and DLN box are thought to play roles in protein-protein interactions or in maintaining the folded structure (Sakamoto et al., 2000). All six genes may function similarly as transcription factors in Arabidopsis plants.
As the zinc-finger motifs of the AZFs and STZ have high structural similarity to those of the ZPT2-related proteins, we thought that these proteins would bind two tandemly repeated AGT cores separated by 10 bp in the EP2 sequence (Takatsuji et al., 1994). These proteins bound to the EP2 sequence, but they showed different DNA-binding preferences from that of petunia ZPT2. All four proteins recognized AGT and ACT cores separated by 3 bp and ACT and AGT cores separated by 4 bp in the EP2 sequence; the consensus sequence was A(G/C)T-X3-4-A(G/C)T. By contrast, ZPT2-1 and ZPT2-5 recognized two tandemly repeated AGT cores separated by 10 bp (Takatsuji et al., 1994). These observations led us to speculate that each ZPT-type zinc-finger domain recognizes tandemly repeated A(G/C)T core sequences and that the spacing between each pair of A(G/C)T may be different among the ZPT2-related proteins. The optimum spacing of the binding sites for these AZFs and STZ was shorter than that of the petunia ZPT2s.
Using transient analysis we showed that the EP2 sequence is a negative cis-element, and the AZFs and STZ act as transcriptional repressors mediated by binding to this negative cis-element in Arabidopsis protoplasts. Ohta et al. (2001) reported that fusion proteins of the GAL4 DNA-binding domain and STZ/ZAT10 or ZAT11 were shown to function as active repressors of transcription and identified the DLN box in the C-terminal region as a repression domain. Initially, they reported that class II ERFs are active repressors of transcription and that conserved C-terminal regions of these proteins function as repression domains. These conserved regions contain the sequence motif L/FDLNL/F(X)P and were designated ERF-associated amphiphilic repression motifs. Recently it was shown that chimeric proteins that include the ERF-associated amphiphilic repression motif act as dominant repressors (Hiratsu et al., 2003). The DLN box in STZ/ZAT10 also functions as an ERF-associated amphiphilic repression motif, and the DLN box in AZF1, AZF2, and AZF3 may have a similar function in transcriptional repression.
Using RNA gel-blot analysis, we showed that AZF2 and STZ were clearly induced at the transcription level by abiotic stresses such as drought, cold, and high salt. These genes were also induced by ABA. These results indicate that these proteins function under abiotic stress conditions. The expression of all four AZF and STZ genes was induced by ethephon. Ethylene is one of the key regulators that mediate a plant's response to biotic and abiotic stresses, such as pathogen infection, wounding, and UV irradiation (Alonso and Ecker, 2001). This indicates that there is cross-talk between ABA and ethylene-signaling pathways under abiotic and biotic stress conditions. These AZF and STZ proteins may play an important role under abiotic and biotic stress conditions.
Both AZF2 and STZ bound to the same cis-acting element, the A(G/C)T repeat, and repressed gene expression, but the different manner of response to each stress suggested that these two proteins function in different signal transduction pathways under stress conditions. Analysis of gene expression in transgenic Arabidopsis plants containing the promoter::GUS fusion genes showed that both the AZF2 and STZ promoter regions are sufficient for drought-stress-responsive gene expression. We searched both promoters to find some putative cis-acting elements involved in gene expression associated with drought, high-salt, and cold stresses. The cis-acting elements we found—DRE (CCGAC), ABRE (RACGTGGC), MYCRS (CANNTG), and MYBRS (RAACYR)—are well known (Shinozaki et al., 2003). The AZF2 promoter region contained all four elements, whereas STZ contained DRE (CCGAC), MYCRS (CANNTG), and MYBRS (RAACYR). We found some notable ABRE-related sequences in the AZF2 promoter. ABRE functions as a cis-acting element for the expression of ABA-inducible genes. Expression of AZF2 was strongly induced by ABA treatment, and induction of AZF2 under drought stress was shown by using ABA mutants to be mediated by ABA (Fig. 5). Expression of AZF2 may be regulated by ABA under drought and high-salt stress conditions. Recently, we analyzed transgenic plants overexpressing DREB1A and found that STZ was overexpressed in them, indicating that STZ is one of the downstream genes of DREB1A (Maruyama et al., 2004). DRE in the STZ promoter may function as a cis-acting element for its expression under drought, high-salt, and cold stress conditions.
We also found many MYBRS and MYCRS in both STZ and AZF2 promoters. In plants, typical examples of the cooperation of MYB and MYC proteins, which bind to MYBRS and MYCRS respectively, are maize C1 and R for anthocyanin biosynthesis (Tuerck and Fromm, 1994). AtMYB2 and AtMYC2 (rd29BP1) cooperatively regulate the induction of the rd29A gene under drought and high-salt stress conditions (Abe et al., 1997, 2003). The promoter regions of MYBRS and MYCRS may also function as cis-acting elements under drought and high-salt conditions. The AZF2 and STZ promoters also contain some A(G/C)T repeats that contain suitable bases between the A(G/C)T repeats for the binding of AZFs and STZ. This means that expression of AZF2 and STZ might be repressed by ZPT2-related proteins. The expression of STZ showed a transient increase under drought stress, so STZ might self-regulate. Similarly, WZF1 bound to its own promoter and repressed its own gene (Sakamoto et al., 1996).
For further understanding of the function of AZF2 and STZ under abiotic stress conditions, we tried to generate transgenic Arabidopsis plants overexpressing AZF2 and STZ under the control of the constitutive CaMV 35S promoter. However, transformants were rare. Moreover, in almost all of the obtained transformants, the expression of transgenes was not enhanced (data not shown). These results suggest that overexpression or ectopic expression of each of AZF2 and STZ causes serious damage to plant development or growth. We obtained two independent lines of STZ overexpressors, but we could not get any overexpressors for AZF2. This may be due to stronger repression activity of AZF2 than of STZ, as shown in the transient assay using Arabidopsis protoplasts.
As both independent lines of the STZ overexpressors showed growth retardation and tolerance to drought stress, the target down-regulated genes might promote plant tolerance and inhibit plant growth. Because the expression of AZF2 was also induced by drought stress, AZF2 might regulate similar target genes to those of STZ. By using microarray analysis, we have shown that many photosynthesis-related genes and genes for carbohydrate metabolism are down-regulated under drought, high-salt, and cold stress conditions (Seki et al., 2002). Under stress conditions, the energy balance changes and photosynthesis is reduced in plants (Huner et al., 1998). The expression of certain photosynthesis-related genes and genes for carbohydrate metabolism would become unnecessary and be reduced at the transcription level under stress conditions. Reduction of these proteins may lead to a suitable energy balance for plants under stress conditions. Therefore, plant growth is inhibited and stress tolerance is increased. These photosynthesis-related genes and genes for carbohydrate metabolism may be target stress-down-regulated genes of STZ and AZF2, and reduction of these proteins may increase stress tolerance in the 35S:STZ transgenic plants. Histochemical analysis in transgenic plants containing the promoter::GUS fusion genes showed that the expression of both AZF2 and STZ was induced mainly in leaves under drought stress. These facts support our hypothesis, as photosynthesis occurs in leaves.
Independently, we tried to generate transgenic Arabidopsis plants that overexpress AZF1 and AZF3 under the control of the CaMV 35S promoter. However, the transformation efficiency of these plants was very low and we could not obtain any transgenic plants overexpressing AZF1 or AZF3 like AZF2. Both AZF1 and AZF3 were expressed specifically in roots at a low level under the control condition and were induced only weakly by stress. As both AZF1 and AZF3 bind to the same cis-acting element as that of STZ and function as repressors, these two proteins may repress the expression of some specific genes in roots such as some kinds of photosynthesis-related genes or genes for carbohydrate metabolism. We are now generating transgenic plants overexpressing AZF1 and AZF3 using a chemical induction system for transcription. The result of this experiment will show us the real target genes of AZF1 and AZF3, and the function of these proteins will be elucidated in roots.
Plant transcription factors were classified on the basis of the conserved DNA-binding domains (Riechman et al., 2000). Interestingly, members of the same family often function in common aspects in the plant life cycle. We have previously described how a subset of the ZPT2-related protein subfamily might be involved in the water stress response at the level of transcriptional regulation (Sakamoto et al., 2000). We suspect that there is a common function of many of these proteins under the control of not only water stress but also other environmental stresses. In petunia, ZPT2-2 was induced by low temperature and dehydration, wounding, and UV-B (van der Krol et al., 1999). In soybean, SCOF-1 was shown to function under low temperature (Kim et al., 2001). In wheat, WZF1 was induced by high salt (H. Sakamoto, H. Araki, T. Meshi, and M. Iwabuchi, unpublished data). Among genes inducible by other than water stress, Arabidopsis RHL41 was induced by high light irradiation (Iida et al., 2000), and alfalfa Mszpt2-1 was induced during nodulation after bacterial inversion (Frugier et al., 2000). Further elucidation of the roles of this gene family in relation to plant stress adaptation, including water stress, will show us a new way of improving plant tolerance to environment stress conditions.
MATERIALS AND METHODS
Plant Materials and Treatments
Plants (Arabidopsis L. Heynh. ecotype Columbia or Landsberg erecta) were grown on GM agar plates for 3 weeks, as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). Dehydration, high-salt, and cold stress treatments and treatment with ABA were performed as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). The plants were subjected to the stress treatments for various periods and then frozen in liquid nitrogen for RNA gel-blot analysis or stained for GUS histochemical assay.
Subcellular Localization Analysis of Transiently Expressed Fusion Proteins
The termination codon of each AZF and STZ coding region was removed with the following primers: 5′-CGGGATCCGAAATCACATCTCACAG-3′ plus 5′-CGGGATCCGAAGTCGTCACTGAGAC-3′ for AZF1, 5′-GCTCTAGAATGGCCCTCGAAGCGATGAAC-3′ plus 5′-CGGATCCAAGATAAATCTTCTTTCTTGATGACTTGG-3′ for AZF2, 5′-GCTCTAGATTTTCTATAGCAATGGCGC-3′ plus 5′-GCTCTAGATTCAGGCGAGGCTTCTTA-3′ for AZF3, and 5′-CGGGATCCCTCAGAATCTTTAACTT-3′ plus 5′-CGGGATCCAGTTGAAGTTTGACCGG-3′ for STZ.
The PCR-amplified fragments were cut with BamHI (AZF1 and STZ), BamHI and XbaI (AZF2), or XbaI (AZF3) and filled in 35S-sGFP to fuse in frame to the GFP gene (Nakashima et al., 1998). Suspension-cultured cells of Arabidopsis (T87) were maintained in Jouanneau and Péaud-Lenoël medium (Axelos et al., 1992) under continuous illumination. Protoplast preparation and polyethylene glycol-mediated transformation were performed essentially according to the method described by Takeuchi et al. (2000). Protoplasts were transfected with 20 μg of the plasmid DNA. Transformed protoplasts were incubated at 22°C in the dark for 2 to 4 d to allow accumulation of sGFP or sGFP fusion proteins. For the nuclear staining, DAPI was added to the culture medium at a rate of 1 μg μL−1, and the mixture was incubated for 10 min.
Preparation of Recombinant Proteins and Gel Mobility Shift Assay
Fragments encoding the truncated proteins containing the two canonical zinc-finger motifs of each AZF and STZ were PCR amplified with the following primers: 5′-CGGGATCCTCACCGTCCGATCACCGAG-3′ plus 5′-GCTCTAGATACCGTTGTTGCCACCGTC-3′ for AZF1, 5′-GCTCTAGAACGCCGCCGCCAGAATCAAAG-3′ plus 5′-GCTCTAGAGGTTGCCTTCGTAGTGACAAC-3′ for AZF2, 5′-CGGGATCCACGGTTGCGGAGAAGCCG-3′ plus 5′-GCTCTAGAAGTTCGAAACGCCACCATC-3′ for AZF3, and 5′-CGGGATCCCCGGCGGTGGAGAAGTTG-3′ plus 5′-GCTCTAGATGTTGTTGTTTCCTTCGT-3′ for STZ. The PCR fragments were cut with BamHI (AZF1, AZF3, and STZ) or XbaI (AZF2) and filled in pMAL-c2 (New England Biolabs, Beverly, MA). The resulting plasmids were transformed to Escherichia coli BL21-Gold. Production and purification of the MBP and gel-shift assays were performed essentially according to the method described by Yoshioka et al. (2001). DNA-binding reactions were carried out in 25 mm HEPES-KOH, pH 7.6, 40 mm KCl, 0.1% Nonidet P-40, 0.01 mm ZnCl2, 10 μg/mL poly(dIdC), and 0.1 mm dithiothreitol (Yoshioka et al., 2001). After incubation for 20 min at room temperature, the mixtures were subjected to electrophoresis in a 0.7% agarose/3% polyacrylamide gel. The 26-bp fragments with or without base substitutions were labeled with 32P-dCTP, as described previously (Urao et al., 1993).
Transient Expression Assay
The 35S:AZFs/STZ effector plasmids were constructed with DNA fragments containing each AZF or STZ coding region cloned into the SmaI site of the plant expression vector pBI35SΩ. The 35S:DREB1A and 35S:AREB2 genes were constructed as described (Liu et al., 1998; Uno et al., 2000). The pBI35SΩ vector was constructed as described (Abe et al., 1997). To construct reporter plasmids, we used the mp:GUS vector, in which the 35S promoter of pBI221 was replaced with the rd29A minimal TATA promoter (Liu et al., 1998). The (29A)3-mp:GUS, (29B)5-mp:GUS, and (EP2)3-mp:GUS genes were created by ligation of (29A)3, (29B)5, and (EP2)3 fragments, respectively, into the HindIII site of mp:GUS. The (EP2)3-(29A)3-mp:GUS, (29A)3-(EP2)3-mp:GUS, (EP2)3-(29B)3-mp:GUS, and (29B)3-(EP2)3-mp:GUS genes were created by ligation of the blunt-ended fragments of (29B)3-(EP2)3, (29B)3-(EP2)3, (29B)3-(EP2)3, and (29B)3-(EP2)3, respectively, into the blunt-ended HindIII site of mp:GUS. The codes (29A)3 and (29B)5 mean the 71-bp fragment of the rd29A promoter with three tandem copies and the 77-bp fragment of the rd29B promoter with five tandem copies, respectively (Liu et al., 1998; Uno et al., 2000). (EP2)3 means the 26-bp EP2 sequence with three tandem copies. To equalize the total amount of plasmids at each transfection, we used the pBI35SΩ vector as a blank plasmid. In each experiment, 20 μg of the CaMV35S promoter-luciferase plasmid, 35S::LUC, was cotransfected to normalize for transfection efficiency (Abe et al., 1997). Isolation of Arabidopsis mesophyll protoplasts and polyethylene glycol-mediated DNA transfection were performed as described (Abe et al., 1997).
RNA Gel-Blot Analysis
RNA gel-blot hybridization was performed as described (Yamaguchi-Shinozaki and Shinozaki, 1994). Gene-specific probes for each AZF and STZ were PCR amplified with the following degenerate primers: 5′-CT(C/A)GCTCTTTG(T/C)CTCCT(C/T)ATGCTCGCTCG-3′ plus 5′-(T/C)TT(A/G)TGGCCGCCGAGGGC(T/C)TG-3′, as described (Sakamoto et al., 2000). The DNA fragment corresponding to the coding region was used as a probe for the GUS gene.
Histochemical Localization Assay for GUS Activity
The fragments containing the upstream region of each of the AZF and STZ genes were PCR-amplified with the following primers: 5′-ACGCGTCGACATAAGTTGCATAACGACAGC-3′ plus 5′-CGGGATCCAAGATTTAATTCTGTGAGATG-3′ for AZF1, 5′-ACGCGTCGACGATTCCATAGCCGTCACAGTG-3′ plus 5′-CGGGATCCGATCAGATGAATCTTCTTCTA-3′ for AZF2, 5′-ACGCGTCGACAGGTCCCCCTTCCGCTTGTGA-3′ plus 5′-CGGGATCCCTTCAAGCGCCATTGCTATAG-3′ for AZF3, and 5′-CCCAAGCTTGCATGCACACACAGAGGAGAG-3′ plus 5′-CGGGATCCTAAGTTAAAGATTCTGAGG-3′ for STZ. The resulting fragments were cut with SalI and BamHI (AZF1, AZF2, and AZF3) or HindIII and BamHI (STZ) and filled in pBI101 (CLONTECH, Palo Alto, CA). These constructs were used to transform Arabidopsis plants by using the vacuum infiltration method with Agrobacterium, as described by Bechtold and Pelletier (1998). Mature T3 seeds were used for subsequent experiments. Histochemical localization of GUS activities in the transgenic plants was performed by incubating the plants in 5-bromo-4-chloro-3-indolyl glucuronide (X-gluc) buffer (50 mm sodium phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% v/v Triton X-100, 2% v/v DMSO, 0.5 mm potassium ferrocyanide, 2 mg mL−1 X-gluc) at 37°C for 6 to 12 h.
Transgenic Plants Overexpressing the STZ Gene
To construct the 35S::STZ plasmid, the SmaI fragment of the STZ coding region was cloned into the SmaI site of a binary vector, pBE2113Not, which was constructed as described (Liu et al., 1998). The resulting plasmid was used to transform Arabidopsis plants as above. Mature T2 seeds were used for subsequent experiments.
Drought-Stress Tolerance of Transgenic Plants
Arabidopsis plants were germinated and grown on GM plates containing 20 mg L−1 kanamycin for 10 d. The plants were transferred to 9-cm pots filled with 1:1 perlite:vermiculite. They were grown under continuous illumination of approximately 2,500 lux at 22°C. Drought stress was imposed by withholding water for 2 weeks. They were photographed, and the numbers of plants that survived and continued to grow were counted. The statistical significance of the values was determined by using the χ2 test.
Acknowledgments
We thank Drs. H. Takatsuji and K. Yoshioka for their helpful advice on the gel-shift assay. We thank Dr. M.M. Parvez for critically reading this manuscript. We are also grateful to Mss. E. Ohgawara, K. Murai, M. Yamamoto, and F. Saito for their excellent technical assistance.
This work was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences and by a project grant from the Ministry of Agriculture, Forestry and Fisheries, Japan.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046599.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Ecker JR (2001) The ethylene pathway: a paradigm for plant hormone signaling and interaction. Sci STKE 70:RE1. [DOI] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplast isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bray EA (1997) Plant response to water deficit. Trends Plant Sci 2: 48–64 [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Choi H-I, Hong J-H, Ha J-O, Kang J-Y, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier F, Poirier S, Satiat-Jeunemaitre B, Kondorosi A, Crespi M (2000) A Kruppel-like zinc finger protein is involved in nitrogen-fixing root nodule organogenesis. Genes Dev 14: 475–482 [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ (1998) Low-temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced cor gene-expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan AB, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Huner NPA, Oquist G, Sarhan F (1998) Energy balance and acclimation to light and cold. Trends Plant Sci 3: 224–230 [Google Scholar]
- Iida A, Kazuoka T, Torikai S, Kikuchi H, Oeda K (2000) A zinc finger protein RHL41 mediates the light acclimation response in Arabidopsis. Plant J 24: 191–203 [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Kim JC, Lee SH, Cheong YH, Yoo C-M, Lee SI, Chun HJ, Yun D-J, Hong JC, Lee SY, Lim CO, et al (2001) A novel cold-inducible zinc-finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J 25: 247–259 [DOI] [PubMed] [Google Scholar]
- Klug A, Schwabe JW (1995) Protein motifs 5. Zinc fingers. FASEB J 9: 597–604 [PubMed] [Google Scholar]
- Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Nishino T, Takatsuji H (1998) Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res 26: 608–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V, Cyert MS, Gasser CS (1996) Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J Biol Chem 271: 12859–12866 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Meissner R, Michael AJ (1997) Isolation and characterisation of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs. Plant Mol Biol 33: 615–624 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Michael AJ, Hofer JMI, Ellis THN (1996) Isolation by PCR of a cDNA clone from pea petals with similarity to petunia and wheat zinc finger proteins. Plant Mol Biol 30: 1051–1058 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol 118: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo CO, Peisach E, Grant RA (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 70: 313–340 [DOI] [PubMed] [Google Scholar]
- Riechman JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2109 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Minami M, Huh GH, Iwabuchi M (1993) The putative zinc-finger protein WZF1 interacts with a cis-acting element of wheat histone genes. Eur J Biochem 217: 1049–1056 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Omirulleh S, Nakayama T, Iwabuchi M (1996) A zinc-finger-type transcription factor WZF-1 that binds to a novel cis-acting element of histone gene promoters represses its own promoter. Plant Cell Physiol 37: 557–562 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Araki T, Meshi T, Iwabuchi M (2000) Expression of a subset of the Arabidopsis Cys(2)/His(2)-type zinc-finger protein gene family under water stress. Gene 248: 23–32 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H (1999) Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol 39: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Takatsuji H, Mori M, Benfey PN, Ren L, Chua N-H (1992) Characterization of a zinc finger DNA-binding protein expressed specifically in Petunia petals and seedlings. EMBO J 11: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H, Nakamura N, Katsumoto Y (1994) A new family of zinc finger proteins in petunia: structure, DNA sequence recognition, and floral organ-specific expression. Plant Cell 6: 947–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A (2000) A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 23: 517–525 [DOI] [PubMed] [Google Scholar]
- Tuerck JA, Fromm ME (1994) Elements of the maize A1 promoter required for transactivation by the anthocyanin B/C1 or phlobaphene P regulatory genes. Plant Cell 6: 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37: 115–127 [DOI] [PubMed] [Google Scholar]
- van der Krol AR, van Poecke RM, Vorst OF, Voogt C, van Leeuwen W, Borst-Vrensen TW, Takatsuji H, van der Plas LH (1999) Developmental and wound-, cold-, desiccation-, ultraviolet-B-stress-induced modulations in the expression of the petunia zinc finger transcription factor gene ZPT2-2. Plant Physiol 121: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Fukushima S, Yamazaki T, Yoshida M, Takatsuji H (2001) The plant zinc finger protein ZPT2-2 has a unique mode of DNA interaction. J Biol Chem 276: 35802–35807 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]