Abstract
SPINDLY (SPY) protein from barley (Hordeum vulgare L. cv Himalaya; HvSPY) negatively regulated GA responses in aleurone, and genetic analyses of Arabidopsis thaliana predict that SPY functions in a derepressible GA-signaling pathway. Many, if not all, GA-dependent responses require SPY protein, and to improve our understanding of how the SPY signaling pathway operates, a yeast two-hybrid screen was used to identify both upstream and downstream components that might regulate the activity of the HvSPY protein. A number of proteins from diverse classes were identified using HvSPY as bait and barley cDNA libraries as prey. Two of the HvSPY-interacting (HSI) proteins were transcription factors belonging to the myb and NAC gene families, HSImyb and HSINAC. Interaction occurred via the tetratricopeptide repeat domain of HvSPY and specificity was shown both in vivo and in vitro. Messenger RNAs for these proteins were expressed differentially in many parts of the barley plant but at very low levels. Both HSImyb and HSINAC inhibited the GA3 up-regulation of α-amylase expression in aleurone, both were activators of transcription in yeast, and the green fluorescent protein-HSI fusion proteins were localized in the nucleus. These results are consistent with the model that HSI transcription factors act downstream of HvSPY as negative regulators and that they in turn could activate other negative regulators, forming the HvSPY negative regulator-signaling pathway for GA response.
GAs are essential for a number of processes, including gene expression in cereal aleurones, seed germination, elongation growth, and flowering. Significant advances have been made in understanding how GAs exert these effects, especially in the area of GA biosynthesis. Enzymes of the biosynthetic pathway have been studied by the identification of mutants and by the cloning and the characterization of the genes and enzymes involved, including aspects of regulation (Hedden and Kamiya, 1997; Olszewski et al., 2002). By contrast, the area of GA signaling remains less well characterized. In the aleurone, the receptor is predicted to be located on the plasma membrane (Hooley et al., 1991; Gilroy and Jones, 1994.), and models for signaling pathways have been proposed based on functional and genetic analyses with a number of protein components (Richards et al., 2001; Olszewski et al., 2002). These factors have been identified, cloned, and characterized in both monocotyledonous and dicotyledonous species. These factors could be classified into positive and negative regulators of GA signaling; the functionality of a factor was assigned based on its effects on the final GA-induced responses. The positive regulators include rice (Oryza sativa) D1 (α-subunit of the trimeric G protein complex; Ashikari et al., 1999; Fujisawa et al., 1999), F-box proteins in rice (GID2) and in Arabidopsis (Arabidopsis thaliana; SLEEPY1 or SLY1; McGinnis et al., 2003; Sasaki et al., 2003), and barley (Hordeum vulgare) GAmyb (Gubler et al., 1995).
The negative regulators are Arabidopsis SPINDLY (SPY; Jacobsen and Olszewski, 1993), GA-insensitive and repressor of ga1-3 (GAI and RGA; Peng et al., 1997; Silverstone et al., 1997), SHORT-INTERNODES (SHI; Fridborg et al., 1999), and Hordeum repressor of transcription (HRT; Raventós et al., 1998). The Arabidopsis genome contains three additional members of the GAI/RGA class of proteins (RGL1, RGL2, and RGL3), and they were found to function in both overlapping as well as in distinct GA responses (Lee et al., 2002; Wen and Chang, 2002). Their orthologous genes were identified in other species. The green revolution genes of wheat (Triticum aestivum) were found to be mutant alleles of GAI orthologs Rht-A1a and Rht-D1a (Peng et al., 1999a), where the highly conserved DELLA domain is expected to be absent. As a result, the mutant proteins remain active as negative regulators of GA responses regardless of GA content. These semidominant mutants are dwarfed and have higher grain yields. Dominant dwarf mutants are found in other species: Maize (Zea mays) D8 mutant has a similar mutation to wheat (Peng et al., 1999), while barley Sln1d and grape (Vitis vinifera) dwarf mutants have a point mutation near or in the DELLA sequence (Boss and Thomas, 2002; Chandler et al., 2002). For barley, the classic slender mutant, sln1a, was shown to be a recessive allele of the dominant dwarf mutant, Sln1d, demonstrating the opposite effects resulting from different mutations at the same locus (Foster, 1977; Chandler et al., 2002; Fu et al., 2002). It has been proposed that a signal from GA would inhibit the activity of negative regulators, such as GAI, RGA, and SLN1, resulting in the derepression of GA signaling (Peng et al., 1997; Silverstone et al., 1998; Chandler and Robertson, 1999). This derepression mechanism targets RGA, SLN1, and rice slender proteins to the proteolytic pathway (Silverstone et al., 2001; Gubler et al., 2002; Itoh et al., 2002). Additional genes responsible for GA-insensitive dwarf mutants in Arabidopsis and rice were cloned recently (SLY1 and GID2), and they encode orthologous F-box proteins that are likely to be involved in the 26S proteasome pathway mediating degradation of RGA and slender proteins (McGinnis et al., 2003; Sasaki et al., 2003). In addition, a number of other factors (Ca2+, CaM, cGMP, pHi) have been identified as involved in GA signaling (Bethke et al., 1997).
The first GA-signaling gene cloned was Arabidopsis SPY (Jacobsen et al., 1996). SPY protein is composed of two domains; the amino half contains 10 tetratricopeptide repeats (TPRs), and the carboxyl half (C-half) has no obvious motifs. Recessive spy mutants are affected in all of the known GA responses (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996) and show a constitutive GA response phenotype. Combined, the genetic evidence in Arabidopsis indicates that SPY functions as a negative regulator of GA response. Orthologs of Arabidopsis SPY were isolated in barley (cv Himalaya; HvSPY), in petunia (Petunia hybrida; PhSPY), and in tomato (Lycopersicon esculentum; LeSPY; Robertson et al., 1998; Izhaki et al., 2001; Greb et al., 2002). Cereal aleurone is a model system used to study GA action, especially the induction of high-pI α-amylase gene expression by GA3 (Bethke et al., 1997). Using this system, the specific involvement of HvSPY as a negative regulator of GA response was demonstrated in barley (Robertson et al., 1998).
The cloning of animal UDP-N-acetyl-d-glucosamine:protein β-N-acetyl-d-glucosaminyltransferase (OGT; EC 2.4.1.94; Kreppel et al., 1997; Lubas et al., 1997) indicated that SPY proteins may also have OGT activity, based on their sequence relatedness. Preliminary results supported this conclusion; however, the results were inconclusive due to the assay procedure used, which detected terminal N-acetyl glucosamine (GlcNAc) residues, many of which may not have been a simple O-GlcNAc modification on Ser or Thr residues (Thornton et al., 1999; Hanover, 2001). Arabidopsis SPY was thought to act downstream of GAI in the derepression system based on epistatic analyses using double mutants (Jacobsen and Olszewski, 1993; Jacobsen et al., 1997) and on a suppressor screen of gai (Peng et al., 1999b). It was also proposed that SPY may modify GAI with O-GlcNAc residues, thus acting upstream of GAI. GlcNAc modification of GAI and RGL1 may be GA independent and may account for the stability of these proteins against the GA signal (Fleck and Harberd, 2002; Wen and Chang, 2002), while the modification of RGA and SLN1 may be subject to GA regulation and a GA-induced loss of the modification may target these proteins to the proteasome pathway.
The TPR domain is important for Arabidopsis SPY protein function, since a number of spy alleles contained mutations only in the TPR domain (Tseng et al., 2001). The TPR motif is known to be involved in protein-protein interactions with another TPR protein or a non-TPR protein (Goebl and Yanagida, 1991; Lamb et al., 1995). Repeats within the TPR domain are different for each of the partner proteins, demonstrating that the TPR domain can interact with many proteins, each with specificity (Scheufler et al., 2000; Lyer and Hart, 2003). It is expected that the activity of HvSPY protein would be modified by an upstream component following an interaction through the TPR domain. The domain may also be used to recognize downstream components, perhaps using different TPR repeats.
Protein factors in the GA-signaling pathway have been identified by genetic screens and subsequent cloning in Arabidopsis and in other species using the Arabidopsis sequences. Further genetic screens identified mainly alleles of known mutants, suggesting that the identification of additional factors may not be accomplished by genetic screens that look for a limited number of obvious GA response phenotypes resulting from mutations. In order to overcome the limitations of genetic screens, an alternative approach was applied in this study, which used the yeast two-hybrid (Y2H) screen to identify proteins that would interact with the HvSPY protein. The GA-signaling pathway would require protein components to interact for the signal to be transduced from the receptor to downstream components. The presence of the TPR domain in the HvSPY protein and the pleiotropic effects of spy mutations would suggest that there may be many HvSPY-interacting (HSI) proteins for a number of SPY pathways. Some of these proteins would function in the GA-signaling pathway, as upstream components which may regulate HvSPY activity or as downstream components which may receive signals from HvSPY. This article reports the construction and screening of Y2H libraries in barley, the identification of HvSPY interacting proteins, and the functional and molecular characterization of two transcription factors. This study shows that these transcription factors are novel negative regulators of GA response in barley aleurone.
RESULTS
Identification of HvSPY-Interacting Proteins by Y2H Screen
To clone proteins that specifically interact with HvSPY, a Y2H screen was carried out using the whole HvSPY protein as bait and barley two-hybrid cDNA libraries as prey (see “Materials and Methods”). In total, 8.3 million colony-forming units were screened to identify 41 clones corresponding to 15 cDNAs (Supplemental Tables I and II, available at www.plantphysiol.org).
Novel Myb and NAC Transcription Factors Interact with HvSPY
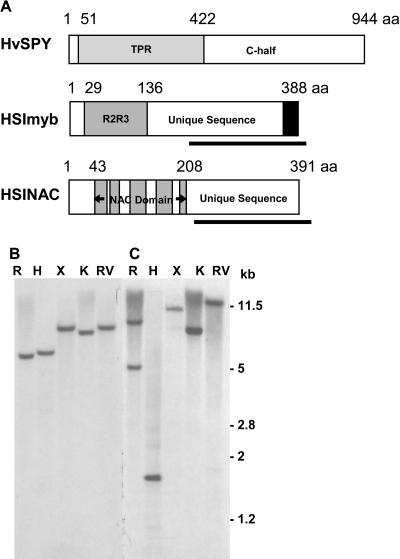
Five cDNA clones (clones 354, 597, 1132, 1342, and 1356) encoded the same overlapping sequence, with clone 1132 having the longest sequence. The database search using the deduced amino acid sequence showed that it belonged to the R2R3 class of myb transcription factors (Fig. 1A). The sequence, HSImyb for HvSPY-interacting myb, encodes a 388-amino acid polypeptide with the R2R3 myb domain located toward the amino end of the protein. The database search identified many plant R2R3 myb sequences with the highest sequence identity for a homologous rice gene at 97.2% using the myb domain alone; however, no closely related sequence could be identified using the C-half of the protein sequence except for the rice homolog. The absence of monocot expressed sequence tags (ESTs) suggests a low abundance of expression for this class of myb genes (see below). When the Arabidopsis genome database was searched with the amino acid sequences from the myb domain and non-myb domain separately, AtMyb83 was identified with the highest identity score for the myb domain. Significantly, it identified a motif at the carboxyl end that is conserved in myb genes between cereals (barley and rice) and dicotyledonous plants (Arabidopsis and soybean [Glycine max]), which is likely to reflect functional conservation (Fig. 1A).
Figure 1.
HSINAC and HSImyb are unique sequences in the barley genome. A, Schematic diagrams of HvSPY, HSINAC, and HSImyb (not to scale). The rectangular boxes are open reading frames with the numbers above referring to amino acid residues. HvSPY has the TPR domain and the C-half. The R2R3 myb domain in HSImyb and the five NAC subdomains in HSINAC are shown in shaded boxes. The thick bars under HSImyb and HSINAC indicate the region used for the Southern-blot analyses. The black box in HSImyb shows the region with a signature motif specific to HSImyb and its homologs. B and C, Genomic Southern-blot analyses for HSImyb (B) and HSINAC (C). Himalaya barley DNA was digested with EcoRI (R), HindIII (H), XbaI (X), KpnI (K), and EcoRV (RV). The numbers on the right refer to the sizes of markers in kilobases.
Clone 531 encodes a member of the NAC gene family of transcription factors (Fig. 1A). The N-terminal half of the open reading frame encodes the highly conserved NAC domain. There is no in-frame stop codon before the proposed start codon in the fusion clone, thus we cannot be certain that this clone contains the complete open reading frame. However, comparison with a number of sequences in the NAC gene family suggests that clone 531 contains a complete open reading frame, since the number of amino acid residues before the subdomain A of the NAC domain is relatively short for NACs, which is less than 20 amino acid residues. It is even possible that the translation start may be from the second ATG, since there are 43 amino acid residues before the subdomain A for the proposed open reading frame. Based on these comparisons, it is proposed that clone 531 encodes a polypeptide of 391 amino acid residues in length (Fig. 1A) and that the clone be named as HSINAC (HvSPY-interacting NAC). The amino-half is the well-defined NAC domain with 166 amino acid residues, and it consists of five (A–E) subdomains (Aida et al., 1997; Kikuchi et al., 2000). The domain is highly conserved among a number of genes across genera, but it is found only in plants (Riechmann et al., 2000). BLAST searches have identified many sequences with the NAC domain from both dicotyledonous and monocotyledonous plant species such as Arabidopsis and rice. When the search was carried out using the sequence unique to HSINAC (see below), no gene was identified, but 12 cereal ESTs were found. It thus appears that this part of the sequence may be unique to monocots. The identification of relatively few ESTs suggests that the transcript may also be expressed at very low levels, and this was later confirmed (see below).
To estimate gene copy numbers of HSImyb and HSINAC, genomic Southern-blot experiments were carried out. When a filter containing Himalaya genomic DNA digested with five restriction enzymes was hybridized with the non-MYB fragment of HSImyb, a single band in each of the digests was detected (Fig. 1B). With the non-NAC sequence from HSINAC used as a probe, single hybridizing bands were detected in all lanes except for EcoRI (Fig. 1C). The presence of two bands for the EcoRI-digested DNA is in agreement with the presence of an internal EcoRI site in the non-NAC region of HSINAC. There are also two HindIII sites in the non-NAC region, however, two smaller fragments are most likely to have run off the gel and the largest 5′ fragment is the hybridizing band. Therefore, these results suggest that both HSImyb and HSINAC are unique genes in the barley genome.
The Interaction Is through the TPR Domain
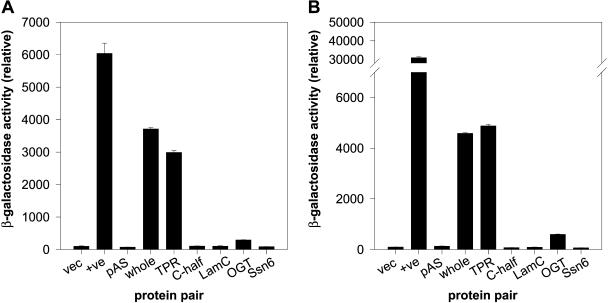
To investigate the specificity of the interaction between HvSPY and the HSI proteins, and to examine which domain of the HvSPY protein was responsible for the interaction, additional interaction analyses were carried out with the Y2H system. The relative strengths of interactions were quantified using the lacZ reporter gene system with o-nitrophenyl β-d-galactopyranoside as a substrate (Fig. 2). The interaction between the two vectors was at the background level, while the interaction between positive control proteins, p53 and T-antigen, was very high. The interaction between the GAL4 DNA-binding protein-HvSPY fusion and GAL4 activation domain (pAS) was again background level as shown before. For HSImyb (Fig. 2A), the interaction with HvSPY was highly significant, and this interaction was through the TPR domain of HvSPY, since the reporter gene activities were similar for the HvSPY protein (whole) and the TPR domain (TPR), while the activity was at the background level for the C-half of HvSPY. The interaction with the TPR domain was shown to be specific with the motif in HvSPY when the interaction with two other proteins containing 10 TPR repeat motifs was tested. HSImyb interacted very weakly with OGT and did not show interaction with Ssn6. When laminC was used as a control bait protein, the interaction between HSImyb and laminC was at the background level, demonstrating that the interaction with HvSPY was specific. For HSINAC (Fig. 2B), similar results were obtained for the interaction assays; it also interacted specifically with HvSPY, and this interaction was through the TPR domain. In addition, the same results were obtained using HIS3 as a reporter gene for both HSImyb and HSINAC, where yeast cells grew only when HvSPY (whole) or HvSPY (TPR) was present along with the HSI proteins (data not shown).
Figure 2.
Specific interaction between HvSPY and HSImyb or HSINAC. Y2H interaction assays for HSImyb (A) and HSINAC (B). The interactions were assayed in the GAL4 Y2H system to test the interactions of HSImyb and HSINAC proteins with GAL4DNABD alone (pAS), HvSPY (whole), the TPR domain of HvSPY (TPR), the C-half of HvSPY (C-half), LamC control (LamC), rat OGT (OGT), or yeast Ssn6 (SSn6) measured by β-galactosidase (lacZ) reporter gene activity. As a control, reporter gene activities for the pACT2 vector alone (vec) and a positive interaction between p53 and T-antigen (+ve) are also shown.
HvSPY Interacts with HSImyb and HSINAC in Vitro
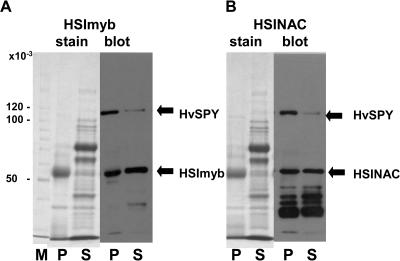
To demonstrate that HvSPY interacts with HSImyb and HSINAC in another system, their interactions were tested in vitro using recombinant proteins. Recombinant HvSPY, HSImyb, and HSINAC were expressed in Escherichia coli with 6× His tag, and the expression conditions were optimized to produce small but soluble amounts of HvSPY, HSImyb, and HSINAC. HvSPY was purified from soluble fractions of E. coli protein extract using the His tag. The interaction was tested by incubating the purified HvSPY and the soluble E. coli proteins containing the recombinant HSImyb or HSINAC. The protein complex was immunoprecipitated with a polyclonal antibody raised against the HvSPY TPR domain. The immunoprecipitated complex was then examined by SDS-PAGE, followed by staining to show the protein profile, as well as by anti-His western-blot analysis for the recombinant proteins (Fig. 3). For both HSImyb and HSINAC, the stained gel showed that there were very few proteins in the immunoprecipitated complex, except for the major band of rabbit IgG, which was isolated by the Protein A. Most of the soluble E. coli proteins were in the supernatant fraction. Anti-His western-blot analysis showed that HvSPY coprecipitated with HSImyb (Fig. 3A) and HSINAC (Fig. 3B) and there was little HvSPY in the supernatant. The excess HSImyb and HSINAC were also present in the supernatant. The many smaller bands in the HSINAC western blot were shorter peptides of the HSINAC. These results show that in addition to the interactions in vivo as hybrid proteins in yeast cells, both HSImyb and HSINAC interacted with HvSPY in vitro.
Figure 3.
HvSPY protein interacts with HSImyb or HSINAC protein in vitro. A, Gel analysis of immunoprecipitation products of recombinant HvSPY in the presence of HSImyb. The top arrow indicates HvSPY and the bottom arrow indicates HSImyb. B, Gel analysis of the immunoprecipitation products of recombinant HvSPY in the presence of HSINAC. The top arrow indicates HvSPY and the bottom arrow indicates HSINAC. For both A and B, the left section is a stained gel for immunoprecipitated proteins (P) or its supernatant (S). The right section is a western blot probed with anti-His MAb to detect the recombinant proteins. The lane M contains Mr markers. The numbers on the left are Mr × 10−3.
The Two Transcription Factors Are Activators of Transcription
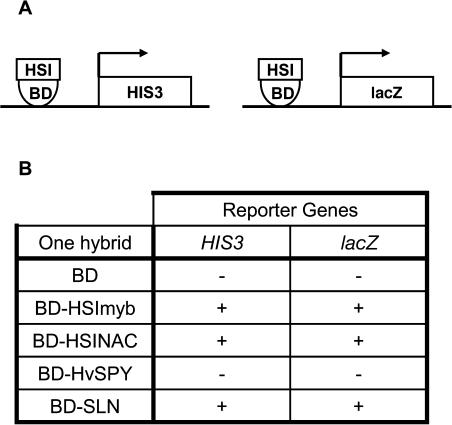
In order to investigate whether the two transcription factors functioned as activators or repressors of transcription, HSImyb and HSINAC proteins were tested in the yeast one-hybrid transcriptional activation assay. These proteins were isolated as fusion proteins with the GAL4 activation domain (AD), so the activation of the reporter genes in Y2H could have been by the GAL4 AD or an AD of the transcription factors. When HSI proteins were fused to the GAL4 DNA binding domain (BD), they would be placed proximal to the transcription start site of the reporter genes. If the HSI proteins had transcriptional activation activity, the activity could be detected by the activities of the reporter genes HIS3 and lacZ (Fig. 4A).
Figure 4.
HSImyb, HSINAC, and SLN are transcription activators. A, Schematic representation of reporter gene activation by transcription activators. If the protein in question is an activator, it should activate both reporter genes, HIS3 and lacZ. B, Reporter gene activities for GAL4DNABD alone (BD), the BD with HSImyb (BD-HSImyb), HSINAC (BD-HSINAC), HvSPY (BD-HvSPY), or SLN (BD-SLN). −, Absence of reporter gene activity; +. presence of reporter gene activity.
When the yeast cells were transformed with the vector containing the GAL4 DNA BD alone, the cells failed to grow in the absence of His and there was no blue color in the lacZ assay (Fig. 4B). By contrast, when HSImyb was fused to the GAL4 DNA BD, cells containing the plasmid grew without His and also developed a blue color from the β-galactosidase encoded by lacZ. The GAL4 DNA BD-HSINAC fusion protein was also capable of activating reporter genes HIS3 and lacZ. Thus, both transcription factors, HSImyb ad HSINAC, were transcription activators in yeast. By contrast, the GAL4 DNA BD-HvSPY fusion protein did not activate the reporter genes as previously shown when HvSPY was tested for its ability to autoactivate. One of the HSImyb clones (clone 597) was isolated, even though it was out of frame with the GAL4 AD protein. The original clone must have been able to activate the reporter genes causing its isolation, and this provides additional evidence that HSImyb is an activator of transcription.
Barley SLN1 is a negative regulator of GA signaling and is a member of the DELLA class of proteins that includes Arabidopsis GAI and RGA. SLN1 was tested for its role in transcriptional regulation in the yeast system. Similarly to HSImyb and HSINAC, SLN1 was also able to activate both reporter genes, HIS3 and lacZ, showing that it too was a transcriptional activator.
Differential Expression of HSImyb and HSINAC
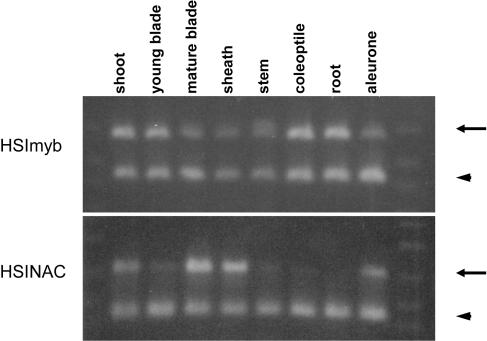
To investigate where HSIs are expressed in barley, northern-blot experiments were carried out using those cDNA fragments containing sequences unique in detecting HSImyb and HSINAC (Fig. 1A). However, no signal could be detected in total RNA samples from different parts of the plant (data not shown), presumably due to a low abundance of mRNAs. Alternatively, semiquantitative duplex reverse transcription (RT)-PCR analyses were carried out with a pair of gene-specific primers and an internal standard, EF1α (Fig. 5). HSImyb was expressed in all the organs examined at similar abundance. When the primer concentration of HSImyb was at a 2-fold molar excess of EF1α, the band intensities of HSImyb amplification products were about the same as the intensities of the internal standard EF1α. By contrast, expression of HSINAC varied more between various plant parts. The expression was higher in shoot, mature blade, sheath, and mature aleurone; lower in young blade and stem; and was barely detectable in coleoptile and root under the conditions shown with 45 cycles. HSINAC was indeed expressed in coleoptile and root, as the amplification products were detectable with more PCR cycles (data not shown).
Figure 5.
HSImyb and HSINAC mRNAs are expressed differently in barley plant parts. HSImyb (top) and HSINAC (bottom) mRNA expression was analyzed by RT-PCR. For each panel, the top band (arrow) is the gene of interest and the bottom band (arrowhead) is the internal standard EF1α. Their expression was analyzed for the plant parts shown at top. The experiments were carried out with at least two biologically replicated samples and more than 10 experimental replications and a representative result is shown.
HSImyb and HSINAC Are Negative Regulators of GA Response in Barley Aleurone
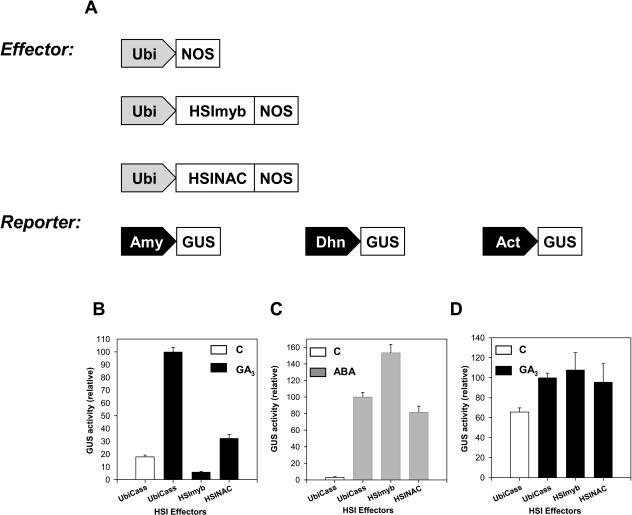
The bait protein, HvSPY, is a negative regulator of GA signaling. The HSI proteins could function as negative or positive regulators, or perhaps not be involved in GA signaling at all. To investigate these possibilities, the HSI proteins were functionally tested in the barley aleurone transient expression system. In these assays, the effects of HSI proteins on the GA3-induction of high-pI α-amylase promoter activity were examined. Effects of the HSI proteins were tested by overexpressing them from effector plasmid constructs (UbiHSImyb or UbiHSINAC) and measuring the promoter activity. The activity was quantified by the reporter gene activity, β-glucuronidase (GUS), in cobombardment experiments with a reporter plasmid, Amy-GUS, and one of the effector plasmids, UbiCass (empty vector) or UbiHSI (Fig. 6A).
Figure 6.
HSImyb and HSINAC are negative regulators of GA response in barley aleurone. A, Schematic diagrams of effector and reporter gene constructs. Effector constructs were under the control of an ubiquitin promoter (Ubi) with a NOS termination sequence. The vector cassette, UbiNOS, does not contain any open reading frame. Effector constructs overexpressing HSImyb and HSINAC are Ubi::HSImybNos and Ubi::HSINACNos. Reporter constructs are under the control of a high-pI α-amylase promoter (AmyGUS), a dehydrin promoter (DhnGUS), or an actin promoter (ActGUS) to express the reporter gene GUS. B to D, Effects of overexpression of HSImyb and HSINAC proteins in barley aleurone on promoter activities of the amylase (B), dehydrin (C), or actin (D) genes. The promoter activities were expressed relative to the GA3 induction of amylase, to the ABA induction of dehydrin or to the GA3-treated actin promoter activities without effector protein overexpression, UbiCass. The mean and se from 24 to 30 independent transfections per treatment are shown.
When no effector protein was expressed (UbiCass), the promoter activity increased 5.6-fold in response to GA3 (Fig. 6B), showing the GA responsiveness of the system. When HSImyb protein was overexpressed under the control of a maize ubiquitin promoter, this increase was completely abolished and the activity was lower than the control background value. Similarly, the overexpression of HSINAC also blocked the GA3-induced promoter activity, but not as effectively as the overexpression of HSImyb. These results show that both HSImyb and HSINAC can inhibit the GA response in aleurone.
The inhibitory effect may have been specific to the GA response or may have been a general effect on any gene. To differentiate between these two possibilities, further functional assays were carried out using promoters from two genes, an abscisic acid (ABA)-induced gene, barley dehydrin (Dhn), and a constitutively expressed gene, rice actin (Act). Effects on the promoters were measured using the reporter gene GUS (Fig. 6A). The promoter activity of the Dhn gene increased more than 30-fold in response to ABA (Fig. 6C). When HSImyb was expressed, the activity increased significantly by 50%, while it decreased slightly from the overexpression of HSINAC, but this decrease was not significant at 99% confidence level. The promoter activity of the Act gene increased by 35% in response to GA3, and the increase was significant (Fig. 6D). These results show that the Act gene expression is not completely constitutive. The promoter activities did not increase in additional control experiments when HSImyb and HSINAC were overexpressed in the absence of hormones (data not shown). Together, these results show that the inhibitory function of HSImyb and HSINAC protein is specific to the GA up-regulated α-amylase and that these transcription factors are not a general inhibitory factor.
HSImyb and HSINAC Are Localized in the Nucleus
Transcription activators are expected to be localized in the nucleus in order to function. To investigate if these HSI transcription activators were localized in the nucleus, and also to determine whether their protein abundance or their localization changed in response to hormones, the expression of green fluorescent protein (GFP)-HSI fusion proteins was examined in barley aleurone protoplasts that had been transfected with GFP-HSI constructs under the control of a maize ubiquitin promoter (Fig. 7A). The expression of GFP alone and of the GFP-HvSPY fusion protein were also examined. These GFP-fusion HSI proteins were functionally active in the barley aleurone layer transient assays, which showed results similar to those obtained with nonfusion proteins. The promoter activity of high-pI α-amylase increased 6.7-fold in response to GA3 when GFP alone was overexpressed, while the overexpression of GFP-HSImyb or GFP-HSINAC decreased the promoter activity significantly to around the control level (data not shown).
Figure 7.
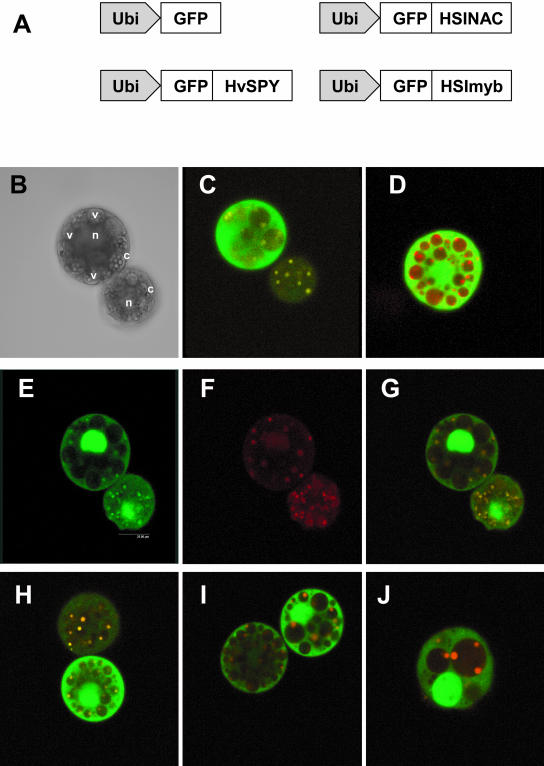
HSINAC and HSImyb are localized in the nucleus of barley aleurone protoplasts. A, Schematic diagrams of GFP fusion protein genes under the control of ubiquitin promoter. They are genes for overexpressing GFP alone (Ubi::GFP) and three fusions (Ubi::GFP-HvSPY, Ubi::GFP-HSImyb, and Ubi::GFP-HSINAC). B, Phase contrast image of aleurone protoplasts that show protein storage vacuoles (v), the nuclei (n), and the cytoplasm (c). C to J, Confocal laser scanning microscope images of protoplasts. C, HvSPY fusion; D, GFP alone; E to G, HSINAC fusion; and H to J, HSImyb fusion. E, GFP fluorescence at 500 to 600 nm; F, autofluorescence at 600 to 720 nm; and G, an overlay of E and F to show GFP-specific fluorescence (green) and autofluorescence (orange). C, D, G, H, I, and J, Overlay images. H, Localization in stage I protoplasts; I, localization in stage II protoplasts; and G, localization in a stage III protoplast.
Barley aleurone protoplasts contain three major organelles, which are the nuclei, protein storage vacuoles containing smaller protein storage bodies, and the cytoplasm in between (Fig. 7B). Protoplasts were examined for GFP expression 18 h after the transfection. In the emission range between 500 and 600 nm, strong signals were detectable in the nucleus when GFP-HSINAC was overexpressed. There were also smaller spots of fluorescence in the protein storage vacuoles (Fig. 7E). The background autofluorescence was detected in the emission range between 600 and 770 nm (Fig. 7F), and small fluorescent spots were present in the protein storage vacuoles. These two images were overlaid to identify the GFP-specific signal. The signal from the nucleus was the only GFP fluorescence and the signal from the protein storage vacuoles was the autofluorescence signal (Fig. 7G). The analysis of many transfected protoplasts showed that GFP-HSINAC was expressed exclusively in the nucleus.
When GFP-HSImyb was overexpressed in the protoplasts, the protein was also localized in the nucleus, but not exclusively (Fig. 7, H–J). GFP fluorescence was also present in the cytoplasm. When a large number of protoplasts expressing GFP-HSImyb were examined in replicated transfection experiments, there was a correlation between the localization and the stages of protoplast aging (Bush et al., 1986). In addition to the presence in the nucleus, GFP fluorescence was detected in the cytoplasm of stages I and II protoplasts (Fig. 7, H and I) and was almost absent in the cytoplasm of stage III protoplasts that were more vacuolated (Fig. 7J).
GFP-HvSPY was present in the nucleus and in the cytoplasm at very low levels (Fig. 7C). No changes were detected in cellular localization or abundance of expression by hormone treatments (data not shown). When GFP alone was expressed, it was expressed to very high levels in the nucleus and in the cytoplasm at all times (Fig. 7D).
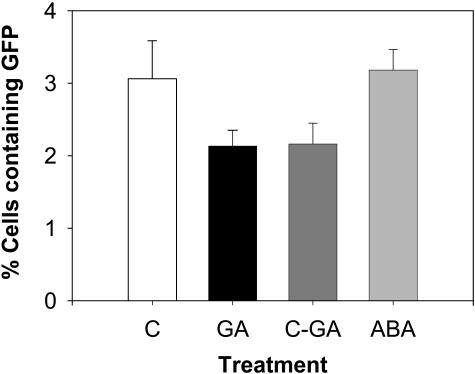
To investigate whether the localization and/or the abundance of HSI proteins are regulated by GA3 and/or ABA, the expression of GFP-HSI fusion protein was examined following various hormone treatments. For GFP-HSINAC, when the transfected protoplasts were incubated without any hormone (C), 3.06% ± 0.52% of the cells showed detectable GFP fluorescence in the nucleus (Fig. 8). When the cells were incubated in 1 μm GA3, 2.13% ± 0.21% of the cells showed detectable GFP, a significant decrease compared to the control cells. The decrease in the number of cells with detectable GFP fluorescence was quite rapid, as cells incubated without hormone for 15 h followed by 3 h in GA3 (C-GA) also had significantly fewer cells expressing GFP, 2.16% ± 0.28%. When treated with ABA, the proportion of cells with the GFP fluorescence was 3.18% ± 0.28%, and this was similar to the control cells. The GFP expression was exclusively in the nucleus in all treatments. By contrast, the proportion of cells expressing GFP-HSImyb did not change significantly in response to hormone treatments. The results with GFP-HSImyb also show that the decrease in the expression of HSINAC in GA3-treated cells is not a general proteolysis response. Were this the case, we would have seen a decreased expression of several GFP-fusion proteins.
Figure 8.
GA3 treatment decreased the expression of HSINAC in aleurone protoplasts. The relative abundance of the expression is shown as the percentage of cells that show detectable GFP fluorescence among the total protoplasts. A pool of transfected protoplasts were treated with no hormone (c), with 1 μm GA3 (GA), or 1 μm ABA (ABA) for 18 h, or no hormone for 15 h followed by 3 h with GA3 (C-GA). The results are from 1,500 to 4,000 protoplasts per treatment from two transfection experiments.
DISCUSSION
SPY and HvSPY proteins are negative regulators of GA signaling based on genetic evidence and functional analyses in Arabidopsis and in barley (Jacobsen and Olszewski, 1993; Robertson et al., 1998; Richards et al., 2001; Olszewski et al., 2002). Arabidopsis spy mutants show GA-independent seed germination and growth, faster transition to flower, and partial male sterility (Jacobsen and Olszewski, 1993). Unlike other GA-signaling components such as GAI and RGA, SPY is the only factor to influence all known GA-dependent processes as observed in spy mutants. In addition, spy mutants also show phenotypes that may not be GA related, suggesting that SPY proteins may be involved in regulating response pathways other than the GA pathway (Swain et al., 2002). Consistent with this, ABA up-regulated dehydrin promoter activity was increased by an overexpression of HvSPY in barley aleurone; however, the action of HvSPY was independent of the ABA pathway (Robertson, 2003). Mutations at the SPY locus have pleiotropic effects in Arabidopsis, and these observations are consistent with the idea that the protein is essential for many aspects of normal plant development. The diversity of affected responses suggests that SPY protein is a hub for many signaling pathways, some involving GA, others not. Such a signaling hub would be receiving and transmitting signals between many upstream and downstream factors as a part of SPY function. In agreement with the role of SPY in signaling, many HSI factors were identified in the Y2H. These HSI proteins are expected to define components upstream and downstream of HvSPY, which could be receiving signals from diverse input ligands and conveying the signal downstream. Most of the identified factors, including the two transcription factors described in this article, were previously unknown. Therefore, the use of the Y2H screen has proved a useful method, complementary to mutant screens, in isolating novel components.
SPY proteins are members of a protein family with TPR motifs that are repeats of 34 semiconserved amino acid residues (Goebl and Yanagida, 1991; Lamb et al., 1995). The TPRs form amphipathic α-helices and the motif is the site for protein-protein interactions (Das et al., 1998; Scheufler et al., 2000). For HvSPY, the role of the TPR motif was verified with the finding that the interaction between HvSPY, and every one of the HSI proteins was through the TPR domain and the C-half of the protein was not involved in the interaction (this study and data not shown). The interaction is unlikely to be a nonspecific interaction with any TPR domain, since a number of HSI proteins did not interact with a different TPR protein having 10 repeats, yeast Ssn6. HSI proteins did interact with rat OGT, but these interactions were much weaker than with HvSPY in the Y2H system. The TPR domain is essential for SPY protein function as demonstrated by the sequence analysis of many spy alleles that have mutations only in the TPR domain (Tseng et al., 2001). For animal OGTs, not all of the repeats are required for the interaction between the OGTs and their substrates (Lubas and Hanover, 2000; Lyer and Hart, 2003). Thus, the spy alleles with mutations only in the TPR domain may result from an inability to interact with a subset of partners. The identification of many HSI proteins will enable the testing of the specificity of repeats that are required for HvSPY interaction with the interacting partners.
GA-signaling components include enzymes (SPY, SLY/GID), heterotrimeric G proteins (Gα), and transcription factors (GAI/RGA/SLN1, GAmyb, HRT). In this screen, two of the HSI proteins are transcription factors, HSImyb and HSINAC. HSImyb is a member of the R2R3 myb family, and HSINAC belongs to the NAC family of transcription factors based on the presence of the highly conserved DNA BDs. The myb and NAC classes of transcription factors are two of the more abundant classes of plant transcription factors, and NAC proteins are found only in plants (Riechmann et al., 2000). The myb family of transcription factors has more members in plant genomes than other genomes that have been sequenced, particularly the R2R3 class. The large number of R2R3 mybs is thought to carry out many plant-specific functions (Stracke et al., 2001).
Several myb transcription factors have been shown to be involved in regulating GA response in cereal aleurone. The myb factor described in this article, HSImyb, is a R2R3 myb, and functional assays have shown that it acts as a negative regulator of GA response. The well-known positive regulator of aleurone GA response is GAmyb with a R2R3 DNA BD (Gubler et al., 1995). GAmyb binds the TAACAA motif in the GA-response elements, which are found in the promoters of many GA-induced hydrolases, and it functions as a transcriptional activator (Gubler et al., 1995, 1999). Thus, it has been proposed that GAmyb protein binds the GA-response element to transactivate α-amylase and other hydrolase genes and that this is the basis of GA-induced α-amylase expression. GAmyb activation of low-pI α-amylase promoter was further enhanced by OsMYB transcription factors. These OsMYB proteins have only one DNA-binding myb domain (1R), and upon binding the TATCCA element in the GA-response complex within the promoter, OsMYB1 and OsMYB2 transactivated the promoter (Lu et al., 2002). As DNA binding is essential for the function of these myb factors and the sequence conservation of the myb domain is high, there is a possibility that HSImyb may block the GA3-induced α-amylase promoter activity by blocking GAmyb binding. However, this is unlikely for three reasons. First, HSImyb is a strong transcription activator; it transactivated two reporter genes in yeast when brought proximal to the transcription starts site via the GAL4 DNA BD. If HSImyb had bound the GAmyb binding site in the α-amylase promoter, it would have activated the transcription of the reporter gene GUS in the transient assays, resulting in increased GUS activity. Secondly, the myb class of transcription factors binds DNA via the R2R3 DNA BD, and the sequence conservation among the R2R3 class myb is variable, with a sequence identity from 40% to over 95% (Stracke et al., 2001). Mybs with similar functions are thought to have a higher sequence identity in the myb domain, even across taxa, to ensure correct binding to the target in a promoter. Amino acid sequence identity between the R2R3 myb domains of HSImyb and GAmyb is only 58%. The relatively low sequence identity suggests that HSImyb is unlikely to bind the TAACAAA GAmyb binding site of the amylase promoter. Thirdly, another R2R3 myb protein, C1 myb from maize, was partly inhibitory to GA3-induced α-amylase promoter activity (Gubler et al., 1995). However, C1 was much less effective in inhibiting the GA3-induced promoter activation than HSImyb, despite the similar sequence identity of the myb domains of C1 to GAmyb (57%) and despite the fact that 30 times more C1 effector plasmid construct was used in the experiment than HSImyb. Taken together, the inhibitory effect of HSImyb is unlikely to have resulted from the binding of HSImyb protein to block GAmyb binding in transfected layers. Similarly, it is unlikely that HSImyb blocks OsMYBs (or their orthologs in barley) from binding, especially when the OsMYBs are 1R mybs.
The fact that these transcription factors were identified by protein-protein interaction would strongly suggest that they are downstream factors of HvSPY. If they were upstream factors regulating HvSPY activity, the interaction between the transcription factors and HvSPY would have been through the promoter of HvSPY gene. The sequence similarity of HvSPY and animal OGTs indicates that HvSPY is likely to be a plant OGT, and it is known that transcription factors are substrates for GlcNAc modification. Recent studies have shown that GlcNAc modification regulated the activities of transcription factors. For example, GlcNAc modification of transcription factors has been found both to decrease transcriptional activities of mER-β (Cheng and Hart, 2001) and Sp1 (Roos et al., 1997; Yang et al., 2001) and to increase activities of p53 (Shaw et al., 1996), YY1 (as a repressor; Hiromura et al., 2003), and PDX-1(Gao et al., 2003). At several sites p53 is modified with GlcNAc; however, the modification at the carboxyl end appears to be important in regulating the activity. When the p53 protein is localized in the nucleus, it is O-GlcNAc-modified at the carboxyl end and with this modification has an increased DNA-binding ability. These results indicate the regulatory role of the modification on protein activity by localization and by biochemical capability. By contrast, the modification inhibited Sp1 from transactivating. When Sp1 was O-GlcNAc modified at terminal GlcNAc residues, its transcriptional activation was repressed. It was further shown that the modification controls its transcriptional activity by affecting its ability for protein-protein interaction (Roos et al., 1997; Yang et al., 2001). Therefore, the interaction between HvSPY and HSITFs may be an enzyme-substrate interaction, with GlcNAc modification of the transcription factors forming the basis of signaling. The identification of these transcription factors would enable the investigation of these possibilities in future research.
Transcription factors regulate gene expression through modulating transcription. They can be repressors or activators of transcription. R2R3 myb proteins are shown to be both transcriptional activators and repressors (Stracke et al., 2001), while only the activator function has been shown for NACs so far (Souer et al., 1996). Yeast one-hybrid transcription assays were used to examine whether HSImyb and HSINAC are transcriptional activators or repressors. The results in this article clearly show that both HSImyb and HSINAC are strong transcriptional activators in yeast. Target proteins downstream of HSI transcription activators could function in one of two possibilities: The downstream factor activated by the HSI transcription factors may be either a positive or negative regulator of GA responses. Functional analysis examining these possibilities has shown that overexpression of HSImyb and HSINAC specifically inhibited GA-induced α-amylase expression, as had HvSPY. These results show that HSImyb and HSINAC are also negative regulators of GA response in barley aleurone and suggest that they could activate the transcription of further downstream negative regulators. Taken together, it is proposed that HvSPY, HSI transcription factors, and unknown targets of the HSI transcription factors define three steps in the HvSPY negative regulator-signaling pathway.
How is it that the two transcription factors both function as negative regulators of GA response? It is possible that different downstream negative regulators may be the targets of these transcription factors, thus they would define steps of divergence in signaling. This possibility is consistent with our observation of no change in α-amylase expression using RNAi constructs of HSImyb and HSINAC in our transient assays (data not shown). Silencing a negative regulator in one of the pathways would leave the other negative regulator pathway active, thus there would be little or no change in the level of expression. It is also likely that these two transcription factors have different roles spatially within the barley plant. HvSPY is ubiquitously expressed in barley plants at similar mRNA abundance (Robertson et al., 1998). By contrast, HSImyb and HSINAC show different abundances. HSImyb may play a more significant role in young blade, coleoptile, and root, while HSINAC may function more in mature blade and sheaths. Their involvement in the negative signaling pathway is determined by the differential expression of the two factors. When both factors are expressed in the same plant part or tissue, their expression appears to be regulated differently. In the aleurone, both HSImyb and HSINAC are expressed, yet HSINAC expression is regulated by GA3, at least in part, while HSImyb is not. Thus, the presence and regulation of various factors with different spatial expression may be the means of diversifying the signal from HvSPY.
In addition to HvSPY and HSI transcription factors, two other negative regulators of α-amylase expression had been identified in barley: SLN1 and HRT. SLN1 is a member of the DELLA class of negative regulators in GA signaling, which includes GAI and RGA. Although SLN1 is a repressor of GA response, SLN1 is also a transcriptional activator (Ogawa et al., 2000), thus its role in inhibiting α-amylase expression is indirect (Chandler et al., 2002). Therefore, predicted models for SLN1 and HSI transcription factors are quite similar. Whether they are part of the same negative regulator-signaling pathway or not remains to be examined.
Transcription factors are active only when they are in the nucleus, and these proteins are expected to contain the nuclear localization signal (NLS) enabling their transport into the nucleus, following translation in the cytoplasm. With HSImyb, NLS is predicted to be KR.{45}KKRL, by the PredictNLS program (Cokol et al., 2000; http://cubic.bioc.columbia.edu/predictNLS) and located in the R3 DNA BD. For HSINAC, the NLS is predicted to be in subdomains C and D within the highly conserved NAC domain (Kikuchi et al., 2000). In support of the predictions, these proteins were localized in the nucleus in barley aleurone protoplasts using GFP-HSI fusion proteins. It is interesting to note that the DNA BDs and the NLS are colocalized. Perhaps, it is a functional coevolution to have the two motifs within the same sequence. In addition to HSImyb and HSINAC, it is reported that the DNA BD and the NLSs overlap for many transcription factors (Cokol et al., 2000).
Using the same approach, GFP-HvSPY was localized both in the nucleus and the cytoplasm, in agreement with the animal OGTs (Kreppel et al., 1997; Lubas et al., 1997) and the nuclear and cytoplasmic-localized Arabidopsis SPY (Swain et al., 2002). The proposed NLS in animal OGTs is located just downstream of the TPR domain (Hanover, 2001), and plant SPY proteins were localized in the nucleus despite the absence of this motif in plant SPY proteins. The colocalization of HvSPY and HSI proteins in the nucleus would suggest that the interaction would occur in the nucleus, after their transport into the nucleus by their NLSs. However, it is also possible that the interaction could occur in the cytoplasm and, following the proposed GlcNAc modification of the transcription factors by HvSPY, HSI proteins are marked for translocation. In this situation, there would be two levels of regulation for the nuclear import: one is the presence of NLS and the other is GlcNAc modification. As discussed earlier, the nuclear-localized p53 is GlcNAc modified at the carboxyl end (Shaw et al., 1996). Also, this model of cytoplasmic interaction is in agreement with the observation that there was a slight but significant decrease in the GFP-HSINAC fusion protein abundance in protoplasts when they were treated with GA3. Here, it is proposed that the GA3 treatment would decrease the activities of the negative regulators; HvSPY would become less active and result in the reduced GlcNAc modification of HSINAC, which would lead to decreased protein abundance in the nucleus, hence the decreased HSINAC activity.
In conclusion, the Y2H screen identified a number of candidate proteins for inclusion in the HvSPY-signaling pathway. The identification of many novel components confirms the advantage of the Y2H approach, which would complement the genetic-screening approach, in advancing our understanding of GA signaling through the identification of its components. This article also describes two HSI transcription factors regulating GA response in aleurone. The results predict signaling pathways composed of negative regulators and that this type of pathway may be an important characteristic of the regulation of GA responses.
MATERIALS AND METHODS
Y2H cDNA Library Construction
Total RNA was extracted from the first leaf blade of 5- to 7-d-old seedlings of barley (Hordeum vulgare L. cv Himalaya) that were germinated in petri dishes. The blade was frozen and stored in liquid N2. Poly(A)-RNA was isolated using Poly(A) Quik (Stratagene, La Jolla, CA) according to the manufacturer's instructions. cDNA was synthesized from 5 μg poly(A)-RNA using a cDNA synthesis kit (Stratagene) according to the manufacturer's instructions, except that double-stranded cDNA was size fractionated on a gel and cDNAs larger than 0.6 or 0.8 kb were purified from the gel to make two cDNA libraries, V1 and V2, respectively.
Lambda ACT2 vector was obtained from Dr. S. Elledge (Baylor College of Medicine, Houston) and propagated in Escherichia coli LE392. Lambda DNA was isolated using Qiagen tip 100 columns (Qiagen, Hilden, Germany), digested with EcoRI and XhoI, and treated with calf intestine phosphatase. Following the ligation of lambda arms and the cDNA, the libraries were packaged using MaxPlax packaging extract (Epicentre Technologies, Madison, WI) according to the manufacturer's instructions. Primary barley Y2H libraries in λACT2 of 1.8 million plaque-forming units (pfu; V1) and 1.9 million pfu (V2) were constructed. The size of the cDNAs was examined by PCR amplification of the inserts using plate lysates, and the inserts of the V1 library ranged from 0.55 to 3 kb and the V2 from 0.8 to 4 kb (data not shown). The libraries were titred and amplified in E. coli LE392MP once. The amplified libraries were frozen at −80°C with 9% (v/v) dimethyl sulfoxide.
About 5 × 107 pfu of each of the two Y2H libraries were mass excised from the λ libraries in E. coli BNN132 by cre-loxp recombination (Elledge et al., 1991). The excised colonies were grown in Terrific Broth (Tartof and Hobbs, 1987), and the plasmid DNA was purified by Qiagen plasmid maxi columns, yielding about 1.5 mg each of the barley prey cDNA libraries in pACT2.
Yeast Two-Hybrid Screen
Prior to screening for HSI proteins, the bait constructs and GAL4 DNA BD vector plasmid (pAS2-1) in the Matchmaker 2 system (CLONTECH, Palo Alto, CA) were transformed into Y187 and Y190 yeast hosts according to the TRAFO procedure (Agatep et al., 1998) to be tested for autoactivation. Bait constructs tested were pAS2-1::HvSPY (whole) with the whole open reading frame of HvSPY fused in frame with the GAL4 DNA BD, pAS2-1::HvSPY (TPR) with amino acid residues 1 to 421 (the TPR domain) fused, and pAS2-1::HvSPY (C-half) with residues 422 to 944 (C-half) fused. The fusion bait proteins alone [pAS::HvSPY (whole)/Y190 and pAS::HvSPY (C-half)/Y190] did not have any autoactivating activities, while pAS::HvSPY (TPR)/Y190 was mildly autoactivating. All three bait proteins, HvSPY (whole, TPR, and C-half), did not interact with GAL4 AD protein carried in pACT2 plasmid (data not shown). Based on these results, pAS2-1::HvSPY (whole) was chosen to be used as a bait to screen the libraries.
The yeast cells Y190 carrying HvSPY (whole) as a bait [pAS::HvSPY (whole)/Y190] were transformed with the cDNA library in pACT2 by the TRAFO procedure (Agatep et al., 1998) and a small fraction of the transformation reaction was plated on synthetic dropout (SD) medium −Leu, -Trp to estimate the total number of transformants. The remainder was plated out on SD −Leu, -Trp, -His, +3-AT (40 mm) to identify HIS3 reporter gene positive colonies. The concentration of 3-AT was optimized at 40 mm, since a large number of colonies were able to grow at 20 mm. A similar number of colonies grew at 40 and 60 mm, but the growth was faster at 40 mm than 60 mm.
HIS3 positive colonies were tested for lacZ reporter gene activity by colony-lift filter β-galactosidase assay (Yeast Protocols Handbook [YPH]; CLONTECH). Those colonies that grew on SD −Leu, -Trp, -His, +3AT and blue on filters (HIS3 and lacZ positives) were grown on SD −Leu, -Trp plates for further analyses. First, they were streaked out on SD −Leu, cycloheximide plates to eliminate the bait plasmid, and the prey cDNA clones were tested for autoactivation (YPH).
Preliminary sequence analysis showed that the majority of the inserts encoded Rubisco small subunit (RSU) isozymes. These sequences were in frame but had a nonspecific interaction for the lacZ reporter gene (data not shown). Therefore, it was necessary to identify clones encoding RSU among the HIS3 and lacZ positive colonies and this was carried out by duplex PCR reactions using yeast cell lysates as a template, which contained only the prey plasmid. Briefly, yeast cells carrying the prey plasmid were grown on SD −Leu plates and were lysed in water by freeze/thaw in liquid N2. An aliquot of the lysate was directly used in plasmid PCR reactions to amplify inserts and RSU, if present, using vector primers and an RSU-specific forward primer. When two bands were produced, with the smaller one around 400 to 800 bp, these clones were designated as RSU and discarded. The remaining clones that produced single bands, now designated as HvSPY-HSI clones, were studied further.
The prey cDNA plasmids were isolated from yeast cells, transformed into E. coli XL1-blue MRF′ and plasmids were isolated from E. coli for sequence analysis. Those clones with long open reading frames, which were in frame with the GAL4 AD protein, were tested for interaction specificity. HSI clones in Y190 were mated with Y187 cells containing different bait plasmids, which would encode negative control proteins [pAS2-1/Y187 and pLAM5′1 (laminC)/Y187], the positive control protein [pAS::HvSPY (whole)/Y187] and test proteins [pAS::HvSPY (TPR)/Y187 and pAS::HvSPY (C-half)/Y187]. The mated samples were plated on SD −Leu, -Trp plates to select for diploid cells carrying both the prey (cDNA) and bait plasmids. At least three diploid cells from each mating were tested for HIS3 and lacZ reporter gene activities. Those clones with specific interaction were selected and grouped into sequence family groups when they had overlapping sequences. A quantitative β-galactosidase assay was carried out using o-nitrophenyl β-d-galactopyranoside according to YPH.
Yeast One-Hybrid Assays
The vector plasmid pAS2-1 and plasmid constructs carrying GAL4 DNA BD-HSI protein fusions (pAS2-1::HSImyb and pAS2-1::HSINAC) and GAL4DNABD-HvSPY (pAS2-1::HvSPY) were transformed into yeast Y190 cells (TRAFO) and the transformants were tested on SD −Trp, -His, +3-AT (40 mm) plates for the HIS3 reporter gene, and by lacZ filter assays (YPH) using colonies growing on SD −Trp plates.
Functional Transient Assays in Barley Aleurone
The transient assays were carried out essentially as described (Robertson et al., 1998) except using 1.6 μm Biolistic gold (Bio-Rad, Hercules, CA). Each transfection used 0.45 mg of gold particles carrying 150 ng of AmyGUS or DhnGUS reporter construct with 15 ng of UbiCass or UbiHSI effector construct, or 75 ng of ActinGUS with 7.5 ng of UbiCass or UbiHSI construct. The promoter activity was quantified by GUS assays using 4-methyl umbelliferyl glucuronide, and the activities were normalized after subtracting the blank background activities and were expressed as a percentage of the value for the aleurone that were transfected with UbiCass and treated with GA3 for AmyGUS and ActinGUS or ABA for DhnGUS.
Protein Localization Studies in Barley Aleurone Protoplasts
Barley aleurone protoplasts were prepared from Himalaya barley (1998 harvest at Pullman, Washington University) as described (Robertson et al., 1995). Protoplasts were transfected with 20 μg of UbiGFP-HSI fusion constructs and incubated for 18 h at 25°C. For hormone treatments, transfected protoplasts were aliquotted into four or eight flasks that received no hormone, GA3, or ABA. The protoplasts were examined for protein localization using an SP2 confocal laser scanning microscope (Leica, Wetzlar, Germany). GFP fluorescence was examined at 500 to 600 nm and autofluorescence at 600 to 720 nm, following an excitation at 488 nm.
Southern Analysis
Barley genomic Southern experiments were carried out as described (Robertson et al., 1998). Since HSImyb and HSINAC are members of multigene families with well-conserved myb and NAC domains, respectively, sequences 3′ of myb (PvuII-EcoRI fragment) and NAC (KpnI-XhoI, in the multiple cloning site, fragment) domains were used in hybridization experiments. The fragments used in the hybridization experiments are indicated in Figure 1A.
Expression Analysis by RT-PCR
Barley plant parts were harvested from immature seedlings and mature Himalaya plants. They were stored frozen in liquid N2 and total RNA was isolated as described (Robertson et al., 1998). RT-PCR analysis was carried out in duplex reactions with a pair of gene-specific primers and a pair of internal-standard primers using a SuperScriptII Platinum Taq long-range one-step RT-PCR kit (Invitrogen, Carlsbad, CA). The internal standard used was barley elongation factor 1α, with primers 5′-TGTAACAAGATGGACGCC-3′ and 5′-GAAGCCACCATTGTCACC-3′. The HSImyb transcript was reverse transcribed and PCR amplified using the gene-specific primers 5′-CAGGAAACAGCAATGTGG-3′ and 5′-ATGGTCTTGGAAGAACGG-3′. The primers used for amplifying the HSINAC transcript were 5′-GTGAAACCGAACGGTACC-3′ and 5′-AAGTTACTTGAAGCTTGTGCA-3′. One of the primers in each of the pairs was designed over an intron, so that all of the amplification products were from RNA. The primer concentrations and the number of cycles were adjusted for each of the duplex reactions so that both the internal standard and the gene of interest were in the linear range of amplification at the time of end point analysis.
Recombinant Protein Production and Immunoprecipitation
To produce recombinant proteins, bacterial cells carrying His-tagged HvSPY, HSImyb, and HSINAC (HIS-HvSPY/BL21-RIL, HIS-HSImyb/BL21-RP, and HIS-HSINAC/BL21-RP) were grown at 37°C to mid-log phase. The cultures were then incubated at 20°C and the expression was induced with 0.6 mm isopropylthio-β-galactoside for 3 h. The cells were harvested by centrifugation and frozen at −80°C until purification. Soluble protein was prepared as per QIAexpressionist by native lysis (Qiagen) for HvSPY, HSImyb, and HSINAC, and the preparations were stored at −80°C. The HIS-tagged HvSPY was purified from the soluble protein extract using Ni-NTA according to the manufacturer's instructions (Qiagen). The purified protein was stored at −80°C with 10% (w/v) Suc.
For immunoprecipitation, the purified HIS-HvSPY was incubated with soluble protein extracts from E. coli containing the recombinant HIS-HSImyb or HIS-HSINAC in TBS-NP40 (50 mm Tris-Cl, pH 8.0, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and 1% NP-40) at 4°C for 1 h. HvSPY and associated proteins were precipitated with an antibody raised against the TPR domain of HvSPY and then with protein-A Sepharose (Pharmacia, Uppsala) for 1 h each at 4°C. The precipitates were washed three times with TBS-NP40. Both the precipitates and the supernatant were analyzed by SDS-PAGE and stain or western blot using anti-HIS MAb (Qiagen).
Plasmid Constructs
Bait Constructs
HvSPY (whole), HvSPY (TPR), and HvSPY (C-half) sequences were cloned into pAS2-1 in frame with the GAL4 DNA BD at NcoI and EcoRI sites. The fragments were amplified using Pfu Turbo (Stratagene) and primers F13 and R11 for HvSPY (whole), F13 and R12 for HvSPY (TPR), and F14 and R11 for HvSPY (C-half), where F13 was 5′-TTCCATGGAGTCCCTCCAAGGG-3′; F14, 5′-TTCCATGGATTCACGAAATGCCGGT-3′; R11, 5′-TGAATTCATCGGCTGCTGTGCCC-3′; and R12, 5′-CGAATTCATGAATCAGGATCTATTTGAAG-3′. The HvSPY inserts did not contain any PCR errors when these sequences were analyzed.
GATEWAY Vectors
In order to streamline the cloning of various sequences into expression vectors, a number of expression cassettes were modified for the GATEWAY cloning technology (Invitrogen). For the Y2H prey and bait vectors pACT2 and pAS2-1, the region flanked by attR1 and attR2 sites in pDEST17 was amplified using vector primers 5′-AGGATATCGTACTACCATCACCAT-3′ and 5′-TTGAGATCTCGAATCAACCACTTTG-3′, and Pfx DNA polymerase (Invitrogen). The fragment with ccdB and chloramphenicol resistance (Cmr) genes with the attR1 and attR2 sites was digested with EcoRV and cloned into pACT2 or pAS2-1 digested with SmaI and treated with calf intestine phosphatase. Vectors with the correct frame were identified by sequence analysis, and they were named pDEST-ACT2 and pDEST-AS2-1.
For recombinant protein productions, the region flanked by aatR1 and aatR2 sites in pDEST17 was amplified with primers NarR1 5′-ATTGGCGCCCACCTCGAATCAACAAGT-3′ and KpnR2 5′-AAGGTACCTGTTAGCCTCGAATC-3′ and Pfx DNA polymerase (Invitrogen). The fragment with ccdB and Cmr genes with attR1 and attR2 sites was cloned into the EheI and KpnI sites of pProEx-1 (Invitrogen) to produce pDEST-ProEx.
To modify the effector construct vector UbiCassNotI for the transient assays, the NotI site of pDEST26 was eliminated by digesting the plasmid with NotI, the overhang was filled in using Klenow DNA polymerase (Perkin-Elmer, Foster City, CA), and the resulting blunt ends were ligated using T4 ligase to produce pDEST26 (no Not). The region flanked by attR1 and attR2 sites in pDEST26 (no Not) was amplified using vector primers 5′-ATGGTACCGCGGACCATGGCGTAC-3′ and 5′-ATATGAGCTCTCAACCACTTTGTACAAGAA-3′, and Pfx DNA polymerase (Invitrogen). The fragment with ccdB and Cmr genes with attR1 and attR2 sites was cloned into the KpnI and SacI sites of Ubi1CassNotI (Z. Li, Plant Industry, Commonwealth Scientific and Industrial Research Organisation), to produce pDEST-Ubi1CassNotI.
For the protein localization study in aleurone protoplasts, a vector was constructed to clone GFP fusion proteins under the control of a constitutive promoter, maize ubiquitin. An enhanced GFP sequence (Chui et al., 1996) was PCR amplified with Pfx (Invitrogen) using a forward primer with a BglII site (5′-AAAGATCTCCATGGTGAGCAAGGGCGAGGA-3′) and a reverse primer with a BamHI site (5′-AAGGATCCCTTGTACAGCTCGTCCATGCCG-3′). The GFP fragment was digested with BglII and ligated into the BamHI and SmaI sites in the UbiCassNotI. The resulting UbiGFPCassNotI was sequenced to confirm that there was no error from the PCR reaction. A fragment containing attR1, Cmr, ccdB, and attR2 in pDEST26 (no Not) was PCR amplified with Pfx using a forward primer with a BclI site (5′-TTTTTGATCACACTCTAGATCAACAAGTTTG-3′) and a reverse primer with a SacI site (5′-ATATGAGCTCTCAACCACTTTGTACAAGAAAG-3′). The attR fragment was digested with BclI and SacI and ligated into the BamHI and SacI sites in UbiGFPCassNotI. The resulting pDEST-UbiGFPCassNotI was sequenced to confirm the attR1 and attR2 sites and Cmr and ccdB genes were functionally tested by growing the transformants in DB3.1 cells on chloramphenicol plates and transforming the plasmid into DH5α cells, respectively.
pENTR Constructs
Open reading frames of HvSPY, HSImyb, and HSINAC were amplified with GATEWAY gene-specific primers (Invitrogen): B1HvSPY 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGAGTCCCTCCAAGGGA-3′, B2HvSPY 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATCGGCTGCTGTGCCC-3′, B1HSImyb 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGACGGAAGCCCGTGGAGT-3′, B2HSImyb 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCACTCAACTTGGAAATCAAG-3′, B1HSINAC 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGACTCGATCAAGGCCGAC-3′, and B2HSINAC 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTACTTGAAGCTTGTGCAAAC-3′ using Pfx DNA polymerase. The products were cloned into pDONR201 by BP clonase (Invitrogen) to produce pENTR-HvSPY, pENTR-HSImyb, and pENTR-HSINAC, respectively. The open reading frames in the entry constructs were confirmed not to contain any PCR errors by sequence analysis.
Expression Constructs
For recombinant protein production, HvSPY, HSImyb, and HSINAC open reading frames were cloned from pENTR constructs into pDEST-ProEx by LR clonase (Invitrogen) to produce pProEx::HvSPY, pProEx::HSImyb, and pProEx::HSINAC, respectively. Plasmid DNAs were prepared by Qiagen mini-spin columns for restriction enzyme analysis and transformation into a codon-optimized host, BL21-RIL or BL21-RP.
For one-hybrid analysis, HvSPY, HSImyb, and HSINAC open reading frames in pENTR constructs were cloned into pDEST-AS2-1 by LR clonase (Invitrogen) to produce pAS2-1::HvSPY, pAS2-1::HSImyb, and pAS2-1::HSINAC, respectively. Plasmid DNAs were prepared by Qiagen mini-spin columns for restriction enzyme analysis and transformation into yeast.
For transient assay effector constructs, open reading frames of HSImyb and HSINAC in pENTR constructs were cloned into pDEST-Ubi1Cass by LR clonase (Invitrogen) to produce Ubi::HSImyb and Ubi::HSINAC, respectively. Effector plasmid DNAs for transfection were prepared by Qiagen maxi column.
For protein localization studies, open reading frames of HvSPY, HSImyb, and HSINAC in pENTR constructs were cloned into pDEST-UbiGFPCassNotI by LR clonase (Invitrogen) to produce Ubi::GFP-HvSPY, Ubi::GFP-HSImyb, and Ubi::GFP-HSINAC, respectively. The resulting expression constructs were prepared using Qiagen mini-spin columns for restriction enzyme analysis. Plasmids used for transfection into aleurone layers and protoplasts were prepared by Qiagen maxi columns.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY672068 and AY672069 for HSImyb and HSINAC, respectively.
Supplementary Material
Acknowledgments
We thank Dr. S. Elledge (Baylor College of Medicine) for λACT2 vector, Dr. Gietz for yeast Y190 and advice on yeast transformation, Drs. F. Gubler and Z. Li for SLN cDNA and the UbiCassNotI, Jan van de Velde for results in Figure 6, and Judy Radik and Robyn East for technical assistance.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.041665.
References
- Agatep R, Kirkpatrick RD, Parchaliuk DL, Woods RA, Gietz RD (1998) Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Technical Tips Online. http://tto.trends.com (January 1999)
- Aida M, Ishida T, Fukaki H, Fujusawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci USA 96: 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Schuurink R, Jones RL (1997) Hormonal signalling in cereal aleurone. J Exp Bot 48: 1337–1356 [Google Scholar]
- Boss PK, Thomas MR (2002) Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416: 847–850 [DOI] [PubMed] [Google Scholar]
- Bush DS, Cornejo M-J, Huang C-N, Jones RL (1986) Ca2+-stimulated secretion of α-amylase during development in barley aleurone protoplasts. Plant Physiol 82: 566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002) Mutants at the Slender1 locus of barley cv Himalaya: molecular and physiological characterization. Plant Physiol 129: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Robertson M (1999) Gibberellin dose-response curves and the characterization of dwarf mutants in barley. Plant Physiol 120: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Hart GW (2001) Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor. Post-translational regulation of turnover and transactivation activity. J Biol Chem 276: 10570–10575 [DOI] [PubMed] [Google Scholar]
- Chui W-L, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter gene in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B (2000) Finding nuclear localization signals. EMBO Rep 1: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AK, Cohen PTW, Barford D (1998) The structure of the tetretricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J 17: 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Mulligan JT, Ramer SW, Spottswood M, Davis RW (1991) λYES: a multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Echerichia coli mutations. Proc Natl Acad Sci USA 88: 1731–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck B, Harberd NP (2002) Evidence that the Arabidopsis nuclear gibberellin signaling protein GAI is not destabilised by gibberellin. Plant J 32: 935–947 [DOI] [PubMed] [Google Scholar]
- Foster CA (1977) Slender: an accelerated extension growth mutant of barley. Barley Genet Newsl 7: 24–27 [Google Scholar]
- Fridborg I, Kuusk S, Moritz T, Sundberg E (1999) The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 11: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Ait-ali T, Hynes LW, Ougham H, Peng J, Harberd NP (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14: 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y (1999) Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA 96: 7575–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Miyazaki J-I, Hart GW (2003) The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 β–cells. Arch Biochem Biophys 415: 155–163 [DOI] [PubMed] [Google Scholar]
- Gilroy S, Jones RL (1994) Perception of gibberellin and abscisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L.) aleurone protoplasts. Plant Physiol 104: 1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M, Yanagida M (1991) The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci 16: 173–175 [DOI] [PubMed] [Google Scholar]
- Greb T, Schmitz G, Theres K (2002) Isolation and characterization of the Spindly homologue from tomato. J Exp Bot 53: 1829–1830 [DOI] [PubMed] [Google Scholar]
- Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV (2002) Gibberellin signaling in barley aleurone cells: control of SLN1 and GAMYB expression. Plant Physiol 129: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV (1995) Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV (1999) Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J 17: 1–9 [DOI] [PubMed] [Google Scholar]
- Hanover JA (2001) Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J 15: 1865–1876 [DOI] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Hiromura M, Choi CH, Sabourin NA, Jones H, Bachvarov D, Usheva A (2003) YY1 is regulated by O-linked N-acetylglucosaminylation (O-GlcNAcylation). J Biol Chem 278: 14046–14052 [DOI] [PubMed] [Google Scholar]
- Hooley R, Beale MH, Smith SJ (1991) Gibberellin perception at the plasma membrane of Avena fatua aleurone protoplasts. Planta 183: 274–280 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki A, Swain SM, Tseng T-s, Borochov A, Olszewski NE, Weiss D (2001) The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J 28: 181–190 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93: 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE, Meyerowitz EM (1997) SPINDLY's role in the gibberellin response pathway. Symp Soc Exp Biol 51: 73–78 [PubMed] [Google Scholar]
- Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano H-Y (2000) Molecular analysis of the NAC gene family in rice. Mol Gen Genet 262: 1047–1051 [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Blomberg MA, Hart GW (1997) Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterisation of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem 272: 9308–9315 [DOI] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P (1995) Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci 20: 257–259 [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulated Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C-A, Ho T-hD, Ho S-L, Yu S-M (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14: 1963–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas WA, Frank DW, Krause M, Hanover JA (1997) O-Linked GlcNAc transferase is a conserved nucleocytoplasmid protein containing tetratricopeptide repeats. J Biol Chem 272: 9316–9324 [DOI] [PubMed] [Google Scholar]
- Lubas WA, Hanover JA (2000) Functional expression of O-linked GlcNAc transferase: domain structure and substrate specificity. J Biol Chem 275: 10983–10988 [DOI] [PubMed] [Google Scholar]
- Lyer SPN, Hart GW (2003) Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J Biol Chem 278: 24608–24616 [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-p, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Kusano T, Katsumi M, Sano H (2000) Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 245: 21–29 [DOI] [PubMed] [Google Scholar]
- Olszewski N, Sun T-p, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism and response pathways. Plant Cell 14 (Suppl): S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, et al (1999. a) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Moritz T, Cano-Delgado A, Harberd NP (1999. b) Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin responses. Plant Physiol 119: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J (1998) HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem 273: 23313–23320 [DOI] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-ali T, Harberd NP (2001) How gibberellin regulated plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52: 67–68 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Robertson M (2003) Increased dehydrin promoter activity caused by HvSPY is independent of the ABA response pathway. Plant J 34: 39–46 [DOI] [PubMed] [Google Scholar]
- Robertson M, Cuming AC, Chandler PM (1995) Sequence analysis and hormonal regulation of a dehydrin promoter from barley, Hordeum vulgare. Physiol Plant 94: 470–478 [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE (1998) Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 10: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos MD, Su K, Baker JR, Kudlow JE (1997) O-glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol Cell Biol 17: 6472–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong D-H, An G, Kitano H, Ashikari M, et al (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegorano S, Moroder L, Bartunik H, Hartl FU, Moarefi S (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101: 199–210 [DOI] [PubMed] [Google Scholar]
- Shaw P, Freeman J, Bovey R, Iggo R (1996) Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene 12: 921–930 [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T-p (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transcription pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun T-p (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martínez EC, Sun T-p (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The No Apical Meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordial boundaries. Cell 85: 159–170 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Swain SM, Tseng T-S, Thornton TM, Gopalraj M, Olszewski NE (2002) SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol 129: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof KD, Hobbs CA (1987) Improved media for growing plasmid and cosmid clones. Bethesda Res Lab Focus 9: 12 [Google Scholar]
- Thornton T, Kreppel L, Hart G, Olszewski N (1999) Genetic and biochemical analysis of arabidopsis SPY. In Altman A, Ziv M, Izhar S, eds, Plant Biotechnology and In Vitro Biology in the 21st Century. Kluwer Academic Publishers, Jerusalem, pp 445–448
- Tseng T-S, Swain SM, Olszewski NE (2001) Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol 126: 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C-K, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Su K, Roos MD, Chang Q, Paterson AJ, Kudlow JE (2001) O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc Natl Acad Sci USA 98: 6611–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.