Summary
Hippocalcin (HPCA) is a calcium-binding protein that is restricted to nervous tissue and contributes to neuronal activity. Here we report that, in addition to inducing neurogenesis, HPCA inhibits astrocytic differentiation of neural stem cells. It promotes neurogenesis by regulating protein kinase Cα (PKCα) activation by translocating to the membrane and binding to phosphoinositide-dependent protein kinase 1 (PDK1), which induces PKCα phosphorylation. We also found that phospholipase D1 (PLD1) is implicated in the HPCA-mediated neurogenesis pathway; this enzyme promotes dephosphorylation of signal transducer and activator of transcription 3 (STAT3[Y705]), which is necessary for astrocytic differentiation. Moreover, we found that the SH2-domain-containing tyrosine phosphatase 1 (SHP-1) acts upstream of STAT3. Importantly, this SHP-1-dependent STAT3-inhibitory mechanism is closely involved in neurogenesis and suppression of gliogenesis by HPCA. Taken together, these observations suggest that HPCA promotes neuronal differentiation through activation of the PKCα/PLD1 cascade followed by activation of SHP-1, which dephosphorylates STAT3(Y705), leading to inhibition of astrocytic differentiation.
Keywords: hippocalcin (HPCA), neural stem cells, neurogenesis, gliogenesis, phospholipase D1 (PLD1), signal transducers and activator of transcription 3 (STAT3), phosphoinositide-dependent protein kinase-1 (PDK1), SH2-domain-containing tyrosine phosphatase-1 (SHP-1)
Highlights
-
•
Hippocalcin is required for neuronal differentiation in neural stem cells
-
•
PKCα/PLD1 activation is required for hippocalcin-mediated neuronal differentiation
-
•
Blocking of STAT3(Y705) activity by hippocalcin decreases astrocytic differentiation
-
•
Hippocalcin promotes neurogenesis by inhibiting gliogenesis in neural stem cells
In this article, Han and colleagues investigate the role of hippocalcin in neuronal differentiation of neural stem cells. They show that hippocalcin promotes neuronal differentiation through activation of the PKCα/PLD1 cascade followed by activation of SHP-1, which dephosphorylates STAT3(Y705), leading to inhibition of astrocytic differentiation. These findings suggest that hippocalcin functions as an on-off switch and a main regulator for neuronal differentiation of neural stem cells.
Introduction
Neural stem cells (NSCs), defined as neural precursor cells that can self-renew, give rise to three major cell types: neurons, astrocytes, and oligodendrocytes (Miller and Gauthier, 2007, Qian et al., 2000). Multipotent NSCs can be isolated and cultured from primary cortical or hippocampal cultures after passage in the presence of mitogenic growth factors (Gage et al., 1995). In cultures of rat embryonic day 14 (E14) cells, cortical NSCs give rise primarily to neurons and dividing precursor cells; astrocytes are generated only when cells are cultured for several days (Ghosh and Greenberg, 1995). The molecular mechanisms by which NSCs undergo differentiation into neurons versus astrocytes are not well understood.
Hippocalcin (HPCA) is a high-affinity calcium-binding protein restricted to the CNS and is most abundant in pyramidal cells of the hippocampus CA1 region (Kobayashi et al., 2012). During brain development HPCA expression increases sharply, concurrent with synapse formation (Saitoh et al., 1994). HPCA belongs to the family of EF-hand-containing neuronal calcium sensor proteins, which possess a Ca2+/myristoyl switch that allows translocation to membranes in response to increased cytosolic Ca2+ concentrations. It also regulates mixed-lineage kinase 2, phospholipase D (PLD), and neuronal apoptosis inhibitory protein (Burgoyne and Weiss, 2001, Kobayashi et al., 1993, Lindholm et al., 2002, Nagata et al., 1998, O'Callaghan et al., 2002, O'Callaghan et al., 2003). We have reported that HPCA is a major regulatory protein in the Ca2+-mediated PLD signaling pathway (Oh et al., 2006) and that it induces expression of Neuro-D, leading to neurite outgrowth in H19-7 cells during differentiation (Oh et al., 2008). It is possible that HPCA regulates neurogenesis through these molecules; however, molecular mechanisms by which HPCA affects neuronal/astrocyte cell-fate decision have not been studied.
PLD is a ubiquitous enzyme that catalyzes the hydrolysis of phosphatidylcholine (PC) to phosphatidic acid and choline (Exton, 1997). Phosphatidic acid (PA) itself acts as a cellular messenger or can be transformed by PA phosphohydrolase into diacylglycerol, which is essential for activation of protein kinase C (PKC) (Manifava et al., 2001, Zhao et al., 2007). PKCα regulates Ca2+-dependent differentiation in several cell lines and primary cells (Kopach et al., 2013, Park et al., 2015) and plays an essential role in synaptic plasticity by raising intracellular Ca2+ levels in neurons (Kopach et al., 2013). Activation and phosphorylation of PLD1 are regulated by PKCα in phorbol myristate acetate-treated COS-7 cells, and a similar interrelationship between PLD and PKC isoforms is seen in a variety of cell types (Kim et al., 2005). Recent studies have reported that PLD plays a key role in neuronal differentiation of cells such as PC12 cells (Banno et al., 2008) and cerebellar granule neurons (Watanabe et al., 2004). We have also reported that it has a critical role in the neuronal differentiation of rat NSCs (Yoon et al., 2005) and H19-7 cells (Yoon et al., 2012). However, the relevance of PLD1 signaling in neuronal differentiation remains unclear.
The signal transducer and activator of transcription (STAT) family participates in the regulation of genes involved in the acute phase of inflammatory response, cell growth, and cell differentiation (Park et al., 2010). Among them, STAT3 is an important transcription factor for the regulation of glial fibrillary acidic protein (GFAP) expression, and the DNA binding of STAT3 was shown to be affected by phosphorylation of the Ser727 or/and Tyr705 site (Yokogami et al., 2000). STAT3 binds to different domains of CBP/p300, and the STAT/p300/Smad complex, acting at the STAT-binding element in the astrocyte-specific GFAP promoter, is particularly effective at inducing astrocyte differentiation in NSCs (Nakashima et al., 1999). Moreover, STAT3 signaling is upregulated in certain neurodegenerative diseases (Shibata et al., 2009), and in vitro suppression of Stat3 or its conditional deletion in vivo promoted neurogenesis and inhibited astrogliogenesis (Cao et al., 2010, Gu et al., 2005). Thus, STAT3 is considered an attractive target for promoting neurogenesis. In our previous study, STAT3 activation is associated with PLD2 through the S6K1-ERK pathway in lipopolysaccharide (LPS)-induced inflammation mechanism (Park et al., 2010), but the relationship between PLD1 signaling and STAT3 function is not yet defined. Thus, the present study showed that PLD1 is required for HPCA-mediated STAT3 activation of neuronal differentiation. In addition, a number of protein tyrosine phosphatases negatively regulate STAT3 signaling through direct dephosphorylation of p-STAT3(Y705); these include members of the SH2-domain-containing tyrosine phosphatase family (SHP-1 and SHP-2) and protein tyrosine phosphatase 1B (PTP-1B) (Han et al., 2006). More specifically, SHP-1 regulates STAT3(Y705) phosphorylation in Huh-7 HCC, PLC5, and HepG2 cells (Chen et al., 2012). Thus, activity of SHP-1 may be critical for regulating STAT3 phosphorylation in neuronal differentiation.
In this study, we aimed to clarify the role of HPCA in the neuronal differentiation of NSCs. Our findings indicate that HPCA is essential for neurogenesis of NSCs, and that it promotes neuronal differentiation and inhibits astrocytic differentiation.
Results
HPCA Is Required for Neuronal Differentiation in NSCs
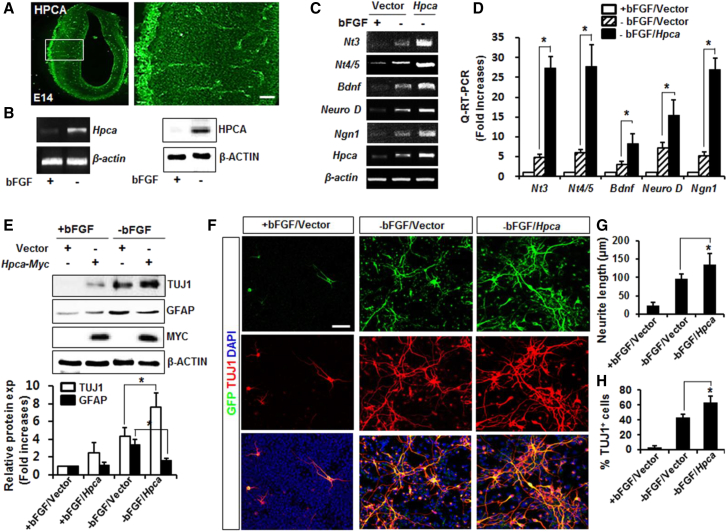
Many studies of the neurogenic-to-gliogenic switch have focused on the developing neocortex (Qian et al., 2000, Shen et al., 2006). We show here that HPCA is expressed in the cerebral neocortex of the E14 rat brain (Figure 1A) and examine its possible role in neuronal differentiation using cortical NSCs. During growth of these cells, basic fibroblast growth factor (bFGF) was present to prevent differentiation and promote proliferation. To investigate the role of HPCA in neuronal differentiation, we removed bFGF for 24 hr. As shown in Figure 1B, mRNA expression of Hpca and the protein level of HPCA were markedly increased under differentiation conditions. Nerve growth factors such as NT-3, NT4/5, and BDNF, together with the basic helix-loop-helix transcription factors Neuro-D and neurogenin-1 (NGN1), are closely associated with neuronal differentiation and can be used as markers of this process (Markus et al., 2002, Shin-young et al., 2007). Therefore, we generated NSCs that overexpressed Hpca and monitored the levels of neuronal differentiation markers. As shown in Figures 1C and 1D, the expression levels of Nt3, Nt4/5, Bdnf, Neuro-D, and Ngn1 were significantly enhanced by Hpca overexpression compared with the vector control in the absence of bFGF. NSCs are considered as the primary progenitor cells for neuronal and glial cell lineages during development (Rietze et al., 2001). We examined the effects of HPCA on the expression of neuronal and glial markers during neuronal differentiation. In the absence of bFGF, Hpca overexpression resulted in markedly enhanced expression of neuron-specific class III β-tubulin (TUJ1, a neuronal marker), while GFAP expression was significantly decreased by Hpca when compared with the vector control (Figure 1E). These data suggest that HPCA promotes neuronal differentiation and suppresses astrocyte differentiation in NSCs.
Figure 1.
Effect of HPCA Expression during Neuronal Differentiation of NSCs
(A) HPCA immunostaining of coronal sections of E14 rat brain cortex. The boxed area is magnified. Scale bar, 20 μm.
(B) Neuronal differentiation was induced by removal of bFGF for 1 day, and Hpca mRNA expression was analyzed by RT-PCR. Twenty micrograms of protein was analyzed by western blotting with anti-HPCA and anti-β-ACTIN.
(C and D) NSCs were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and allowed to differentiate for 24 hr. mRNA levels of neuronal factors were analyzed by RT-PCR (C) and real-time RT-PCR (D). ∗p < 0.05 compared with the −bFGF/vector control (mean ± SD; n = 3).
(E) Cells were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and induced to differentiate for 24 hr. Levels of TUJ1, GFAP, MYC, and β-ACTIN were determined by western blot. ∗p < 0.05 compared with the −bFGF/vector control (mean ± SD; n = 3).
(F) Cells were transduced with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc and induced to differentiate by withdrawal of bFGF. After 3 days, GFP-positive cells were examined by fluorescence microscopy and stained with anti-EGFP (green) and anti-TUJ1 (red). Scale bar, 20 μm.
(G and H) Neurite lengths were measured and the proportions of TUJ1-positive cells and total cells were determined in randomly selected areas from at least three slides of each condition. ∗p < 0.05 compared with the −bFGF/vector control (mean ± SD; n = 3).
We reported previously that HPCA leads to neurite outgrowth of H19-7 cells (Oh et al., 2008). To confirm its role in neurite outgrowth in NSCs, we exposed cells to EGFP-tagged Hpca for 2 days. After 3 days of differentiation, neurite outgrowth was measured under a fluorescence microscope (EGFP-transfected cells fluoresce green, TUJ1-stained cells fluoresce red, and cells transfected with both EGFP and TUJ1 fluoresce yellow) (Figure 1F). Neurons and astrocytes were quantified by normalizing the total number of TUJ1- and GFAP-positive cells to the total number of DAPI-labeled cell nuclei. When Hpca was introduced, neurite outgrowth (97.1 ± 8.3 μm versus 135.2 ± 14.1 μm, p < 0.05, Figure 1G) and the number of TUJ1-positive cells (43.2% ± 5.5% versus 64.1% ± 9.9%, p < 0.05, Figure 1H) were enhanced, indicating that HPCA plays an important role in NSC neurite outgrowth.
PKCα Is Involved in HPCA-Mediated Expression of TUJ1 in NSCs
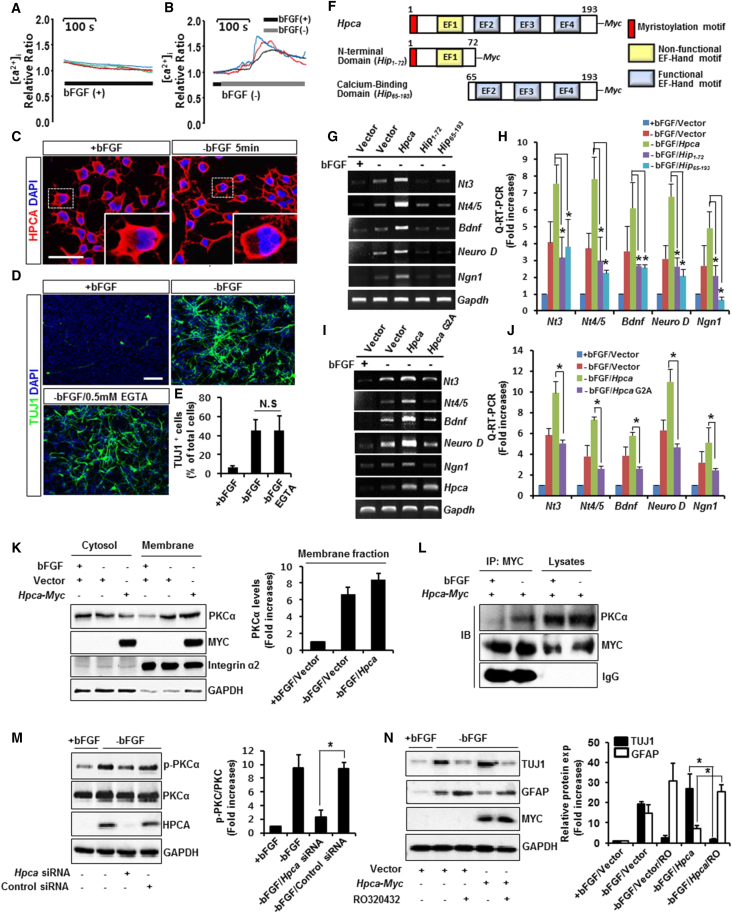
HPCA is a Ca2+-binding protein in the neuronal Ca2+ sensor family of proteins, which possess a Ca2+/myristoyl switch that allows translocation to membranes in response to increased cytosolic Ca2+ levels (Palmer et al., 2005). Therefore, we analyzed intracellular Ca2+ mobilization in fluo-3-loaded NSCs. When the cells were induced to differentiate by removal of bFGF, intracellular Ca2+ levels increased (Figures 2A and 2B). We also observed translocation of HPCA from the cytosol to the membrane after bFGF removal (Figure 2C), suggesting that binding of Ca2+ to HPCA induces a conformational change that promotes membrane association.
Figure 2.
Increased Intracellular Ca2+ Affects HPCA-PKCα Activation during Neuronal Differentiation of NSCs
(A and B) NSCs were used for fluo-3/AM Ca2+ imaging 1 day after plating on poly-L-lysine-coated glass coverslips (see Experimental Procedures). We collected images every 1 s at 470 nm excitation/514 nm emission using Metafluor software. The threshold of increased intracellular Ca2+ was defined as a 20% increase from the basal level.
(C) Cells were depleted with bFGF for 5 min, fixed, and immunostained with anti-HPCA (red). The boxed area is magnified. Scale bar, 5 μm.
(D and E) Cells were pretreated with 500 μM EGTA for 1 day and incubated for 3 days after removal of bFGF. After immunostaining with anti-TUJ1, TUJ1-positive cells were counted under fluorescence microscopy. The proportion of TUJ1-positive cells and total cells was determined in random areas from at least three slides of each condition. Data are shown as mean ± SD (n = 3). N.S, not significant (p = 0.245). Scale bar, 20 μm.
(F) Schematic of Hpca (wild-type) and its deletion mutants.
(G and H) NSCs were transfected with pMSCV-IRES-EGFP, pMSCV-IRES-EGFP-Hpca-Myc, pMSCV-IRES-EGFP-Hip1-72-Myc, or pMSCV-IRES-EGFP-Hip65-193-Myc for 48 hr and induced to differentiate for 24 hr. mRNA levels of neuronal factors were analyzed by RT-PCR (G) and real-time RT-PCR (H). ∗p < 0.05 compared with −bFGF/Hpca (mean ± SD; n = 3).
(I and J) G2 is a key residue for myristoylation of HPCA. NSCs were transfected with pMSCV-IRES-EGFP, pMSCV-IRES-EGFP-Hpca-Myc, or pMSCV-IRES-EGFP-Hpca G2A-Myc for 48 hr and induced to differentiate for 24 hr. mRNA levels of neuronal factors were analyzed by RT-PCR (I) and real-time RT-PCR (J). ∗p < 0.05 compared with −bFGF/Hpca (mean ± SD; n = 3).
(K) Cells were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and induced to differentiate by withdrawal of bFGF for 24 hr. Cells were then lysed and centrifuged at 100,000 × g to separate cytosolic and membrane fractions. Samples of protein (30 μg) from each fraction were analyzed by western blotting with anti-PKCα, anti-MYC, anti-integrin α2, and anti-GAPDH. Data are shown as mean ± SD (n = 3).
(L) Cells were transfected with p-MSCV-IRES-EGFP-Hpca-Myc for 48 hr, and induced differentiate for 24 hr. Immunoprecipitates (IP) from the cell lysates with anti-MYC were analyzed by western blotting with anti-PKCα and anti-MYC.
(M) Cells were transiently transfected with control siRNA or Hpca siRNA for 48 hr, then incubated for 1 day after removal of bFGF. Proteins were analyzed by western blotting with anti-p-PKCα, anti-PKCα, anti-HPCA, and anti-GAPDH. ∗p < 0.05 compared with the −bFGF/control siRNA (mean ± SD; n = 3).
(N) Cells were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and then treated with 10 μM RO320432 for 30 min before differentiation for 1 day. Cells were lysed and analyzed by western blotting with anti-TUJ1, anti-GFAP, anti-MYC, and anti-GAPDH. ∗p < 0.05 compared with −bFGF/Hpca (mean ± SD; n = 3).
To confirm that increased Ca2+ levels represent intracellular Ca2+, we pretreated cells with EGTA, an extracellular Ca2+chelator, before inducing differentiation. The EGTA had no effect on neurite outgrowth (Figure 2D) or TUJ1-positive cells (Figure 2E), indicating that HPCA is responsible for the locational changes induced by the increase of intracellular Ca2+. HPCA has three putative Ca2+-binding domains of the EF-hand structure and a possible NH2-terminal myristoylation site, which is referred to as a “calcium/myristoyl switch” and considered to be primarily a mechanism by which the protein can be translocated from the cytosol to the membrane. Both structural features are known to regulate the intracellular Ca2+-dependent activation of HPCA. To determine whether Ca2+ binding is required for the expressions of HPCA-mediated nerve growth factors, we constructed Hip1-72 (N-terminal domain without all three functional EF hands [EF2–4]) and Hip65-193 (calcium-binding domain without myristoylation motif) deletion mutants using the pMSCV-IRES-EGFP-Hpca-Myc (Figures 2F and S1). As shown in Figures 2G and 2H, Hpca-induced expression levels of Nt3, Nt4/5, Bdnf, Neuro-D, and Ngn1 were significantly decreased by the Hip1–72 (N-terminal domain) or Hip65–193 (calcium-binding domain). This result indicated that HPCA-mediated expressions of nerve growth factors are dependent on Ca2+ binding as well as the myristoylation of HPCA during neuronal differentiation. To further investigate whether HPCA-mediated expressions of nerve growth factors depend on myristoylation, we made constructs with the second glycine residue, essential for myristoylation, mutated to an alanine (G2A) in HPCA protein. As shown in Figures 2I and 2J, G2A decreased Hpca-induced expression levels of Nt3, Nt45, Bdnf, Neuro-D, and Ngn1 in the absence of bFGF. This result suggests that myristoylation is essential for expression of nerve growth factors during neuronal differentiation of NSCs.
Interestingly, increased Ca2+ levels regulate PKCα activation and translocation to the membrane from the cytosol in various processes (Boncoeur et al., 2013, Champion and Kass, 2004). Therefore, we analyzed translocation of PKCα after depleting bFGF. Translocation of PKCα from the cytosol to the membrane occurred when neuronal differentiation was induced (Figure 2K). In addition, the translocation was further enhanced by overexpression of Hpca (Figure 2K). Based on our results, we hypothesized that Ca2+-bound HPCA binds PKCα after removal of bFGF. To test this hypothesis, we used immunoprecipitation to determine whether HPCA directly binds to PKCα during neuronal differentiation. As shown in Figure 2L, HPCA binds to PKCα after removal of bFGF, indicating that Ca2+-binding HPCA directly binds to PKCα. Next, to investigate whether HPCA affects PKCα activation, we knocked down Hpca in NSCs using Hpca siRNA. This resulted in decreased phosphorylation of PKCα (Figure 2M). To further investigate whether PKCα regulates HPCA-mediated neuronal differentiation, we pretreated NSCs with the PKCα inhibitor RO320432 for 30 min before inducing differentiation by removing bFGF. Inhibition of PKCα markedly reduced Hpca-induced expression of TUJ1, whereas GFAP increased (Figure 2N), demonstrating that PKCα is implicated in HPCA-mediated neuronal differentiation.
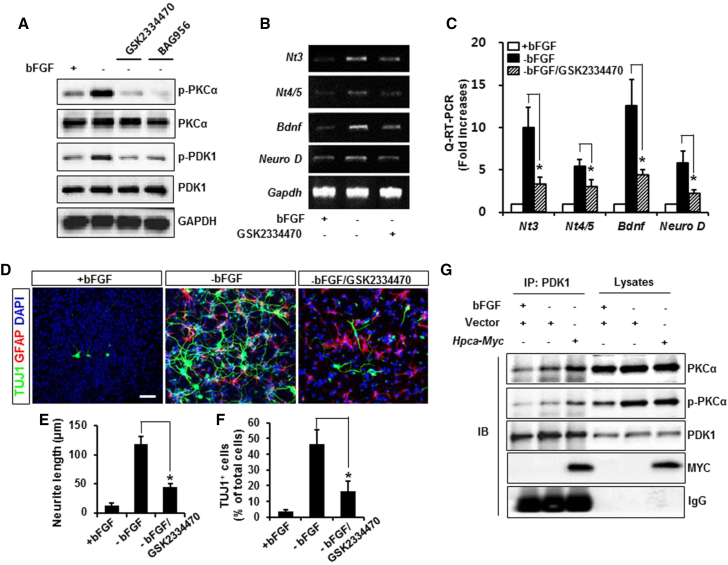
We next asked which signaling pathway was responsible for PKCα activation during neuronal differentiation in NSCs. PDK1 plays a central role in many signal transduction pathways by phosphorylating a site in the activation loop of PKC isoforms and initiating their maturation (Dutil et al., 1998, Williams et al., 2000). To determine whether PDK1 could enhance phosphorylation of PKCα, we pretreated cells with the PDK1 inhibitors GSK2334470 or BAG956 for 30 min. Inhibition of PDK1 abolished phosphorylation of PKCα, indicating that PDK1 regulates PKCα activation during neuronal differentiation (Figure 3A). In addition, GSK2334470 not only decreased transcripts of Nt3, Nt4/5, Bdnf, and Neuro-D (Figures 3B and 3C) but also significantly reduced neurite length (125 ± 14.3 μm versus 45.3 ± 3.3 μm, p < 0.05, Figures 3D and 3E) and the number of TUJ1-positive cells (47.2% ± 10% versus 18.5% ± 4%, p < 0.05, Figure 3F) when compared with removal of bFGF. Taken together, these results demonstrate that PDK1 signals upstream of PKCα trigger neurite outgrowth, leading to expression of neuronal factors in NSCs. However, the mechanism by which PDK1 affects HPCA-mediated PKCα activation is still unknown. We hypothesized that HPCA brings PKCα to PDK1, after which PDK1 activates PKCα. To test this hypothesis, we used co-immunoprecipitation to determine whether HPCA contributes to binding between PDK1 and PKCα. HPCA stimulated this binding and promoted PKCα activation (Figure 3G), supporting the idea that HPCA induces the formation of a PDK1-PKCα complex that promotes PKCα phosphorylation during neuronal differentiation.
Figure 3.
PDK1 Is Involved in HPCA-Mediated PKCα Activation and Neuronal Differentiation of NSCs
(A) NSCs were pretreated with 5 μM GSK2334470 or 10 μM BAG956 for 30 min before differentiation for 1 day. Cells were lysed and analyzed by western blotting with anti-p-PKCα, anti-PKCα, anti-p-PDK1, anti-PDK1, and anti-GAPDH.
(B and C) Cells were pretreated with 5 μM GSK 2334470 and allowed to differentiate for 1 day. mRNA levels of neuronal factors were analyzed by RT-PCR (B) and real-time RT-PCR (C). ∗p < 0.05 compared with the −bFGF control (mean ± SD; n = 3).
(D) Cells were pretreated with 5 μM GSK2334470 and induced to differentiate by withdrawal of bFGF. After differentiation for 3 days, fixed cells were immunostained with anti-TUJ1 (green) and anti-GFAP (red). Scale bar, 20 μm.
(E and F) Neurite lengths were measured and the numbers of TUJ1-positive cells and total cells were determined in randomly selected areas from at least three slides of each condition. ∗p < 0.05 compared with the −bFGF control (mean ± SD; n = 3).
(G) Cells were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr, and induced to differentiate for 24 hr. Immunoprecipitates from the cell lysates with anti-PDK1 were analyzed by western blotting with anti-p-PKCα, anti-PKCα, anti-PDK1, and anti-MYC.
Effect of PLD1 Activation on HPCA-Mediated Neuronal Differentiation
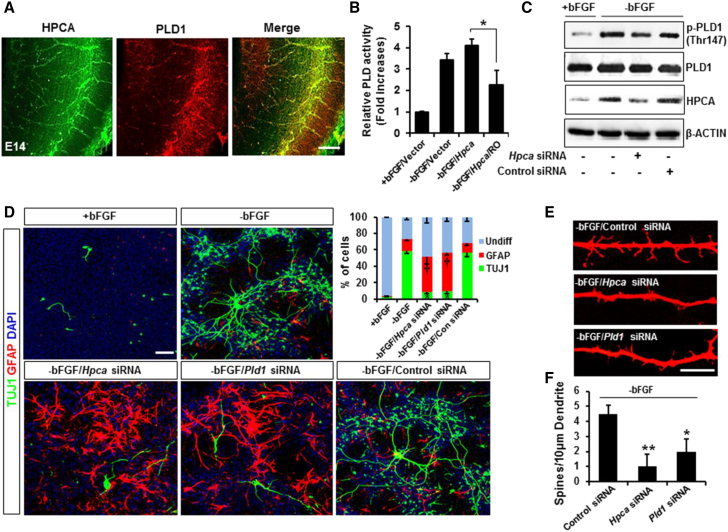
We previously reported that PLD1 is involved in various neuronal signaling pathways and is an important regulator of neuron function (Yoon et al., 2006, Yoon et al., 2012). We also showed that HPCA increased the activity and expression of PLD in various cell types (Hyun et al., 2000, Oh et al., 2006). In the present study, we found that PLD1 co-localized with HPCA in the cerebral neocortex of the E14 rat brain (Figure 4A) and therefore hypothesized that PLD1 was implicated in the HPCA-mediated neuronal differentiation of NSCs. Since PLD1 is regulated by PKCα in NSCs (Park et al., 2015), we examined whether HPCA-activated PKCα affects PLD activation during neuronal differentiation. We found that in the absence of bFGF, Hpca-induced PLD activation was decreased by the PKCα inhibitor RO320432 (Figure 4B). To further elucidate the role of HPCA in PLD activation, we transfected cells with Hpca small interfering RNA (siRNA). Knockdown of Hpca markedly inhibited PLD1 phosphorylation, which is related to PLD1 activation (Figure 4C). To determine the role of HPCA-mediated PLD1 activation in neuronal differentiation, we knocked down Hpca or Pld1 using siRNA and found that the number of TUJ1-positive cells significantly decreased due to knockdown of Hpca or Pld1, whereas the number of GFAP-positive cells increased (Figure 4D). These results suggest that HPCA-dependent PLD1 activation is required for neuronal differentiation of NSCs.
Figure 4.
PLD1 Is Required for Hpca-Induced Neuronal Differentiation of NSCs
(A) Coronal sections of E14 rat brain cortex immunostained with anti-HPCA and anti-PLD1. Scale bar, 10 μm.
(B) NSCs were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and labeled with 2 μCi/mL [3H]palmitic acid. They were pretreated with 10 μM RO320432 for 30 min and allowed to differentiate for 1 day. PLD activities were determined by measuring the formation of [3H]PBt in the presence of 1-butanol. ∗p < 0.05 compared with −bFGF/Hpca (mean ± SD; n = 5).
(C) Cells were transfected with control siRNA or Hpca siRNA for 48 hr and then incubated for 1 day after removal of bFGF. Proteins were analyzed by western blotting with anti-p-PLD1 (T147), anti-PLD1, anti-HPCA, and anti-β-ACTIN.
(D) Cells were transfected with control siRNA, Hpca siRNA, or Pld1 siRNA for 48 hr and induced to differentiate through withdrawal of bFGF. After differentiation for 3 days, cells were stained for immunocytochemical analysis of neuronal (TUJ1, green) and astrocytic (GFAP, red) markers. Scale bar, 10 μm. Graphs show the percentages of TUJ1- and GFAP-positive cells. Data are shown as mean ± SD (n = 3). ∗Significantly different from −bFGF/control siRNA at p < 0.05 (for TUJ1). †Significantly different from −bFGF/control siRNA at p < 0.05 (for GFAP).
(E) Cells were transfected with control siRNA, Hpca siRNA, or Pld1 siRNA for 48 hr and induced to differentiate by withdrawal of bFGF. After differentiation for 3 days, fixed cells were immunostained with anti-TUJ1. Scale bar, 10 μm.
(F) The numbers of spines per 10-μm dendrite were counted in randomly selected areas from at least three slides of each condition. Data are shown as mean ± SD (n = 5). ∗p < 0.05 compared with the −bFGF/control siRNA. ∗∗p < 0.01 compared with the −bFGF/control siRNA.
To test whether HPCA and PLD1 promote spine formation, which implies synapse formation during neuronal differentiation, we examined dendritic spine formation in cells transfected with siRNA for Hpca or Pld1. Dendritic spines were significantly decreased by knockdown of Hpca or Pld1 (Figures 4E and 4F). Taken together, these results indicate that PLD1 plays an important role in HPCA-mediated neuronal differentiation of NSCs.
Inhibition of STAT3(Y705) Activation Increases Neuronal Differentiation in NSCs
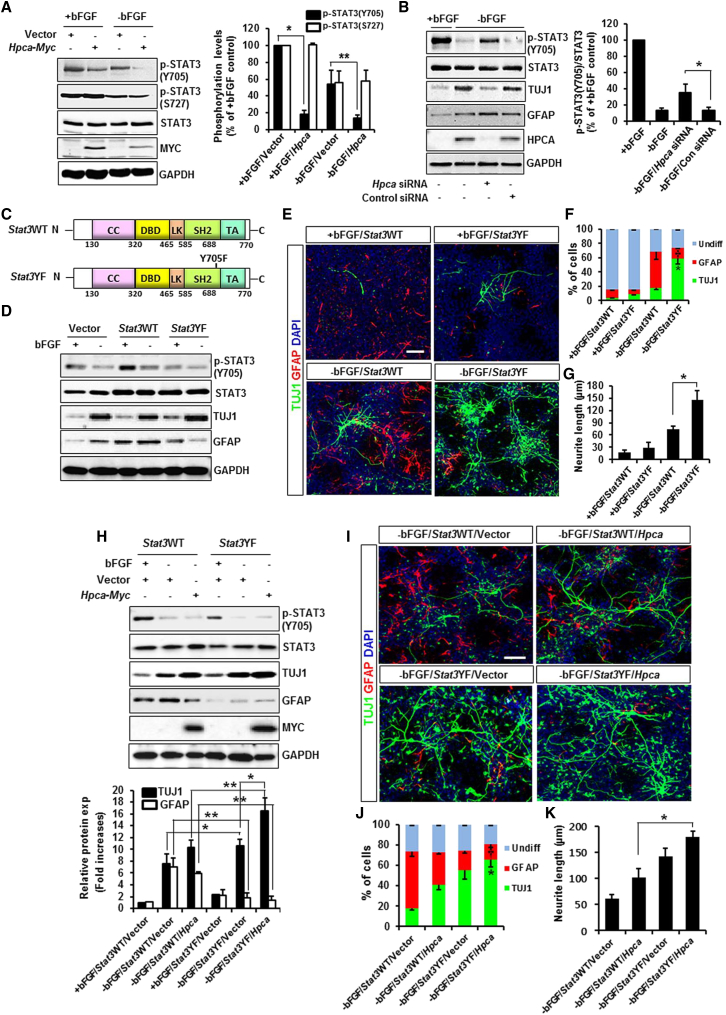
The determination of neuronal/glial fate during CNS development involves complex interactions between intrinsic signals that act through many transcription factors (Edlund and Jessell, 1999). STAT3 inhibition enhances differentiation of human NSCs to motor neurons and reduces astrocytosis, and similarly induces neurogenesis and inhibits gliogenesis (Cao et al., 2010, Natarajan et al., 2014), supporting our hypothesis that STAT3 activation inhibits neuronal differentiation and promotes astrocytic differentiation. We therefore examined whether HPCA affects STAT3 activation. Hpca was transfected into NSCs for 2 days and differentiation was induced for 1 day. As shown in Figure 5A, Hpca increased the dephosphorylation of p-STAT3(Y705) compared with the vector control, even in the absence of bFGF, but did not affect p-STAT3(S727). Moreover, bFGF removal-induced p-STAT3(Y705) dephosphorylation and TUJ1 expression were abolished by Hpca siRNA while GFAP expression was enhanced (Figures 5B and S2), suggesting that Hpca-induced dephosphorylation of p-STAT3(Y705) is crucial for the promotion of neuronal differentiation. To further investigate whether dephosphorylation of p-STAT3(Y705) is essential for the neuronal differentiation of NSCs, we prepared Stat3YF mutant (point mutation of tyrosine 705 to phenylalanine 705). When Stat3WT (wild-type) and Stat3YF were transfected into HEK293 cells, respectively, STAT3 was successfully overexpressed (Figure S3). We validated Stat3YF mutant by showing the disappearance of LPS-induced tyrosine phosphorylation of STAT3(Y705) (Figure S3). Next, we determined the effects of Stat3YF on the expression of neuronal and glial markers during differentiation of NSCs. Stat3YF resulted in markedly enhanced TUJ1 expression while GFAP expression was significantly decreased by removal of bFGF when compared with the Stat3WT (Figure 5D). In addition, Stat3YF significantly increased the number of TUJ1-positive cells (63.4% ± 9% versus 19.2% ± 2%, p < 0.05, Figures 5E and 5F) and neurite length (147.2 ± 20.3 μm versus 75.8 ± 6.9 μm, p < 0.05, Figures 5E and 5G), whereas it decreased the number of GFAP-positive cells (18.8% ± 1.2% versus 52.5% ± 10.1%, Figures 5E and 5F) when compared with Stat3WT with bFGF removal. Consistent with these results, Stat3YF markedly enhanced expression levels of Nt3, Nt4/5, Bdnf, Neuro-D, and Ngn1 by withdrawal of bFGF when compared with the Stat3WT (Figure S4). These results indicate that inhibition of STAT3(Y705) activity promotes neuronal differentiation accompanied by suppressed astrocytic differentiation.
Figure 5.
Functional Analysis of STAT3(Y705) Activity in HPCA-Mediated Neuronal Differentiation
(A) NSCs were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and induced to differentiate for 1 day. Cell lysates were analyzed by western blotting with anti-p-STAT3(Y705), anti-p-STAT3 (S727), anti-STAT3, anti-MYC, and anti-GAPDH. Data are shown as mean ± SD (n = 3). ∗p < 0.05 compared with the +bFGF/vector control. ∗∗p < 0.05 compared with the −bFGF/vector control.
(B) Cells were transfected with control siRNA or Hpca siRNA for 48 hr and allowed to differentiate for 1 day. Proteins were analyzed by western blotting with anti-p-STAT3(Y705), anti-STAT3, anti-TUJ1, anti-GFAP, and anti-GAPDH. ∗p < 0.05 compared with the −bFGF/control siRNA (mean ± SD; n = 3).
(C) Diagrams of Stat3WT (wild-type) and Stat3YF (mutant). CC, coiled-coil domain; DBD, DNA-binding domain; LK, linker domain; SH2, Src homology 2; TA, transcriptional activation domain.
(D) Vector (pBOS-FLAG), Stat3WT, or Stat3YF were transfected into NSCs for 48 hr, and the cells were induced to differentiate for 2 days. Cells were lysed and analyzed by western blotting with anti-p-STAT3(Y705), anti-STAT3, anti-TUJ1, anti-GFAP, and anti-GAPDH.
(E and F) Cells were transfected with Stat3WT or Stat3YF for 48 hr. After differentiation for 3 days, cells were stained for neuronal (TUJ1, green) and astrocytic (GFAP, red) markers. Scale bar, 10 μm. Graphs show the percentages of TUJ1- and GFAP-positive cells. Data are shown as mean ± SD (n = 3). ∗Significantly different from −bFGF/Stat3WT at p < 0.05 (for TUJ1). †Significantly different from −bFGF/Stat3WT at p < 0.05 (for GFAP).
(G) Neurite lengths were measured in randomly selected areas from at least three slides of each condition. ∗p < 0.05 compared with −bFGF/Stat3WT (mean ± SD; n = 3).
(H) Cells infected with retroviruses expressing pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc were transiently transfected with Stat3WT or Stat3YF for 48 hr. The cells were induced to differentiate for 2 days. Levels of TUJ1 and GFAP were determined by western blotting. Data are shown as mean ± SD (n = 3). ∗p < 0.05, ∗∗p < 0.01.
(I and J) Cells were treated as in (H) and induced to differentiate for 3 days. Cells were stained for immunocytochemical analysis of neuronal (TUJ1, green) and astrocytic (GFAP, red) markers. Scale bar, 10 μm. Graphs show the percentages of TUJ1- and GFAP-positive cells. Data are shown as mean ± SD (n = 3). ∗Significantly different from −bFGF/Stat3WT/Hpca at p < 0.05 (for TUJ1). †Significantly different from −bFGF/Stat3WT/Hpca at p < 0.05 (for GFAP).
(K) Neurite lengths were measured in randomly selected areas from at least three slides of each condition. ∗p < 0.05 compared with −bFGF/Stat3WT/Hpca (mean ± SD; n = 3).
Next, we asked whether p-STAT3(Y705) played a role in HPCA-mediated neuronal differentiation in NSCs. Hpca-induced TUJ1 expression was increased by Stat3YF, while GFAP expression was significantly decreased when compared with Stat3WT with bFGF removal (Figure 5H). In addition, Stat3YF increased Hpca-induced TUJ1-positive cells (55.4% ± 9% versus 18.3% ± 1.2%, p < 0.05, Figures 5I and 5J) and neurite length (178.6 ± 11.3 μm versus 101.2 ± 18 μm, p < 0.05, Figures 5I and 5K) compared with Stat3WT/Hpca with bFGF removal. By contrast, Stat3YF decreased GFAP-positive cells (14.9% ± 2% versus 32.2% ± 2%, p < 0.05, Figures 5I and 5J) compared with Stat3WT with bFGF removal in Hpca-transduced cells. These results suggest that p-STAT3(Y705) inhibits Hpca-induced neuronal differentiation, leading to astrocyte differentiation in NSCs.
SHP-1 Is Essential for Neuronal Differentiation of NSCs
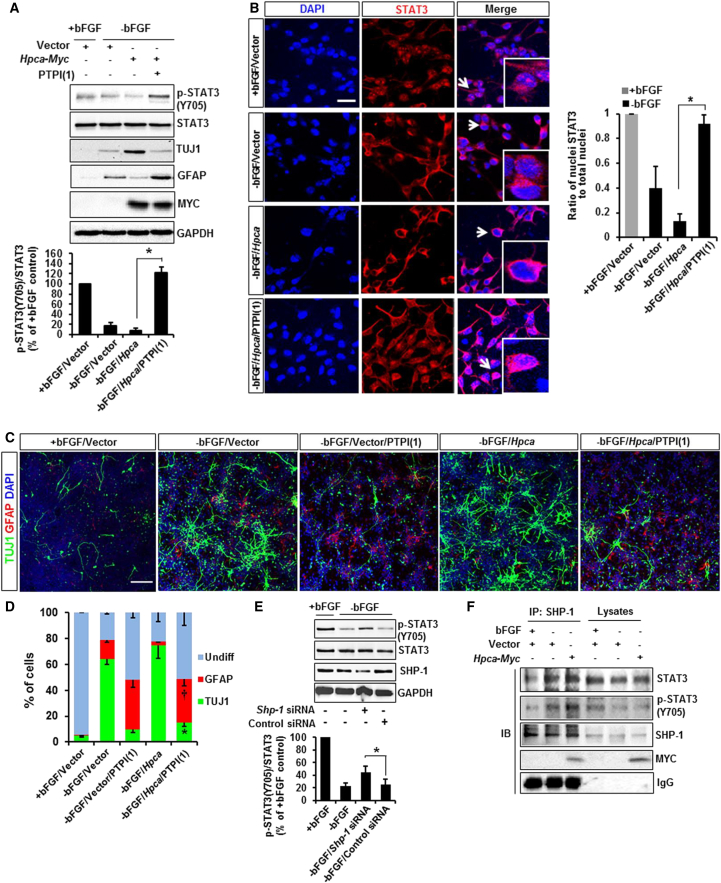
Protein tyrosine phosphatases, such as SHP-1, SHP-2, and PTP-1B, negatively regulate STAT3 signaling through direct dephosphorylation of p-STAT3(Y705) (Pandey et al., 2010, Witkiewicz et al., 2007). Thus, the activity of protein tyrosine phosphatases may be critical for the regulation of STAT3 phosphorylation in NSCs. SHP-1 is a negative regulator of p-STAT3(Y705) (Chen et al., 2012). Therefore, we examined the participation of SHP-1 in dephosphorylation of p-STAT3(Y705) during neuronal differentiation in NSCs. First, we showed that PTPI(1), an inhibitor of SHP-1, increased STAT3(Y705) phosphorylation and GFAP expression, while TUJ1 expression decreased (Figures 6A and S5) in Hpca-induced NSCs during differentiation. Translocation of STAT3 to the nucleus from the cytosol is known to reflect STAT3 activation (Ihle, 2001). Therefore, we determined whether SHP-1 affected the translocation of STAT3 to the nucleus during neuronal differentiation. We found that PTPI(1) indeed enhanced its translocation to the nucleus (Figure 6B), indicating that SHP-1 is sufficient to reduce STAT3 activation, most likely by lowering nuclear STAT3. Furthermore, the number of TUJ1-positive cells was decreased by PTPI(1), whereas the number of GFAP-positive cells was increased (Figures 6C and 6D), indicating that HPCA-mediated SHP-1 activation is critical for neuronal differentiation by promoting neurogenesis and inhibiting gliogenesis. In addition, the role of SHP-1 in STAT3 activation was confirmed in a Shp-1 knockdown study. Shp-1 siRNA abolished dephosphorylation of p-STAT3(Y705), suggesting a crucial role of SHP-1 in STAT3(Y705) phosphorylation (Figure 6E). We also found that SHP-1 bound to p-STAT3(Y705) during differentiation, and that this binding was promoted by Hpca (Figure 6F). This suggests that HPCA promotes the formation of an SHP-1/p-STAT3(Y705) complex, leading to dephosphorylation of p-STAT3(Y705) during neuronal differentiation.
Figure 6.
Dephosphorylation of STAT3(Y705) by SHP-1 Is Related to the Neuronal Differentiation of NSCs
(A) NSCs were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr before pretreatment with 1 μM PTPI(1) for 30 min. Cells were then allowed to differentiate for 1 day before lysis and analysis by western blotting with anti-p-STAT3(Y705), anti-STAT3, anti-TUJ1, anti-GFAP, anti-MYC, and anti-GAPDH. ∗p < 0.05 compared with −bFGF/Hpca (mean ± SD; n = 3).
(B) Cells were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and then treated with 1 μM PTPI(1) for 30 min before differentiation for 1 day. Fixed cells were immunostained with anti-STAT3 (red) and anti-DAPI (blue); STAT3 nuclei were counted. Arrows indicate magnified cells. Graphs show the ratio of STAT3 nuclei to total nuclei in random areas from at least three slides of each condition. ∗p < 0.05 compared with −bFGF/Hpca (mean ± SD; n = 3).
(C and D) Cells were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and pretreated with 1 μM PTPI(1) for 30 min. After differentiation for 3 days, cells were analyzed immunocytochemically using neuronal (TUJ1, green) and astrocytic (GFAP, red) markers. Scale bar, 50 μm. Graphs show the percentages of TUJ1- and GFAP-positive cells. Data are shown as mean ± SD (n = 3). ∗Significantly different from −bFGF/Hpca at p < 0.05 (for TUJ1). †Significantly different from −bFGF/Hpca at p < 0.05 (for GFAP).
(E) Cells were transfected with control siRNA or Shp-1 siRNA for 48 hr and allowed to differentiate for 1 day. Proteins were analyzed by western blotting with anti-p-STAT3(Y705), anti-STAT3, anti-SHP-1, and anti-GAPDH. ∗p < 0.05 compared with the −bFGF/control siRNA (mean ± SD; n = 3).
(F) Cells were transfected with pMSCV-IRES-EGFP or pMSCV-IRES-EGFP-Hpca-Myc for 48 hr and induced to differentiate for 24 hr. Anti-SHP-1 immunoprecipitates from the cell lysates were analyzed by western blotting with anti-STAT3, anti-p-STAT3, anti-SHP-1, and anti-MYC.
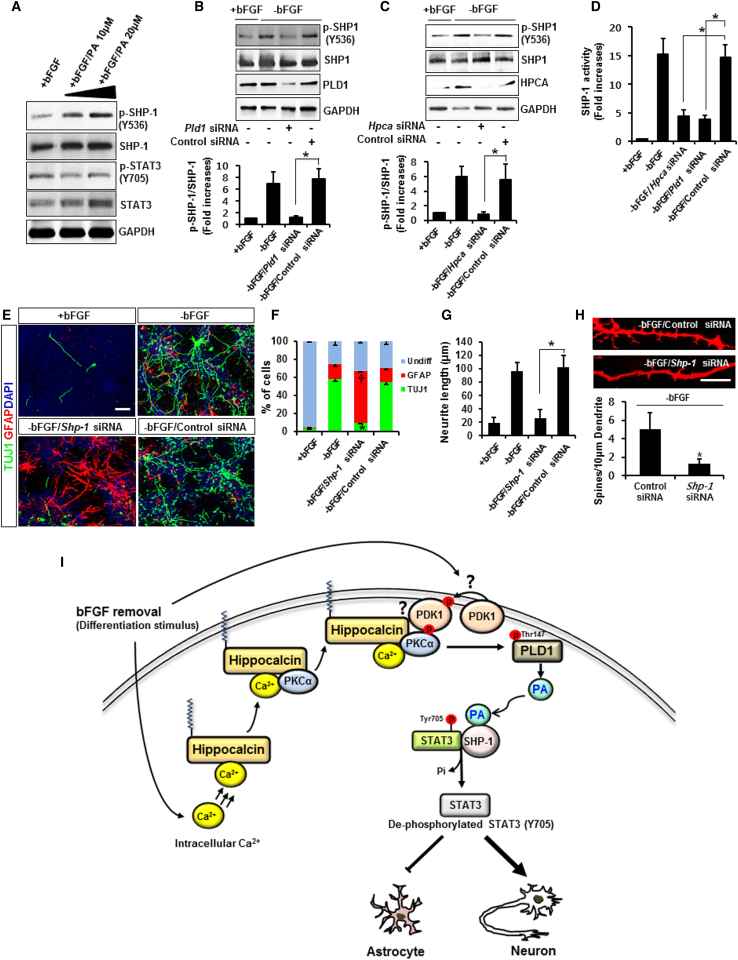
Activation of SHP-1 by acidic phospholipids such as PA may help to regulate SHP-1 activity (Frank et al., 1999). PA is the functional enzymatic product of PLD activity (Exton, 2002). As SHP-1 activity can be influenced by tyrosine phosphorylation, we determined whether tyrosine phosphorylation of SHP-1 is regulated by PA. Exogenously added PA induced phosphorylation of SHP-1 and dephosphorylation of STAT3 in a dose-dependent manner (Figure 7A). We next determined whether PLD1, the major source of PA, affects SHP-1 phosphorylation. Pld1 knockdown inhibited SHP-1 phosphorylation (Figure 7B), and Pld1 siRNA affected dephosphorylation of STAT3 (Figure S6). These results suggest that SHP-1/STAT3 activation is regulated by PLD1 activation. We further showed that phosphorylation of SHP-1 was decreased by Hpca knockdown (Figure 7C). In addition, silencing Hpca or Pld1 with siRNA decreased SHP-1 phosphatase activity (Figure 7D), as expected.
Figure 7.
SHP-1 Is Required for Neuronal Differentiation of NSCs
(A) NSCs were treated with 10 μM or 20 μM phosphatidic acid (PA) for 1 day in the presence of bFGF. Cell lysates were analyzed by western blotting with anti-p-SHP-1, anti-SHP-1, anti-p-STAT3(Y705), anti-STAT3, and anti-GAPDH.
(B) Cells were transfected with control siRNA or Pld1 siRNA for 48 hr and allowed to differentiate for 1 day. Proteins were analyzed by western blotting with anti-p-SHP1 (Y536), anti-SHP-1, anti-PLD1, and anti-GAPDH. ∗p < 0.05 compared with the −bFGF/control siRNA (mean ± SD; n = 3).
(C) Cells were transfected with control siRNA or Hpca siRNA for 48 hr and allowed to differentiate for 1 day. Cell lysates were analyzed by western blotting with anti-p-SHP1 (Y536), anti-SHP-1, anti-HPCA, and anti-GAPDH. ∗p < 0.05 compared with the −bFGF/control siRNA (mean ± SD; n = 3).
(D) Cells were transfected with control siRNA, Hpca siRNA, or Pld1 siRNA for 48 hr and allowed to differentiate for 1 day. SHP-1 activity was measured by phosphatase activity assay (see Experimental Procedures). ∗p < 0.05 compared with the −bFGF/control siRNA (mean ± SD; n = 5).
(E and F) Cells were transfected with control siRNA or Shp-1 siRNA for 48 hr and allowed to differentiate for 3 days. Fixed cells were immunostained with anti-TUJ1 and anti-GFAP, and TUJ1- and GFAP-positive cells were counted. Scale bar, 20 μm. Graphs show the percentages of TUJ1- and GFAP-positive cells. Data are shown as mean ± SD (n = 5). ∗Significantly different from −bFGF/control siRNA at p < 0.05 (for TUJ1). †Significantly different from −bFGF/control siRNA at p < 0.05 (for GFAP).
(G) Neurite lengths were measured in randomly selected areas from at least three slides of each condition. ∗p < 0.05 compared with the −bFGF/control siRNA (mean ± SD; n = 3).
(H) Cells were transfected with control siRNA or Shp-1 siRNA for 48 hr and induced to differentiate by withdrawal of bFGF. After differentiation for 3 days, fixed cells were immunostained with anti-TUJ1. Scale bar, 10 μm. The numbers of spines per 10-μm dendrite were counted in randomly selected areas from at least three slides of each condition. ∗p < 0.05 compared with the −bFGF/control siRNA (mean ± SD; n = 3).
(I) The model suggests that HPCA plays a crucial role in neurogenesis through modulation of the PKCα/PLD1/SHP-1/STAT3 signaling pathway.
To confirm the role of SHP-1 in neuronal differentiation, we knocked down Shp-1, which decreased TUJ1-positive cells, increased GFAP-positive cells (Figures 7E and 7F), and significantly reduced neurite length compared with control siRNA (24.5 ± 6 μm versus 103.2 ± 8 μm, p < 0.05) (Figure 7G). Furthermore, the number of dendritic spines was greatly decreased by Shp-1 siRNA (Figure 7H). These results confirm that SHP-1 is an important regulator of neurogenesis.
Discussion
In the present study, we have shown that HPCA is an important regulator of the neuronal differentiation of NSCs and that it promotes neurogenesis and inhibits gliogenesis. HPCA expression markedly increased during differentiation of NSCs isolated from the cerebral cortex in the E14 rat brain. In addition, neurite outgrowth was enhanced by overexpression of Hpca, indicating that HPCA could be an important player in neuronal differentiation in the brain. Studies of the signal transduction pathways that lead to neuronal differentiation have been impeded by limitations in the genetic manipulation and biochemical analysis of primary neuronal cells, including NSCs. Nevertheless, we have identified a mechanism of HPCA action that promotes neurogenesis. HPCA recruits PKCα to PDK1, which facilitates the PKCα-regulated kinase cascade; PKCα-dependent PLD1 activation is required for HPCA-mediated neurite outgrowth. PA, a functional product of PLD1, affects tyrosine phosphatase SHP-1 activation during neuronal differentiation, and SHP-1 blocks STAT3(Y705) activation, thereby inhibiting gliogenesis in NSCs (Figure 7I). Thus, this signaling pathway provides insight into HPCA-mediated neuronal function in rat NSCs and its potential contribution to cell-fate signaling.
HPCA Promotes Neurogenesis by Inhibiting Gliogenesis
The specification of cell fate involves the commutative activation of genes related to a particular cell fate and the inhibition of genes related to alternative fates. Cell-fate-specifying transcription factors can both positively and negatively regulate alternative fates (Scully et al., 2000, Tajbakhsh et al., 1996), and a recent study reported that NGN1 has dual functions in activating neuronal differentiation genes and suppressing glial differentiation genes (Sun et al., 2001). Similarly, our study suggests that HPCA positively regulates neurogenesis and negatively regulates gliogenesis in NSCs. This is supported by our findings that: (1) knockdown of Hpca strongly decreased neurite outgrowth and dendritic spinogenesis, whereas it increased expression of the astrocyte marker GFAP and GFAP-positive cells; and (2) overexpression of Hpca significantly stimulated neurite outgrowth and expression of the neuron marker TUJ1, even in the presence of bFGF. These results are consistent with the concept that a specific gene product such as Ngn-1 inhibits glial differentiation (Nieto et al., 2001).
The Role of HPCA in PKCα/PLD1 Activation during Neuronal Differentiation
HPCA has a Ca2+/myristoyl switch, allowing membrane translocation from the cytosol in response to increased free Ca2+ concentrations (O'Callaghan et al., 2003). In neurons, HPCA is located in the cytoplasm and plasma membrane of cell bodies, dendrites, and axons (Burgoyne et al., 2004). HPCA exerts a neuroprotective action by blocking the formation of Ca2+-induced cell death stimuli (Masuo et al., 2007), and infusion of a mutant Hpca lacking Ca2+-binding sites prevents long-term depression in hippocampal neurons (Jo et al., 2010). Since HPCA has a crucial role in Ca2+-mediated neuronal activity in the brain, we investigated its potential role in Ca2+-dependent signaling pathways in NSCs. When cells were induced to differentiate by removal of bFGF, Ca2+ levels increased and HPCA translocated to the membrane. Furthermore, HPCA assisted the formation of a PDK1-PKCα complex, which induces PKCα phosphorylation. As summarized in Figure 7I, it is not known whether HPCA has direct or indirect effects on PDK1 activation or binds to PDK1. Moreover, the molecular mechanism responsible for the phosphorylation of PDK1 by differentiation stimulus has not yet been defined. Thus, further studies are needed to clarify the relationship between HPCA and PDK1 signaling during neuronal differentiation in NSCs; this could be important for a complete understanding of HPCA-mediated signaling cascade in neurogenesis of NSCs. Our previous studies showed that HPCA played a role in Ca2+-dependent PLD activation (Hyun et al., 2000), and PLD1 promotes neurite outgrowth in H19-7 cells (Yoon et al., 2012). As expected, PLD1 is involved in HPCA-mediated neurogenesis and its activation is regulated by HPCA. Taken together, our results suggest that HPCA is required for optimal formation of a complex between PDK1 and PKCα, resulting in PLD1 activation during neuronal differentiation.
Blocking of STAT3(Y705) Activation by HPCA Increases Neuronal Differentiation and Decreases Astrocytic Differentiation of NSCs
STAT3 plays essential roles in determining the fate of NSCs (Cao et al., 2010). In the present study, NSC differentiation induced by removal of bFGF resulted in a dramatic reduction in the phosphorylation of STAT3(Y705). Interestingly, HPCA's inhibition of STAT3(Y705) phosphorylation induced neurogenesis and decreased gliogenesis. Furthermore, Stat3YF mutant significantly promotes Hpca-induced TUJ1 expression and TUJ1-positive cells, while it failed to stimulate GFAP expression and GFAP-positive cells. Our results are consistent with earlier reports in which suppression or conditional deletion of Stat3 promotes neuronal differentiation and inhibits glial differentiation (Gu et al., 2005, Natarajan et al., 2014). The activities of intracellular transcription factors are controlled by the competing activities of kinases and phosphatases (Massa et al., 2004). We found that SHP-1 led to loss of STAT3(Y705) activation. Interestingly, SHP-1 binds to phosphorylated STAT3(Y705) under differentiation conditions and this process was promoted by HPCA. Thus, we conclude that HPCA-mediated SHP-1 activity most likely inhibits STAT3 activity by blocking its import to the nucleus and, presumably, by decreasing the nuclear accumulation of phosphorylated STAT3(Y705), resulting in increased neurogenesis.
In summary, HPCA appears to have two distinct roles: promoting neurogenesis and inhibiting gliogenesis. We propose that it has an important role in enhancing the generation of neurons from NSCs. Neurogenesis is tightly controlled because of its critical importance in proper physiological function, and multiple signals control the growth and directionality of the relevant cell-fate decision. However, the mechanism by which HPCA affects Ca2+-dependent signaling during neurogenesis remains to be determined, and a more detailed characterization of the mechanisms is currently under way. HPCA may be implicated in memory formation, neuronal excitability, and activity-dependent plasticity (Kobayashi et al., 2012). Hpca knockout (Hpca−/−) mice have a defect in CREB activation that is associated with impaired spatial and associative memory (Kobayashi et al., 2005) as well as an impaired RAS-regulated kinase cascade in response to RAF activation (Noguchi et al., 2007). It is possible that HPCA plays an important role in long-lasting neural plasticity and promotes spatial and associative memory. Nevertheless, despite recent progress, much remains unknown regarding the contribution of HPCA-mediated signaling mechanisms to brain injury and recovery. Further studies are required to address its possible use as a marker of neurodegenerative disorders and in promoting neuronal regeneration in vivo.
Experimental Procedures
Preparation of Animal Tissue
All experimental animal procedures (Sprague-Dawley rats) were approved by the Institutional Animal Care and Use Committee (IACUC) at Hanyang College of Medicine under approval number 2015-0009. Experiments were performed in accordance with the NIH guidelines.
Primary Culture of Neural Precursor Cells
Embryonic brain cortices from E14 rat embryos were mechanically triturated in Ca2+/Mg2+-free Hank's balanced salt solution (HBSS; Gibco), seeded at 2 × 105 cells in 10-cm culture dishes (Corning Life Sciences), which were precoated with 15 μg/mL poly-L-ornithine (Sigma) and 1 μg/mL fibronectin (Invitrogen) and then cultured for 5–6 days in serum-free N2 medium supplemented with 20 ng/mL bFGF (R&D Systems). Cell clusters generated by precursor cell proliferation were dissociated in HBSS and plated at 2 × 104 cells per well on coated 24-well plates, 2 × 105 cells per well on coated 6-well plates, and 8 × 105 cells per dish on coated 6-cm culture dishes. The plated cells were allowed to proliferate further in N2 + bFGF up to 70%–80% cell confluence before induction of differentiation. All experiments were carried out using the passage 1 (P1) neural precursor cells.
Other Procedures
Full additional information is available in Supplemental Experimental Procedures.
Author Contributions
S.-Y.P.: Conception and design, manuscript writing, collection and assembly of data, data analysis and interpretation, financial support. S.N.Y.: Conception and design, collection and assembly of data; M.-J.K.: Collection and assembly of data; Y.Y.L.: Data analysis and interpretation; S.J.J.: Collection and assembly data, data analysis and interpretation; J.-S.H.: Conception and design, manuscript writing, data analysis and interpretation, financial support.
Acknowledgments
We owe great thanks to Dr. Mi-Ryoung Song (Gwangju Institute of Science and Technology, Gwangju, Republic of Korea) for Stat3 plasmids. This work was supported by a Medical Research Center program (NRF-2012R1A5A2A34671243) of the Ministry of Science and Technology, Republic of Korea, and partly supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning (2015R1C1A1A02037376).
Published: December 22, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.11.009.
Supplemental Information
References
- Banno Y., Nemoto S., Murakami M., Kimura M., Ueno Y., Ohguchi K., Hara A., Okano Y., Kitade Y., Onozuka M. Depolarization-induced differentiation of PC12 cells is mediated by phospholipase D2 through the transcription factor CREB pathway. J. Neurochem. 2008;104:1372–1386. doi: 10.1111/j.1471-4159.2007.05085.x. [DOI] [PubMed] [Google Scholar]
- Boncoeur E., Bouvet G.F., Migneault F., Tardif V., Ferraro P., Radzioch D., de Sanctis J.B., Eidelman D., Govindaraju K., Dagenais A. Induction of nitric oxide synthase expression by lipopolysaccharide is mediated by calcium-dependent PKCalpha-beta1 in alveolar epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2013;305:L175–L184. doi: 10.1152/ajplung.00295.2012. [DOI] [PubMed] [Google Scholar]
- Burgoyne R.D., Weiss J.L. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem. J. 2001;353:1–12. [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R.D., O'Callaghan D.W., Hasdemir B., Haynes L.P., Tepikin A.V. Neuronal Ca2+-sensor proteins: multitalented regulators of neuronal function. Trends Neurosci. 2004;27:203–209. doi: 10.1016/j.tins.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Cao F., Hata R., Zhu P., Nakashiro K., Sakanaka M. Conditional deletion of Stat3 promotes neurogenesis and inhibits astrogliogenesis in neural stem cells. Biochem. Biophys. Res. Commun. 2010;394:843–847. doi: 10.1016/j.bbrc.2010.03.092. [DOI] [PubMed] [Google Scholar]
- Champion H.C., Kass D.A. Calcium handler mishandles heart. Nat. Med. 2004;10:239–240. doi: 10.1038/nm0304-239. [DOI] [PubMed] [Google Scholar]
- Chen K.F., Su J.C., Liu C.Y., Huang J.W., Chen K.C., Chen W.L., Tai W.T., Shiau C.W. A novel obatoclax derivative, SC-2001, induces apoptosis in hepatocellular carcinoma cells through SHP-1-dependent STAT3 inactivation. Cancer Lett. 2012;321:27–35. doi: 10.1016/j.canlet.2012.03.023. [DOI] [PubMed] [Google Scholar]
- Dutil E.M., Toker A., Newton A.C. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1) Curr. Biol. 1998;8:1366–1375. doi: 10.1016/s0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- Edlund T., Jessell T.M. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- Exton J.H. Phospholipase D: enzymology, mechanisms of regulation, and function. Physiol. Rev. 1997;77:303–320. doi: 10.1152/physrev.1997.77.2.303. [DOI] [PubMed] [Google Scholar]
- Exton J.H. Regulation of phospholipase D. FEBS Lett. 2002;531:58–61. doi: 10.1016/s0014-5793(02)03405-1. [DOI] [PubMed] [Google Scholar]
- Frank C., Keilhack H., Opitz F., Zschornig O., Bohmer F.D. Binding of phosphatidic acid to the protein-tyrosine phosphatase SHP-1 as a basis for activity modulation. Biochemistry. 1999;38:11993–12002. doi: 10.1021/bi982586w. [DOI] [PubMed] [Google Scholar]
- Gage F.H., Ray J., Fisher L.J. Isolation, characterization, and use of stem cells from the CNS. Annu. Rev. Neurosci. 1995;18:159–192. doi: 10.1146/annurev.ne.18.030195.001111. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Greenberg M.E. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- Gu F., Hata R., Ma Y.J., Tanaka J., Mitsuda N., Kumon Y., Hanakawa Y., Hashimoto K., Nakajima K., Sakanaka M. Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J. Neurosci. Res. 2005;81:163–171. doi: 10.1002/jnr.20561. [DOI] [PubMed] [Google Scholar]
- Han Y., Amin H.M., Franko B., Frantz C., Shi X., Lai R. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood. 2006;108:2796–2803. doi: 10.1182/blood-2006-04-017434. [DOI] [PubMed] [Google Scholar]
- Hyun J.K., Yon C., Kim Y.S., Noh D.Y., Lee K.H., Han J.S. Role of hippocalcin in Ca2+-induced activation of phospholipase D. Mol. Cells. 2000;10:669–677. doi: 10.1007/s10059-000-0669-1. [DOI] [PubMed] [Google Scholar]
- Ihle J.N. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 2001;13:211–217. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- Jo J., Son G.H., Winters B.L., Kim M.J., Whitcomb D.J., Dickinson B.A., Lee Y.B., Futai K., Amici M., Sheng M. Muscarinic receptors induce LTD of NMDAR EPSCs via a mechanism involving hippocalcin, AP2 and PSD-95. Nat. Neurosci. 2010;13:1216–1224. doi: 10.1038/nn.2636. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kim J.H., Ohba M., Suh P.G., Ryu S.H. Novel functions of the phospholipase D2-Phox homology domain in protein kinase Czeta activation. Mol. Cell. Biol. 2005;25:3194–3208. doi: 10.1128/MCB.25.8.3194-3208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Takamatsu K., Saitoh S., Noguchi T. Myristoylation of hippocalcin is linked to its calcium-dependent membrane association properties. J. Biol. Chem. 1993;268:18898–18904. [PubMed] [Google Scholar]
- Kobayashi M., Masaki T., Hori K., Masuo Y., Miyamoto M., Tsubokawa H., Noguchi H., Nomura M., Takamatsu K. Hippocalcin-deficient mice display a defect in cAMP response element-binding protein activation associated with impaired spatial and associative memory. Neuroscience. 2005;133:471–484. doi: 10.1016/j.neuroscience.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Hamanoue M., Masaki T., Furuta Y., Takamatsu K. Hippocalcin mediates calcium-dependent translocation of brain-type creatine kinase (BB-CK) in hippocampal neurons. Biochem. Biophys. Res. Commun. 2012;429:142–147. doi: 10.1016/j.bbrc.2012.10.125. [DOI] [PubMed] [Google Scholar]
- Kopach O., Viatchenko-Karpinski V., Atianjoh F.E., Belan P., Tao Y.X., Voitenko N. PKCalpha is required for inflammation-induced trafficking of extrasynaptic AMPA receptors in tonically firing lamina II dorsal horn neurons during the maintenance of persistent inflammatory pain. J. Pain. 2013;14:182–192. doi: 10.1016/j.jpain.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D., Mercer E.A., Yu L.Y., Chen Y., Kukkonen J., Korhonen L., Arumae U. Neuronal apoptosis inhibitory protein: structural requirements for hippocalcin binding and effects on survival of NGF-dependent sympathetic neurons. Biochim. Biophys. Acta. 2002;1600:138–147. doi: 10.1016/s1570-9639(02)00454-5. [DOI] [PubMed] [Google Scholar]
- Manifava M., Thuring J.W., Lim Z.Y., Packman L., Holmes A.B., Ktistakis N.T. Differential binding of traffic-related proteins to phosphatidic acid- or phosphatidylinositol (4,5)-bisphosphate-coupled affinity reagents. J. Biol. Chem. 2001;276:8987–8994. doi: 10.1074/jbc.M010308200. [DOI] [PubMed] [Google Scholar]
- Markus A., Patel T.D., Snider W.D. Neurotrophic factors and axonal growth. Curr. Opin. Neurobiol. 2002;12:523–531. doi: 10.1016/s0959-4388(02)00372-0. [DOI] [PubMed] [Google Scholar]
- Massa P.T., Wu C., Fecenko-Tacka K. Dysmyelination and reduced myelin basic protein gene expression by oligodendrocytes of SHP-1-deficient mice. J. Neurosci. Res. 2004;77:15–25. doi: 10.1002/jnr.20155. [DOI] [PubMed] [Google Scholar]
- Masuo Y., Ogura A., Kobayashi M., Masaki T., Furuta Y., Ono T., Takamatsu K. Hippocalcin protects hippocampal neurons against excitotoxin damage by enhancing calcium extrusion. Neuroscience. 2007;145:495–504. doi: 10.1016/j.neuroscience.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Miller F.D., Gauthier A.S. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Nagata K., Puls A., Futter C., Aspenstrom P., Schaefer E., Nakata T., Hirokawa N., Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Kawabata M., Miyazono K., Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Natarajan R., Singal V., Benes R., Gao J., Chan H., Chen H., Yu Y., Zhou J., Wu P. STAT3 modulation to enhance motor neuron differentiation in human neural stem cells. PLoS One. 2014;9:e100405. doi: 10.1371/journal.pone.0100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M., Schuurmans C., Britz O., Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Noguchi H., Kobayashi M., Miwa N., Takamatsu K. Lack of hippocalcin causes impairment in Ras/extracellular signal-regulated kinase cascade via a Raf-mediated activation process. J. Neurosci. Res. 2007;85:837–844. doi: 10.1002/jnr.21180. [DOI] [PubMed] [Google Scholar]
- O'Callaghan D.W., Ivings L., Weiss J.L., Ashby M.C., Tepikin A.V., Burgoyne R.D. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca2+ signal transduction. J. Biol. Chem. 2002;277:14227–14237. doi: 10.1074/jbc.M111750200. [DOI] [PubMed] [Google Scholar]
- O'Callaghan D.W., Tepikin A.V., Burgoyne R.D. Dynamics and calcium sensitivity of the Ca2+/myristoyl switch protein hippocalcin in living cells. J. Cell Biol. 2003;163:715–721. doi: 10.1083/jcb.200306042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.Y., Yon C., Oh K.J., Lee K.S., Han J.S. Hippocalcin increases phospholipase D2 expression through extracellular signal-regulated kinase activation and lysophosphatidic acid potentiates the hippocalcin-induced phospholipase D2 expression. J. Cell Biochem. 2006;97:1052–1065. doi: 10.1002/jcb.20665. [DOI] [PubMed] [Google Scholar]
- Oh D.Y., Cho J.H., Park S.Y., Kim Y.S., Yoon Y.J., Yoon S.H., Chung K.C., Lee K.S., Han J.S. A novel role of hippocalcin in bFGF-induced neurite outgrowth of H19-7 cells. J. Neurosci. Res. 2008;86:1557–1565. doi: 10.1002/jnr.21602. [DOI] [PubMed] [Google Scholar]
- Palmer C.L., Lim W., Hastie P.G., Toward M., Korolchuk V.I., Burbidge S.A., Banting G., Collingridge G.L., Isaac J.T., Henley J.M. Hippocalcin functions as a calcium sensor in hippocampal LTD. Neuron. 2005;47:487–494. doi: 10.1016/j.neuron.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M.K., Sung B., Aggarwal B.B. Betulinic acid suppresses STAT3 activation pathway through induction of protein tyrosine phosphatase SHP-1 in human multiple myeloma cells. Int. J. Cancer. 2010;127:282–292. doi: 10.1002/ijc.25059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Cho J.H., Ma W., Choi H.J., Han J.S. Phospholipase D2 acts as an important regulator in LPS-induced nitric oxide synthesis in Raw 264.7 cells. Cell Signal. 2010;22:619–628. doi: 10.1016/j.cellsig.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Ma W., Yoon S.N., Kang M.J., Han J.S. Phospholipase D1 increases Bcl-2 expression during neuronal differentiation of rat neural stem cells. Mol. Neurobiol. 2015;51:1089–1102. doi: 10.1007/s12035-014-8773-y. [DOI] [PubMed] [Google Scholar]
- Qian X., Shen Q., Goderie S.K., He W., Capela A., Davis A.A., Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Rietze R.L., Valcanis H., Brooker G.F., Thomas T., Voss A.K., Bartlett P.F. Purification of a pluripotent neural stem cell from the adult mouse brain. Nature. 2001;412:736–739. doi: 10.1038/35089085. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Takamatsu K., Kobayashi M., Noguchi T. Expression of hippocalcin in the developing rat brain. Brain Res. Dev. Brain Res. 1994;80:199–208. doi: 10.1016/0165-3806(94)90105-8. [DOI] [PubMed] [Google Scholar]
- Scully K.M., Jacobson E.M., Jepsen K., Lunyak V., Viadiu H., Carriere C., Rose D.W., Hooshmand F., Aggarwal A.K., Rosenfeld M.G. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- Shen Q., Wang Y., Dimos J.T., Fasano C.A., Phoenix T.N., Lemischka I.R., Ivanova N.B., Stifani S., Morrisey E.E., Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shibata N., Kakita A., Takahashi H., Ihara Y., Nobukuni K., Fujimura H., Sakoda S., Sasaki S., Iwata M., Morikawa S. Activation of signal transducer and activator of transcription-3 in the spinal cord of sporadic amyotrophic lateral sclerosis patients. Neurodegener. Dis. 2009;6:118–126. doi: 10.1159/000213762. [DOI] [PubMed] [Google Scholar]
- Shin-young P., Koh Y.J., Cho J.H., Oh D.Y., Shin S.A., Lee K.S., Lee H.B., Han J.S. Nicotine inhibits bFGF-induced neurite outgrowth through suppression of NO synthesis in H19-7 cells. Neurochem. Res. 2007;32:481–488. doi: 10.1007/s11064-006-9256-y. [DOI] [PubMed] [Google Scholar]
- Sun Y., Nadal-Vicens M., Misono S., Lin M.Z., Zubiaga A., Hua X., Fan G., Greenberg M.E. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt D., Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996;384:266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Yamazaki M., Miyazaki H., Arikawa C., Itoh K., Sasaki T., Maehama T., Frohman M.A., Kanaho Y. Phospholipase D2 functions as a downstream signaling molecule of MAP kinase pathway in L1-stimulated neurite outgrowth of cerebellar granule neurons. J. Neurochem. 2004;89:142–151. doi: 10.1111/j.1471-4159.2004.02308.x. [DOI] [PubMed] [Google Scholar]
- Williams M.R., Arthur J.S., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D.R. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- Witkiewicz A., Raghunath P., Wasik A., Junkins-Hopkins J.M., Jones D., Zhang Q., Odum N., Wasik M.A. Loss of SHP-1 tyrosine phosphatase expression correlates with the advanced stages of cutaneous T-cell lymphoma. Hum. Pathol. 2007;38:462–467. doi: 10.1016/j.humpath.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Yokogami K., Wakisaka S., Avruch J., Reeves S.A. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- Yoon M.S., Yon C., Park S.Y., Oh D.Y., Han A.H., Kim D.S., Han J.S. Role of phospholipase D1 in neurite outgrowth of neural stem cells. Biochem. Biophys. Res. Commun. 2005;329:804–811. doi: 10.1016/j.bbrc.2005.02.087. [DOI] [PubMed] [Google Scholar]
- Yoon M.S., Cho C.H., Lee K.S., Han J.S. Binding of Cdc42 to phospholipase D1 is important in neurite outgrowth of neural stem cells. Biochem. Biophys. Res. Commun. 2006;347:594–600. doi: 10.1016/j.bbrc.2006.06.111. [DOI] [PubMed] [Google Scholar]
- Yoon S.N., Kim K.S., Cho J.H., Ma W., Choi H.J., Kwon S.J., Han J.S. Phospholipase D1 mediates bFGF-induced Bcl-2 expression leading to neurite outgrowth in H19-7 cells. Biochem. J. 2012;441:407–416. doi: 10.1042/BJ20110302. [DOI] [PubMed] [Google Scholar]
- Zhao C., Du G., Skowronek K., Frohman M.A., Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.