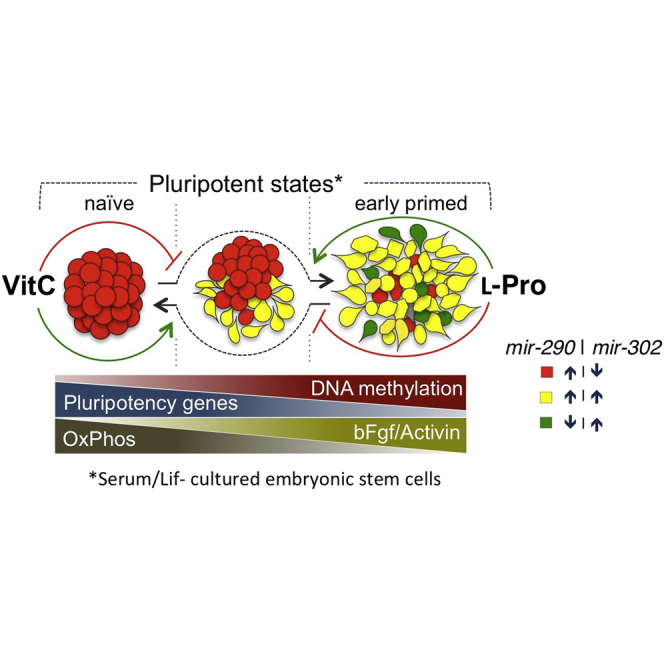
Summary
Metabolites and cofactors are emerging as key regulators of cell plasticity and reprogramming, and their role in the control of pluripotency is just being discovered. Here we provide unprecedented evidence that embryonic stem cell (ESC) pluripotency relies on the relative levels of two physiological metabolites, namely ascorbic acid (vitamin C, VitC) and l-proline (l-Pro), which affect global DNA methylation, transcriptional profile, and energy metabolism. Specifically, while a high VitC/l-Pro ratio drives ESCs toward a naive state, the opposite condition (l-Pro excess) captures a fully reversible early primed pluripotent state, which depends on autocrine fibroblast growth factor and transforming growth factor β signaling pathways. Our findings highlight the pivotal role of metabolites availability in controlling the pluripotency continuum from naive to primed states.
Keywords: vitamin C, l-proline, DNA methylation, naive pluripotency, early primed pluripotency, metabolic reprogramming, metabolomics
Graphical Abstract
Highlights
-
•
Global DNA methylation is modulated by l-Pro and VitC availability in ESCs
-
•
l-Pro and VitC relative levels induce naive and reversible primed pluripotency
-
•
High l-Pro/low VitC levels induce metabolic reprogramming in ESCs
-
•
l-Pro induces and sustains autocrine FGF and TGF-β signaling in ESCs
In this article, Minchiotti, De Cesare, and colleagues show that the relative levels of two physiological metabolites, vitamin C and l-proline, direct ESCs toward naive or reversible primed states of pluripotency, placed between the naive/2i and FGF/activin states, and highlight a pivotal role of metabolic-epigenetic crosstalk in the developmental continuum.
Introduction
Pluripotency is transiently induced during the early stages of mammalian embryo development. Blastocyst stem cells progress from a “naive/ground” state to a “primed” state of pluripotency before lineage commitment (Martinez Arias et al., 2013). Two distinct pluripotent stem cells, embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs), are considered as their in vitro counterparts. ESCs and EpiSCs differ with respect to their morphology, metabolism, DNA methylation levels, transcription profiles, and growth factors requirement (Weinberger et al., 2016). Pluripotency states are unstable and thus difficult to stabilize in vitro. Indeed, ESC cultures consist of heterogeneous cells dynamically fluctuating between different pluripotent states (Hayashi et al., 2008, Toyooka et al., 2008). EpiSCs frequently lose the primed state, acquiring features of late pre-gastrula embryos (Wu and Izpisua Belmonte, 2015). Known molecular determinants of such plasticity are mainly transcription factors, while the role of metabolism has been largely unexplored until recently. Indeed, it has now become evident that metabolites, including amino acids, act as key regulators of pluripotent stem cell plasticity and behavior. For instance, it has been shown that ESC self-renewal depends on l-threonine (Wang et al., 2009), while ESC identity is regulated by l-proline (l-Pro) availability (Casalino et al., 2011, Comes et al., 2013, D'Aniello et al., 2015, Washington et al., 2010). Moreover, several metabolites act as epigenetic signals (Blaschke et al., 2013, Comes et al., 2013, Shyh-Chang et al., 2012), thus defining a regulatory network among metabolism, epigenetic modification, and pluripotency, knowledge of which is still limited (Harvey et al., 2016). Here we provide evidence that pluripotency is finely controlled by the mutual availability of two physiological metabolites, vitamin C (VitC) and l-Pro, and propose that naive and early primed pluripotency states can be captured in vitro by exploiting the epigenetic activity of these metabolites.
Results
Vitamin C and l-Proline Induce Opposite Effects on DNA Methylation in ESCs
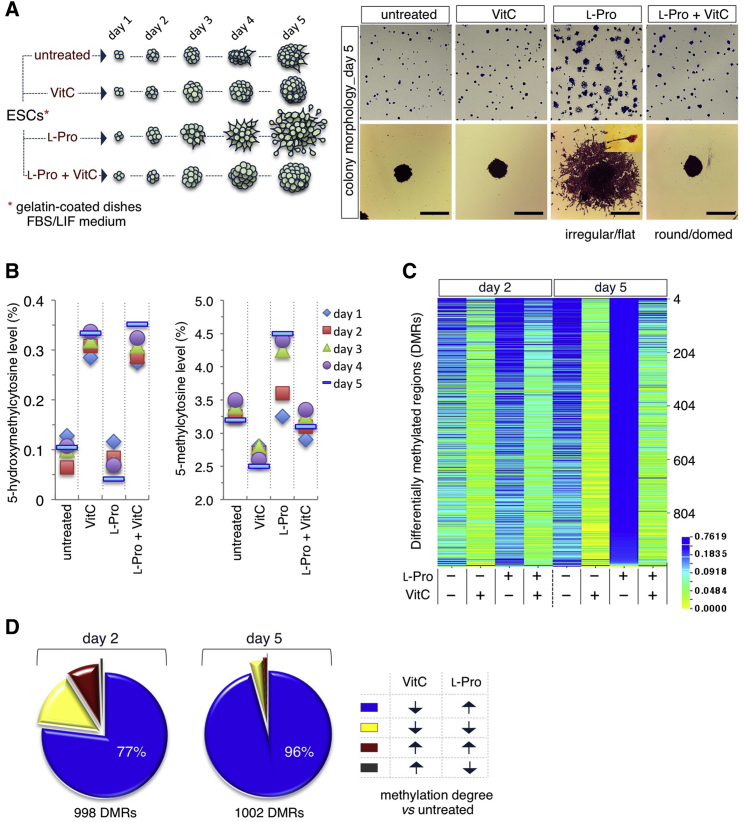
Recent evidence from our laboratory demonstrates that ESCs suffer a highly specific intrinsic shortage of the nonessential amino acid l-Pro, which induces the amino acid stress response pathway (D'Aniello et al., 2015). Exogenously provided l-Pro alleviates this stress condition and converts round-shaped ESCs into flat-shaped pluripotent stem cells. This phenotypic transition is fully reversible, either after l-Pro withdrawal or by addition of VitC (Casalino et al., 2011, Comes et al., 2013), raising the hypothesis that these two metabolites play antagonistic roles in controlling ESC identity. VitC is the most relevant naturally occurring reducing agent and enhances the catalytic activity of 2-oxoglutarate/Fe(II)-dependent dioxygenases, including TET DNA demethylases, and thereby contributes to epigenetic regulation, cell differentiation, and reprogramming (Hore et al., 2016, Krishnakumar and Blelloch, 2013, Monfort and Wutz, 2013). We thus reasoned that VitC and l-Pro might regulate pluripotency by exerting opposite effects on the dynamics of DNA methylation. We first quantified DNA methylation levels in ESCs treated with l-Pro ± VitC at different time points by liquid chromatography followed by mass spectrometry (LC-MS) (Figures 1A and 1B). VitC supplementation led to a rapid and sustained increase of 5-hydroxymethylcytosine (5hmC) and reduced 5-methylcytosine (5mC) levels. Conversely, supplemental l-Pro increased 5mC and reduced 5hmC levels, and its effect was fully counteracted by VitC (Figure 1B). We then compared genome-wide methylation profiling of ESCs ± l-Pro and ESCs, ± VitC, at days 2 and 5 of treatment. Reduced representation bisulfite sequencing (RRBS) analysis identified ∼1,000 differentially methylated regions (DMRs) distributed throughout all chromosomes (Figure 1C and Table S1). The methylation levels of individual CpGs were highly correlated (r > 0.9) between the different groups (Figures S1A and S1B). Interestingly, the majority of DMRs (77% at day 2 and 96% at day 5) that were hypomethylated in VitC-treated ESCs were conversely hypermethylated in l-Pro-treated ESCs (Figures 1D and S1C), thus indicating that VitC and l-Pro modulate methylation at the same DNA regions in an opposite manner. Of note, a high fraction of DMRs (50%) lay in promoters (mostly HCPs) and ∼20% in enhancers (Figures S1D and S1E; Table S1). Moreover, DMR-associated genes are highly enriched in genes related to the developmental process (p = 6.3 × 10−38) and DNA-binding transcriptional regulators (p = 9.7 × 10−34) (Table S1). Our findings provide evidence that l-Pro availability regulates DNA methylation in ESCs. Of note, DNA methylation levels increase in the transition from naive to primed pluripotency (Maruotti et al., 2010), thus leading to the hypothesis that the antagonistic effects of VitC and l-Pro on the DNA methylation pattern might regulate pluripotency.
Figure 1.
Vitamin C and l-Proline Induce Opposite Effects on DNA Methylation in ESCs
(A) Schematic representation of the time-course colony assay. FBS/LIF ESCs plated at low density ± VitC (500 μM)/± l-Pro (150 μM) for 5 days (left panel). Representative pictures of crystal violet-stained colonies (right panels). Scale bars, 250 μm.
(B) Time-course mass spectrometry (MS) analysis of 5hmc and 5mc levels in ESCs ± VitC (500 μM)/± l-Pro (150 μM). Data are relative levels (%) compared with day 1.
(C) Heatmap (CIMminer, http://discover.nci.nih.gov/cimminer/) showing the differentially methylated regions (DMRs) in ESCs ± VitC (500 μM)/± l-Pro (150 μM) at days 2 and 5.
(D) Pie graphs showing the percentage of DMRs shared by VitC- and l-Pro-treated ESCs at days 2 and 5.
See also Figure S1.
Vitamin C and l-Proline Modify mir-290/mir-302 Signature in ESCs
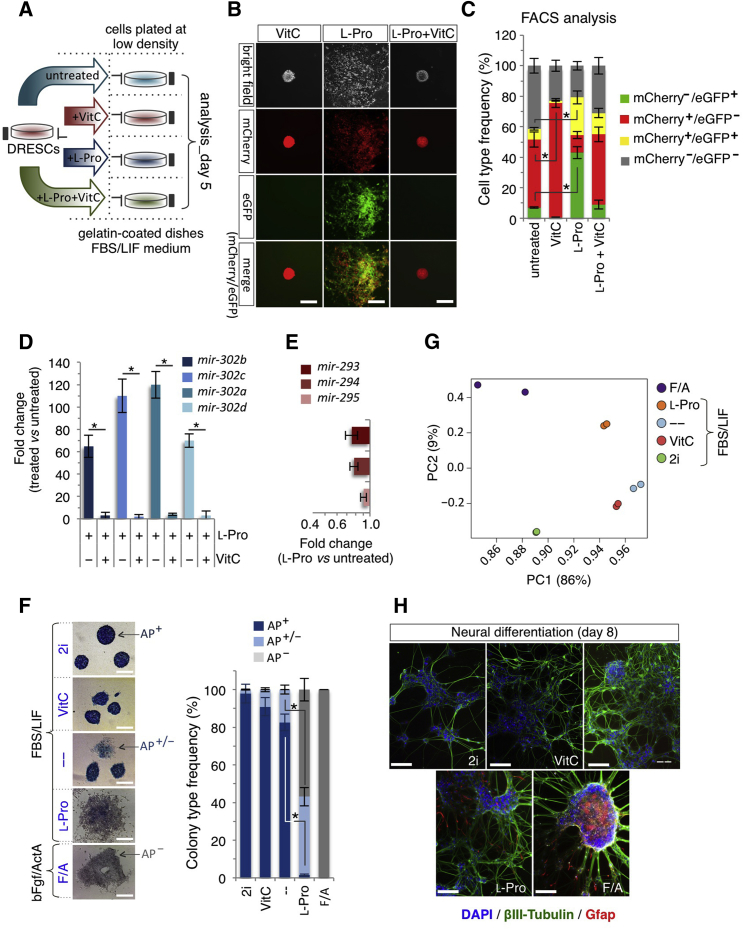
Pluripotent stem cell transition from naive to primed state is marked by changes in the expression of the mir-290 and mir-302 clusters, which correspond to naive (pre-implantation epiblast) and primed (late post-implantation epiblast of the pre-gastrulating embryo) states, respectively; whereas co-expression of the two clusters identifies an intermediate (early post-implantation epiblast) state (Parchem et al., 2014). To assess whether VitC and l-Pro impact on the expression of these microRNA clusters, we exploited the potential of ESCs carrying the reporters for the mir-290 and mir-302 loci (mir-290_mCherry/mir-302_eGFP dual-reporter ESCs, named DRESCs) (Parchem et al., 2014). Addition of VitC to fetal bovine serum (FBS)/leukemia inhibitory factor (LIF) DRESCs increased the fraction of mCherry+/eGFP− (red) cells at the expense of mCherry±/eGFP+ cells (yellow and green) (Figures 2A–2C). A similar effect was observed in feeder-dependent FBS/LIF culture conditions (Figures S2A–S2C). Conversely, l-Pro supplementation increased the frequency of mCherry+/eGFP+ (yellow) and mCherry−/eGFP+ (green) cells (Figures 2A–2C), and its effect was fully reversed by VitC (Figures 2A–2C). Accordingly, the expression of all members of the mir-302-367 cluster, and the pri-mir-302 was strongly induced in l-Pro-treated ESCs (Figures 2D, S2D, and S2E), whereas VitC blocked l-Pro-dependent induction of mir-302 (Figure 2D). Of note, expression of mir-290 cluster persisted, though to a reduced extent in l-Pro-treated cells (Figure 2E). Our findings suggest that FBS/LIF ESCs supplemented with either VitC or l-Pro acquire a naive (mir-290+/mir-302−) and a primed (mir-290±/mir-302+) signature, respectively.
Figure 2.
Molecular and Pluripotency Features of Vitamin C- and l-Proline-Induced Cells
(A) Schematic representation of the experimental strategy. Dual-reporter ESCs (DRESCs) plated at low density ± VitC (500 μM)/± l-Pro (150 μM) and analyzed at day 5.
(B) Representative bright-field and fluorescence images of colonies generated from DRESCs ± VitC (500 μM)/± l-Pro (150 μM) at day 5. Red (mir-290), green (mir-302), and yellow (mir-290 and mir-302) signals indicate expression of the transgenes. Scale bars, 250 μm.
(C) Fluorescence-activated cell sorting (FACS) quantification of mCherry±/eGFP± cells in DRESCs ± VitC (500 μM)/± l-Pro (150 μM). Data are mean ± SEM; ∗p < 0.01 (n = 3 independent experiments).
(D) qPCR analysis of mir-302 expression in l-Pro (150 μM) ± VitC (500 μM)-treated ESCs at day 5. Data are fold change in gene expression compared with control, normalized to Gapdh, and are mean ± SEM; ∗p < 0.005 (n = 3 independent experiments).
(E) qPCR analysis of mir-290 expression in l-Pro-treated ESCs (150 μM) at day 5. Data are fold change versus control, normalized to Gapdh, and are mean ± SEM (n = 3 independent experiments).
(F) Representative pictures (left) and frequency (right) of AP+, AP+/−, and AP− staining on colonies derived from ESCs plated at low density in the presence of the indicated inhibitors, metabolites, and growth factors. Data are mean ± SEM; ∗p < 0.05 (n = 3 independent experiments). Scale bars, 250 μm.
(G) Principal component (PC) analysis of RNA-sequencing data generated from ESCs grown as indicated in (F).
(H) Representative pictures of βIII-tubulin (green) and GFAP (red) immunofluorescence on neurons derived from ESCs grown as in (F). Nuclei are stained with DAPI. Scale bars, 100 μm.
See also Figure S2.
Molecular and Pluripotency Features of Vitamin C- and l-Proline-Induced Cells
To further support our findings, we compared VitC- and l-Pro-treated ESCs with naive/LIF/2i and primed/basic fibroblast growth factor (bFGF)/Activin A (F/A) cells, respectively. Interestingly, the percentage of round/domed-shaped and alkaline phosphatase-positive (AP+) colonies increased in 2i and VitC culture conditions compared with control, which, as expected, showed some irregular colonies with an intermediate AP staining (AP+/−) (Figure 2F). Conversely, flat-shaped F/A EpiSC colonies were AP negative. Interestingly, the large majority (≥90%) of l-Pro-induced flat-shaped colonies were either AP+/− or AP− (Figure 2F). To further investigate this phenotype, we compared the transcriptome profiling of FBS/LIF ± VitC or l-Pro, naive/2i, and F/A EpiSCs, and identified ∼7,900 deregulated genes in the different conditions (fold change [FC] ≥ 2; p < 0.05). Interestingly, principal component analysis placed VitC between 2i and untreated control, and l-Pro between control and F/A (Figure 2G). Accordingly, a set of pluripotency-associated genes was upregulated in 2i and VitC conditions but downregulated in l-Pro and F/A compared with control. Conversely, priming markers showed the opposite trend (Figure S2F and Table S2). Consistent with these findings, the four cell populations significantly differ in their differentiation potential. Indeed, only l-Pro- and F/A-derived neural cell populations already showed complex βIII-tubulin+ structures at an early time, and stained positive for the late glial marker glial fibrillary acidic protein (GFAP) (Figure 2H) (Fico et al., 2008). These data suggest that VitC and l-Pro captured alternative pluripotency states placed between the naive/2i (Habibi et al., 2013) and primed/F/A.
l-Proline and F/A Capture Distinct Temporal Stages of Primed Pluripotency
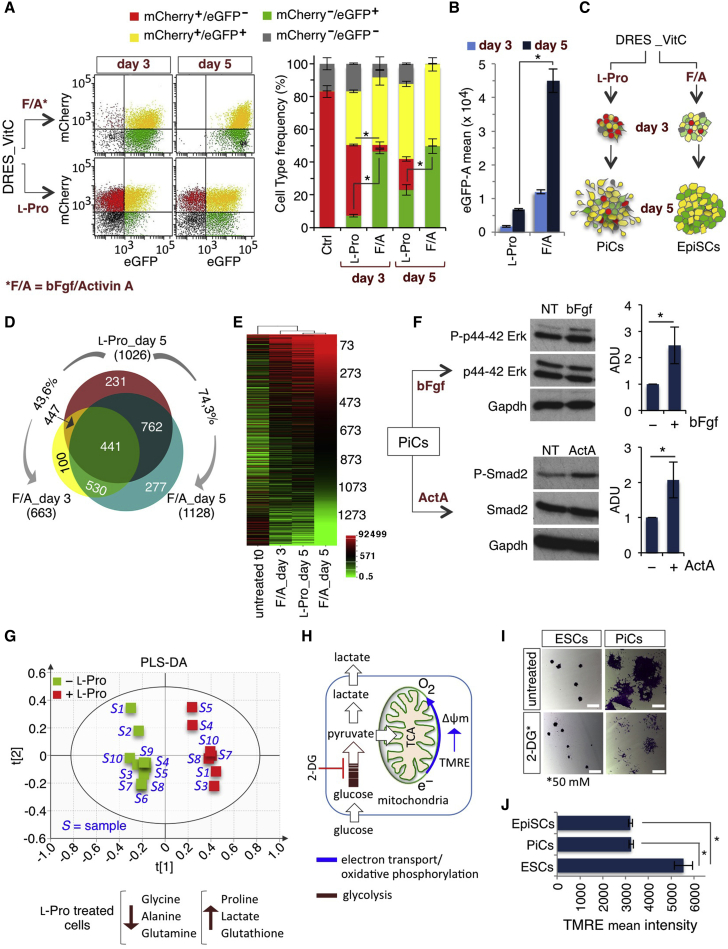
To further compare the effect of l-Pro and F/A, we analyzed the distribution of mCherry±/eGFP± cells in VitC/DRESCs upon treatment with either l-Pro or F/A. Interestingly, mCherry+/eGFP+ (yellow) and mCherry−/eGFP+ (green) cells strongly and progressively increased in both l-Pro- and F/A-treated ESCs at the expense of the mCherry+/eGFP− (red) population (Figure 3A). However, the extent of l-Pro and F/A activity was different. Specifically, the percentage of mCherry−/eGFP+ (green) cells almost doubled in F/A-treated compared with l-Pro-treated cells and mCherry+/eGFP− (red) cells persisted, although at a low level, only in l-Pro-treated DRESCs (day 5, Figure 3A). Furthermore, eGFP fluorescence intensity was higher (∼4-fold) in F/A compared with l-Pro (Figure 3B). In line with these findings, markers of the late epiblast (Brachyury, Cerberus, and Sox17) were induced by l-Pro, although at lower levels compared with F/A (Figure S3A). Finally, although the fraction of mCherry+/eGFP+ (yellow) cells was comparable, the mir-302/mir-290 expression ratio was ∼10-fold higher in F/A compared with l-Pro (Figure S3B). To further explore the possibility that l-Pro and F/A induced distinct temporal stages of primed pluripotency, we compared the transcriptome profiling of the l-Pro and F/A mCherry+/eGFP+ (yellow) population, and found >1,000 genes deregulated compared with mCherry+/eGFP− (red) control cells (NoiSeq; posterior probability > 0.99; FC ≥ 4; Table S3). The two populations shared up to 75% of common genes (Figure 3D) with the same deregulation trend (Figure 3E) but different magnitudes. Specifically, pluripotency-associated genes were downregulated either at similar level (Nanog, Klf2, Klf4, and Gbx2) or at lower levels (up to 10-fold) (Dppa 2, 3, 4, 5a, Rex1, Esrrb) in F/A- compared with l-Pro-treated cells (Figure S3C and Table S3). Furthermore, mesendodermal-related genes (e.g., Brachyury, Cer1, Dkk1, Eomes, Foxa2, and Sox17) were induced at higher levels in F/A than in l-Pro (Figure S3D and Table S3). We then asked whether l-Pro-induced cells (PiCs) were still responsive to F/A. Both Erk and Smad2 phosphorylation was induced in PiCs upon addition of either bFGF or activin (Figure 3F), and mCherry+/eGFP+ (yellow) and mCherry−/eGFP+ (green) cells increased at the expense of mCherry+/eGFP− (red) cells (Figure S3E). In addition, mean eGFP intensity also increased in F/A-treated PiCs (Figure S3E). These data indicate that l-Pro and F/A similarly induce a naive to primed transition and further suggest that l-Pro captures an earlier primed state.
Figure 3.
l-Proline Mimics bFGF/Activin A Treatment in ESCs
(A and B) DRESCs were passaged three times in VitC (100 μM), plated at low density and treated with either l-Pro (1 mM) or F/A and analyzed at days 3 and 5. Representative FACS plots (A, left panels) and quantification (A, right panel) of mCherry±/eGFP± cell distribution. Quantification of eGFP intensity mean (B). Data are mean ± SEM; ∗p < 0.001 (n = 3 independent experiments).
(C) Schematic representation of eGFP/mCherry-positive cells' distribution in colonies from l-Pro- and F/A-treated DRESCs.
(D) Venn diagram showing the overlap between differentially expressed genes in mCherry+/eGFP+ (yellow) cells from l-Pro-treated (day 5) and F/A-treated (days 3 and 5) DRESCs. Relative expression of each gene was normalized to untreated cells.
(E) Heatmap representation of genes deregulated in l-Pro-treated (day 5) and F/A-treated (days 3 and 5) cells compared with control.
(F) Western blot analysis of phospho-Erk/Erk (upper panels) and phospho-Smad2/Smad2 (lower panels) in l-Pro-induced cells (PiCs) treated with ±bFGF (12 ng/mL) and ±Activin A (20 ng/mL) (15 min), respectively. Anti-Gapdh antibody was used as loading control. Densitometric analysis (arbitrary densitometry units [ADU]) is shown as pErk/Erk and pSmad2/Smad2 ratio. Data are mean ± SEM; ∗p < 0.01 (n = 3 independent experiments).
(G) Projection to latent structure-discriminant analysis (PLS-DA) score plot showing the projection of all the spectral classes analyzed (−l-Pro, green boxes; and +l-Pro, red boxes). The model quality is evaluated by the goodness of fit and the goodness of prediction, represented by parameters R2Y = 0.98 and Q2 = 0.96. Classes are separated along the first principal component t[1], which represents interclass variation. The second component t[2] accounts for intraclass variation.
(H) Schematic representation of the oxidative phosphorylation (blue) and glycolysis (red) metabolic pathways.
(I) Representative phase-contrast images of ESC and PiC colonies (day 5), and ±2-DG (50 mM; 16 hr), stained with crystal violet. Scale bars, 200 μm.
(J) Tetramethylrhodamine (TMRE) mean intensity in ESCs, EpiSCs, and PiCs at day 5. Data are mean ± SEM; ∗p < 0.05 (n = 3 independent experiments).
See also Figure S3.
l-Proline Induces a Metabolic Reprogramming in ESCs
One of the earliest events that occur in the transition from naive to primed pluripotency is a switch from a bivalent to exclusively glycolytic metabolism (Zhou et al., 2012). We thus performed a nuclear magnetic resonance (NMR)-based global metabolomic profiling of ESCs ± l-Pro and found strong differences in their metabolite content (Figure 3G and Table S4). In particular, PiCs showed increased levels of l-Pro and reduced levels of different amino acids (l-Gln, l-Tyr, l-Gly, l-Ala l-Val, l-Leu, l-Ser) (Table S4), in line with our previous findings that l-Pro supplementation inactivates the amino acid stress response pathway in ESCs (D'Aniello et al., 2015). Remarkably, lactate levels significantly increased (Figures 3G, 3H, and S3F; Table S4) and PiCs were more susceptible to the glycolysis inhibitor 2-deoxy-D-glucose (2-DG) compared with ESCs (Figures 3H, 3I, and S3G), suggesting a metabolic shift to glycolysis. Moreover, the mitochondrial membrane potential was significantly reduced in PiCs as in F/A EpiSCs compared with ESCs (Figure 3J), indicating that oxidative phosphorylation was reduced, and further supporting the idea that PiCs undergo a metabolic reprogramming, which resembles the one occurring in the naive to primed transition (Zhou et al., 2012).
l-Proline-Induced Primed State Depends on Autocrine FGF and TGF-β Signaling Pathways
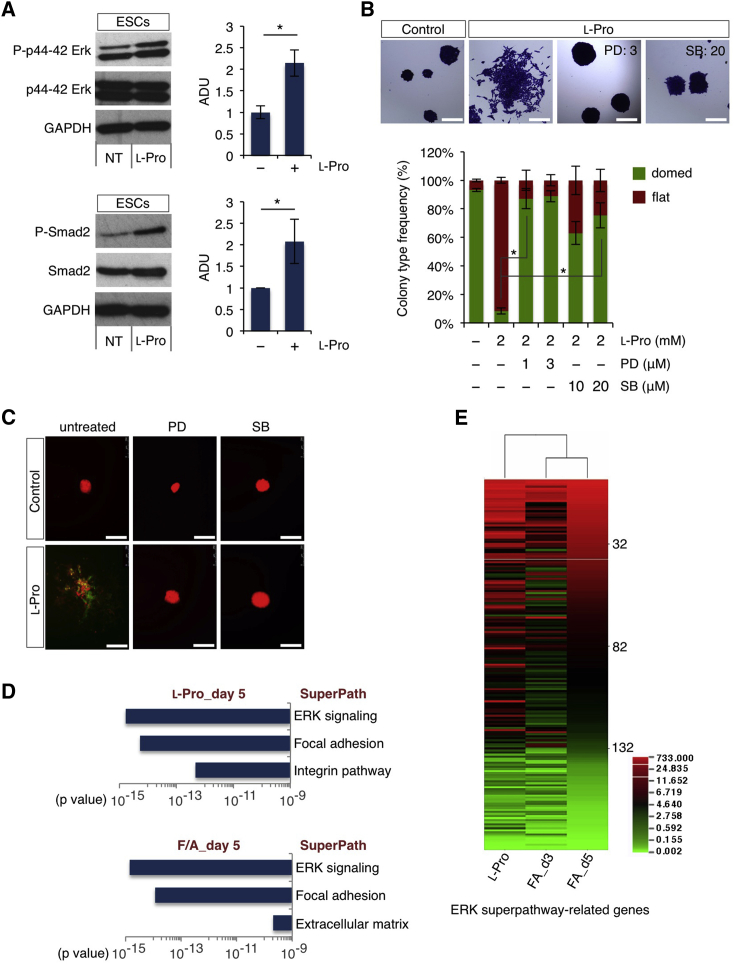
To further explore the notion that l-Pro and F/A exert similar effects on ESCs, we first evaluated whether l-Pro induced Erk (FGF) and Smad2 (transforming growth factor β [TGF-β]) signaling pathways in FBS/LIF ESCs. Interestingly, both Erk and Smad2 phosphorylation were induced by supplemental l-Pro (Figure 4A). Furthermore, l-Pro-induced phenotypic transition was impaired in the presence of FGF and TGF-β chemical inhibitors PD0325901 and SB431542, respectively. Indeed, while l-Pro-treated cells generated only 15% of domed colonies, domed-shaped mCherry+/eGFP− (red) colonies increased up to 70% in the presence of PD0325901 or SB431542, even with high doses of l-Pro (Figures 4B and 4C). Moreover, gene ontology analysis revealed that the transcriptome of both l-Pro- and F/A-derived mCherry+/eGFP+ (yellow) cells was highly enriched in genes of Erk superpathway (Figures 4D and 4E; Table S3). These data suggested that l-Pro drives naive ESCs toward a primed state of pluripotency by inducing and sustaining autocrine FGF and TGF-β signaling.
Figure 4.
l-Proline Induces FGF and TGF-β Signaling in ESCs
(A) Western blot analysis of phospho-Erk/Erk (upper panels) and phospho-Smad2/Smad2 proteins (lower panels) in ESCs ± l-Pro (1 mM; 20 hr). Anti-Gapdh antibody was used as loading control. Densitometric analysis (ADU) is shown as pErk/Erk and pSmad2/Smad2 ratio. Data are mean ± SEM; ∗p < 0.05 (n = 3 independent experiments).
(B) Representative photomicrographs (upper panels) of colonies generated from ESCs ± l-Pro, either alone or ±PD0325901 (PD, 1–3 μM) or SB431542 (SB, 10–20 μM), and stained with crystal violet. Scale bars, 250 μm. Classification of colonies based on their morphology (lower panel). Data are mean ± SEM; ∗p < 0.01 (n = 3 independent experiments).
(C) Representative images of mCherry+ colonies generated from ESCs ± l-Pro (1 mM), either alone or ±PD or SB, at day 5. Scale bars, 250 μm.
(D) Transcriptome analysis using GeneAnalytics software showing the enrichment in Erk pathway-associated genes in l-Pro- and FA-treated cells (day 5).
(E) Heatmap showing Erk-superpathway-related genes deregulated in l-Pro-treated (day 5) and F/A-treated (days 3 and 5) ESCs.
See also Figure S3.
Discussion
The naive to lineage primed transition was proposed as a gradual continuum of pluripotent states with distinct functional and transcriptional signatures and biases (Tesar, 2016). Several stem cell types with different degrees of pluripotency can be established under artificial culture conditions (Wu and Izpisua Belmonte, 2015). Specifically, naive and primed (EpiSCs) cells rely on chemical inhibitors (2i) or growth factors (F/A), respectively, and are largely used as in vitro models of pluripotency. Here we provide evidence that the relative levels of two physiological metabolites, namely VitC and l-Pro, push FBS/LIF ESCs toward naive (low l-Pro/high VitC) or early primed (high l-Pro/low VitC) states, between naive/2i and F/A primed states of the pluripotency continuum. In particular, similarly to the F/A primed state (EpiSCs), the high l-Pro/low VitC condition is associated with a metabolic reprogramming toward glycolysis and is strictly dependent on the activation and maintenance of autocrine bFGF and TGF-β signaling pathways. Interestingly, while F/A EpiSCs show molecular features of late pre-gastrulation epiblast and are difficult to revert to naive ESCs (Guo et al., 2009), PiCs display molecular, metabolic, and functional features (Table S5) of a transitional/reversible primed state in the pluripotency continuum, which has been hypothesized to define the boundary beyond which the transition becomes irreversible (Hackett and Surani, 2014, Martello and Smith, 2014). Of note, our findings indicate that VitC and l-Pro are both limiting in ESCs even in complete FBS/LIF culture conditions. This is in line with our recent findings that an autoregulatory loop limits l-Pro biosynthesis in ESCs (D'Aniello et al., 2015); nevertheless, whether and how intracellular levels of VitC are regulated in ESCs is still unknown and deserves further investigation. VitC enhances epigenetic modifications, including DNA and histone demethylation, and promotes cell reprogramming (Esteban et al., 2010, Stadtfeld et al., 2012, Wang et al., 2011). Here we show that VitC and l-Pro supplementation oppositely modify DNA methylation at genomic regions that normally gain methylation during the blastocyst to epiblast transition (Blaschke et al., 2013). The DNA demethylation effect of VitC is well explained by its ability to positively regulate Fe(II)/oxoglutarate-dependent dioxygenase enzymes of the TET family (5mC to 5hmC conversion) (Blaschke et al., 2013, von Meyenn et al., 2016). Conversely, no data have been reported so far linking l-Pro and DNA methylation. Although the molecular mechanism by which a sudden increase of l-Pro induces DNA methylation in ESCs is still far from being fully elucidated, we suggest that l-Pro may influence VitC homeostasis, reducing its availability/activity for the TET demethylases. First, the vast majority of the genomic regions that are hypomethylated in VitC-treated ESCs are in turn hypermethylated upon l-Pro supplementation. Moreover, at appropriate stoichiometric ratios, VitC fully counteracts l-Pro-induced DNA methylation. How may l-Pro affect VitC availability? l-Pro could alter the redox balance and eventually induce VitC oxidation, i.e., its conversion to dehydroascorbic acid, by generating reactive oxygen species, which are by-product of the activity of mitochondrial l-Pro oxidase enzyme (PRODH/POX) (Phang et al., 2010). However, previous findings argue against a primary role of oxidative stress/redox signaling in this process (Comes et al., 2013, D'Aniello et al., 2015), and further evidence comes from metabolomics analysis, which reveals no sign of global oxidation and even higher levels of reduced glutathione (GSH) in l-Pro-treated cells (Figure 3G and Table S4), suggesting that l-Pro supplementation does not exert an oxidizing effect in these culture conditions. Thus, we propose that the mechanism by which VitC and l-Pro antagonistic effects capture alternative states in the pluripotency continuum does not rely, at least primarily, on altered redox balance, which would favor a reducing (VitC) versus an oxidant (l-Pro) state. Of note, this is consistent with the idea that pluripotency is linked to a reduced state, while activation of oxidation is a metabolic signature of ESC differentiation (Yanes et al., 2010). Indeed, the GSH/glutathione disulfide (GSSG) ratio and VitC levels are inversely correlated in ESC cardiac and neural differentiation, suggesting that VitC compensates for GSSG accumulation to maintain homeostasis (Yanes et al., 2010). Since l-Pro activity depends on protein synthesis (D'Aniello et al., 2015), we hypothesize that a sudden increase of l-Pro could influence VitC homeostasis/availability, at least in part, by inducing the synthesis of l-Pro-rich proteins such as collagens. Indeed, nascent collagens are modified by VitC-dependent Fe(II)/oxoglutarate-dependent prolyl hydroxylase enzymes (Gorres and Raines, 2010). Thus, an abrupt increment of collagen biosynthesis/hydroxylation may reduce VitC availability for TET demethylases as well as for other VitC-dependent enzymes, such as the prolyl hydroxylases that regulate the hypoxia-inducible factor (HIF) steady-state level (Fong and Takeda, 2008). Interestingly, HIF is a key regulator of the metabolic reprogramming and is stabilized in primed but not in naive human ESCs (Sperber et al., 2015, Zhou et al., 2012). Of note, some of the HIF targets are induced in l-Pro-treated cells, such as Ldha, Pfkfb3, Plin2, Gbe1, and Pygl (Table S2). Although future studies are needed to clarify the molecular mechanisms, our findings have important implications for stem cell biology as they provide insights into how the availability of natural metabolites contribute to global epigenetic changes regulating alternative pluripotent states (Carey et al., 2015, Ryall et al., 2015, Wang et al., 2009). Furthermore, they attest to the definition of the optimal culture conditions for the capture in vitro of an early primed state of pluripotency.
Experimental Procedures
Full details of the Experimental Procedure are included in Supplemental Experimental Procedures.
Culture of ESCs, Reagents, and Treatments
Wild-type TBV2 (129/SvP) and dual-reporter (DRESCs) mouse ESCs were maintained on a feeder layer of mitomycin C-treated MEFs according to standard procedures. ESC to PiC transition and ESC to EpiSC transition were performed as previously described (Casalino et al., 2011, Guo et al., 2009).
VitC was dissolved in water and used at the indicated concentrations (100–500 μM). VitC/l-Pro were added concomitantly. PD0325901 and SB431542 (Sigma-Aldrich) were used at concentration ranging from 1 to 3 μM and 10 to 20 μM, respectively. 2-Deoxy-D-glucose (2-DG, Sigma-Aldrich) was dissolved in water and used at concentration ranging from 6.25 to 50 mM.
5mC and 5hmC HPLC-MS/MS Measurement
Genomic DNA was extracted using the Wizard Genomic DNA Purification kit (Promega). 5mC and 5hmC high-performance liquid chromatography (HPLC)-tandem MS measurements were performed as described previously (Kroeze et al., 2014).
RNA Sequencing and Reduced Representation Bisulfite Sequencing
RNA sequencing and RRBS were performed at the Institute of Applied Genomics (Udine, Italy).
NMR-Based Metabolomic Analysis
The metabolic profiles were acquired through NMR spectroscopy. Details on data processing are provided in Supplemental Experimental Procedures.
Statistical Analysis
Statistical significance was determined by a two-tailed paired Student's t test. p Values of <0.05 were considered as statistically significant. Error bars in the figures represent SEM.
Multivariate statistical analysis of NMR data was performed using SIMCA 14 software (Umetrics). Details are provided in Supplemental Experimental Procedures.
Author Contributions
C.D'A., A.F., D.d.C., E.J.P., and G.M. conceived and designed the study. C.D'A., D.d.C., F.C., G.d.N., A.F., E.H., and G.C. performed the experiments. E.H., F.R., and C.A. performed bioinformatics analysis. D.P., D.J.M., and A.M. performed metabolomics analysis. R.B., A.M., and H.G.S. gave conceptual advice and edited the manuscript. C.D'A., E.J.P., and G.M. wrote the manuscript.
Acknowledgments
We are grateful to members of the Integrated Microscopy and FACS Facilities of IGB-ABT, CNR. We thank Gennaro Andolfi for excellent technical assistance. This study was supported by Epigenomics Flagship Project (EPIGEN) MIUR-CNR to G.M. and C.A., and AIRC (grant 11599), Italian Ministry of Education-University-Research (grant CTN01_00177 Cluster ALISEI_IRMI and PRIN) and CARIPLO to G.M.
Published: December 22, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.11.011.
Contributor Information
Dario De Cesare, Email: dario.decesare@igb.cnr.it.
Gabriella Minchiotti, Email: gabriella.minchiotti@igb.cnr.it.
Accession Numbers
The accession number for all RNAseq and RRBS data reported in this paper is GEO: GSE84373.
Supplemental Information
References
- Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martinez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Finley L.W., Cross J.R., Allis C.D., Thompson C.B. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino L., Comes S., Lambazzi G., De Stefano B., Filosa S., De Falco S., De Cesare D., Minchiotti G., Patriarca E.J. Control of embryonic stem cell metastability by L-proline catabolism. J. Mol. Cell Biol. 2011;3:108–122. doi: 10.1093/jmcb/mjr001. [DOI] [PubMed] [Google Scholar]
- Comes S., Gagliardi M., Laprano N., Fico A., Cimmino A., Palamidessi A., De Cesare D., De Falco S., Angelini C., Scita G. L-Proline induces a mesenchymal-like invasive program in embryonic stem cells by remodeling H3K9 and H3K36 methylation. Stem Cell Reports. 2013;1:307–321. doi: 10.1016/j.stemcr.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aniello C., Fico A., Casalino L., Guardiola O., Di Napoli G., Cermola F., De Cesare D., Tate R., Cobellis G., Patriarca E.J. A novel autoregulatory loop between the Gcn2-Atf4 pathway and (L)-Proline [corrected] metabolism controls stem cell identity. Cell Death Differ. 2015;22:1094–1105. doi: 10.1038/cdd.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Fico A., Manganelli G., Simeone M., Guido S., Minchiotti G., Filosa S. High-throughput screening-compatible single-step protocol to differentiate embryonic stem cells in neurons. Stem Cell Dev. 2008;17:573–584. doi: 10.1089/scd.2007.0130. [DOI] [PubMed] [Google Scholar]
- Fong G.H., Takeda K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008;15:635–641. doi: 10.1038/cdd.2008.10. [DOI] [PubMed] [Google Scholar]
- Gorres K.L., Raines R.T. Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 2010;45:106–124. doi: 10.3109/10409231003627991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Surani M.A. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Harvey A.J., Rathjen J., Gardner D.K. Metaboloepigenetic regulation of pluripotent stem cells. Stem Cell Int. 2016;2016:1816525. doi: 10.1155/2016/1816525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hore T.A., von Meyenn F., Ravichandran M., Bachman M., Ficz G., Oxley D., Santos F., Balasubramanian S., Jurkowski T.P., Reik W. Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naive pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. USA. 2016;113:12202–12207. doi: 10.1073/pnas.1608679113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R., Blelloch R.H. Epigenetics of cellular reprogramming. Curr. Opin. Genet. Dev. 2013;23:548–555. doi: 10.1016/j.gde.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeze L.I., Aslanyan M.G., van Rooij A., Koorenhof-Scheele T.N., Massop M., Carell T., Boezeman J.B., Marie J.P., Halkes C.J., de Witte T. Characterization of acute myeloid leukemia based on levels of global hydroxymethylation. Blood. 2014;124:1110–1118. doi: 10.1182/blood-2013-08-518514. [DOI] [PubMed] [Google Scholar]
- Martello G., Smith A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Martinez Arias A., Nichols J., Schroter C. A molecular basis for developmental plasticity in early mammalian embryos. Development. 2013;140:3499–3510. doi: 10.1242/dev.091959. [DOI] [PubMed] [Google Scholar]
- Maruotti J., Dai X.P., Brochard V., Jouneau L., Liu J., Bonnet-Garnier A., Jammes H., Vallier L., Brons I.G., Pedersen R. Nuclear transfer-derived epiblast stem cells are transcriptionally and epigenetically distinguishable from their fertilized-derived counterparts. Stem Cells. 2010;28:743–752. doi: 10.1002/stem.400. [DOI] [PubMed] [Google Scholar]
- Monfort A., Wutz A. Breathing-in epigenetic change with vitamin C. EMBO Rep. 2013;14:337–346. doi: 10.1038/embor.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem R.J., Ye J., Judson R.L., LaRussa M.F., Krishnakumar R., Blelloch A., Oldham M.C., Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14:617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang J.M., Liu W., Zabirnyk O. Proline metabolism and microenvironmental stress. Annu. Rev. Nutr. 2010;30:441–463. doi: 10.1146/annurev.nutr.012809.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryall J.G., Cliff T., Dalton S., Sartorelli V. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell. 2015;17:651–662. doi: 10.1016/j.stem.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Locasale J.W., Lyssiotis C.A., Zheng Y., Teo R.Y., Ratanasirintrawoot S., Zhang J., Onder T., Unternaehrer J.J., Zhu H. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2012;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H., Mathieu J., Wang Y., Ferreccio A., Hesson J., Xu Z., Fischer K.A., Devi A., Detraux D., Gu H. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T., Ooi S.S., Kim S.Y., Bestor T.H., Shioda T. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44:398–405. doi: 10.1038/ng.1110. S1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J. Snapshots of pluripotency. Stem Cell Reports. 2016;6:163–167. doi: 10.1016/j.stemcr.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- von Meyenn F., Iurlaro M., Habibi E., Liu N.Q., Salehzadeh-Yazdi A., Santos F., Petrini E., Milagre I., Yu M., Xie Z. Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol. Cell. 2016;62:848–861. doi: 10.1016/j.molcel.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Alexander P., Wu L., Hammer R., Cleaver O., McKnight S.L. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Chen K., Zeng X., Yang J., Wu Y., Shi X., Qin B., Zeng L., Esteban M.A., Pan G. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Washington J.M., Rathjen J., Felquer F., Lonic A., Bettess M.D., Hamra N., Semendric L., Tan B.S., Lake J.A., Keough R.A. L-Proline induces differentiation of ES cells: a novel role for an amino acid in the regulation of pluripotent cells in culture. Am. J. Physiol. Cell Physiol. 2010;298:C982–C992. doi: 10.1152/ajpcell.00498.2009. [DOI] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Wu J., Izpisua Belmonte J.C. Dynamic pluripotent stem cell states and their applications. Cell Stem Cell. 2015;17:509–525. doi: 10.1016/j.stem.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Yanes O., Clark J., Wong D.M., Patti G.J., Sanchez-Ruiz A., Benton H.P., Trauger S.A., Desponts C., Ding S., Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C.A., Horwitz M.S., Hockenbery D., Ware C. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.