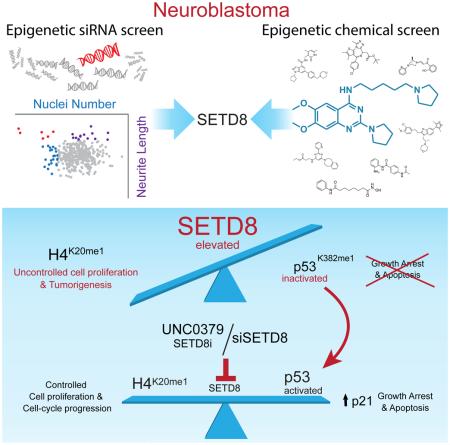
Summary
Given the paucity of druggable mutations in high-risk neuroblastoma (NB), we undertook chromatin-focused siRNA and chemical screens to uncover epigenetic regulators critical for the differentiation block in high-risk NB. High-content Opera imaging identified 53 genes whose loss of expression led to a decrease in NB cell proliferation and 16 also induced differentiation. From these, the secondary chemical screen identified SETD8, the H4K20me1 methyltransferase, as a druggable NB target. Functional studies revealed that SETD8 ablation rescued the proapoptotic and cell-cycle arrest functions of p53 by decreasing p53K382me1, leading to activation of the p53 canonical pathway. In pre-clinical xenograft NB models, genetic or pharmacological (UNC0379) SETD8 inhibition conferred a significant survival advantage, providing evidence for SETD8 as a therapeutic target in NB.
Graphical abstract
Introduction
Alterations in epigenetic machinery, including mutations or overexpression of chromatin remodelers and modifiers as well as DNA hypermethylation, can disrupt normal development, resulting in pathologies, including cancer (Arrowsmith et al., 2012; Jones and Baylin, 2007). Neuroblastoma (NB) is a neural crest-derived tumor and, based on its histology, transcriptome and propensity to differentiate, is thought to arise from a failure of sympathoadrenal progenitors to differentiate (Schulte and Eggert, 2015). Genetic and epigenetic regulators have been implicated in the undifferentiated status of high-risk NB (HR-NB) tumors (Lawlor and Thiele, 2012).
NB accounts for 15% of all deaths in children due to cancer. Despite intense multimodality treatment, current therapy for NB is still inadequate for 50% of HR-NB patients and has significant toxicities and morbidities (Maris, 2010). MYCN amplification is a powerful independent marker of adverse outcome in HR-NB (Bagatell et al., 2009; Canete et al., 2009). While TP53 is rarely (<2%) mutated in primary HR-NB, its functional activity is attenuated in MYCN-amplified NB by a variety of mechanisms (Petroni et al., 2012; Van Maerken et al., 2006; Van Maerken et al., 2009). However, the majority (60-70%) of HR-NBs do not have MYCN amplification and mechanisms that attenuate the activity of p53 in these tumors have not been intensively studied. So the challenge is to identify novel targeted therapies for HR-NB patients.
Aside from the anaplastic lymphoma kinase ALK, NexGen sequencing revealed few druggable, recurrent somatic mutations in NBs (Pugh et al., 2013). In older HR-NB patients, studies identified mutations in the chromatin remodelers ATRX (22%) and ARID1A/ARID1B (11%) (Alexandrov et al., 2013; Cheung et al., 2012; Molenaar et al., 2012; Pugh et al., 2013; Sausen et al., 2013), which are not directly druggable but may exhibit synthetic lethality in drug combinations (Bitler et al., 2015; Wilson et al., 2010). Studies showing overexpression of BMI-1 and KDM1A (LSD1), and our work showing dysregulation of EZH2 and sensitivity to EZH2 inhibitors, indicate a linkage between aberrations in epigenetic regulators and NB (Cui et al., 2006; Lawlor and Thiele, 2012; Schulte et al., 2009; Wang et al., 2012). This led us to hypothesize that epigenetic dysregulation in sympathoadrenal progenitors contributes to the initiation and progression of NB.
The goal of this study is to survey the epigenetic landscape of NB in order to identify those regulators that block neuroblastoma terminal differentiation. By characterizing critical epigenetic pathways regulating cell proliferation and differentiation that are altered in NB tumor cells, we may identify therapeutically relevant targets.
Results
siRNA screen of chromatin modifiers identifies regulators of NB cell proliferation and differentiation
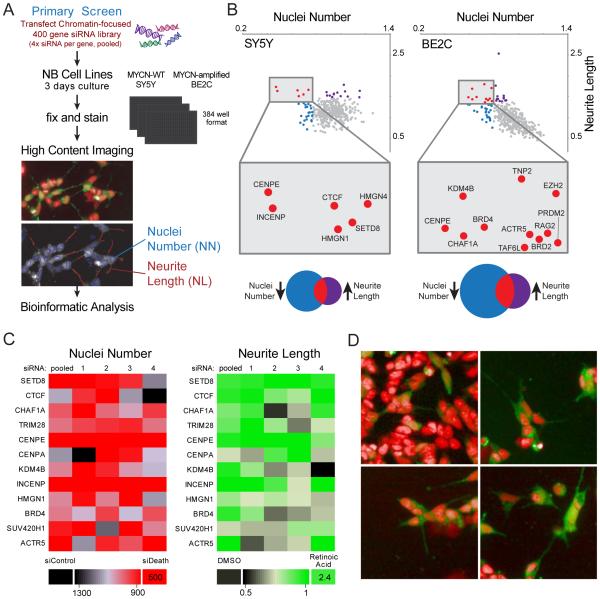
To identify epigenetic regulators of NB cell proliferation and differentiation, we performed a cell-based high-throughput siRNA screen of almost 400 genes (pool of 4 siRNAs per gene) encoding modulators of chromatin structure and function (including ~55% of the known epigenetic enzymes, writers and erasers) in 2 NB cell lines SY5Y (MYCN-WT) and SK-N-BE2C (MYCN-amplified (MYCN-amp)). Nuclei number (NN) and neurite length (NL) were used as measures of cell proliferation and morphologic differentiation, respectively (Figure 1A and Table S1). The primary screen identified 53 genes whose knockdown significantly and substantially reduced cell proliferation in one or both cell lines (Table S2). Of these, 16 genes also induced differentiation (increased NL) (Figure 1B, red dots, and Table S2). The reproducibility of replicates was assessed by Pearson correlation and permutation tests (Figure S1A-D and Supplemental Experimental Procedures). Four of the top candidates in the screen, CENPE (Balamuth et al., 2010), BRD4 (Puissant et al., 2013), CHAF1A (Barbieri et al., 2014) and KDM4B (Yang et al., 2015) were already characterized as required for NB cell survival. We performed a validation screen using individual siRNAs against select candidate genes in SY5Y and BE2C (Figure 1C, for selection and validation criteria, see Supplemental Experimental Procedures). For all selected genes, silencing led to a statistically significant reduction in NN. Silencing of SETD8, CTCF, CENPE, INCENP and ACTR5 also significantly increased NL (Table S3). Representative images showing decreased cell number and increased neurite formation after silencing of selected hits are shown (Figure 1D). Thus, we identified 16 genes whose silencing led to significant reduction in NB cell proliferation and induction of morphologic differentiation, nine of which were validated for NN and five for NL.
Figure 1. Epigenetic focused high-throughput RNAi screen of NB cells reveals 16 vulnerability genes.
(A) A flow chart of the high-throughput imaging assay identified key steps. A library of siRNAs against ~400 chromatin-focused genes was reverse transfected into NB cell lines. After 3 days of culture, the cells were fixed and stained. A high-throughput imaging assay was used to determine nuclei number (NN) and neurite length (NL). NB cells driven to differentiate by retinoic acid treatment (upper panel) were visualized using stably expressed GFP and a nuclear stain (Hoechst 33342). Nuclei (blue outline) and neurite outgrowths (red line segment) were detected by an automated image analysis algorithm (lower panel).
(B) Scatter plots showing statistically significant genes in SY5Y (left panel) and SK-N-BE2C (right panel). siRNAs that met both parameters: decrease of NN and increase of NL (red dots). Venn diagrams showing statistically significant genes for which siRNAs decreased NN (blue) or increased NL (purple) or both (red).
(C) Heatmaps showing the results of the secondary screen performed with a siRNAs library with 4 deconvoluted sequences and smartpool siRNAs (pooled): 12 statistically significant genes were analyzed in SY5Y on the basis of NN (red heatmap) and NL (green heatmap). The positive controls used were an siRNA that induced cell death (NN) and retinoic acid (NL).
(D) Stably expressed GFP and Hoechst 33342 (pseudocolored as red) were used to label SY5Y cells. Representative images show effects on cell number and morphology after silencing of CENPE, SETD8 and BRD4 compared with non-targeting siRNA.
A chemical screen confirms inhibition of SETD8 activity as a vulnerability in NB
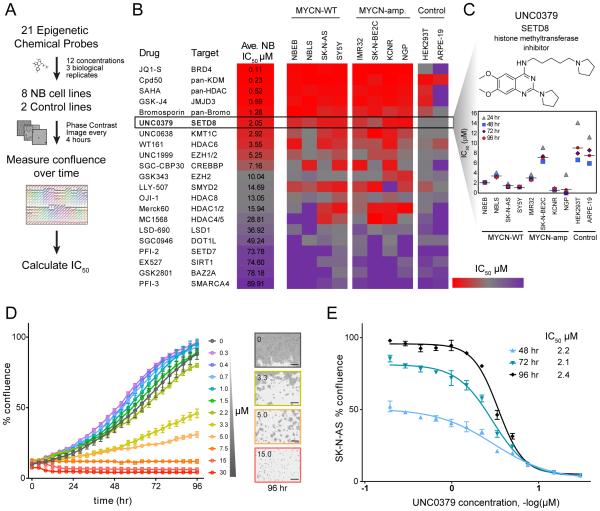
We next performed a chemical screen using 21 epigenetic chemical probes, provided by the Structural Genomics Consortium (SGC) and targeting 90% of proteins with enzymatic activity that were targeted by the siRNA library. This chemical library includes pharmaceutical tool compounds in the drug development pipeline. Each compound was evaluated in 8 NB (4 MYCN-WT and 4 MYCN-amp) and 2 immortal but non-transformed (HEK293T and ARPE-19) cell lines using 12 concentrations (0.1-30 μM). Cell viability was assessed over 7 days (Figures 2A and 2B). JQ1-S, a BRD4 inhibitor (Puissant et al., 2013), was the most potent and showed the expected difference in sensitivity between MYCN-WT and MYCN-amp cells (Table S4). Pan-lysine demethylase, pan-histone acetyltransferase and pan-Bromodomain inhibitors also showed high potency. The targets of JQ1 (BRD4, BRD2) and Cpd50 (KDM4B) were among the genes whose siRNAs inhibited cell growth and induced differentiation in the genetic screen (Figure 1B). Interestingly, a selective, substrate-competitive inhibitor of SETD8, UNC0379 (Ma et al., 2014a) was one of the most active compounds, exhibiting a relatively low average IC50 (2 μM) (Figure 2B) and a highly significant p value for the In Vitro Therapeutic Index (IVTI) (Figure S2 and Table S4) across the 8 NB cell lines compared with control cell lines. Median IC50 values for the NB cell lines following UNC0379 treatment at 24-96 hr are depicted in Figure 2C, with 96 hr IC50 values ranging from 0.64 to 7 μM (95% confidence interval 0.98, 3.82). A representative experiment shows the effects of UNC0379 on the NB cell growth (Figures 2D and 2E). Together, the genetic and chemical-based screens suggest that SETD8 regulates NB cell growth and differentiation.
Figure 2. Chemical screen of 21 epigenetic probes identifies SETD8 inhibitor as one of the top compounds with the lowest IC50.
(A) Flow chart of the chemical screen: 21 epigenetic probes were tested in the indicated cell lines and images were recorded by an IncuCyte Zoom System in order to calculate IC50.
(B) Heatmap showing the average IC50 of the indicated epigenetic chemical probes at 96 hr in 4 MYCN-WT and 4 MYCN-amp NB cell lines compared to 2 control cell lines (ARPE-19 and 293T). The color key represents the average IC50 (μM) values: red color indicates low IC50 whereas purple color represents high IC50. Cell confluence was recorded by an IncuCyte Zoom System. The average IC50 was calculated based on the cell confluence values of 3 biological replicates across 8 NB cell lines at 96 hr using GraphPad Prism. For in vitro therapeutic index (IVTI) and p values, see Figure S2 and Table S4.
(C) In vitro UNC0379 treatment of NB cell lines and control cell lines ARPE-19 and 293T (average of 3 biological replicates).
(D, E) A representative experiment showing effects of in vitro treatment of SK-N-AS cells with UNC0379 at indicated times (D) and concentrations (E). Bars show the average of 3 replicates ± SD. In panel (D) the images were captured by IncuCyte Zoom. Scale bars, 300 μm.
See also Table S4 and Figure S2.
SETD8 is overexpressed in NB cells and high SETD8 expression correlates with poor outcome in MYCN-WT HR-NB patients
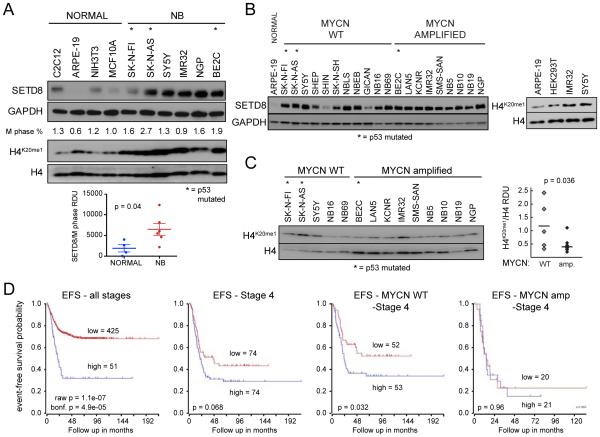
SETD8 is a histone methyltransferase that specifically catalyzes mono-methylation of K20 on histone H4 (H4K20me1). Levels of SETD8 and its target H4K20me1 are increased during mitosis and required for cell proliferation in human cell models (Wu and Rice, 2011; Wu et al., 2010). To assess the expression of SETD8 in NB, we cultured NB cells and harvested them during exponential growth. We found elevated levels of both SETD8 (3.6-fold) and its target H4K20me1 (2.6-fold) in NB cells compared with non-transformed normal cell lines. SETD8 levels were higher when normalized to the
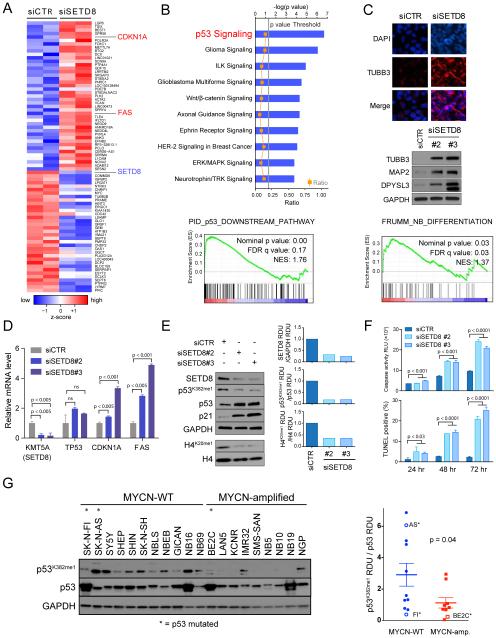
M-phase fraction (Figure 3A). This is consistent with previous studies showing overexpression of SETD8 in various types of cancer (Takawa et al., 2012). Although the levels of SETD8 expression were not significantly different between MYCN-WT and MYCN-amp NB cells (Figures 3B, S3A, and S3B), the levels of H4K20me1 were higher in MYCN-WT compared with MYCN-amplified NB cell lines in vitro (Figures 3C). This may be due to higher levels of PHF8, the demethylase for H4K20me1, in MYCN-amp HR-NBs (database: R2 http://r2.amc.nl/ Kocak Stage 4 tumors p = 1.3×10−4). In primary NB tumors (database: R2 Kocak NB; n = 476 tumors), elevated levels of SETD8 mRNA are significantly correlated with poor prognosis in primary NB tumors (Figure 3D; bonferroni p = 4.9×10−5). In addition, Stage 4 HR-NB patients whose tumors had high levels of SETD8 tend to do worse, although the correlation does not reach statistical significance (p = 0.068) (Figure 3E). However, elevated levels of SETD8 are associated with worse prognoses in Stage 4 MYCN-WT tumors (p = 0.032) (Figure 3F), but not MYCN-amp tumors (p = 0.96) (Figure 3G). The prognostic significance of SETD8 in MYCN-WT primary tumors was replicated in two additional R2 databases: SEQC-498 and Seeger (Figures S3C-S3E). Focal amplifications of Chr12q24 occur in a region that encompasses KMT5A (SETD8) in almost 22% of MYCN-WT and 6% of MYCN-amp HR-NB tumors (Schnepp et al., 2015; Wolf et al., 2010) and may contribute to the association of elevated SETD8 mRNA levels with poor outcome in NBs.
Figure 3. SETD8 is overexpressed in NB cells and high SETD8 expression associated with worse prognosis in MYCN-WT tumors.
(A) Immunoblots show SETD8 and H4K20me1 levels in NB cells compared with control cells. SETD8 protein levels normalized to M phase % in NB compared to normal cells were calculated as RDU (Relative Densitometric Unit, p = 0.04). Bars show the mean ± SEM.
(B) Immunoblot showing SETD8 protein levels in MYCN-WT and MYCN-amp NB cells.
(C) Western blot (left) and densitometric analysis (right) of H4K20me1 protein levels normalized to H4 protein levels in 14 NB cell lines were calculated as RDU (p = 0.036) using Image J Software.
(D) Kaplan-Meier plots based on the expression of SETD8 tumors from NB patients at all stages (left panel) or in tumors from stage 4 (middle left panel) NB patients (R2 database: Kocak raw p = 1.1e-07 bonf p = 4.9e-05; p = 0.068). Kaplan-Meier plots based on the expression of SETD8 in MYCN-WT (middle right panel) or MYCN-amp (right panel) tumors from stage 4 NB patients (R2 database: Kocak n = 105, MYCN-WT, stage 4 NB tumors, p = 0.032; n = 41, MYCN-amp, stage 4 NB tumors, p = 0.96).
See also Figure S3.
SETD8 silencing rescues p53 function by decreasing p53K382me1 levels
Since SETD8 activity has been implicated in multiple biologic processes, including control of cell division and modulation of transcription, RNA-seq transcriptome analyses after SETD8 silencing were performed in order to gain insights into potential mechanisms contributing to the SETD8-mediated growth inhibition and differentiation in NB cells. Preliminary studies established conditions for effective silencing of SETD8 while minimizing effects on cell viability (Figure S4A-G and Supplemental Experimental Procedures). The top differentially up- and down-regulated genes, ranked by statistical significance based on edgeR software analysis and a false discovery rate (FDR) < 0.001, are depicted as a clustered heatmap in Figure 4A (see also Table S5). IPA (Ingenuity Pathway Analysis, www.ingenuity.com) of the differentially expressed genes identified p53 signaling and neuronal differentiation-associated pathways among the most significantly up-regulated (Figure 4B). GSEA (Gene Set Enrichment Analysis) showed enrichment in several p53 pathways (Figure 4B and Table S6). SETD8 silencing resulted in increased expression of biochemical markers of neural differentiation (TUBB3, MAP2, DPYSL3) (Choi et al., 2005) and GSEA showed an enrichment in the FRUMM NB differentiation signature (Frumm et al., 2013) (Figure 4C and Table S7) supporting the morphological differentiation noted in the siRNA screen (Figure 1B-1D). The alteration in the expression of selected genes following silencing of SETD8 was confirmed (Figure 4D). These data suggest that SETD8 ablation leads to increased differentiation and activation of the p53 canonical pathway.
Figure 4. Genetic inhibition of SETD8 leads to activation of the p53 canonical pathway by decreased p53K382me1 levels.
(A) Heatmap showing the top up- and down-regulated genes ranked by statistical significance following 36 hr of SETD8 silencing in SY5Y cells. Data are presented as normalized expression values of 2 biological replicates based on edgeR software analysis and FDR < 0.001. The color key represents the normalized expression values: blue (low) to red (high).
(B) The top ten differentially expressed canonical pathways after SETD8 silencing in SY5Y cells defined by IPA (Ingenuity pathway analysis) based on edgeR software analysis and FDR < 0.001 (upper panel). GSEA (Gene Set Enrichment Analysis) of the p53 downstream pathway (lower panel, nominal p value = 0.00, FDR = 0.17, NES = 1.76) after SETD8 silencing.
(C) Immunofluorescence analysis showing the expression of TUBB3 (red) after SETD8 silencing for 72 hr in SY5Y cells counter-stained with DAPI (blue). Scale bars, 100 μm (upper panel). Immunoblot of proteins from SY5Y cells 72 hr after transfection with control siRNA and SETD8 siRNAs and blotted with antibodies detecting TUBB3, MAP2 and DPYSL3 proteins (middle panel). GSEA of the FRUMM NB differentiation signature (lower panel, nominal p value = 0.03, FDR = 0.03, NES = 1.37) after SETD8 silencing.
(D) qRT-PCR analysis showing relative mRNA levels of the indicated genes after SETD8 silencing for 36 hr. Bars show the mean ± SEM of 3 replicates.
(E) Immunoblot analysis of SY5Y (MYCN-WT) cells upon treatment with either of 2 different siRNAs targeting SETD8 for 72 hr. Densitometric analysis of SETD8 protein levels normalized to GAPDH (upper right panel), of p53K382me1 protein levels normalized to p53 (middle right panel) and of H4K20me1 protein levels normalized to H4 (lower right panel) after SETD8 silencing calculated as RDU using Image J Software.
(F) Caspase 3/7 activity and number of TUNEL positive cells upon SETD8 silencing in SY5Y cells. Bars show the average of 3 replicates ± SD.
(G) Immunoblot (left) and densitometric analysis (right) of p53K382me1 protein levels normalized to p53 protein levels in 20 NB cell lines were calculated as RDU (p = 0.04) using Image J Software. Bars show the mean ± SEM.
In tumor and normal cell lines, SETD8 attenuates the pro-apoptotic and cell-cycle arrest functions of p53 through methylation of lysine 382 (p53K382me1) (Shi et al., 2007). To test whether the elevated levels of SETD8 in NB may inhibit the activity of p53 through increased p53K382me1 levels, we evaluated p53 expression and activity in SETD8-silenced SY5Y cells. Decreased expression of SETD8 was associated with reduced H4K20me1 and p53K382me1 levels, increased p53 and p21 protein levels (Figures 4E and S4A, D). Depletion of SETD8 led to increased apoptosis (Figure 4F).
Analysis of p53 methylation levels across a panel of NB cell lines showed higher p53K382me1 levels in MYCN-WT compared with MYCN-amp NB cells (Figure 4G). Among the p53 mutated NB cell lines, SK-N-BE2C and SK-N-FI were less sensitive to the SETD8 inhibitor (UNC0379) and showed relatively low p53K382me1 levels compared with SK-N-AS, which was sensitive to UNC0379 (Figure 4G). These observations demonstrate that SETD8 silencing decreased p53K382me1 levels and rescued the proapoptotic and growth arrest functions of p53 through activation of the p53 canonical pathway.
Comparison of pharmacologic and genetic inhibition of SETD8 in MYCN-WT NB
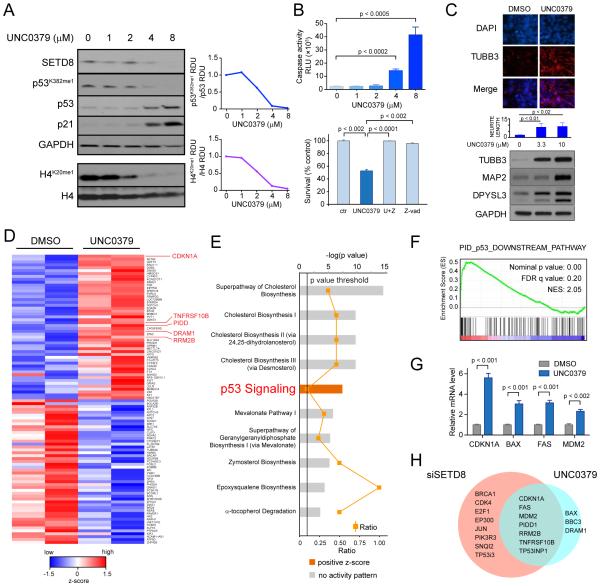
To determine the molecular mechanisms of NB cell growth inhibition by a small molecule SETD8 inhibitor, we treated SY5Y cells with UNC0379 and examined effects on target levels, cell cycle, and cell growth for up to 48 hr (Figure S5A-C and Supplemental Experimental Procedures). UNC0379 is a substrate-competitive inhibitor selective for SETD8 (Ma et al., 2014a) compared with 5 other methyltransferases in SY5Y cells (Figure S5D). Analysis of protein lysates harvested after 12 hr indicated a dose-dependent decrease in the SETD8 targets H4K20me1 and p53K382me1 (Figure 5A). The decrease in p53K382me1 levels was accompanied by an increase in p53 levels and induction of p21 (Figures 5A and S5A). Activation of the p53 pathway was accompanied by a dose-dependent increase in apoptotic cells (Figure 5B and S5E). UNC0379 induced cell death was blocked by pre-incubation with the pan-caspase inhibitor Z-VAD-FMK, indicating a caspase-dependent mode of cell death (Figure 5B). UNC0379 treatment led to increased expression of neural differentiation markers and increased neurite length (Figure 5C). These functional studies suggest that the pharmacologic inhibition of SETD8 led to a similar decrease in p53K382me1 levels, activation of the p53 pathway and increased differentiation as had been detected after genetic silencing of SETD8.
Figure 5. SETD8 pharmacological inhibition reduces cell growth and activates p53 proapoptotic and growth arrest functions in SY5Y cells.
(A) Western blot of SY5Y (MYCN-WT) cells treated with UNC0379 at the indicated concentrations (left panel). Densitometric analysis of p53K382me1 levels normalized to p53 protein levels (upper right) and of H4K20me1 levels normalized to H4 protein levels (lower right) after treatment with the SETD8 inhibitor, UNC0379, for 12 hr calculated as RDU using Image J Software.
(B) Caspase 3/7 activity upon SETD8 pharmacological inhibition for 12 hr in SY5Y cells. Bars show the mean ± SD of 3 replicates (upper graph). The effects of caspase inhibitor Z-VAD-FMK on UNC0379 treated SY5Y cells (lower graph). SY5Y cells were pretreated with 10 μM Z-VAD for 3 hr, followed by treatment with 10 μM UNC0379 for 48 hr, or treated with UNC0379 or Z-VAD alone for 48 hr. MTS assay was used to detect cell survival. Data represent mean ± SD of 2 independent experiments (p < 0.001).
(C) Immunofluorescence analysis showing the expression of TUBB3 (red) after treatment with 4 μM UNC0379 for 72 hr in SY5Y cells. Nuclei are stained with DAPI (blue). Scale bars, 100 μm (upper panel). Neurite length in SY5Y treated with 3.3 μM and 10 μM UNC0379 for 72 hr, measured by the Opera microscope. Bars represent the mean ± SD of 3 replicates (middle panel). Immunoblot of proteins from SY5Y cells treated with 3.3 μM and 10 μM UNC0379 for 72 hr and immunoblotted with antibodies to TUBB3, MAP2 and DPYSL3 (lower panel).
(D) Heatmap of the top 50 up- and down-regulated genes in SY5Y cells ranked by statistical significance following 12 hr of treatment with 4 μM (IC80) UNC0379. Data are presented as normalized expression values of 2 biological replicates based on edgeR software analysis and FDR < 0.001. The color key represents the normalized expression values: blue (high) to red (low).
(E) The top ten differentially expressed canonical pathways after SETD8 pharmacological inhibition defined by IPA (Ingenuity pathway analysis) based on edgeR software analysis and FDR < 0.001. Pathways related to p53 signaling are among top differentially expressed pathways.
(F) GSEA of the p53 downstream pathway (Nominal p value = 0.00, FDR = 0.20, NES = 2.05) after UNC0379 treatment.
(G) qRT-PCR analysis showing relative mRNA levels of the indicated genes after SETD8 pharmacological inhibition. Bars show mean ± SEM of 3 replicates.
(H) Venn diagram showing numbers of differentially expressed p53 target genes from genetic and pharmacological inhibition of SETD8.
Transcriptional changes in SY5Y cells treated with 4 μM UNC0379 for 12 hr were investigated using RNA-seq as both histone and non-histone targets were inhibited with minimal cell death. The top 50 differentially upregulated and downregulated genes are depicted as a clustered heatmap (Figure 5D and Table S5). At least 10% of the upregulated genes were involved in the p53 pathway, including CDKN1A (p21) as the most upregulated p53 target gene (Figure 5D, red marked genes). IPA of the RNA-seq data revealed that among the top ten pathways, p53 signaling was the most significantly upregulated (Figures 5E and S5F). Interestingly, cholesterol biosynthetic and mevalonate pathways were the most differentially regulated (Figures 5E and S5F). GSEA showed enrichment in several p53 pathways (Figure 5F, Table S6 and Figure S5G) after SETD8 pharmacological inhibition. Quantitative RT-PCR analysis confirmed upregulation of several of the identified p53 target genes (Figure 5G).
Activation of the p53 pathway was common to both the genetic and pharmacologic inhibition of SETD8: approximately half of the differentially expressed p53 target genes identified following SETD8 silencing overlapped with those affected by UNC0379 treatment (Figure 5H). Interestingly, the eight p53 target genes differentially expressed only after genetic silencing of SETD8 included oncosuppressors and oncogenes such as BRCA1, E2F1 and JUN, suggesting that UNC0379 treatment only partially recapitulates the effects of SETD8-specific genetic ablation (Figure 5H). These observations indicate that either genetic or pharmacological inhibition of SETD8 leads to activation of p53 canonical transcriptional pathways.
SETD8 inhibition induces p53 dependent cell death in NB cells
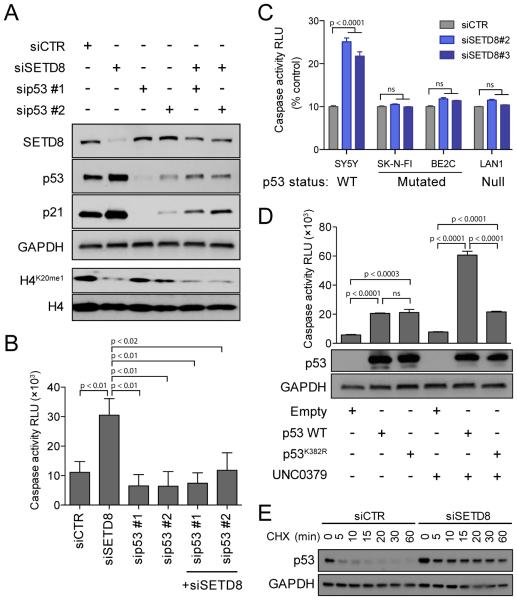
To evaluate whether SETD8 inhibition-mediated cell death was p53 dependent, we silenced SETD8 alone or in combination with different siRNAs targeting p53 (Figure 6A and S6A). The silencing of SETD8 induced increases in caspase-dependent cell death that were blocked upon p53 inhibition (Figure 6B and S6A). SETD8 silencing led to increased caspase 3/7 activity in p53 WT NB cells (SY5Y) while the levels were relatively unchanged in p53 mutant (SK-N-FI and BE2C) or p53 null (LAN1) NB cell lines (Figure 6C and S6B). Additionally, the p53 mutant and null NB cell lines were less sensitive to UNC0379 compared with p53 WT SY5Y cells (Figure S6C). Genetic rescue experiments indicated that WT p53 but not mutant p53K382R mediates increased UNC0379-induced cell death in p53 null LAN1 cells (Figure 6D). SETD8 silencing increased p53 half-life approximately 10-fold (Figure 6E and Supplemental Experimental Procedures). These results indicate that SETD8 inhibition leads to increases in p53 stability and SETD8-induced cell death is p53 dependent.
Figure 6. SETD8 inhibition-mediated cell death is p53 dependent.
(A) Immunoblot analysis showing p53 and/or SETD8 protein levels upon treatment with 2 different siRNAs targeting p53 alone or in combination with siSETD8 #3 for 48 hr in SY5Y cells.
(B) Caspase 3/7 activity calculated as RLU (Relative Luminescence Unit) upon treatment with 2 different siRNAs targeting p53 alone or in combination with siSETD8 #3 for 72 hr in SY5Y cells. Data represent mean ± SD of 2 independent experiments (p < 0.001).
(C) Caspase 3/7 activity calculated as RLU after SETD8 knockdown for 72 hr in two p53 mutated (SK-N-FI and BE2C) and in one p53 null (LAN1) NB cells compared with p53 WT SY5Y. Data represent mean ± SD of 2 independent experiments (p <0.001).
(D) Caspase 3/7 activity calculated as RLU with overexpression of p53 WT or p53K382R and treatment with UNC0379 8 μM for 24 hr in LAN1, p53 null NB cells. Data represent mean ± SD of 2 independent experiments (p < 0.001) (upper panel). Immunoblot analysis showing p53 levels under indicated conditions (lower panel).
(E) Immunoblot analysis for p53 with siCTR or siSETD8 in SY5Y cells. Cells were collected after treatment with 50 μg/ml CHX at the indicated time.
See also Figure S6.
Genetic inhibition of SETD8 and ex-vivo treatment with SETD8 inhibitor impairs tumor growth and significantly prolongs murine survival in in vivo models of NB
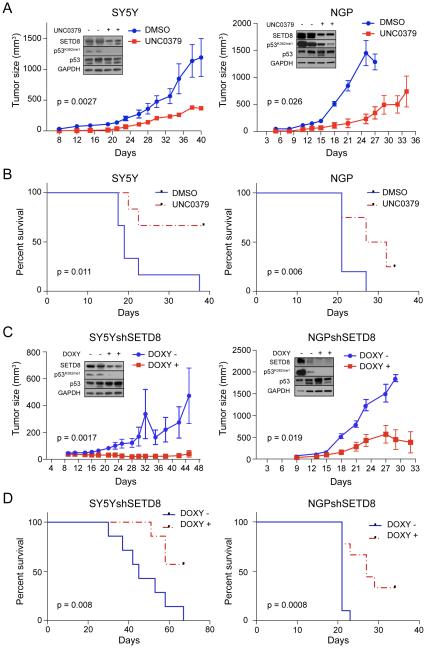
We evaluated effects of SETD8 inhibition on in vivo tumor growth. Bioavailability and animal toxicology precluded treatment of mice with the SETD8 inhibitor UNC0379 (Ma et al., 2014a). We therefore performed an ex-vivo tumorigenicity assay by exposing NB cells to 2 μM UNC0379 or control solvent for 24 hr in vitro, then subcutaneously implanting treated NB cells into nude mice and monitoring tumor growth. Pharmacological inhibition of SETD8 showed decreased NB tumor growth in vivo and significantly reduced SY5Y (p ≤ 0.01) and NGP (p = 0.026) tumor growth (Figure 7A). Assessment of tumor lysates indicated that UNC0379 treatment reduced levels of p53K382me1 modification in xenografts (Figure 7A, insets). Kaplan-Meier survival curves revealed a statistically significant survival advantage for mice bearing ex-vivo UNC0379-treated NGP or SY5Y NB cells compared with control-treated NB cells (Figure 7B). These results suggest that a small molecule inhibitor of SETD8 may have therapeutic relevance.
Figure 7. Genetic and pharmacological inhibition of SETD8 impairs tumor growth and prolongs murine survival in in vivo model of NB.
(A) SY5Y (left) and NGP (right) cells were treated ex-vivo for 24 hr with 2 μM UNC0379, SETD8 inhibitor, and then injected into nude mice. Day 0 indicates the day of cell implantation. Immunoblot of proteins from 2 tumors randomly chosen from each group (untreated and ex-vivo UNC0379 treated) collected 20 days after the injection and blotted with antibodies to SETD8, p53K382me1, p53 and GAPDH (insets). Bars show the tumor size average of 15 mice/group ± SEM. Slopes of the growth rate were compared by t test.
(B) Kaplan-Meier graphs showing the murine survival upon ex-vivo treatment of SY5Y (left) and NGP (right) with UNC0379. The statistical significance between 2 treatment groups was evaluated using a log-rank test.
(C) SY5Y-NB (left) and NGP-NB (right) xenograft tumor size in mice treated with doxycycline (DOXY) after tumors reached 75-100 mm3. Day 0 indicates the day of cell implantation. After 9 days, mice were divided in 2 groups: untreated and doxy-treated. Immunoblot of proteins from 2 tumors randomly chosen from each group 20 days after the injection and blotted with antibodies to SETD8, p53K382me1, p53 and GAPDH (insets). Bars show the tumor size average of 15 mice/group ± SEM. Slopes of the growth rate were compared by t test.
(D) Kaplan-Meier graphs showing the murine survival upon SETD8 silencing in SY5Y-NB (left) and NGP-NB (right) tumor bearing mice. The statistical significance between 2 treatment groups was evaluated using a log-rank test.
See also Figure S7.
To determine whether inhibition of SETD8 affected the growth of established NB tumors, we generated stable TET-inducible shRNAs targeting SETD8 in SY5Y (SY5Y-Tet-shSETD8) and NGP (NGP-Tet-shSETD8) NB cell lines. SY5Y-TET-shSETD8 or NGP-TET-shSETD8 cells were implanted subcutaneously into nude mice. SY5Y-Tet-TRIPZ control and NGP-Tet-TRIPZ control cell lines showed that doxycycline (DOXY)-containing chow did not significantly affect tumor growth (Figures S7A and S7B). When tumors reached ~100 mm3, animals were stratified into cohorts that received DOXY-chow or regular chow. Tumor growth was monitored at least three times a week. Assessment of tumor protein lysates showed reduced levels of SETD8 protein, decreased levels of the p53K382me1 (Figure 7C, insets) and increases in neural differentiation markers (Figure S7C) in tumors taken from mice receiving DOXY-chow compared with those receiving normal chow. Tumor RNA (qRT-PCR) showed up-regulated p53 and its targets after SETD8 genetic inhibition (Figure S7D). Mice receiving DOXY-chow had significantly reduced tumor size in both the SY5Y (p ≤ 0.001) and the NGP cell lines (p ≤ 0.01) (Figure 7C) and increased overall survival (Figure 7D) compared with those receiving normal chow.
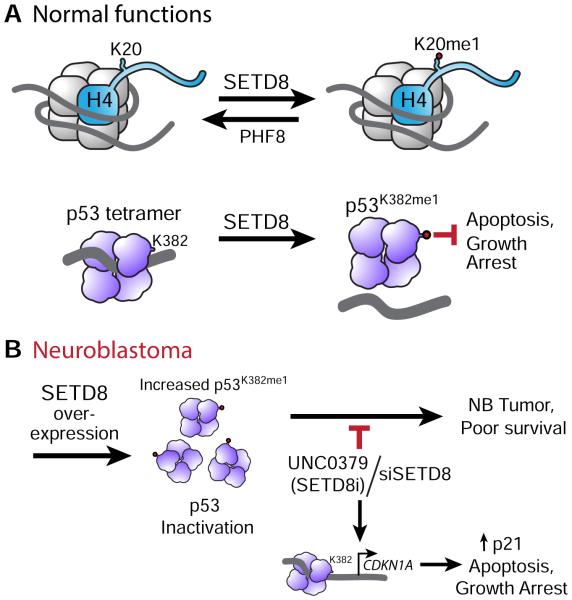
We propose a model in which in NB, SETD8 may be overexpressed, resulting in increased levels of p53K382me1 and inactivation of p53. Genetic or pharmacologic inhibition of SETD8 rescues p53 function by decreasing p53K382me1 and activating p53 canonical pathways (Figure 8A-B). These studies demonstrate that targeting SETD8 expression in NB cells significantly inhibited tumor xenograft growth and prolonged survival of mice. Thus, the biochemical, cell and animal model data support SETD8 as a bona fide target in NB.
Figure 8. Model of SETD8 normal function and role in NB.
(A) Model showing H4K20me1 and p53K382me1 as major targets of SETD8. Monomethylation of p53 K382 by SETD8 attenuates p53 proapoptotic and growth arrest function.
(B) In NB, SETD8 may be overexpressed, resulting in increased levels of p53K382me1 and inactivation of p53. Genetic or pharmacologic inhibition of SETD8 rescues p53 function by decreasing p53K382me1 and activating p53 canonical pathways.
Discussion
Our genetic and chemical screens revealed vulnerabilities in NB tumor cells that are relevant to HR-NB patients and are potentially druggable. The siRNA screen identified 53 genes whose silencing reduced NB tumor cell growth, and 16 of which also induced differentiation. We focused on the role of SETD8 based on the siRNA screen and the results of the chemical screen involving the recently developed small molecule inhibitor of SETD8, UNC0379. Mechanistically, our studies show that genetic or pharmacological inhibition of SETD8 results in activation of the p53 pathway in NB cell lines. Moreover, pre-clinical tumor models showed pharmacologic or genetic ablation of SETD8 inhibits tumor xenograft growth and extends murine survival.
DNA replication, recombination, repair and cell cycle were the major pathways (IPA) implicated after down-regulation of the 53 growth-inhibiting genes. Some 60% of these genes had never before been implicated in limiting NB tumor cell growth and importantly more than 30% have enzymatic activities that are potentially druggable. Of the 16 genes whose silencing also induced NB differentiation, over half have not been described in NB, with TAF6L and SETD8 having enzymatic activities. The potential relevance of these genes is supported by the findings that expression of high levels of 9 out of the 16 genes (EZH2, ACTR5, CTCF, BRD4, TAF6L, CHAF1A, CENPE, INCENP, SETD8) are associated with poor prognosis in primary NB tumors (database: R2 http://r2.amc.nl, Kocak). Furthermore, high-level expression of TAF6L, CENPE, INCENP and SETD8 is significantly associated with poor prognosis in MYCN-WT, Stage 4 patients. Our finding that high-level expression of SETD8 is associated with poor prognosis in NB may be due in part to focal DNA copy number gains at chr12q24, in the region including SETD8 (Schnepp et al., 2015; Wang et al., 2011; Wolf et al., 2010). Chr12q24 gain is significantly associated with the high-risk MYCN-WT subset and several genes in this region, including SETD8, are significantly differentially expressed when compared to MYCN-amp NB tumors (Schnepp et al., 2015). However, only 33% of MYCN-WT HR-NB tumors harbor Chr12q24 gains (Schnepp et al., 2015; Wolf et al., 2010), indicating that other mechanisms contribute to elevated levels of SETD8. Increased SETD8 expression has been found in lung cancers and this may reflect the tight regulation of SETD8 during the cell cycle (Takawa et al., 2012). During anaphase through early G1, SETD8 protein levels are negatively regulated by the APCcdh1 E3-ubiquitin ligase (Beck et al., 2012). In NB, DNA hypermethylation of CDH1 (Hoebeeck et al., 2009) and low-level expression of CDH1 are associated with poor prognosis (database: R2 Kocak NB). As CDH1 is required for recognition and recruitment of SETD8 to the APC E3-ubiquitin ligase complex, low levels of CDH1 in NB may impair appropriate cell cycle-regulation of SETD8. CDH1 is implicated as a tumor and metastasis suppressor as well as a regulator of genomic stability (Garcia-Higuera et al., 2008). Interestingly other APCcdh1 E3-ubiquitin ligase substrates (AuroraA, -B kinase and BARD1) have been shown to have a role in NB susceptibility or tumorigenesis (Bosse et al., 2012; Schnepp et al., 2015).
Our functional siRNA and chemical screens indicated that inhibition of SETD8 led to decreased NB cell growth and induction of differentiation. The transcriptome data indicate a major driver is SETD8-mediated methylation and inactivation of the p53 pathway. SETD8 is the sole mono-methyltransferase of lysine 20 on histone H4 (H4K20me1) and this histone mark is critical in regulating DNA replication and chromosome condensation during mitosis. In many studies (Beck et al., 2012), loss of SETD8 leads to DNA damage, which may also contribute to the p53 activation we detect in NB cells. SETD8 also methylates non-histone proteins including proliferating cell nuclear antigen (PCNA), the p53-stabilizing factor NUMB as well as the tumor suppressor p53 (Dhami et al., 2013; Takawa et al., 2012), which are critical for cell cycle progression. Dysregulation of SETD8 mostly has been implicated as a driver of oncogenesis. SETD8 mediated methylation of PCNA promotes tumor cell proliferation (Takawa et al., 2012) and its interaction with TWIST leads to H4K20me1 of the N-cadherin promoter, a TWIST target stimulating invasiveness (Yang et al., 2012). SETD8 methyltransferase decreases activity of the p53 tumor suppressor by two distinct mechanisms: monomethylation of NUMB, which inhibits NUMB-dependent stabilization of p53, and the K382 monomethylation of p53, which decreases the transcriptional activity of p53 in non-damaged cells (Shi et al., 2007). In NB, our findings are consistent with a model in which SETD8-mediated cell death is p53-dependent and p53K382 is important for this activity.
In primary NBs, TP53 is rarely mutated (<2%) and this study identifies SETD8 methylation as a mechanism that attenuates the activity of p53 in NB cells. Previous studies, primarily in MYCN-amp NB cells, have implicated cytosolic sequestration and deregulation of the p14 ARF/MDM2/p53 axis as mechanisms contributing to p53 inactivation (Tweddle et al., 2003). Aside from mutations in p14ARF, mechanisms that attenuate the activity of p53 in MYCN-WT NB have not been well described, even though MYCN-WT tumors comprise almost 60% of Stage 4 HR-NB patients (Van Maerken et al., 2009). Here, we find that SETD8 inhibition leads to decreased levels of p53K382me1, decreased cell growth and increased p53 activity in both MYCN-WT and MYCN-amp cell lines in vitro and in vivo. Since SETD8 methylation activity is part of the normal cell cycle progression, it is expected that SETD8 would play a role in both HR-NB subsets. However, SETD8 mediated mechanisms of p53 inactivation may be more relevant in MYCN-WT NB tumors as: 1) MYCN-WT NB cell lines have relatively higher levels of p53K382me1 compared with those in MYCN-amp NB; 2) the levels of SETD8 are associated with a poor prognosis in both the subsets, but this association is statistically significant only in Stage 4 MYCN-WT tumors; 3) Chr12q24 gain is significantly associated with the MYCN-WT HR-NB (Schnepp et al., 2015); and 4) low mRNA levels of SETD8-ubiquitin ligases, CHD1 and BTRC (Wang et al., 2015), are associated with poor prognosis in Stage 4 MYCN-WT tumors (database: R2 Kocak).
Pharmacologic approaches that lead to activation of p53 have been a long term goal of cancer therapeutics ever since its tumor suppressor function was identified. Most studies have been focused on Nutlin-3 in NB (Van Maerken et al., 2011; Zawacka-Pankau and Selivanova, 2015). Here, we identify that UNC0379, a small molecule inhibitor of SETD8, leads to activation of p53 in NB tumor cells. UNC0379 is a recently discovered substrate-competitive SETD8 inhibitor with preferential selectivity for SETD8 compared with 15 other methyltransferases (Ma et al., 2014a). In our study, UNC0379 demonstrated in vitro and in vivo efficacy at low micromolar concentrations in both MYCN-WT and -amp NB cell lines, although p53 mutant cell lines exhibited lower sensitivity compared with p53 WT cell lines. The IVTI of UNC0379 in NB cells was highly significant when compared with normal cells, suggesting that its characteristics warrant further development. The recent elucidation of structure-activity relationships among SETD8 inhibitors (Ma et al., 2014b) and the identification of other small-molecule inhibitors of SETD8 (Blum et al., 2014) should lead to compounds with increased activity and selectivity (A. Ma unpublished data). As with many targeted compounds, combination studies may prove more efficacious and further studies of inhibitors of SETD8 with Nutlin-3 or DNA damaging conventional cytotoxic agents will be explored.
By integrating a high-content imaging siRNA screen with a chemical screen we identified SETD8 as a crucial regulator of NB proliferation and differentiation and UNC0379 as a small molecule inhibitor with anti-tumor growth activity. This study reveals inhibition of SETD8 as a mechanism in NB to activate p53 by decreasing p53K382me1. A few studies have implicated SETD8 as a putative target, but this study also shows that genetic targeting of SETD8 in pre-clinical xenograft tumor models affects in vivo growth and confers a significant survival advantage. Collectively, our work sheds light on the chromatin modifier SETD8 and its role in NB tumorigenesis providing evidence for a therapeutic strategy to activate p53 function in HR-NBs.
Experimental procedures
Cell culture
Neuroblastoma cell lines were cultured in RPMI-containing media; ARPE-19 and HEK293T were grown in DMEM/F12. Further details are provided in the Supplemental Experimental Procedures.
Real-Time PCR analysis
Total RNA extraction was carried out using an RNeasy Plus Kit (Qiagen Inc., Hilden, Germany) and quantitative reverse transcription-PCR (qRT-PCR) was performed as described (Veschi et al., 2014). For primer sequences, see Supplemental Experimental Procedures.
siRNA transfection in a 384-well format and automated imaging
siGenome pools and OnTargetPlus pools of 4 siRNA oligos per gene were purchased from Dharmacon. The epigenetic focused siRNA library targets 395 known modulators of chromatin structure and function. siRNA oligos targeting genes in the primary screening are described in Table S1. Automated imaging was performed using an Opera high-throughput confocal microscope (PerkinElmer, Waltham, MA, USA). The methodology is detailed in the Supplemental Experimental Procedures.
Animal experiments
All xenograft studies were approved by the Animal Care and Use Committee of the National Cancer Institute, and all mouse treatments, including their housing, were in accordance with the institutional guidelines (PB-023). The methodology used is detailed in the Supplemental Experimental Procedures.
Statistical analysis
Statistical analyses were performed using Microsoft Excel, standard two-tailed Student’s t test and the software GraphPad Prism 6.0. Statistical methodology for gene expression profiling, for siRNA and chemical screens and for the animal experiments is described in the Supplemental Experimental Procedures. Image J software was used for quantification of selected immunoblots.
Supplementary Material
Highlights.
We identified 53 epigenetic regulators that control NB growth and/or differentiation
SETD8 inhibition activates the p53 pathway by decreasing p53K382me1 levels
UNC0379 is a small molecule inhibitor of SETD8 that activates the p53 pathway
SETD8 is validated as a druggable therapeutic target in NB
Significance.
The functional significance of alterations in epigenetic enzymes is incompletely understood in NB. Here, we identify epigenetic regulators critical for NB proliferation and differentiation. Specifically, combined genetic and chemical screens reveal the protein lysine methyltransferase SETD8 as a crucial regulator of growth and differentiation in NB. The role of SETD8 has not been intensively studied in tumorigenesis. We demonstrate that inhibition of SETD8 activates the p53 pathway particularly in MYCN-WT NBs, which account for the majority of high-risk NBs (60-70%), and we show that SETD8 ablation impairs tumor growth in preclinical xenograft NB models.
Acknowledgments
We are grateful to Drs. G. Pegoraro (High-Throughput Imaging Facility, CCR) and D. Wei (Urologic Oncology Branch, CCR) for technical assistance, to Dr. N. Caplen for her critical review, and to Drs. M. Aladjem and D. Banerjee for insightful discussions. Studies on the epigenetic siRNA screen were funded by a grant from the St. Baldrick’s Foundation to CJT and ZL. SGC is a registered charity (1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck & Co, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. Dr. G. Giannini was funded by AIRC IG17734; MIUR PRIN 2015. This work is supported by the Center for Cancer Research, NCI as part of the Intramural Research Program, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
The NCBI Gene Expression Omnibus accession number of the RNAseq data is GEO: GSE81626.
Author Contributions
V.V., C.J.T. and Z.L. designed experiments. V.V. and C.J.T. drafted the manuscript. C.J.T., V.V, T.C.V., L.O., G.H., Z.L. developed and/or performed the High-content Imaging screen. V.V. and B.G. performed the chemical screen and figure generation. A.M., J.J. and C.A. provided chemical probes and technical advice. V.V. and N.L. generated stable shSETD8 NB lines and qRT-PCR. V.V. and B. K. S. performed Western blotting, experiments for RNA-seq analysis and animal studies. C.Y., Y.H., Z. L. and B. G. performed bioinformatic analyses. S.J.M., E.A., provided unique reagents and with C.A., G.G., and J.K. critically edited the manuscript. All authors reviewed the manuscript.
In Brief
Veschi et al. perform both genetic and chemical screening to identify histone methyltransferase SETD8 as a potential target in neuroblastoma (NB). Chemical or genetic inhibition of SETD8 in NB leads to increased p53 activity and reduced tumor cell growth, resulting in prolonged survival in mouse models of NB.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Bagatell R, Beck-Popovic M, London WB, Zhang Y, Pearson AD, Matthay KK, Monclair T, Ambros PF, Cohn SL. Significance of MYCN amplification in international neuroblastoma staging system stage 1 and 2 neuroblastoma: a report from the International Neuroblastoma Risk Group database. J Clin Oncol. 2009;27:365–370. doi: 10.1200/JCO.2008.17.9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamuth NJ, Wood A, Wang Q, Jagannathan J, Mayes P, Zhang Z, Chen Z, Rappaport E, Courtright J, Pawel B, et al. Serial transcriptome analysis and cross-species integration identifies centromere-associated protein E as a novel neuroblastoma target. Cancer Res. 2010;70:2749–2758. doi: 10.1158/0008-5472.CAN-09-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri E, De Preter K, Capasso M, Chen Z, Hsu DM, Tonini GP, Lefever S, Hicks J, Versteeg R, Pession A, et al. Histone chaperone CHAF1A inhibits differentiation and promotes aggressive neuroblastoma. Cancer Res. 2014;74:765–774. doi: 10.1158/0008-5472.CAN-13-1315. [DOI] [PubMed] [Google Scholar]
- Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih Ie M, Conejo-Garcia JR, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, Ibanez G, Rao X, Shum D, Radu C, Djaballah H, Rice JC, Luo M. Small-molecule inhibitors of SETD8 with cellular activity. ACS Chem Biol. 2014;9:2471–2478. doi: 10.1021/cb500515r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse KR, Diskin SJ, Cole KA, Wood AC, Schnepp RW, Norris G, Nguyen le B, Jagannathan J, Laquaglia M, Winter C, et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;72:2068–2078. doi: 10.1158/0008-5472.CAN-11-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canete A, Gerrard M, Rubie H, Castel V, Di Cataldo A, Munzer C, Ladenstein R, Brichard B, Bermudez JD, Couturier J, et al. Poor survival for infants with MYCN-amplified metastatic neuroblastoma despite intensified treatment: the International Society of Paediatric Oncology European Neuroblastoma Experience. J Clin Oncol. 2009;27:1014–1019. doi: 10.1200/JCO.2007.14.5839. [DOI] [PubMed] [Google Scholar]
- Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. J Amer Med Assoc. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YL, Kim CJ, Matsuo T, Gaetano C, Falconi R, Suh YL, Kim SH, Shin YK, Park SH, Chi JG, Thiele CJ. HUlip, a human homologue of unc-33-like phosphoprotein of Caenorhabditis elegans; Immunohistochemical localization in the developing human brain and patterns of expression in nervous system tumors. J Neuro-Oncol. 2005;73:19–27. doi: 10.1007/s11060-004-3013-3. [DOI] [PubMed] [Google Scholar]
- Cui H, Ma J, Ding J, Li T, Alam G, Ding HF. Bmi-1 regulates the differentiation and clonogenic self-renewal of I-type neuroblastoma cells in a concentration-dependent manner. J Biol Chem. 2006;281:34696–34704. doi: 10.1074/jbc.M604009200. [DOI] [PubMed] [Google Scholar]
- Dhami GK, Liu H, Galka M, Voss C, Wei R, Muranko K, Kaneko T, Cregan SP, Li L, Li SS. Dynamic methylation of Numb by Set8 regulates its binding to p53 and apoptosis. Mol Cell. 2013;50:565–576. doi: 10.1016/j.molcel.2013.04.028. [DOI] [PubMed] [Google Scholar]
- Frumm SM, Fan ZP, Ross KN, Duvall JR, Gupta S, VerPlank L, Suh BC, Holson E, Wagner FF, Smith WB, et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem Biol. 2013;20:713–725. doi: 10.1016/j.chembiol.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- Hoebeeck J, Michels E, Pattyn F, Combaret V, Vermeulen J, Yigit N, Hoyoux C, Laureys G, De Paepe A, Speleman F, Vandesompele J. Aberrant methylation of candidate tumor suppressor genes in neuroblastoma. Cancer Lett. 2009;273:336–346. doi: 10.1016/j.canlet.2008.08.019. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor ER, Thiele CJ. Epigenetic changes in pediatric solid tumors: promising new targets. Clin Cancer Res. 2012;18:2768–2779. doi: 10.1158/1078-0432.CCR-11-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A, Yu W, Li F, Bleich RM, Herold JM, Butler KV, Norris JL, Korboukh V, Tripathy A, Janzen WP, et al. Discovery of a selective, substrate-competitive inhibitor of the lysine methyltransferase SETD8. J Med Chem. 2014a;57:6822–6833. doi: 10.1021/jm500871s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A, Yu W, Xiong Y, Butler KV, Brown PJ, Jin J. Structure-activity relationship studies of SETD8 inhibitors. MedChemComm. 2014b;5:1892–1898. doi: 10.1039/C4MD00317A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM. Recent advances in neuroblastoma. New Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- Petroni M, Veschi V, Gulino A, Giannini G. Molecular mechanisms of MYCN-dependent apoptosis and the MDM2-p53 pathway: an Achille's heel to be exploited for the therapy of MYCN-amplified neuroblastoma. Front Oncol. 2012;2:141. doi: 10.3389/fonc.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH, Nekritz EA, Zeid R, Gustafson WC, Greninger P, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Jr., Papadopoulos N, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepp RW, Khurana P, Attiyeh EF, Raman P, Chodosh SE, Oldridge DA, Gagliardi ME, Conkrite KL, Asgharzadeh S, Seeger RC, et al. A LIN28B-RAN-AURKA Signaling Network Promotes Neuroblastoma Tumorigenesis. Cancer Cell. 2015;28:599–609. doi: 10.1016/j.ccell.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Eggert A. Neuroblastoma. Crit Rev Oncogenesis. 2015;20:245–270. doi: 10.1615/critrevoncog.2015014033. [DOI] [PubMed] [Google Scholar]
- Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69:2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takawa M, Cho HS, Hayami S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Maejima K, Ueda K, Masuda A, et al. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 2012;72:3217–3227. doi: 10.1158/0008-5472.CAN-11-3701. [DOI] [PubMed] [Google Scholar]
- Tweddle DA, Pearson AD, Haber M, Norris MD, Xue C, Flemming C, Lunec J. The p53 pathway and its inactivation in neuroblastoma. Cancer Lett. 2003;197:93–98. doi: 10.1016/s0304-3835(03)00088-0. [DOI] [PubMed] [Google Scholar]
- Van Maerken T, Rihani A, Dreidax D, De Clercq S, Yigit N, Marine JC, Westermann F, De Paepe A, Vandesompele J, Speleman F. Functional analysis of the p53 pathway in neuroblastoma cells using the small-molecule MDM2 antagonist nutlin-3. Mol Cancer Ther. 2011;10:983–993. doi: 10.1158/1535-7163.MCT-10-1090. [DOI] [PubMed] [Google Scholar]
- Van Maerken T, Speleman F, Vermeulen J, Lambertz I, De Clercq S, De Smet E, Yigit N, Coppens V, Philippe J, De Paepe A, et al. Small-molecule MDM2 antagonists as a new therapy concept for neuroblastoma. Cancer Res. 2006;66:9646–9655. doi: 10.1158/0008-5472.CAN-06-0792. [DOI] [PubMed] [Google Scholar]
- Van Maerken T, Vandesompele J, Rihani A, De Paepe A, Speleman F. Escape from p53-mediated tumor surveillance in neuroblastoma: switching off the p14(ARF)-MDM2-p53 axis. Cell Death Differ. 2009;16:1563–1572. doi: 10.1038/cdd.2009.138. [DOI] [PubMed] [Google Scholar]
- Veschi V, Petroni M, Bartolazzi A, Altavista P, Dominici C, Capalbo C, Boldrini R, Castellano A, McDowell HP, Pizer B, et al. Galectin-3 is a marker of favorable prognosis and a biologically relevant molecule in neuroblastic tumors. Cell Death Dis. 2014;5:e1100. doi: 10.1038/cddis.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei JS, Marquez VE, Bates SE, Jin Q, Khan J, et al. EZH2 Mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer Res. 2012;72:315–324. doi: 10.1158/0008-5472.CAN-11-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Dai X, Zhong J, Inuzuka H, Wan L, Li X, Wang L, Ye X, Sun L, Gao D, et al. SCF(beta-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat Commun. 2015;6:10185. doi: 10.1038/ncomms10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Korja M, Karhu R, Edgren H, Kilpinen S, Ojala K, Mousses S, Kallioniemi A, Haapasalo H. Array-based gene expression, CGH and tissue data defines a 12q24 gain in neuroblastic tumors with prognostic implication. BMC Cancer. 2010;10 doi: 10.1186/1471-2407-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rice JC. A new regulator of the cell cycle: the PR-Set7 histone methyltransferase. Cell Cycle. 2011;10:68–72. doi: 10.4161/cc.10.1.14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 2010;24:2531–2542. doi: 10.1101/gad.1984210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. The EMBO J. 2012;31:110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, AlTahan AM, Hu D, Wang Y, Cheng PH, Morton CL, Qu C, Nathwani AC, Shohet JM, Fotsis T, et al. The role of histone demethylase KDM4B in Myc signaling in neuroblastoma. J Natl Cancer Inst. 2015;107:djv080. doi: 10.1093/jnci/djv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawacka-Pankau J, Selivanova G. Pharmacological reactivation of p53 as a strategy to treat cancer. J Intern Med. 2015;277:248–259. doi: 10.1111/joim.12336. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hou W, Chai M, Zhao H, Jia J, Sun X, Zhao B, Wang R. MicroRNA-127-3p inhibits proliferation and invasion by targeting SETD8 in human osteosarcoma cells. Biochem Biophys Res Commun. 2016;469:1006–1011. doi: 10.1016/j.bbrc.2015.12.067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.