Abstract
Plant root growth is affected by both gravity and mechanical stimulation (Massa GD, Gilroy S [2003] Plant J 33: 435–445). A coordinated response to both stimuli requires specific and common elements. To delineate the transcriptional response mechanisms, we carried out whole-genome microarray analysis of Arabidopsis root apices after gravity stimulation (reorientation) and mechanical stimulation and monitored transcript levels of 22,744 genes in a time course during the first hour after either stimulus. Rapid, transient changes in the relative abundance of specific transcripts occurred in response to gravity or mechanical stimulation, and these transcript level changes reveal clusters of coordinated events. Transcriptional regulation occurs in the root apices within less than 2 min after either stimulus. We identified genes responding specifically to each stimulus as well as transcripts regulated in both signal transduction pathways. Several unknown genes were specifically induced only during gravitropic stimulation (gravity induced genes). We also analyzed the network of transcriptional regulation during the early stages of gravitropism and mechanical stimulation.
Plants adapt their growth in response to environmental cues. Gravity is a constant force that guides the direction of plant growth. Mechanical stimuli such as wind, rain, and obstacles in the soil trigger changes in growth patterns (Braam and Davis, 1990; Massa and Gilroy, 2003). Gravitropic and mechanical stimulation in the root are interactive processes, mutually influencing differential growth (Mullen et al., 2000; Fasano et al., 2002; Massa and Gilroy, 2003). The mechanisms of sensing and signal transduction for either stimulus are not well understood, but it has been shown that the transcript levels of specific genes are regulated early during signal transduction after either stimulus (McClure and Guilfoyle, 1989; Braam and Davis, 1990).
It is widely accepted that gravitropic stimulation by reorientation is perceived by sedimentation of starch-containing plastids (statoliths) in the columella cells of the root tip (Kiss et al., 1989, 1996; Juniper et al., 1966; Blancaflor et al., 1998). Columella cells show differences in their contribution toward gravity perception and in the velocity of sedimentation of their statoliths (Blancaflor et al., 1998). While complete sedimentation of the statoliths requires at least 5 min (Blancaflor et al., 1998; MacCleery and Kiss, 1999), the presentation time (duration of the stimulus that is required to establish a response) was estimated at only approximately 1 min for wild-type Arabidopsis root tips (Blancaflor et al., 1998). Statolith sedimentation is postulated to disrupt the actin-based cytoskeletal network and its links to plasma membrane receptors in some regions of the cell cortex (Yoder et al., 2001). Other groups hypothesized that statoliths are directly connected to the cytoskeleton, activating membrane proteins anchored to the filaments upon dislocation or activation of mechanosensitive ion channels (Evans et al., 1986; Pickard and Ding, 1993; Collings et al., 2001; Blancaflor, 2002).
The transduction of the physical alterations within the cell (statolith settling) into a biochemical signal is not understood. Several different second messengers of the very early signal transduction events have been identified, including inositol-1,4,5-trisphosphate (IP3), pH, and Ca2+. The fastest responses were measured for phospholipase C-mediated changes of IP3 levels in the cereal pulvini of oat and maize. A transient 5-fold increase in IP3 within 10 s of gravity stimulation in the lower half of the pulvinus was detected (Perera et al., 1999, 2001). Within less than a minute of gravity stimulation, changes in the cytosolic pH in the columella cells of the Arabidopsis root cap were evident (Scott and Allen, 1999; Fasano et al., 2001). In response to the cytosolic pH changes, the Arabidopsis root cap apoplast acidified within 2 min of gravity stimulation. These pH changes precede by 10 min detectable tropic growth or growth-related changes in the elongation zone cell wall (Fasano et al., 2001). Within 90 s of gravity stimulation, distinct cytosolic Ca2+ signaling occurs in Arabidopsis seedlings with kinetics very different from cytosolic Ca2+ transients evoked by other mechanical stimuli (Plieth and Trewavas, 2002).
In addition to intracellular signal transduction, the signal must be transmitted intercellularly. In roots, the gravistimulus must be transferred from the sensing cells (columella) to the responding cells elsewhere in the plant. The classic theory holds that auxin is transported laterally and accumulates on the lower side of graviresponding tissue (Cholodny, 1926; Went and Thimann, 1937). Recent results using modern techniques have supported this theory, including asymmetrical distribution of polar auxin transport carriers (Bennett et al., 1996; Chen et al., 1998; Friml et al., 2002) and molecular evidence for auxin asymmetry following gravity stimulation (McClure and Guilfoyle, 1989; Muday et al., 1995; Lomax, 1997; Long et al., 2002; Ottenschlager et al., 2003). Lateral redistribution of radiolabeled indole-3-acetic acid (IAA) has been measured in both shoots (Parker and Briggs, 1990) and roots (Young et al., 1990; Young and Evans, 1996), and the redistribution of IAA has been shown to precede differential growth (Parker and Briggs, 1990). Other plant hormones, specifically brassinosteroids (Kim et al., 2000), ethylene (Lee et al., 1990; Philosoph-Hadas et al., 1996; Madlung et al., 1999), cytokinin (Golan et al., 1996), and gibberellic acid (Moore and Dickey, 1985; Rood et al., 1987; Brock and Kaufman, 1988; Chaban et al., 1999) have been shown to also play a role in gravitropism. Reactive oxygen species may function as a downstream component in auxin-mediated signal transduction (Joo et al., 2001). How all of these perception and signal transduction events are linked and how asymmetric growth is regulated is unknown.
The mechanical stimulation of the root tip affects its gravity sensing and response (Massa and Gilroy, 2003). A sensing or signal transduction pathway for mechanical stimulation is unknown, but fast and transient changes in Ca2+ have been observed after touch stimulation (Knight et al., 1991; Legue et al., 1997). In cell suspension cultures of parsley, a mechanical stimulus alone induced the translocation of cytoplasm and nucleus to the site of stimulation, the generation of intracellular reactive oxygen intermediates, and the expression of some, but not all, elicitor-responsive genes (Gus-Mayer et al., 1998). In shoots, touch stimulates specifically the fast and transient expression of calmodulin (TCH1), calmodulin-like isoforms (TCH2, TCH3), and a xyloglucan endoglycosyltransferase (TCH4; Braam and Davis, 1990; Sistrunk et al., 1994; Xu et al., 1995). Transcriptional changes in roots after reorientation have been shown for individual auxin-regulated genes (McClure and Guilfoyle, 1989; Guilfoyle, 1995; Philippar et al., 1999).
Previous microarray analysis of Arabidopsis transcript abundance compared gravitropic stimulation to mechanical stimulation (Moseyko et al., 2002). Three-week-old seedlings were subjected to gravitropic stimulation (135° reorientation) or mechanical stimulation (a single 360° rotation within 10 s). The authors used Affymetrix (Santa Clara, CA) 8 K GeneChips and compared whole seedling changes in gene expression 15 and 30 min after each stimulus. They identified several stress-specific regulated genes as well as genes regulated by both stimuli (Moseyko et al., 2002).
We selectively monitored changes in the Arabidopsis root apex to eliminate potential masking of changes in the hypocotyl or tissue not directly involved in root gravitropism. Using whole-genome Arabidopsis GeneChips (Affymetrix), we monitored the time course of transcript abundance of 22,744 genes after gravity and mechanical stimulation. Here, we report on the earliest detected changes in transcript levels in the Arabidopsis root apices due to gravity and mechanical stimulation compared to stationary control plants. Some transcriptional changes in Arabidopsis apical root tissues occur prior to changes in auxin concentrations, while statolith sedimentation is still occurring (MacCleery and Kiss, 1999). Regulation of transcript levels for most genes was transient during the first hour after either stimulus. We are now able to distinguish, on a transcriptional level, the differences between the signal transduction of mechanical versus gravitational stimuli in roots.
RESULTS
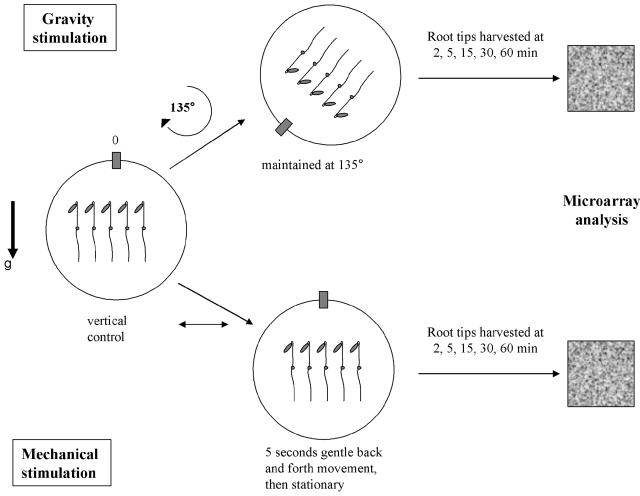
Arabidopsis root apices (7.5 mm, 7-d-old dark-grown seedlings) were harvested to analyze their changes in transcript abundance over the entire genome (Affymetrix GeneChip ATH1) in response to gravity stimulation (135° reorientation) and transient mechanical stimulation (5 s mild movement in the horizontal plane) as shown in Figure 1. The 5 s mechanical stimulation in the horizontal plane was chosen to mimic the mechanical component of our gravity stimulation by reorientation. Root apices were harvested before (control, 0 time point) and 2, 5, 15, 30, and 60 min after the respective stimulus. RNA extraction of approximately 150 root apices (per experiment per time point) yielded approximately 2 μg of mRNA. We amplified the mRNA by two cycles of reverse transcription/in vitro transcription according to the Affymetrix protocol for small samples (GeneChip Eukaryotic Small Sample Target Labeling Assay Version II) prior to labeling, fragmentation, and hybridization. To halt transcription and other cellular activities at the precise time points and preserve RNA profile of each sample, we submerged plants in RNAlater (Ambion, Austin, TX; Paul et al., 2004). RNAlater penetrated the roots in less than 3 s as demonstrated by trypan blue viability staining (Fig. 2). This method allowed for fast preservation of RNA profiles and meticulous dissection of the roots. Root apices for each time point were dissected and pooled immediately following the treatment. Each time point (2, 5, 15, 30, and 60 min) is represented by two biological replicates (150 pooled root apices per sample). The vertical controls (0 time point, 4 biological replicates) were used as references for each time point.
Figure 1.
Schematic representation of experimental design. Gravity stimulation was imposed as a continuous stimulus by 135° reorientation from the vertical. The mechanical stimulation was a transient 5 s movement in the horizontal plane.
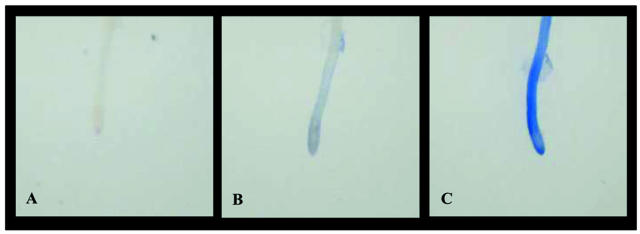
Figure 2.
Root cell death and preservation of RNA profile occurs within 3 s of treatment with RNAlater as shown by trypan blue permeability. Arabidopsis root apex in water (A), after staining in trypan blue for 10 s and rinsing in water (B), and root apex after 3 s in RNAlater immediately followed by rinsing in water prior to trypan blue staining (C).
Gene Expression in the Arabidopsis Root Apex
Fluorescence intensities are a measure of relative abundance of transcripts present in a tissue. Using an arbitrary noise cutoff at a fluorescence intensity of ≥75 signal intensity units to indicate the presence of a transcript, we found that 14,883 genes (approximately 65% of all 22,744 genes presented on the GeneChip) were transcribed in the vertical root apices prior to stimulation (Supplemental Table I, available at www.plantphysiol.org). This high percentage of overall expression corresponds to the findings of Yamada et al. (2003) using an Affymetrix tiling genome array to analyze transcriptional activity in Arabidopsis roots. The highest abundance of transcripts based on normalized fluorescence intensity data was found for genes involved in protein synthesis (ribosomal proteins, elongation factor 1-α, HSP70) and degradation (aspartic proteinase, E2, ubiquitin-conjugating enzyme), detoxification (glutathione transferases, metallothioneins), and maintenance of reduced proteinacious Cys (thioredoxin H). These genes were also identified with highest transcript abundance based on fluorescence intensity in Arabidopsis root tips by Birnbaum et al. (2003) and showed high abundance as a function of frequency of sequenced tags in SAGE libraries of Arabidopsis root tips (Fizames et al., 2004), which validates our overall expression data.
Common and Stress-Specific Changes in Transcript Levels
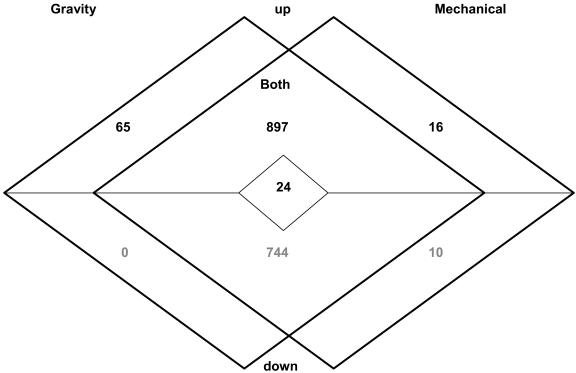
Relative transcript abundance, as reflected in average fluorescence intensities, was used to identify transcripts that showed significant changes (P < 0.01) in their abundance compared to vertical controls. We found 1,730 genes (7.6%) altered in transcript abundance after gravity stimulation, while 1,691 genes (7.5%) were altered in response to mechanical stimulation (Fig. 3; Supplemental Table II). Comparing the two lists of regulated genes, we found transcript levels of 65 genes exclusively up-regulated by gravity.
Figure 3.
Summary of transcripts regulated by either stress specifically or by both stresses. Categorization into “up” or “down” was determined as predominant tendency (majority of time points) during the first hour after stimulation.
Mechanical stimulation exclusively affected the transcript levels of 26 genes. Sixteen of those showed increased transcript levels, while the abundance of 10 transcripts was reduced. Transcript levels of 1,641 genes showed similar changes during gravity and mechanical stimulation. A small set of 24 genes responded to both stimuli but with different behavior, i.e. up-regulated after gravity stimulation while down-regulated after mechanical stimulation. Following either stimulus, transcription of 111 genes that were below the detection limit (75 cutoff) in vertical control root apices showed significant increases in transcript abundance and are therefore considered stress-induced genes.
Gravity-Specific Gene Expression
A functionally diverse group of 65 genes showed increases in their transcript abundance during the first hour after gravity stimulation (Fig. 4; Supplemental Table II). We identified transcription factors (MYB, RingH2, KNAT1, HFR1, etc.), cell wall modifying enzymes (xyloglucan endotransglycosylases [XETs], pectinesterases), transporters (ions, sugar, purines), and genes involved in the response to other environmental stresses (dehydration, cold, pathogen response). Unknown genes (“expressed,” “hypothetical”) absent in vertical control samples that showed up-regulation after gravity stimulation were categorized as gravity induced genes. Genes showing transcripts present in the vertical control conditions, followed by significant increases in relative abundance, were categorized as gravity regulated genes. Genes down-regulated in a gravity-specific manner were not observed in any of the experiments.
Figure 4.
Time-resolved cluster analysis showing ratios of gravity-specific increases in transcript abundances. xG, Minutes after gravity stimulation; xM, minutes after mechanical stimulation. Intensities of red reflect degree of up-regulation while intensities of green reflect the degree of down-regulation compared to the vertical control (0G).
Time-resolved cluster analysis identified groups of genes with distinct temporal expression patterns (Fig. 4). Transcript levels of several genes were strongly increased by 2 min, while for other genes, increases in transcript abundance were not detectable until 30 min after gravity stimulation. A cluster of five genes coding for a pentacyclic triterpene synthase (AtPEN1; At5g48010), an expressed protein (At2g16005), a Cys protease (At4g11310), S-adenosyl-l-Met:carboxymethyltransferase (SAMT)-homolog (At5g38020), and a major latex related protein (At4g23670) were up-regulated more than 3-fold within 2 min and remained at high transcript levels until 30 min. These transcripts were thereafter down-regulated to control levels by 60 min after gravity stimulation. During mechanical stimulation their transcript abundance showed a delayed and transient increase with smaller amplitude (Fig. 4). AtPEN1 is known to be involved in sterol biosynthesis, and its fast up-regulation during gravitropism might be important for the synthesis of brassinosteroids, which are involved in gravitropic signal transduction in maize roots (Kim et al., 2000). SAMT enzymes methylate secondary metabolites, and some of them, such as methylsalicylate and methyljasmonate, are known to be in several signal transduction pathways that lead to systemic and local defense responses (Shulaev et al., 1997; Zubieta et al., 2003; Hendricks et al., 2004). At least one SAMT isoform in Arabidopsis has high affinity for IAA, catalyzing the methylation of IAA (Zubieta et al., 2003). The specific functions/targets of this SAMT and the other fast up-regulated genes are unknown.
Most of the transcripts classified here as gravity specific in their regulation showed transient increases in their abundance after 5 to 15 min of reorientation. Among those gravity-specific regulated genes classified as transcription factors, the basic HLH protein HFR1 has been shown to be required for activation of a group of phytochrome A-mediated photoresponses in Arabidopsis (Kim et al., 2002; Yang et al., 2003). The homeobox transcription factor AtHB-12 has been shown to be expressed in root tips, and its expression can be induced by abscisic acid (Lee et al., 2001). Interestingly, the knotted1-like homeobox gene KNAT1 showed no detectable transcripts in the vertical controls but showed a significant (more than 3 times over threshold) transient transcript increase in abundance between 5 and 30 min after reorientation. Expression studies of KNAT genes in different tissues were not able to detect expression in nonstimulated roots (Lincoln et al., 1994). KNAT1 is a non-cell-autonomous transcriptional activator and has been shown to be involved in shoot meristem cell fate determination (Chuck et al., 1996; Kim et al., 2003). After 30 min of gravity stimulation, but not after mechanical stimulation, the transcript levels of NO APICAL MERISTEM family (NAM) transcription factors were up-regulated as well. NAM transcription factors are required for floral organ development, cotyledon separation, and embryonic shoot apical meristem formation (Souer et al., 1996; Aida et al., 1997; Takada et al., 2001). There is no known function for NAM or KNAT1 in root tissue.
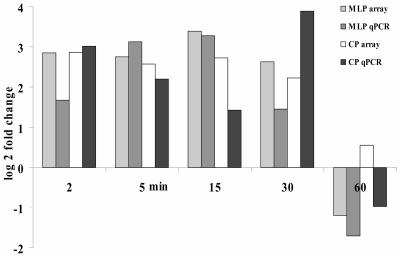
To validate the microarray data, specifically those fast and transient changes in gravity-specific gene expression, we generated sequence-specific primers and performed real-time reverse transcription (RT)-PCR on a third independent biological replicate. Real-time RT-PCR with isoform specific primers for AtPEN1 (At5g48010), major latex protein (MLP; At4g23670), Cys proteinase (CP; At4g11310), the homeobox-Leu zipper protein ATHB-12 (At3g61890), bet vI protein family (At1g70830), and a cyclic nucleotide-regulated ion channel (CNGC2; At5g15410) confirmed the relative abundance and temporal changes for the transcript levels of these genes following gravity stimulation (Fig. 5, only MLP and CP shown).
Figure 5.
Relative transcript abundance changes after gravity stimulation as compared to the vertical control were analyzed by microarray and real-time PCR using SYBR Green for detection. Microarray data for MLP (At4g23670) and CP (At4g11310) are shown as an average of two independent biological replicates. Real-time PCR with isoform-specific primers for those genes was performed on a third independent experiment.
Specific Responses to Mechanical Stimulation
Mechanical stimulation was sensed by the root apex and led to significant changes in transcript abundance of 1,696 genes, of which 26 were specifically regulated under mechanical stimulation and not by gravity (Fig. 6; Supplemental Table II). In addition to nine transcripts with unknown function, we found a mitochondrial proton-ATPase (At1g78020), a putative sulfate transporter (At3g15990), a putative cytochrome P450 (At3g14650), and a wound-induced protein (At4g10270) to be up-regulated within 2 min after mechanical stimulation. Rapid decreases in the abundance of mRNA after mechanical stimulation were observed for genes encoding a cyclic nucleotide regulated K+-channel (CNGC14; At2g24610), an expansin (EXP18; At1g62980), a RING-finger transcription factor (At2g20030), a sugar transporter (ERD6; At1g08900), and a member of the thaumatin family, which are involved in pathogen response (At5g40020). None of those genes have been shown to be involved in the response to any thigmotropic stimulus before. Several genes we expected to find in this category, i.e. “touch” genes (Braam and Davis, 1990), did not show specificity for mechanical stimulation but exhibited changes in transcript abundance in both stimuli conditions (Supplemental Table II). TCH4 encodes a XET that is responsive to mechanical stimulation of leaf tissue. Changes in TCH4 expression were not significantly reproducible in our microarray experiments or in additional real-time PCR measurements (data not shown).
Figure 6.
Cluster analysis of genes showing changes in transcript abundance after mechanical stimulation. xG, minutes after gravity stimulation; xM, minutes after mechanical stimulation. Intensities of red reflect degree of up-regulation while intensities of green reflect the degree of down-regulation compared to the vertical control (0G).
A Transcriptional Network Common to Gravitational and Mechanical Stimulation
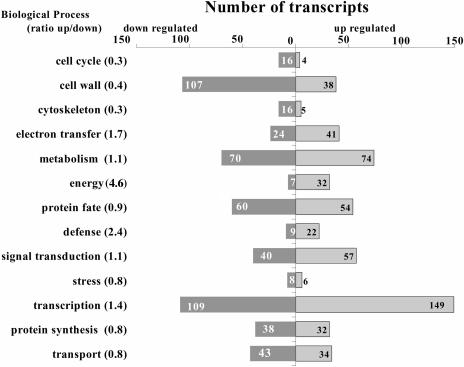
Considering the similarity in growth responses and signaling events (Sistrunk et al., 1994; Sedbrook et al., 1998; Massa et al., 2003; Massa and Gilroy, 2003) in roots, it is not surprising that gravitropic and mechanical stimulation of roots show the majority of genes regulated under both conditions (Fig. 3). Transcript levels of 897 genes were up-regulated after gravity and mechanical stimulation, while the abundance of 744 transcripts decreased. We classified this set of 1,641 genes according to their known or predicted molecular function, localization, temporal pattern of regulation, and biological process. Thirty-five percent of regulated transcript levels have no known molecular function or sequence similarity to a gene with known function. This portion is fairly representative of the entire genome, where 30% of genomic sequence (Arabidopsis Genome Initiative, 2000) to 36% of the protein encoding annotated genes show no sequence similarity to genes with known function (Yamada et al., 2003; TAIR, 2004). Most of the genes regulated were functionally categorized to be involved in transcription (15%), cell wall synthesis and modification (8.4%), metabolism (8.4%), protein fate (6.6%), and signal transduction (5.6%). However, all of these functional groups contain genes with increased and decreased transcript levels in very different ratios. Of those genes involved in cell cycle regulation, only 4 were up-regulated while 16 showed a decrease in transcript abundance (Fig. 7). Overall, the ratio of up- versus down-regulated genes in a category of biological function can indicate the relative activation and contribution to a response. For the transcriptional activities associated with the network of responses common to gravitropic and mechanical stimulation, the data imply down-regulation of cell wall synthesis, cell cycle activity, and cytoskeletal elements occurred at the same time as there was an up-regulation of energy metabolism, electron transfer, defense mechanisms, and transcriptional activity (Fig. 7). We found many more transcripts of genes with unknown function up-regulated (384) than down-regulated (201). This difference is similar to the number of genes we found to be not transcribed in the vertical controls, but present after either stimulus, which requires up-regulation. A time-resolved analysis of up- and down-regulation of those different functional categories did not reveal any significant differences (data not shown).
Figure 7.
Comparison of genes according to functional classification regulated by either gravitational or mechanical stimulation.
In addition to their biological process, we analyzed the subcellular localization of those genes commonly regulated by both stimuli using the Gene Ontology for cellular component (http://www.Arabidopsis.org/tools/bulk/go/index.jsp). Approximately one-third of the genes were predicted to be membrane or membrane-associated proteins. In addition to the 20% of regulated genes whose proteins were predicted to be localized to the plastid, we found 29 and 13 genes to be significantly increased in their transcript abundance that are encoded on the plastid and mitochondrial chromosomes, respectively. These genes code for subunits of the ATPase (atpB), NADH-dehydrogenase, cytochrome b6, several subunits of PSII as well as some small and large ribosomal subunits, RNA polymerase (rpoA and C2), and three hypothetical proteins (ycf1, ycf2) on the plastid chromosome. On the mitochondrial genome, only the NADH-dehydrogenase subunits 1, 2, 3, and 5 were up-regulated in addition to 9 open reading frames.
Same Genes, Different Regulation
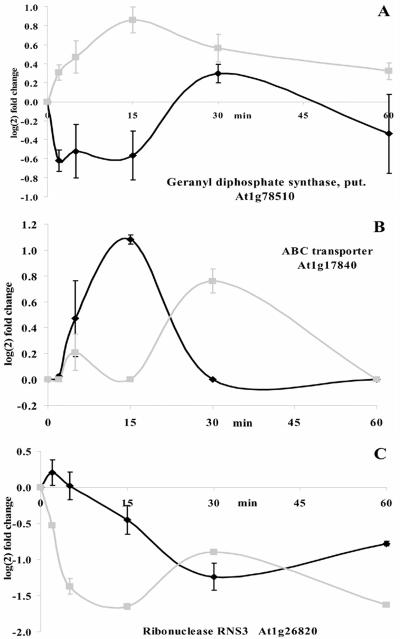
In addition to those genes that exhibited specific changes only after gravity or mechanical stimulation, we found 24 transcripts that were regulated by either stimulus but with different effects on their transcript abundance (Fig. 8; Supplemental Table II). Opposite regulation of transcript levels was identified, where the same gene shows increases in transcript abundance after one stimulus and a decrease after the other stimulus. An example of this is for a putative isoform of geranyl diphosphate synthase (At1g78510; Fig. 8A), which is transiently down-regulated after gravity stimulation while transiently up-regulated after mechanical stimulation. Transcript levels of a member of the Leu-rich repeat disease resistance protein family (At3g20820; Supplemental Table II) showed the reverse regulation with a transient increase after gravity stimulation and a decrease after mechanical stimulation. Some genes exhibited a different temporal sequence in transcript regulation between both stimuli. For example, the transcript level of an ATP-binding cassette transporter (At1g17840) showed a transient increase within the first 30 min after reorientation and showed a significantly delayed response to mechanical stimulation (Fig. 8B). By contrast, transcript levels for ribonuclease RNS3 (At1g26820) were immediately down-regulated upon mechanical stimulation, while a much smaller change in transcript levels after gravity stimulation was only observed after 15 min (Fig. 8C). These differentially responding genes provide insight into early signal transduction events in either stimulus.
Figure 8.
Examples of genes with stress-specific differences in transcript abundance changes. Differentially regulated transcript abundance of three genes after gravity stimulation (black line) or mechanical stimulation (gray line).
Regulators of Gene Expression Involved in Mechanical and Gravity Sensing
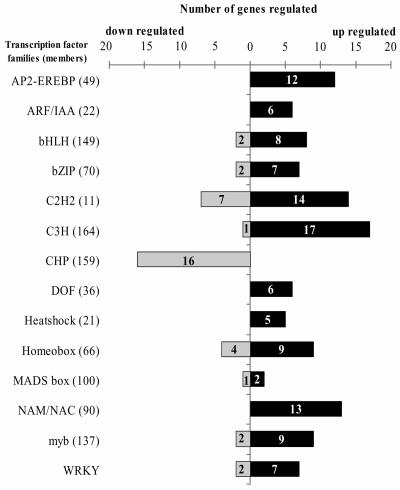
Changes in gene expression comprise changes in the relative rates of transcription, translation, and degradation of its mRNA. Approximately 15% of all transcripts regulated by either stress were classified as transcriptional regulators according to their molecular function. Proteins involved in activation or repression of transcription (chromatin modification, histones, transcription factors, RNA-polymerases) were found to be regulated. Only a few proteins are known to function in RNA stability and degradation. The mRNA levels of ribonuclease RNS3 (At1g26820) decreased after either stimulus with different kinetics (Fig. 8C). Most genes involved in chromatin organization and modification, such as DNA helicase, chromomethylase, chromosome condensation protein, histones H2A and H3, DNA gyrase, SNF2 domain helicases, SET-domain regulators, and replication factors, were down-regulated. Increased abundance was detected for some specific histone isoforms (H1, H2B, and H3.2), RNA polymerase subunits (nuclear and plastid isoforms), a sigma-related factor, a homolog to chromodomain-helicases, and a high mobility group 2-related protein. For most of these proteins, the specific functions are unknown. Based on the homology, it can be speculated that some chromatin modification takes place after either stimulus to regulate gene expression on the chromosomal level. The largest group of transcripts regulated after either stimulus are transcription factors. Transcription factors have been classified into families according to their structure and domain composition. Only specific isoforms of each family showed regulation during either or both stresses (Fig. 9). Most isoforms of those families showed increases in their transcript abundance. Only one group of zinc-finger transcription factors containing a Cys-His-rich domain was identified for which all regulated isoforms showed a decrease in their abundance. These Cys-His-rich domain zinc-finger proteins contain short domains rich in Cys and His and probably bind one or two zinc ions (renamed DC1 for divergent D1, IPR004146). This plant-specific Cys-His-rich domain is found in 159 Arabidopsis proteins and its function is unknown.
Figure 9.
Differential regulation of transcription factor gene families by either gravitational or mechanical stimulation.
Regulation of Cell Wall Loosening
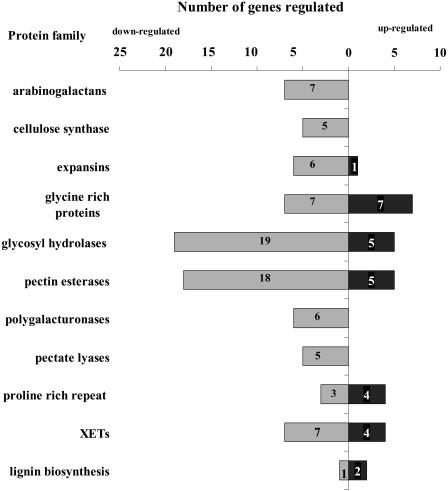
Cell wall composition and extensibility change during gravitropism, and genes regulating cell wall loosening are activated after touch stimulation (Gibeaut et al., 1990; Xu et al., 1995). We found that many transcripts regulated after either stimulus are involved in cell wall synthesis or modification (Fig. 10). Those transcripts represent members of a few families of cell wall proteins. During the first hour of gravitropic and mechanical stimulation in the root apex, regulated transcripts that belong to the protein families of arabinogalactans, cellulose synthases, polygalacturonases, or pectate lyases showed a decrease in their abundance (Fig. 10). While we did not find a family where transcripts were only up-regulated, different ratios of up- versus down-regulated isoforms within a family were observed for expansins, Gly-rich proteins, glycosyl hydrolases, pectin esterases, Pro-rich repeat proteins, XETs, and three isoforms of cinnamoyl-CoA reductases known to be involved in lignin biosynthesis. Some XETs (meri5B and TCH4) are known to be regulated by brassinosteroids under different environmental stimuli, especially touch stimulation of the shoot and leaves (Xu et al., 1995).
Figure 10.
Differential regulation of transcript abundance of genes corresponding to cell wall protein gene families.
Involvement and Temporal Sequence of Hormonal Regulation
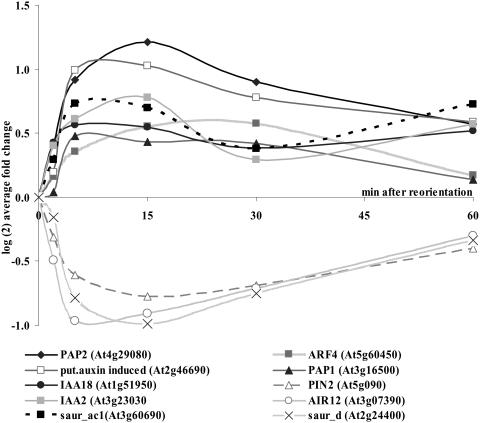
Several of the genes regulated by reorientation and mechanical stimulation have been assigned to functional categories relating to hormone synthesis or hormone regulated gene expression. Auxin-responsive genes (small auxin up-regulated RNA [SAUR], IAAs) were regulated (up and down) immediately and exhibited changes in transcript abundance within 2 min after either stimulus (Fig. 11). Very rapid (2–5 min) induction of transcription had been shown previously for exogenously applied auxin (McClure and Guilfoyle, 1989), and redistribution of SAUR occurs in gravity stimulated soybean hypocotyls within 20 min (McClure and Guilfoyle, 1987, 1989). All auxin-responsive genes showed a similar trend with either stimulus. Interestingly, we found the root-specific auxin efflux carrier EIR1 (or PIN2, At5g57090) to be down-regulated after either stimulus. Changes in 1-aminocyclopropane-1-carboxylate (ACC) synthase gene expression occurred within 2 to 5 min, with an increase in two ACC synthase transcripts, while expression of an ACC oxidase gene was repressed. Changes in the transcript levels of ethylene-responsive elements were not significantly detectable within 15 min of either stimulus.
Figure 11.
Auxin-regulated genes during gravitropic stimulation. All genes regulated by gravitropic stimulation that are known to be affected in their transcript abundance regulation by auxin showed changes of their transcript abundance within 2 to 5 min after reorientation.
Notably, the abundance of 28 transcripts encoding proteins involved in brassinosteroid synthesis or gene regulation was shown to respond to both stimuli (Supplemental Table II). Pentacyclic triterpene synthase (ATPEN1) is a known key step in steroid biosynthesis (Benveniste, 2004) and was induced 4-fold within 2 min after reorientation, while no significant increase was measured in the mechanically stimulated plants (Fig. 4). We found meri5B to be induced greater than 2-fold as early as 15 min after gravity stimulation. Meri5B is an XET involved in cell wall loosening and a known downstream target in the brassinosteroid receptor BRI1 mediated cell elongation pathway (Zurek and Clouse, 1994; Xu et al., 1995; Iliev et al., 2002).
Analysis of Regulatory Elements in Noncoding Regions of Coregulated Genes
To detect potential cis-acting elements among a cluster of coregulated genes, we performed analyses using MotifSampler to find overrepresented motifs in the 2-kb upstream sequence of a set of gravity induced transcripts that were greatly up-regulated prior to 2 min (Thijs et al., 2002). The five coregulated genes in this cluster correspond to the following genes: At4g23670 (major latex protein), At5g38020 (SAMT), At5g48010 (pentacyclic triterpene synthase), At2g16005 (expressed protein), and At4g11310 (Cys proteinase). To distinguish motifs found by chance and those that were meaningful, we included only motifs that were detected at least 5 times in 10 runs and where the motif length was set to either 8 or 12 bp using the third order Arabidopsis background model. We found that these five genes share one common potential regulatory element (TCATTAA). Four of the promoter sequences contained at least two copies of the consensus sequence in both the forward and reverse strand. In the case of At4g23670 (major latex protein-related), the consensus sequence was detected once in both orientations. The motif did not have a significant match to any known plant transcription factor binding sites when queried against the plant cis-acting regulatory DNA elements database (Higo et al., 1999; Lescot et al., 2002) or TRANSFAC (http://transfac.gbf.de/TRANSFAC). Although this novel potential cis-regulatory element is overrepresented in these five coregulated genes, it will require functional characterization to assign to the gravitropic response.
DISCUSSION
We have analyzed the effects of gravity and mechanical stimulation on transcript abundance in Arabidopsis root apices. We observed fast and transient changes in transcript levels throughout the first hour of gravity and mechanical stimulation in functionally diverse groups, such as transcription factors, cell wall modifying enzymes, transporters, kinases, phosphatases, hormone metabolism, and cell cycle. The results suggest both the relative abundance and specific timing of events are important for establishment of gene expression cascades for either stimulus since most of the transcripts abundance changes we observed were transient.
We identified 1,665 transcripts whose abundance was significantly altered as a result of both the gravity and mechanical stimulation. This reflects the large degree (96% of regulated transcripts) of overlap between responses to the two abiotic stimuli. Many transcripts we observed to be regulated by gravity or mechanical stimulation are regulated by other stresses, such as drought, cold, light, and biotic stimulation by pathogens as well. Many of these events, along with thigmotropism and gravitropism, involve cell wall modification and cell elongation. This overlapping response between and within biotic and abiotic stresses shows there is a core set of genes whose expression is regulated in response to a wide array of specific stress stimuli. Also, a smaller specific subset of genes accounting for only 4% of regulated genes is shown to be induced exclusively through gravitropic stimulation, while only approximately 2% of all regulated transcripts are shown to be regulated exclusively through mechanical stimulation. We hypothesize that this small number of stimuli-specific regulated genes are necessary in the establishment of the first events in complex gene cascades that then branch out and act through a general set of stress-regulated genes to result in cell elongation and wall modification.
We focused in this study on characterizing differences and mutuality of early transcriptional responses to gravity and mechanical stimulation in Arabidopsis root apical tissue by analyzing steady-state transcript abundances and their temporal changes. Steady-state transcript abundance of each mRNA is determined by the rate of its transcription and the rate of its degradation. Therefore, the fast and transient regulation of several clusters of genes after either stimulus would require tight control for the regulation of both transcription and degradation. The analysis of changes in transcript abundance during gravitropic and mechanical signal transduction does not allow us to distinguish between the relative changes of synthesis versus stability of specific transcripts in our experimental design. Instability of mRNA facilitates fast changes in mRNA levels that result in transient and tightly controlled regulation of gene expression. Sequence elements like the downstream element in the 3′ untranslated region of SAUR have been shown to determine fast degradation of its mRNA (McClure et al., 1989; Newman et al., 1993). About 1% of all Arabidopsis genes have been estimated to be genes with unstable transcripts (AtGUTs) with a half-life of less than an hour. Specifically touch-induced and auxin-repressed genes have transcripts with low stability (Gutierrez et al., 2002).
Interpretation of previous studies of gravity-induced changes in transcript abundance are limited by the GeneChip (8 K) available at the time, the tissue sample (whole seedlings), the mode of mechanical stimulation (360° rotation within 10 s), and the time points (15 and 30 min) analyzed (Moseyko et al., 2002). We compared our results to those of Moseyko et al. (2002) and found similar responses for three gravity-specific genes, glutathione transferase (At2g29450), glycosyl hydrolase (At4g19810), and a trypsin-related inhibitor (At2g43510). Because they used whole seedlings to extract RNA, it is not known whether the changes are due to differences in the response of the hypocotyl or the root or resulted from a common response of all tissues. Mutant analysis indicates that gravitropism in roots and shoots have both common and separate steps (Lomax, 1997; Tasaka et al., 1999, 2001). The temporal responses of root and inflorescence stem to gravity stimulation are very different. All these differences are likely to be reflected in transcriptional regulation of gravitropism and could be responsible for the differences between the results of Moseyko et al. (2002) and this study.
One of the mysteries of gravitropic signal perception and transduction is how the cell distinguishes its position within the organ and with respect to the vector of gravity. The differential growth response with faster elongation rates on the upper versus the lower side of the tissue, which causes downward bending (in roots), is transduced by a concentration gradient of auxin (Went and Thimann, 1937; Muday et al., 1995; Lomax, 1997; Philippar et al., 1999; Ottenschlager et al., 2003). Cell type-specific transcript abundance in the Arabidopsis root apex was recently analyzed by Birnbaum et al. (2003). Cell type-specific expression of green fluorescent protein was used to sort Arabidopsis root protoplasts and analyze their transcript levels. Our experiments do not provide spatial resolution of transcript abundance changes on a cellular level. This could mask differential transcript abundance changes of genes within the root apex during gravity stimulation, as it has been shown for auxin-induced genes (McClure and Guilfoyle, 1989).
More detailed molecular and biochemical analyses are in progress on many of the stimuli-specific regulated transcripts in an effort to elucidate their function in the response mechanism of gravity and mechanical stimulation. Localization and mutant analyses will allow us to determine whether these particular genes are necessary for gravity sensing and/or response. In addition, identification of the effectors regulating fast and transient gravity-specific gene expression will aid us in determining the initial signal transduction events responsible for gravity sensing in plant root tips.
CONCLUSION
We have shown here that a fast and transient network of transcriptional regulation controls the response to gravitropic and mechanical stimulation in Arabidopsis root apices. Transcript abundance of several genes was specifically up- or down-regulated within less than 2 min after stimulation. Only a small number of transcripts showed stimulus-specific changes in their abundance, while the majority (approximately 96%) of all regulated transcripts responded to both stimuli. Except for some auxin-induced genes, nothing is known about the mechanisms of regulation of transcript abundance after gravity or mechanical stimulation. Second messengers known to be involved in gravitropic signal transduction like IP3, Ca2+, pH, reactive oxygen species, and the cytoskeleton could be directly or indirectly involved in the transcriptional regulation. Our future research will focus on higher temporal and spatial resolution of gravity-induced transcript abundance changes and identify the mechanisms of regulation for gravity-specific regulated gene expression.
MATERIAL AND METHODS
Plant Growth and Experimental Design
Wild-type (Col-O) Arabidopsis seeds (Lehle, Round Rock, TX) were presterilized in 30% bleach and washed five times in sterile water. Sterile seeds were sown to square (100 mm × 100 mm × 15 mm) petri dishes (Becton-Dickinson, Franklin Lakes, NJ) containing 4.3 g/L Murashige and Skoog salts (Invitrogen, Carlsbad, CA), 0.5 g/L MES salt (Sigma, St. Louis), 10 g/L Suc (Sigma), and 8 g/L agar type M (Sigma), pH 5.7. Plates and seeds were wrapped in aluminum foil, vernalized horizontally at 4°C for 48 h and placed vertically in a 22°C growth chamber for 7 d. Approximately 18 h prior to the experiment, plants were removed from foil, randomized, divided into groups (mechanical and gravity stimulation), and allowed to acclimate to dark room conditions (22°C, dim green light; bandpass, 525 ± 15 nm). Plants were subjected to gravity stimulation by slowly rotating plates into a device designed to hold plates in 135° orientation. Mechanical stimulation of plants was performed by gentle vertical oscillation of a set of plates in each time point for a period of 5 s within the gravity vector plane. Harvesting prior to (0 min) and 2 min, 5 min, 15 min, 30 min, and 60 min after either stimulus was carried out by pouring approximately 15 mL of RNAlater (Ambion) directly onto the plates. Approximately 0.75 cm of each root was cut on a chilled microscope slide from root tip toward stem and stored in approximately 40 μL of RNAlater at −20°C. Microarray analysis was carried out on two independent experiments, and the results were confirmed by real-time PCR on a third independent experiment.
Trypan Blue Staining
Root apices of 7-d-old dark grown Arabidopsis seedlings were submerged for approximately 3 s in RNAlater (Ambion), immediately rinsed with water, stained for approximately 10 s in trypan blue (0.4% solution in phosphate buffer saline, Mediatech CELLGRO; Herndon, VA), and again rinsed in water. We used a Carl Zeiss microscope (AXIOSKOP 40; Jena, Germany) and the program Auxio Vision 3.1 to process and analyze all the samples.
RNA Profile Preservation and Isolation
Homogenization of tissue was performed using FastRNA (Qbiogen, Carlsbad, CA) lysing matrix and tube to which 400 μL of RLT extraction buffer and apical root tissue was added (settings: 2 cycles × 30 s on speed 6). The resulting lysis buffer and matrix were poured directly into a Qiashredder column and processed according to the Plant + Fungi protocol in the RNeasy plant minikit manual (Qiagen, Valencia, CA). Purified RNA was eluted from the column using 20 μL of water.
Sample Processing and Analysis
Amplified complementary RNA was synthesized according to Affymetrix GeneChip eukaryotic small sample target labeling assay II (Affymetrix; https://www.affymetrix.com/support/technical/technotes/smallv2_technote.pdf). Double-stranded cDNAs were synthesized from 100 ng of total RNA using oligo(dT)24 containing T7 RNA polymerase promoter sequence with SuperScript II RT (Invitrogen). Complementary RNAs were transcribed from entire cDNA pellet using Megascript T7 RNA polymerase (Ambion). A second cycle of cDNA synthesis was performed to amplify cRNA where the first strand of cDNA was synthesized using random primers with SuperScript II RT (Invitrogen). The RNA template was removed by treatment with RNaseH, and T7 labeled-oligo(dT)24 was annealed to the cDNA. Second strand of the cDNA was synthesized, and the entire cDNA pellet was used to synthesize biotin-labeled cRNA using T7 RNA polymerase (ENZO Biochem, Farmingdale, NY). Ten micrograms of fragmented biotin labeled cRNA containing hybridization controls (BioB, BioC, BioD, cre1-1) was hybridized to an Affymetrix ATH1 whole-genome array for 16 h at 45°C. The arrays were washed and stained with a streptavidin R phycoerythrin conjugate (Molecular Probes, Eugene, OR) on a GeneChip Fluidics Station 400 (Affymetrix) and scanned with an argon-ion laser with a 488-nm emission and a 570-nm detection (GeneArray scanner; Agilent, Palo Alto, CA).
Analysis of Microarray Data
Microarray data were analyzed using MAS5 (Affymetrix). Scanned arrays were normalized to baseline array with median overall expression using dCHIP (Li and Wong, 2001). Probe sets with signal values below the detectable range were adjusted to 75, and probe sets with values of 75 for all conditions were removed from subsequent analyses. sd of the vertical controls (four biological replicates) were used to identify genes with significant changes over the vertical controls (P < 0.01). Only genes that showed transcript level changes in at least two consecutive time points and with the same tendency in both biological replicates were considered as relevant for either stress. Hierarchical clustering was performed on the log(2) fold changes of normalized expression values using CLUSTER and visualized with TREEVIEW (Eisen et al., 1998).
Real-Time RT-PCR
Total RNA was extracted from roots using Qiashredder and RNeasy plant mini kit (Qiagen). Residual DNA was removed by performing an on-column digestion using a DNA-free kit (Ambion). cDNA was synthesized according to the Taqman RT kit protocol (ABI, Foster City, CA) on 300 ng of RNA and dT16-oligonucleotide as primer. Following synthesis, cDNA samples were diluted to 1,000 pg/μL with DNase/RNase-free water. PCR primers were designed using OligoAnalyzer 3.0 (http://biotools.idtdna.com/biotools) to create amplicons 80 to 120 bp in length from regions within 500 bp from the 3′ end of cDNA. Primers used had the following characteristics: melting temperature between 55°C to 60°C, 3′ G/C clamp, 40% to 60% G/C content overall, and matching only desired tentative consensus sequence in The Institute for Genomic Research tgi database (http://tigrblast.tigr.org/tgi/). The primers are F_At2g40880_GTTATGCAAAGGATCCAGTGAGCC, R_At2g40880_CAAACATTGCCCTCCATACAAATC, F_At4g23670_TACAAGAGCTGGAAGAGCGAGAAC, and R_At4g23670_GTCATGTTCGCCTTCGTGAACAAC.
No-RT controls were used on each 96-well plate to ensure that no genomic DNA was carried over. To ensure consistent tissue collection, RNA extraction, and RT, we used the housekeeping control Actin8 primers designed around the 3′ end of the cDNA (5′-CTTTCCGGTTACAGCGTTTG-3′ and 5′-GAAACGCGGATTAGTGCCT-3′).
PCR was performed using an ABI7900HT sequence detection system set to the following conditions: activation of Taq −95°C for 10 min, denaturing −40 cycles at 95°C of 15 s, and annealing and extension −60° for 1 min. The following reagents were combined with each sample and control: 15.5 μL of SYBR Green PCR master mix (ABI), 2.5 μL of primers (0.2 μm), and 4.5 μL of cDNA (0.001 μm). Data were analyzed using ABI SDS software (ABI), and gene expression data were calculated using the 2^ΔC′T method (Livak and Schmittgen, 2001).
Cis-Regulatory Elements
Base sequences (DNA) 2,000 bp upstream of transcription start site corresponding to very early gravity induced genes, At2g16005, At4g11310, At4g23670, At5g38020, and At5g48010, were queried from The Arabidopsis Information Resource (TAIR), and a search for overrepresented sequences was performed according to Thijs et al. (2002). Overrepresented motifs were searched against the plant cis-acting regulatory DNA elements database (Higo et al., 1999; Lescot et al., 2002) and TRANSFAC (http://transfac.gbf.de/TRANSFAC) to determine the novelty of the potential cis-regulatory element.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Supplementary Material
Acknowledgments
We thank Nina Allen, Eric Davies, Gloria Muday, Imara Perera, Niki Robertson, and Ron Sederoff for stimulating discussions.
This work was supported by the National Aeronautics and Space Administration (grant no. NAG 2–1566).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044594.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Benveniste P (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Physiol Plant Mol Biol 55: 429–457 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB (2002) The cytoskeleton and gravitropism in higher plants. Plant Growth Regul 21: 120–136 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Fasano JM, Gilroy S (1998) Mapping the functional roles of cap cells in the response of Arabidopsis primary roots to gravity. Plant Physiol 116: 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J, Davis RW (1990) Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 60: 357–364 [DOI] [PubMed] [Google Scholar]
- Brock TG, Kaufman PB (1988) Altered growth response to exogenous auxin and gibberellic acid by gravistimulation in pulvini of Avena sativa. Plant Physiol 87: 130–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban CI, Kordyum EL, Demkiv OT, Khorkavtsiv OY, Khorkavtsiv YD (1999) The gravireaction of ceratodon protonemata treated with gibberellic acid. Adv Space Res 24: 717–721 [DOI] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH (1998) The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA 95: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholodny N (1926) Beitraege zur Analyse der geotropischen Reaktion. Jahrb Wiss Bot 65: 447–459 [Google Scholar]
- Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings DA, Zsuppan G, Allen NS, Blancaflor EB (2001) Demonstration of prominent actin filaments in the root columella. Planta 212: 392–403 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ML, Moore R, Hasenstein KH (1986) How roots respond to gravity. Sci Am 255: 112–119 [DOI] [PubMed] [Google Scholar]
- Fasano JM, Massa GD, Gilroy S (2002) Ionic signaling in plant responses to gravity and touch. J Plant Growth Regul 21: 71–88 [DOI] [PubMed] [Google Scholar]
- Fasano JM, Swanson SJ, Blancaflor EB, Dowd PE, Kao T-H, Gilroy S (2001) Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell 13: 907–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fizames C, Munos S, Cazettes C, Nacry P, Boucherez J, Gaymard F, Piquemal D, Delorme V, Commes T, Doumas P, et al (2004) The Arabidopsis root transcriptome by serial analysis of gene expression. Gene identification using the genome sequence. Plant Physiol 134: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Karuppiah N, Chang SR, Brock TG, Vadlamudi B, Kim D, Ghosheh NS, Rayle DL, Carpita NC, Kaufman PB (1990) Cell wall and enzyme changes during the graviresponse of the leaf-sheath pulvinus of oat (Avena sativa). Plant Physiol 94: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan A, Tepper M, Soudry E, Horwitz BA, Gepstein S (1996) Cytokinin, acting through ethylene, restores gravitropism to Arabidopsis seedlings grown under red light. Plant Physiol 112: 901–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ (1995) Auxin regulated gene expression and gravitropism in plants. ASGSB Bull 8: 39–45 [PubMed] [Google Scholar]
- Gus-Mayer S, Naton B, Hahlbrock K, Schmelzer E (1998) Local mechanical stimulation induces components of the pathogen defense response in parsley. Proc Natl Acad Sci USA 95: 8398–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS (2004) An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem 326: 100–105 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev EA, Xu W, Polisensky DH, Oh MH, Torisky RS, Clouse SD, Braam J (2002) Transcriptional and posttranscriptional regulation of Arabidopsis TCH4 expression by diverse stimuli. Roles of cis regions and brassinosteroids. Plant Physiol 130: 770–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Bae YS, Lee JS (2001) Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol 126: 1055–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juniper BE, Groves S, Landausc B, Audus LJ (1966) Root cap and perception of gravity. Nature 209: 93 [Google Scholar]
- Kim JY, Yuan Z, Jackson D (2003) Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362 [DOI] [PubMed] [Google Scholar]
- Kim SK, Chang SC, Lee EJ, Chung WS, Kim YS, Hwang S, Lee JS (2000) Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol 123: 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Woo JC, Song PS, Soh MS (2002) HFR1, a phytochrome A-signalling component, acts in a separate pathway from HY5, downstream of COP1 in Arabidopsis thaliana. Plant J 30: 711–719 [DOI] [PubMed] [Google Scholar]
- Kiss JZ, Hertel R, Sack FD (1989) Amyloplasts are necessary for full gravitropic sensitivity in roots of Arabidopsis thaliana. Planta 177: 198–206 [PubMed] [Google Scholar]
- Kiss JZ, Wright JB, Caspar T (1996) Gravitropism in roots of intermediate-starch mutants of Arabidopsis. Physiol Plant 97: 237–244 [DOI] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Lee JS, Chang WK, Evans ML (1990) Effects of ethylene on the kinetics of curvature and auxin redistribution in gravistimulated roots of Zea mays. Plant Physiol 94: 1770–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Oh HS, Cheon CI, Hwang IT, Kim YJ, Chun JY (2001) Structure and expression of the Arabidopsis thaliana homeobox gene Athb-12. Biochem Biophys Res Commun 284: 133–141 [DOI] [PubMed] [Google Scholar]
- Legue V, Blancaflor E, Wymer C, Perbal G, Fantin D, Gilroy S (1997) Cytoplasmic free Ca2+ in Arabidopsis roots changes in response to touch but not gravity. Plant Physiol 114: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lomax TL (1997) Molecular genetic analysis of plant gravitropism. Gravit Space Biol Bull 10: 75–82 [PubMed] [Google Scholar]
- Long JC, Zhao W, Rashotte AM, Muday GK, Huber SC (2002) Gravity-stimulated changes in auxin and invertase gene expression in maize pulvinal cells. Plant Physiol 128: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCleery SA, Kiss JZ (1999) Plastid sedimentation kinetics in roots of wild-type and starch-deficient mutants of Arabidopsis. Plant Physiol 120: 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A, Behringer FJ, Lomax TL (1999) Ethylene plays multiple nonprimary roles in modulating the gravitropic response in tomato. Plant Physiol 120: 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa GD, Fasano JM, Gilroy S (2003) Ionic signaling in plant gravity and touch responses. Gravit Space Biol Bull 16: 71–82 [PubMed] [Google Scholar]
- Massa GD, Gilroy S (2003) Touch modulates gravity sensing to regulate the growth of primary roots of Arabidopsis thaliana. Plant J 33: 435–445 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1987) Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1989) Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 243: 91–93 [DOI] [PubMed] [Google Scholar]
- McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ (1989) Transcription, organization, and sequence of an auxin-regulated gene-cluster in soybean. Plant Cell 1: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R, Dickey K (1985) Growth and graviresponsiveness of primary roots of Zea mays seedlings deficient in abscisic acid and gibberellic acid. J Exp Bot 36: 1793–1798 [DOI] [PubMed] [Google Scholar]
- Moseyko N, Zhu T, Chang HS, Wang X, Feldman LJ (2002) Transcription profiling of the early gravitropic response in Arabidopsis using high-density oligonucleotide probe microarrays. Plant Physiol 130: 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Lomax TL, Rayle DL (1995) Characterization of the growth and auxin physiology of roots of the tomato mutant, diageotropica. Planta 195: 548–553 [DOI] [PubMed] [Google Scholar]
- Mullen JL, Wolverton C, Ishikawa H, Evans ML (2000) Kinetics of constant gravitropic stimulus responses in Arabidopsis roots using a feedback system. Plant Physiol 123: 665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ (1993) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5: 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschlager I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K (2003) From the cover: gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker KE, Briggs WR (1990) Transport of IAA during gravitropism in intact maize coleoptiles. Plant Physiol 94: 1763–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Schuerger AC, Popp MP, Richards JT, Manak MS, Ferl RJ (2004) Hypobaric biology: Arabidopsis gene expression at low atmospheric pressure. Plant Physiol 134: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Boss WF (1999) Transient and sustained increases in inositol 1,4,5-trisphosphate precede the differential growth response in gravistimulated maize pulvini. Proc Natl Acad Sci USA 96: 5838–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Chang SC, Boss WF, Kaufman PB (2001) A role for inositol 1,4,5-trisphosphate in gravitropic signaling and the retention of cold-perceived gravistimulation of oat shoot pulvini. Plant Physiol 125: 1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar K, Fuchs I, Luthen H, Hoth S, Bauer CS, Haga K, Thiel G, Ljung K, Sandberg G, Bottger M, et al (1999) Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc Natl Acad Sci USA 96: 12186–12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philosoph-Hadas S, Meir S, Rosenberger I, Halevy AH (1996) Regulation of the gravitropic response and ethylene biosynthesis in gravistimulated snapdragon spikes by calcium chelators and ethylene inhibitors. Plant Physiol 110: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard BG, Ding JP (1993) The mechanosensory calcium-selective ion channel: key component of a plasmalemmal control centre? Aust J Plant Physiol 20: 439–459 [DOI] [PubMed] [Google Scholar]
- Plieth C, Trewavas AJ (2002) Reorientation of seedlings in the earth's gravitational field induces cytosolic calcium transients. Plant Physiol 129: 786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood SB, Kaufman PB, Abe H, Pharis RP (1987) Gibberellins and gravitropism in maize shoots: endogenous gibberellin-like substances and movement and metabolism of [3H]Gibberellin A20. Plant Physiol 83: 645–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AC, Allen NS (1999) Changes in cytosolic pH within Arabidopsis root columella cells play a key role in the early signaling pathway for root gravitropism. Plant Physiol 121: 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J, Boonsirichai K, Chen R, Hilson P, Pearlman R, Rosen E, Rutherford R, Batiza A, Carroll K, Schulz T, et al (1998) Molecular genetics of root gravitropism and waving in Arabidopsis thaliana. Gravit Space Biol Bull 11: 71–78 [PubMed] [Google Scholar]
- Shulaev V, Silverman P, Raskin I (1997) Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385: 718–721 [Google Scholar]
- Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J (1994) Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell 6: 1553–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85: 159–170 [DOI] [PubMed] [Google Scholar]
- TAIR (2004) The Arabidopsis Information Resource. http://www.arabidopsis.org (February 25, 2004)
- Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135 [DOI] [PubMed] [Google Scholar]
- Tasaka M, Kato T, Fukaki H (1999) The endodermis and shoot gravitropism. Trends Plant Sci 4: 103–107 [DOI] [PubMed] [Google Scholar]
- Tasaka M, Kato T, Fukaki H (2001) Genetic regulation of gravitropism in higher plants. Int Rev Cytol 206: 135–154 [DOI] [PubMed] [Google Scholar]
- Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouze P, Moreau Y (2002) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Comput Biol 9: 447–464 [DOI] [PubMed] [Google Scholar]
- Went FW, Thimann KV (1937) Phytohormones. Macmillan, New York
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yang KY, Kim YM, Lee S, Song PS, Soh MS (2003) Overexpression of a mutant basic helix-loop-helix protein HFR1, HFR1-DeltaN105, activates a branch pathway of light signaling in Arabidopsis. Plant Physiol 133: 1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder TL, Zheng H-Q, Todd P, Staehelin LA (2001) Amyloplast sedimentation dynamics in maize columella cells support a new model for the gravity-sensing apparatus of roots. Plant Physiol 125: 1045–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LM, Evans ML (1996) Patterns of auxin and abscisic acid movement in the tips of gravistimulated primary roots of maize. Plant Growth Regul 20: 253–258 [DOI] [PubMed] [Google Scholar]
- Young LM, Evans ML, Hertel R (1990) Correlations between gravitropic curvature and auxin movement across gravistimulated roots of Zea mays. Plant Physiol 92: 792–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP (2003) Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15: 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Clouse SD (1994) Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol 104: 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.