Abstract
Phytophthora sojae Kaufmann and Gerdemann, which causes Phytophthora root rot, is a widespread pathogen that limits soybean production worldwide. Development of Phytophthora resistant cultivars carrying Phytophthora resistance Rps genes is a cost-effective approach in controlling this disease. For this mapping study of a novel Rps gene, 290 recombinant inbred lines (RILs) (F7 families) were developed by crossing the P. sojae resistant cultivar PI399036 with the P. sojae susceptible AR2 line, and were phenotyped for responses to a mixture of three P. sojae isolates that overcome most of the known Rps genes. Of these 290 RILs, 130 were homozygous resistant, 12 heterzygous and segregating for Phytophthora resistance, and 148 were recessive homozygous and susceptible. From this population, 59 RILs homozygous for Phytophthora sojae resistance and 61 susceptible to a mixture of P. sojae isolates R17 and Val12-11 or P7074 that overcome resistance encoded by known Rps genes mapped to Chromosome 18 were selected for mapping novel Rps gene. A single gene accounted for the 1:1 segregation of resistance and susceptibility among the RILs. The gene encoding the Phytophthora resistance mapped to a 5.8 cM interval between the SSR markers BARCSOYSSR_18_1840 and Sat_064 located in the lower arm of Chromosome 18. The gene is mapped 2.2 cM proximal to the NBSRps4/6-like sequence that was reported to co-segregate with the Phytophthora resistance genes Rps4 and Rps6. The gene is mapped to a highly recombinogenic, gene-rich genomic region carrying several nucleotide binding site-leucine rich repeat (NBS-LRR)-like genes. We named this novel gene as Rps12, which is expected to be an invaluable resource in breeding soybeans for Phytophthora resistance.
Introduction
Phytophthora root and stem rot (PRR), caused by Phytophthora sojae Kaufmann and Gerdemann, is one of the most devastating diseases in soybean [Glycine max (L.) Merr.] [1]. The disease was first reported in Indiana in 1948, in Ohio in 1951, and subsequently spread to all soybean-growing regions of the United States (US) [2]. It is most prevalent in the North Central region where the environmental conditions favor disease development [3]. P. sojae has also been reported in other soybean-growing countries, including Argentina, Brazil, China, Japan, Indonesia, Australia, Canada, and Europe [4–9]. The estimated annual yield suppression from the disease has been valued at $200 million in the North Central United States, and approximately $1–2 billion worldwide [10–11].
Though the soil-borne oomycete P. sojae primarily attacks soybean seedlings prior to emergence [1], disease can occur at any stage of plant development and throughout the growing season. Disease symptoms include brown stem lesions that develop in the roots and gradually progress to the stems, followed by wilting, chlorosis, and plant death [12]. In addition, plants infected with P. sojae may become more vulnerable to infection by other soil-borne pathogens. P. sojae can survive as mycelia or as oospores in soil or soybean plant debris for many years without a host. Under saturated soil conditions, especially during warm and wet weather, oospores germinate and produce sporangia containing hundreds of small, mobile spores called zoospores, which swim through the water-filled soil pores and infect soybean roots [1, 8, 13]. Epidemics of PRR usually occur in poorly drained fields because flooded fields or saturated soil favor sporulation and dissemination of zoospores [1].
Soybean cultivars and germplasm accessions differ in their responses to isolates of P. sojae [2]. The use of resistant soybean cultivars is the most economical and effective method of controlling this pathogen. Two distinct types of host resistance to P. sojae have been described: (i) race-specific resistance conditioned by single dominant genes (Rps); and (ii) broad-spectrum partial non-race-specific resistance conferred by several minor genes [14–15].
When novel Rps genes are introduced through the release of new cultivars P. sojae isolates evolve to overcome the introduced resistance genes [16–17]. Over 200 known pathotypes of this pathogen have been reported [18–19], presumably due to selection pressure on the P. sojae population for new pathotypes that can overcome Rps genes [20]. The rapid evolution of new P. sojae virulent pathotypes limits the effectiveness of an Rps gene to 8–15 years [1]. Consequently, there is a constant need for novel Rps genes that can effectively manage the disease.
The first Rps gene was identified in the 1950s [21]. To date, 27 Rps genes have been identified and mapped to eight chromosomes (S1 Table). The Rps genes encode receptors that presumably recognize P. sojae effectors and induce effector-triggered immunity [22]. The Rps genes mapped to Chromosome 3 include Rps1, Rps7, Rps9, RpsYu25, RpsYD29, RpsYD25, RpsUN1 and Rps1? [14, 23–31]. The Rps1 locus is complex and contains at least five functional alleles, Rps1a, 1b, 1c and 1d and 1k [28, 32–33]. High resolution genetic and physical maps were constructed for the Rps1-k region and two functional nucleotide binding site-leucine rich repeat (NBS-LRR) containing Rps genes, Rps1-k-1 and Rps1-k-2, were cloned from the Rps1-k locus [29, 34–37]. Recent studies have revealed that additional alleles may be present in the Rps1 locus. For example, Rps1? gene in Waseshiroge, RpsYu25 and RpsYD25 in the Chinese cultivar ‘Yudou 25’, and Rps9 in the Chinese cultivar ‘Ludou 4’ have been considered to be either allelic to Rps1 or Rps1-linked genes [14, 38–39]. The Rps2 gene and RpsUN2 have been mapped to Chromosome 16 [27, 40–41]. Three Rps3 alleles, Rps3a, Rps3b and Rps3c, Rps8 and RpsSN10 have been mapped to Chromosome 13 [27, 42–48]. Although earlier studies suggested no linkage between Rps4 and Rps6 [49], Rps4, Rps5, Rps6 and RpsJS are tightly linked genes that are located on the lower arm of Chromosome 18 [27, 50–54]. In fact, Rps4 and Rps6 could be allelic [50]. Rps10 has been mapped to Chromosome 17 [55], RpsYB30 and RpsZS18 [56–57] to Chromosome 19 and Chromosome 2, respectively, and Rps11 to Chromosome 7 [58].
An earlier study [59] suggested that PI399036 contains multiple Rps genes including at least one novel Rps gene. Our recent mapping study of quantitative trait loci underlying partial resistance to P. sojae [60] using a mixture of three P. sojae isolates suggested the presence of a putative novel Rps gene on the lower arm of Chromosome 18. The present study was undertaken to map this potential novel Rps gene. We observed that a single dominant Phytophthora resistance gene, named Rps12, is tightly linked to the proximal side of the Rps4/6 locus in a 5.4 cM region between the SSR marker BARCSOYSSR_18_1840 and the NBSRps4/6-130/533 sequence.
Materials and Methods
Plant genetic material
The AX20925 recombinant inbred line (RIL) population was developed by crossing PI399036 (USDA-ARS National Soybean Germplasm Collection) with the germplasm line AR2, released by Iowa State University (S.R. Cianzio, D.R. Charlson, G. Gebhart, N. Rivera, P. Lundeen, and R. Shoemaker, unpublished). The cross was made at the Iowa State University research site at the University of Puerto Rico’s Isabela Substation (ISU-PR) [60].
The individual F2 plants were advanced to the F6 generation by applying single-seed descent breeding method. One hundred seeds of each individual F6 plant were planted and harvested in bulk to obtain F7 seeds [recombinant inbred line (RILs)] used in this study [60]. In this study, 290 F7 families [recombinant inbred lines (RILs)] were phenotyped for responses to a mixture of three P. sojae isolates [60] that overcome most of the known Rps genes. Of these 290 RILs, 130 were homozygous resistant, 12 heterzygous for Phytophthora resistance and 148 were recessive homozygous and susceptible. In this molecular mapping study, 120 RILs of the 290 RILs were investigated. Eleven plants each from selected 120 RILs were scored again for responses to the P. sojae isolates in each of the three independent experiments. Among these 120 RILs, 59 were homozygous resistant and 61 were susceptible to the pathogen.
Phytophthora sojae isolates
Phytophthora sojae R17 (vir 1b, 1d, 3a, 3b, 3c, 5, 6), Val 12–11 (vir 1a, 1b, 1c, 1d, 1k, 2, 4, 7), and P7074 (vir 1b, 1d, 2, 3a, 3b, 3c, 4, 5, 6, 7, 8) isolates were used in this study (Table 1). Phytophthora sojae isolate R17 was obtained from Dr. Anne Dorrance (Ohio State University, OH), Val 12–11 from Dr. Martin Chilvers (Michigan State University, MI) and strain P7074 from Dr. Alison E. Robertson (Iowa State University). All isolates were grown on half strength V8 agar plates amended with neomycin sulfate and chloramphenicol antibiotics for 5–7 days under room temperature in the dark as described by Dorrance et al. [12].
Table 1. Reactions of soybean differentials carrying Rps1a, 1b, 1c, 1d, 1k, 2, 3a, 3b, 3c, 4, 5, 6, 7, and 8 genes to Phytophthora sojae isolates.
| Differential Line | Rps gene | R17 | Val12-11 | R17 & Val12-11 | P7074 |
|---|---|---|---|---|---|
| L88-8470 | 1a | 0 | 100 | 80–100 | 0–5 |
| L77-1863 | 1b | 83–100 | 100 | 86–100 | 96–100 |
| Williams 79 | 1c | 0–13 | 100 | 86–100 | 0 |
| L93-3312 | 1d | 100 | 100 | 88–100 | 100 |
| Williams 82 | 1k | 0 | 100 | 80–100 | 0–10 |
| L82-1449 | 2 | 33 | 90–100 | 71–100 | 80–100 |
| L83-570 | 3a | 100 | 0 | 100 | 93–100 |
| L91-8347 | 3b | 100 | 0 | 80–100 | 96–100 |
| L92-7857 | 3c | 100 | 17 | 100 | 88–100 |
| L85-2352 | 4 | 17 | 100 | 100 | 90–100 |
| L85-3059 | 5 | 100 | 11 | 88–100 | 95–100 |
| L89-1581 | 6 | 100 | 0 | 86–100 | 85–100 |
| L93-3258 | 7 | 50–67 | 100 | 100 | 93–100 |
| PI 399073 | 8 | 33 | 13 | 31–67 | 77–100 |
| Sloan | 100 | 100 | 100 | 100 |
R17, P. sojae R17 isolate; Val12-11, P. sojae Val12-11 isolate; R17+Val12-11, a mixture of P. sojae R17 and Val12-11 isolates; P7074, P. sojae strain P7074 alone, Data are in % dead seedlings.
Evaluation of genetic materials for phytophthora resistance
The 120 RILs, the parents PI399036 and AR2 along with 14 differential lines and the susceptible cultivar ‘Sloan’ [12, 61] with no known Rps genes were planted in vermiculite filled 237 mL Styrofoam cups (11 seeds per cup) and watered once a day. The differential lines include lines that carry Rps1a, Rps1b, Rps1c, Rps1d, Rps1k, Rps2, Rps3a, Rps3b, Rps3c, Rps4, Rps5, Rps6, Rps7, and Rps8 genes [19, 62]. Seedlings were grown in the greenhouse for a week. Hypocotyls of seven-day old seedlings were inoculated using the wounded-hypocotyl inoculation technique [18–20, 59–63]. An approximately 1 cm long slit was made with the needle tip in each hypocotyl, 1 cm below the cotyledonary node, and 0.2 to 0.4 mL of the culture slurry was placed into the slit using the syringe. Plants were kept in a dew chamber at 25°C for 24 h in the dark after inoculations and then moved to a growth chamber at 25°C with a 12 h photoperiod with light intensity 580 ± 75 μ mol PAR m-2 s-1. The experiment was repeated two more times. Plants were rated seven days after inoculation as either R (resistant, <30% seedling death) or S (susceptible, ≥70% seedling death).
Inocula were prepared using a modified version of the protocol described by Dorrance et al. [12]. Isolates were grown on soft V8 juice agar (12 g agar/liter) at 22°C under dark conditions until the mycelia covered the entire plate. The colonized agar was cut in strips, placed in a 10-mL syringe and forced out through the syringe to prepare inoculum pulp. The macerated culture was placed in a syringe for a second time and a #18 needle was used to further macerate the culture. Macerated R17 and Val 12–11 cultures were mixed in a 1:1 ratio to prepare the mixed inoculum [63], which is virulent to soybean cultivars carrying Rps genes mapped to any of the Rps1 to 7 loci (Fig 1, Table 1). P. sojae strain P7074 [22, 64–65] was also used as a separate source of inoculum as it is virulent to soybean lines carrying Rps4, 5 and 6 (Fig 1).
Fig 1.
Reactions of soybean differentials carrying Rps4, Rps5, and Rps6 genes and RILs with or without the novel Rps gene to (A) a mixture of P. sojae of R17 and Val12-11 isolates, and (B) the P. sojae P7074 isolate. The presence of a dying or expanded lesion indicates a susceptible response or compatible interaction. Resistance response is expressed as hypersensitive cell death at the inoculation sites and healthy nature of infected plants. AR2, susceptible parent AR2; PI, resistant parent PI399036 containing the novel Rps gene; RIL (R), randomly selected recombinant inbred lines resistant to P. sojae isolates; RIL (S), randomly selected recombinant inbred lines susceptible to P. sojae isolates; Sloan, the susceptible cultivar Sloan with no known Rps genes used as the susceptible control.
DNA preparation and bulked segregant analysis (BSA)
Prior to inoculation, one unifoliate leaf from each of 11 random plants per RIL was harvested, bulked and frozen in liquid nitrogen, and stored at -80°C. The genomic DNA was extracted from the bulked leaf samples using the CTAB (cetyl trimethyl-ammonium bromide) method [66]. To identify microsatellite and molecular markers, we conducted bulked segregant analysis (BSA) [67] using pooled DNA samples of 10 homozygous resistant (Resistant Bulk) or 10 susceptible (Susceptible Bulk) RILs. One μg DNA from each selected RIL was used for pooling. Each DNA bulk was diluted to a final concentration of 50 ng DNA/μL.
Molecular marker analyses
Microsatellite (simple sequence repeats, SSR) and molecular markers were used to construct a linkage map of the genomic region carrying the putative novel Rps gene locus. Molecular markers based on previously reported NBSRps4/6 sequence [50] were developed for mapping the novel Rps gene (S2 Table). SSR primers were synthesized using the sequence data available at SoyBase (http://soybase.org/) (S2 Table). Primer sequences for SSR markers linked to RpsJS were obtained from a published report [54] (S2 Table). For SSR analysis, 50 ng DNA extracted from leaf samples of each resistant or susceptible RIL was used as the template in a 25 μL reaction containing 1X PCR reaction buffer (10 mM Tris–HCl, 50 mM KCl, pH 8.3), 2.0 mM MgCl2; 0.25 μM of each primer, 200 μM of each dNTP, and 1 U Taq DNA polymerase. The polymerase chain reaction (PCR) conditions were as follows: 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at primer-specific annealing temperature (S2 Table), 1 min extension at 72°C; followed by 10 min at 72°C. The amplification products were separated on a 4% NuSieveTM 3:1 agarose (Lonza, USA) gel, stained with EtBr and then visualized under UV light using FOTO/Analyst Express Systems (FOTODYNE Incorporated, USA). Thirty-four SSR markers covering the novel Rps gene region on Chromosome 18 (S2 Table) were evaluated for possible polymorphisms between the AR2 (susceptible), and PI399036 (resistant) parents, and resistant and susceptible bulks of BSA.
Linkage map construction and statistical analysis
The Chi square (χ2) analysis was performed to check the phenotypic data for goodness-of-fit to a Mendelian segregation ratio using Graphpad (http://www.graphpad.com/quickcalcs). To determine genetic distances, Mapmaker version 3.0 [68] and the Kosambi mapping function [69] were used. Marker order was determined using the log-likelihood (LOD) method with threshold 3.0. The linkage map of molecular markers and the Rps12 locus was constructed using MapChart 2.3 [70].
Results
Identification of a putative novel Rps gene
PI399036 has been suggested to carry multiple Rps genes including known and unknown Rps genes [59, 60, 71]. Our previous study of two independent segregating populations suggested that there is a major Phytophthora resistance gene in the Rps4/6 region of this accession [60]. Here we determine the inheritance of the putative novel gene by evaluating F2 and RILs for segregation of Phytophthora resistance against an inoculum mixture of Val 12–11 and R17 isolates, which together are virulent on soybean lines carrying all Phytophthora resistance genes mapped to the Rps1 to 7 loci. We also used the P7074 isolate in screening the RILs because this isolate can overcome the resistance encoded by Rps4, 5, and 6 mapped tightly to the Rps4/6 region (Fig 1; Table 1).
Phenotypic evaluation of the 25 F2 plants obtained from the cross between PI399036 x AR2 following inoculation with the mixture of the Val 12–11 and R17 isolates resulted in 19 resistant (R) and six susceptible (S) plants. The F2 segregation ratio fits the expected 3:1 (R:S) ratio for a single dominant gene for resistance (χ2df = 1 = 0.013, p = 0.908). The screening of the 290 RILs of the AX20925 population with the mixture of the P. sojae isolates, PT2004 C2.S1 (vir 1a, 1b, 1c, 1d, 1k, 2, 3c, 4, 6,7), 1005–2.9 (vir 1a, 1b, 1c, 1k, 3b, 7), and R7-2a (vir 1d, 2, 3a, 5, 6, 7) [60] resulted in a 130:12:148::R:H(heterozygous):S segregation ratio, which fits the expected 140.5:9:140.5 (R:S) ratio for a single gene segregation among the homozygous RILs (χ2df = 2 = 2.125, p = 0.346).
Putative mapping of the novel Rps gene by BSA
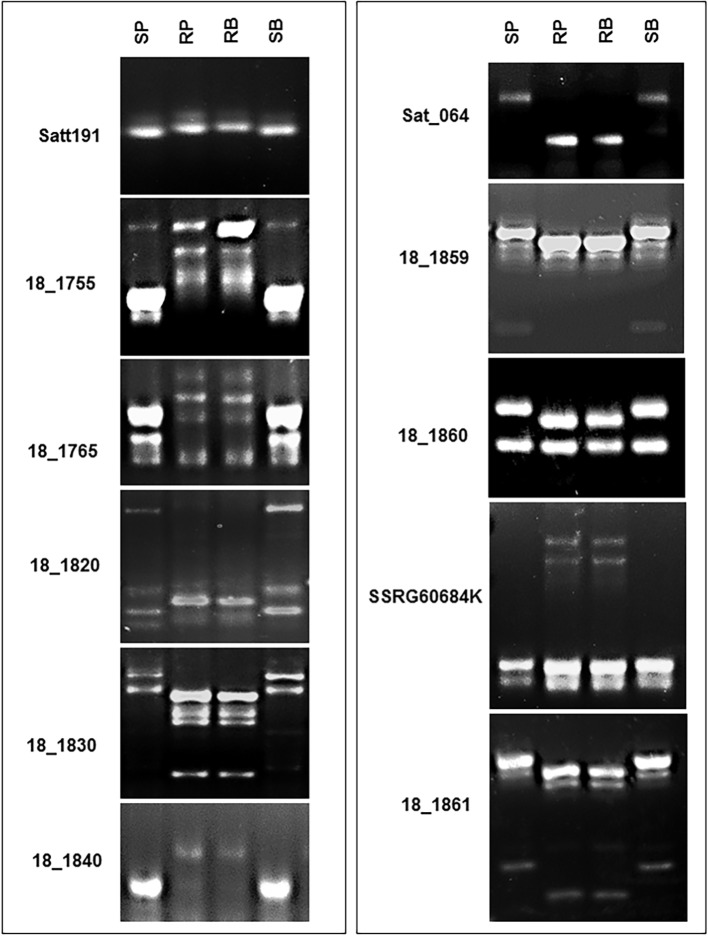
To putatively map the novel Rps gene, we evaluated 34 SSR markers from the Rps4/6 region for polymorphisms among the parents of the population, PI399036 and AR2 (S2 Table). The selected SSR markers encompass the genomic region that includes the RpsJS, Rps4, and Rps6 genes [27, 56–57]. Of the 34 SSR markers evaluated, 14 were polymorphic between PI399036 and AR2. These SSR markers were then used to putatively determine map location of the Rps gene by conducting BSA [67]. The BSA analysis revealed that the novel Rps gene was located in the Rps4/6 region. Of the 14 SSR markers, 11 showing close association to the novel Rps gene were further considered for mapping the 120 RILs (Fig 2).
Fig 2. Eleven polymorphic SSR markers linked to Rps12.
SP, susceptible parent AR2; RP, resistant parent PI399036; RB, bulk of 10 resistant homozygous RILs; SB, bulk of 10 susceptible RILs. Satt191, BARCSOYSSR_18_1750; 18_1755, BARCSOYSSR_18_1755; 18_1765, BARCSOYSSR_18_1765; 18_1820, BARCSOYSSR_18_1820; 18_1830, BARCSOYSSR_18_1830; 18_1840, BARCSOYSSR_18_1840; Sat_064, BARCSOYSSR_18_1858; 18_1859, BARCSOYSSR_18_1859; 18_1860, BARCSOYSSR_18_1860; SSRG60684K, SSRG60684K marker; 18_1861, BARCSOYSSR_18_1861.
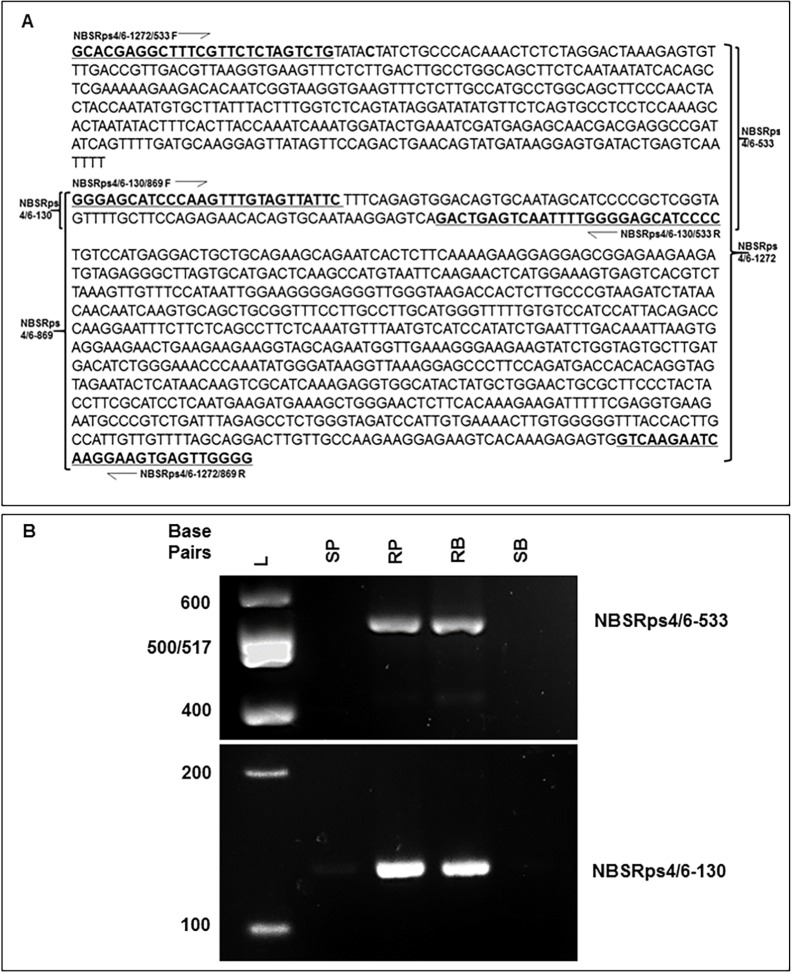
In addition to the 14 polymorphic SSR markers, we determined if the NBSRps4/6 sequence previously reported to be the candidate for the Rps4 gene is polymorphic between the two parents, PI399036 and AR2 [50]. We designed NBSRps4/6 sequence-specific primers and amplified two PCR products of 130 and 533 bp in length, from both PI399036 and the resistant bulked DNA sample, but not from either AR2 or the susceptible bulked DNA sample (Fig 3). BSA analysis suggested that the amplified NBSRps4/6-like sequences co-segregate with the genomic region containing a putative novel Rps gene (Fig 3). The 130 bp and 533 bp PCR fragments showed 93% and 99% nucleic acid sequence identity, respectively, to the NBSRps4/6 sequence reported earlier [50]. The 130 and 533 bp NBSRps4/6-type fragments were named as NBSRps4/6-130 and NBSRps4/6-533, respectively.
Fig 3. Analysis of NBSRps4/6-specific molecular markers linked to a novel Phytophthora resistance gene.
(A) The NBSRps4/6 specific sequence (GenBank accession no. AY258630 [50]) used for developing molecular markers. Primer sequences used for PCR are underlined and marked with half arrows. The PCR primers for amplified targets, NBSRps4/6-1272, NBSRps4/6-869, NBSRps4/6-533 and NBSRps4/6-130, are shown along the primers (S2 Table). (B) The NBSRps4/6 specific molecular markers linked to the novel Rps gene. L, 100 bp DNA Ladder (New England Biolabs, USA); SP, susceptible parent AR2; RP, resistant parent PI399036; RB, bulk of 10 resistant homozygous RILs; SB, bulk of 10 susceptible RILs. NBSRps4/6-533, NBSRps4/6 specific NBSRps4/6-533 marker; NBSRps4/6-130, NBSRps4/6 specific NBSRps4/6-130 marker.
Genetic mapping of the novel Rps gene
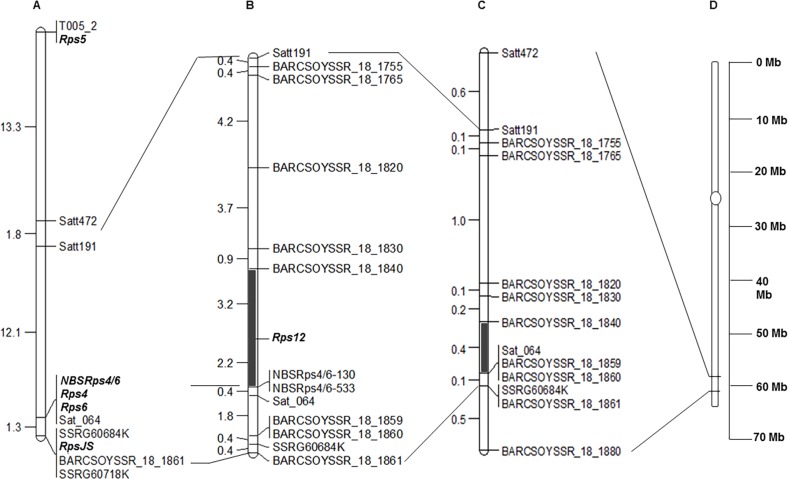
Two dominant markers, NBSRps4/6-130 and NBSRps4/6-533, and 11 co-dominant SSR markers (Figs 2 and 3 and S2 Table) from a genomic region of ~3 Mb containing the novel Rps gene were used to construct a linkage map. We genotyped all 120 RILs (59 R and 61 S) for the 13 molecular markers (S3 Table). With the genotypic and phenotypic data of the mapping population, a genetic map consisting of the 11 SSR markers, the two dominant markers, NBSRps4/6-130 and NBSRps4/6-533, and the novel Rps gene locus was constructed. The new gene was mapped between the SSR markers, BARCSOYSSR_18_1840 and Sat_064 (BARCSOYSSR_18_1858) (Fig 4). Both the NBSRps4/6-130 and NBSRps4/6-533 markers were mapped 2.2 cM distal to the novel Rps locus, suggesting that the new Rps gene is unlikely to be allelic to Rps4. Based on the map positions of the molecular markers linked to previously reported Rps genes, it appears that Rps12 is mapped to a new locus which is distinct from the previously mapped Rps loci of the lower arm of Chromosome 18 (Fig 4A and 4B; S1 Fig).
Fig 4. Genetic and physical map of the Rps12 region.
(A) Molecular genetic map of the Rps loci of the lower arm of Chromosome 18 (S1D Fig). (B) Genetic map of the Rps12 region. SSR markers are shown on the right side of the map and corresponding genetic distances between two adjacent loci are shown on the left side of the map in centi-Morgan (cM). The Rps12 region is shown with a solid line. (C) Physical map of SSR markers on Chromosome 18 according to the soybean reference genome sequence (Glycine max v1.1: http://soybase.org). The corresponding physical distances between two adjacent loci are shown on the left side of the map in mega base pairs (Mb). The Rps12 physical region defined by two molecular markers (BARCSOYSSR_18_1820 and Sat_064) is shown by a solid dark line. (D) Physical location of the Rps12 region on Chromosome 18 (Glycine max v1.1: http://soybase.org). The two long bars indicate two arms of Chromosome 18, the circle indicates the approximate position of the centromeric region, and the marked portion indicates the region containing Rps12.
The RpsJS gene has also been mapped to the Rps4/6 region between the molecular markers, BARCSOYSSR_18_1859 and SSRG60752K [54] (S1 Fig). Both of these markers mapped distal to Sat_064, which co-segregated with the Rps4/6 locus carrying Rps4 and Rps6 genes (S1 Fig). Our mapping data suggest that the novel Rps gene is located in the genomic region proximal to the Rps4, 6 and JS genes and distal to Rps5. We conclude that the gene is novel and named the Phytophthora resistance gene as Rps12 (Fig 4).
Discussion
It has been suggested that PI399036 contains multiple Rps genes including known and unknown Rps genes [59, 60, 71]. Several major and minor QTL for partial resistance to P. sojae have also been identified from this accession [60]. Our previous study indicated the presence of a novel Rps gene in the Rps4/6 region. Responses of the segregating RILs and parents to a P. sojae isolate and a mixture of two isolates established that the gene is distinct from Rps4, 5, and 6. In addition to these three Rps genes, RpsJS was mapped to the lower arm of Chromosome 18 [54]. To determine if the putative novel gene is distinct from Rps4, 5, 6 and JS, we investigated the molecular markers that were shown to be linked to these Phytophthora resistance genes. The Rps4 and Rps6 genes were shown to co-segregate and Rps4 was tightly linked to Sat_064 [50]. Rps5 was shown to co-segregate with the RFLP marker T005_2, which is proximal to both the Satt191 and Satt472 SSR markers (Fig 4 and S1 Fig) [53,72]. Therefore, we conclude that the novel Phytophtora resistance gene identified in this investigation mapped to the novel locus, Rps12.
Rps12 is located in between two SSR markers, BARCSOYSSR_18_1840 and Sat_064, which span a region of 372 kb DNA. The genetic distance between these two loci is 5.8 cM (Fig 4). These results suggest that the Rps12 region is highly recombinogenic, with only 64 kb DNA/cM. Thus, introgression of the gene using the BARCSOYSSR_18_1840 and Sat_064 to elite soybean lines would require molecular analyses of a relatively small segregating population (Fig 4). The Rps12 region contains 45 predicted genes, with on the average one gene in every 8 kb DNA. This means that the highly recombinogenic Rps12 region is gene-rich (S4 Table). It will therefore be feasible to map the candidate Rps12 genes to a small genetic interval through use of molecular markers and a large recombinant inbred line population.
Considering the fact that most identified disease resistance genes encode nucleotide binding site-leucine rich repeat (NBS_LRR) containing proteins, we investigated if there are any NBS-LRR-type genes in the Rps12 region [73]. We identified four clusters of eight NBS-LRR-type genes from this genomic region of the cultivar Williams 82, which has been sequenced (S4 Table) [74]. We observed that although NBSRps4/6 is closer to Rps12 as compared to Sat_064 in the genetic map (Fig 4B), in the Williams 82 genome its physical distance to Rps12 is larger than the distance between Rps12 and Sat_064. This could be due to a micro inversion in the Sat_064 region. Alternatively, this could be just an artifact resulting from misassembling of sequences in the Sat_064 region.
The highly recombinogenic nature of the Rps12 region suggests that positional cloning of the gene could be facilitated through high density mapping of the Rps12 region using a large segregating population. It is expected that a few of the homozygous RILs for Rps12 contain QTL conditioned by minor genes for partial Phytophthora resistance reported earlier [60] and could be an invaluable germplasm for breeding soybeans.
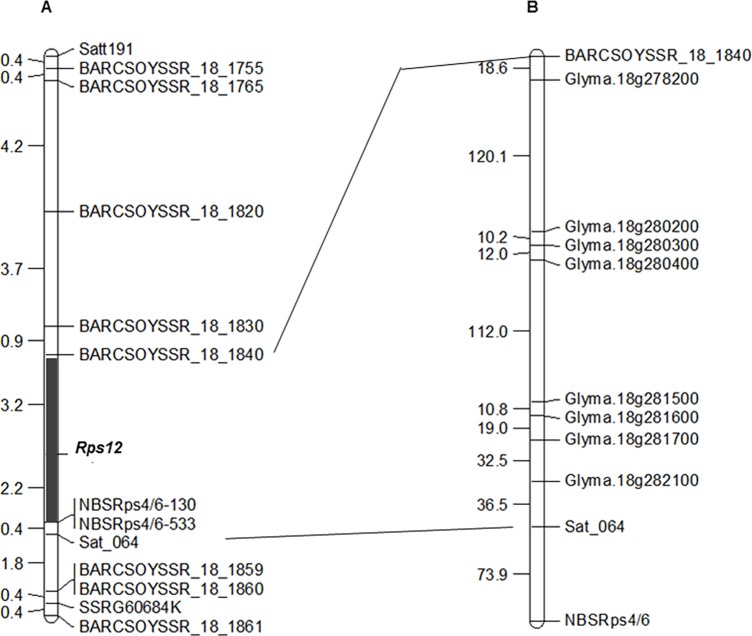
In this study, we have demonstrated that the previously identified Rps4/6 locus is 2.2 cM distal to the Rps12 locus. To date, Rps1-k has been cloned and a strong candidate gene for Rps4 has been identified. Both encode NBS-LRR genes [29, 50]. Several Rps loci have been shown to harbor NBS-LRR sequences, although their functional relevance to Rps genes is yet to be established [75–76]. Our data suggest that Rps12 could be an NBS-LRR-type sequence (Fig 5).
Fig 5. Physical map of NBS-LRR like genes present on the Rps12 region.
(A) Genetic map of the Rps12 region (Fig 4B). (B) Physical map of eight NBS-LRR-like genes identified from the Rps12 region of the Williams 82 genome.
We have evaluated RILs carrying Rps12 against only three important P. sojae isolates, which can overcome resistance encoded by most known Rps genes (Table 1). The study of RILs for their responses to three isolates indicate that the Rps12 gene is expected to have agronomical importance in conferring resistance to most P. sojae isolates that can defeat the Phytophthora resistance encoded by currently available Rps genes.
Supporting Information
(A) The genetic map of the Rps4/6 region from the study by Sandhu et al. (2004) [50]. (B) The genetic map of the Rps4 and Rps5 region from Diers et al. (1992) [53]. (C) The genetic linkage map of the RpsJS region from the study of Sun et al. (2014) [54]. (D) The composite genetic map of the Rps loci located in the lower arm of Chromosome 18. The map was developed from three maps shown in A, B and C, and the co-segregation of Rps4 and Rps6 was from the study of Sandhu et al. (2004) [50].
(TIF)
(DOCX)
(DOC)
(DOC)
(XLS)
Acknowledgments
We are thankful to Drs. Anne Dorrance and Martin Chilvers for kindly providing us the P. sojae isolates that were used in this study. We also thank Dr. Dorrance for constructive discussion. We thank Mr. Peter Lundeen for technical assistance and Dr. Clarice L. Schmidt for technical guidance during hypocotyl inoculation technique. We also thank Ms. Jordan Baumbach for technical guidance for operating Mapmaker and MapChart programs and Dr. David Grant for critically reviewing the manuscript. This work was supported by an Iowa Soybean Association grant.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by an Iowa Soybean Association grant. No role played by the granting agency.
References
- 1.Schmitthenner AF. Problems and progress in control of Phytophthora root-rot of soybean. Plant Dis 1985; 69: 362–368. [Google Scholar]
- 2.Kaufmann MJ, Gerdemann JW. Root and stem rot of soybean caused by Phytophthora sojae n sp. Phytopathology 1958; 48: 201–208. [Google Scholar]
- 3.Dorrance AE, Schmitthenner AF. New sources of resistance to Phytophthora sojae in the soybean plant introductions. Plant Dis 2000; 84: 1303–1308. [DOI] [PubMed] [Google Scholar]
- 4.Anderson TR, Buzzell RI. Diversity and frequency of races of Phytophthora megasperma f sp glycinea in soybean fields in Essex County, Ontario, 1980–1989. Plant Dis 1992; 76:587–589. [Google Scholar]
- 5.Irwin JAG, Cahill DM, Drenth A. Phytophthora in Australia. Aust J Agric Res 1995; 46: 1311–1337. [Google Scholar]
- 6.Drenth A, Whisson SC, Maclean DJ, Irwin JAG, Obst NR, Ryley MJ. The evolution of races of Phytophthora sojae in Australia. Phytopathology 1996; 86: 163–16. [Google Scholar]
- 7.Ryley MJ, Obst NR, Irwin JAG, Drenth A. Changes in the racial composition of Phytophthora sojae in Australia between 1979 and 1996. Plant Dis 1998; 82: 1048–1054. [DOI] [PubMed] [Google Scholar]
- 8.Schmitthenner AF. Phytophthora rot of soybean In: Hartman GL, Sinclair JB, Rupe JC, editors. Compendium of Soybean Diseases. 4th edn The American Phytopathological Society Press; St. Paul, Minnesota; 1999. Pp. 39–42. [Google Scholar]
- 9.Wrather JA, Stienstra WC, Koenning SR (2001) Soybean disease loss estimates for the United States from 1996 to 1998. Canadian Journal of Plant Pathology-Revue Canadienne De Phytopathologie 2001; 23: 122–131. [Google Scholar]
- 10.Tyler BM. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol 2007; 8: 1–8. 10.1111/j.1364-3703.2006.00373.x [DOI] [PubMed] [Google Scholar]
- 11.Wrather J, Koenning S. Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Prog. 2009. [PMC free article] [PubMed] [Google Scholar]
- 12.Dorrance AE, Berry SA, Anderson TR, Meharg C. Isolation, storage, pathotype characterization, and evaluation of resistance for Phytophthora sojae in soybean. Plant Health Prog. 2008. [Google Scholar]
- 13.Drenth A, Janssen EM, Govers F. Formation and survival of oospores of Phytophthora infestans under natural conditions. Plant Pathol 1995; 44: 86–94. [Google Scholar]
- 14.Sugimoto T, Yoshida S, Kaga A, Hajika M, Watanabe K, Aino M, et al. Genetic analysis and identification of DNA markers linked to a novel Phytophthora sojae resistance gene in the Japanese soybean cultivar Waseshiroge. Euphytica 2011; 182: 133–145. [Google Scholar]
- 15.Sugimoto T, Kato M, Yoshida S, Matsumoto I, Kobayashi T, Kaga A, et al. Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed Sci 2012; 61:511–522.16. 10.1270/jsbbs.61.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorrance AE, McClure SA, deSilva A. Pathogenic diversity of Phytophthora sojae in Ohio soybean fields. Plant Dis 2003; 87: 139–146 [DOI] [PubMed] [Google Scholar]
- 17.Grau CR, Dorrance AE, Bond J, Russin J. Fungal diseases In: Boerma HR and Specht JE (eds.) Soybeans: Improvement, production and uses. 3rd ed., Agronomy Monogr. American Soc. Agron; Madison, WI; 2004. pp. 679–763. [Google Scholar]
- 18.Dorrance AE, Jia H, Abney TS. Evaluation of soybean differentials for their interaction with Phytophthora sojae. Plant Health Prog 2004. [Google Scholar]
- 19.Stewart S, Abeysekara N, Robertson AE. Pathotype and genetic shifts in a population of Phytophthora sojae Under Soybean Cultivar Rotation. Plant Dis 2014; 98 (5): 614–624. [DOI] [PubMed] [Google Scholar]
- 20.MacGregor T, Bhattacharyya M, Tyler B, Bhat R, Schmitthenner AF, Gijzen M. Genetic and physical mapping of Avr1a in Phytophthora sojae. Genetics 2002; 160: 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernard RL, Smith PE, Kaufmann MJ, Schmitthenner AF. Inheritance of resistance to Phytophthora root and stem rot in the soybean. Agron Jour 1957; 49: 391–391. [Google Scholar]
- 22.Dong S, Yu D, Cui L, Qutob D, Tedman-Jones J, Kale SD, et al. Sequence variants of the Phytophthora sojae RXLR effector Avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. Plos One 2011; 6: e20172 10.1371/journal.pone.0020172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Zhou B, Zhao J, Guo N, Zhang B, Yang F, et al. Identification of quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Plant Breed 2011; 130: 144–149. [Google Scholar]
- 24.Buzzell RI, Anderson TR. Inheritance and race reaction of a new soybean Rps1 allele. Plant Dis 1992; 76: 600–601 [Google Scholar]
- 25.Xiao-ling W, Bao-qiang Z, Shi S, Jin-ming Z, Feng Y, Na G, et al. Identification, Genetic analysis and mapping of resistance to Phytophthora sojae of pm28 in soybean[J]. Journal of Integrative Agriculture 2011; 10: 1506–1511 [Google Scholar]
- 26.Zhang J, Xia C, Wang X, Duan C, Sun S, Wu X, et al. Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar. Theor Appl Genet 2013; 126: 1555–156 10.1007/s00122-013-2073-1 [DOI] [PubMed] [Google Scholar]
- 27.Demirbas A, Rector BG, Lohnes DG, Fioritto RJ, Graef GL, Cregan PB, et al. Simple sequence repeat markers linked to the soybean Rps genes for Phytophthora resistance. Crop Sci 2001; 41: 1220–1227. [Google Scholar]
- 28.Weng C, Yu K, Anderson TR, Poysa V. Mapping genes conferring resistance to Phytophthora root rot of soybean, Rps1a and Rps7. J Hered 2001; 92: 442–446. [DOI] [PubMed] [Google Scholar]
- 29.Gao HY, Narayanan NN, Ellison L, Bhattacharyya MK. Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol Plant Microbe Interact 2005; 18: 1035–1045. 10.1094/MPMI-18-1035 [DOI] [PubMed] [Google Scholar]
- 30.Sun S, Wu XL, Zhao JM, Wang YC, Tang QH, Yu DY, Gai JY, Xing H. Characterization and mapping of RpsYu25, a novel resistance gene to Phytophthora sojae. Plant Breed 2011; 130: 139–143. [Google Scholar]
- 31.Wu XL, Zhang BQ, Sun S, Zhao JM, Yang F, Guo N, et al. Identification, Genetic analysis and mapping of resistance to Phytophthora sojae of Pm28 in soybean. Agric Sci China 2011; 10: 1506–1511. [Google Scholar]
- 32.Bernard RL and Cremeens CR. An allele at the Rps1 locus from the variety ‘Kingwa’. Soybean Genet. Newsl. 1981; 8: 40–42. [Google Scholar]
- 33.Lohnes DG, Schmitthenner AF. Position of the Phytophthora resistance gene Rps7 on the soybean molecular map. Crop Sci 1997; 37: 555–556. [Google Scholar]
- 34.Bhattacharyya MK, Gonzales RA, Kraft M, Buzzell RI. A copia-like retrotransposon Tgmr closely linked to the Rps1-k allele that confers race-specific resistance of soybean to Phytophthora sojae. Plant Mol Biol 1997; 34: 255–264. [DOI] [PubMed] [Google Scholar]
- 35.Kasuga T, Salimath SS, Shi J, Gijzen M, Buzzell RI, Bhattacharyya MK. High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. Mol Plant Microbe Interact 1997; 10: 1035–1044. [Google Scholar]
- 36.Bhattacharyya MK, Narayanan NN, Gao H, Santra DK, Salimath SS, Kasuga T, et al. Identification of a large cluster of coiled coil-nucleotide binding site-leucine rich repeat-type genes from the Rps1 region containing Phytophthora resistance genes in soybean. Theor Appl Genet 2005; 111: 75–86. 10.1007/s00122-005-1993-9 [DOI] [PubMed] [Google Scholar]
- 37.Gao H, Bhattacharyya MK. The soybean Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol 2008; 8: 29 10.1186/1471-2229-8-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen VT, Vuong TD, VanToai T, Lee JD, Wu X, Rouf Mian MA, et al. Mapping of quantitative trait loci associated with resistance to Phytophthora sojae and flooding tolerance in soybean. Crop Sci 2012; 52: 2481–2493. [Google Scholar]
- 39.Ai-Ying F, Xiao-Ming W, Xiao-Ping F, Xiao-Fei W, Zhen-Dong Z. Molecular identification of Phytophthora resistance gene in soybean cultivar Yudou 25. Acta Agronomica Cinica 2009; 35: 1844–1850 (in Chinese). [Google Scholar]
- 40.Lin F, Zhao M, Ping J, Johnson A, Zhang B, Abney TS, et al. Molecular mapping of two genes conferring resistance to Phytophthora sojae in a soybean landrace PI 567139B. Theor Appl Genet 2013; 126: 2177–2185. 10.1007/s00122-013-2127-4 [DOI] [PubMed] [Google Scholar]
- 41.Kilen TC, Hartwig EE, Keeling BL. Inheritance of a second major gene for resistance to Phytophthora rot in soybeans. Crop Sci 1974; 14: 260–262 [Google Scholar]
- 42.Gordon SG, St. Martin SK, Dorrance AE. Mapping Rps8, a gene for resistance to Phytophthora root and stem rot in soybean. Page 320 in: Crop Sci. Soc. America Annu. Mtg., ASA-CSA-SS, Madison, WI; 2004.
- 43.Sandhu D, Schallock KG, Rivera-Velez N, Lundeen P, Cianzio S, Bhattacharyya MK. Soybean Phytophthora resistance gene Rps8 maps closely to the Rps3 region. J Hered 2005; 96: 536–541. 10.1093/jhered/esi081 [DOI] [PubMed] [Google Scholar]
- 44.Yu AL, Xu PF, Wang JS, Zhang SZ, Wu JJ, Li WB, et al. Genetic analysis and SSR mapping of gene resistance to Phytophthora sojae race 1 in soybean cv. Suinong 10. Chin J Oil Crop Sci 2010; 32: 462–466 [Google Scholar]
- 45.Mueller EH, Athow KL, Laviolette FA. Inheritance of resistance to four physiologic races of Phytophthora megasperma var sojae. Phytopathology 1978; 68: 1318–1322. [Google Scholar]
- 46.Ploper LD, Athow KL, Laviolette FA. A new allele at Rps3 locus for resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathology 1985; 75: 690–694. [Google Scholar]
- 47.Burnham KD, Dorrance AE, Francis DM, Fioritto RJ, Martin SKS. Rps8, a new locus in soybean for resistance to Phytophthora sojae. Crop Sci 2003; 43: 101–105. [Google Scholar]
- 48.Gordon SG, Martin SKS, Dorrance AE. Rps8 maps to a resistance gene rich region on soybean molecular linkage group F. Crop Sci 2006; 46: 168–173. [Google Scholar]
- 49.Athow KL, Laviolette FA. Rps6, a major gene for resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathology 1982; 72: 1564–1567. [Google Scholar]
- 50.Sandhu D, Gao HY, Cianzio S, Bhattacharyya MK. Deletion of a disease resistance nucleotide-binding-site leucine-rich-repeat-like sequence is associated with the loss of the Phytophthora resistance gene Rps4 in soybean. Genetics 2004; 168: 2157–2167. 10.1534/genetics.104.032037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Athow KL, Laviolette FA, Mueller EH. A new major gene for resistance to Phytophthora megasperma var. sojae in soybean. Phytopathology 1980; 70: 977–980 [Google Scholar]
- 52.Buzzell RI, Anderson TR. Another major gene for resistance to Phytophthora megasperma var. sojae in soybean. Soybean Genet Newslett 1981; 18: 30–33 [Google Scholar]
- 53.Diers BW, Mansur L, Imsande J, Shoemaker RC. Mapping Phytophthora resistance loci in soybean with restriction-fragment-length-polymorphism markers. Crop Sci 1992; 32: 377–383. [Google Scholar]
- 54.Sun J, Li L, Zhao J, Huang J, Yan Q, Xing H, Guo N. Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthora sojae in soybean Glycine max (L.) Merr. Theor Appl Genet 2014; 127: 913–919. 10.1007/s00122-014-2266-2 [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Xia C, Duan C, Sun S, Wang X, Wu X, et al. Identification and candidate gene analysis of a novel Phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PLoS ONE 2013; 8: e69799 10.1371/journal.pone.0069799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao HY, Wang XM, Wu XF, Xiao YN, Zhu ZD. Molecular mapping of Phytophthora resistance gene in soybean cultivar Zaoshu18. J Plant Genet Resour 2010; 11: 213–217. [Google Scholar]
- 57.Zhu ZD, Huo YL, Wang XM, Huang JB, Wu XF. Molecular identification of a novel Phytophthora resistance gene in soybean. Acta Agron Sinica 2007; 33: 154–157 [Google Scholar]
- 58.Ping J, Fitzgerald JC, Zhang C, Lin F, Bai Y, Wang D, et al. Identification and molecular mapping of Rps11, a novel gene conferring resistance to Phytophthora sojae in soybean. Theor Appl Genet 2016;129: 445–451. 10.1007/s00122-015-2638-2 [DOI] [PubMed] [Google Scholar]
- 59.Gordon SG, Kowitwanich K, Pipatpongpinyo W, Martin SKS, Dorrance AE. Molecular marker analysis of soybean plant introductions with resistance to Phytophthora sojae. Phytopathology 2007; 97: 113–118. 10.1094/PHYTO-97-0113 [DOI] [PubMed] [Google Scholar]
- 60.Abeysekara NS, Matthiesen RL, Cianzio SR, Bhattacharyya MK, Robertson AE. Novel sources of partial resistance against Phytophthora sojae in soybean PI 399036. Crop Sci 2016; 56: 1–14. [Google Scholar]
- 61.Dorrance AE, Robertson AE, Cianzio S, Giesler LJ, Gran CR, Draper MA, et al. Integrated management strategies for Phytophthora sojae combining host resistance and seed treatments. Plant Dis 2009; 93: 875–882. [DOI] [PubMed] [Google Scholar]
- 62.Stewart S, Robertson AE. A modified method to screen for partial resistance to Phytophthora sojae in soybean. Crop Sci 2012; 52: 1181–1186. [Google Scholar]
- 63.Matthiesen R, Abeysekara N, Ruiz-Rojas J, Biyashev R, Saghai-Maroof M, Robertson AE. A method for combining isolates of Phytophthora sojae to screen for novel sources of resistance to Phytophthora stem and root rot in soybean. Plant Dis 2016; 100 (7): 1424–1428. [DOI] [PubMed] [Google Scholar]
- 64.Dong S, Qutob D, Tedman-Jones J, Kuflu K, Wang Y, Tyler BM, et al. The Phytophthora sojae avirulence locus Avr3c encodes a multi-copy RXLR effector with sequence polymorphisms among pathogen strains. PLoS ONE 2009; 4 (5): e5556 10.1371/journal.pone.0005556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Na R, Yu D, Chapman BP, Zhang Y, Kuflu K, Austin R, et al. Genome re-sequencing and functional analysis places the Phytophthora sojae avirulence genes Avr1c and Avr1a in a tandem repeat at a single locus. PLoS ONE 2014; 9: e89738 10.1371/journal.pone.0089738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allen GC, Flores-Vergara MA, Krasnyanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 2006; 1: 2320–2325. 10.1038/nprot.2006.384 [DOI] [PubMed] [Google Scholar]
- 67.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis—a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 1991; 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, et al. Mapmaker an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987; 1: 174–181. [DOI] [PubMed] [Google Scholar]
- 69.Kosambi D. The estimation of map distances from recombination values. Ann Eugen 1944; 12: 172–175. [Google Scholar]
- 70.Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered 2002; 93: 77–78. [DOI] [PubMed] [Google Scholar]
- 71.Gordon SG, Berry SA, St. Martin SK, Dorrance AE. Genetic analysis of soybean plant introductions with resistance to Phytophthora sojae. Phytopathology 2007; 97: 106–112. 10.1094/PHYTO-97-0106 [DOI] [PubMed] [Google Scholar]
- 72.Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, et al. A new integrated genetic linkage map of the soybean. Theor Appl Genet 2004; 109: 122–128. 10.1007/s00122-004-1602-3 [DOI] [PubMed] [Google Scholar]
- 73.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biology 2006; 7:212 10.1186/gb-2006-7-4-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmutz J, Cannon SB, Schlueter J, Ma J, Hyten D, Song Q, et al. Genome sequence of the paleopolyploid soybean (Glycine max (L.) Merr.). Nature 2010; 463: 178–83. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- 75.Kanazin V, Marek LF, Shoemaker RC. Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci U S A. 1996; 93(21): 11746–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu YG, Buss GR, Saghai Maroof MA. Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 1996; 93: 11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The genetic map of the Rps4/6 region from the study by Sandhu et al. (2004) [50]. (B) The genetic map of the Rps4 and Rps5 region from Diers et al. (1992) [53]. (C) The genetic linkage map of the RpsJS region from the study of Sun et al. (2014) [54]. (D) The composite genetic map of the Rps loci located in the lower arm of Chromosome 18. The map was developed from three maps shown in A, B and C, and the co-segregation of Rps4 and Rps6 was from the study of Sandhu et al. (2004) [50].
(TIF)
(DOCX)
(DOC)
(DOC)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.