Abstract
Salinity is a major limiting factor for the proliferation of plants and inhibits central metabolic activities such as photosynthesis. The halotolerant green alga Dunaliella can adapt to hypersaline environments and is considered a model photosynthetic organism for salinity tolerance. To clarify the molecular basis for salinity tolerance, a proteomic approach has been applied for identification of salt-induced proteins in Dunaliella. Seventy-six salt-induced proteins were selected from two-dimensional gel separations of different subcellular fractions and analyzed by mass spectrometry (MS). Application of nanoelectrospray mass spectrometry, combined with sequence-similarity database-searching algorithms, MS BLAST and MultiTag, enabled identification of 80% of the salt-induced proteins. Salinity stress up-regulated key enzymes in the Calvin cycle, starch mobilization, and redox energy production; regulatory factors in protein biosynthesis and degradation; and a homolog of a bacterial Na+-redox transporters. The results indicate that Dunaliella responds to high salinity by enhancement of photosynthetic CO2 assimilation and by diversion of carbon and energy resources for synthesis of glycerol, the osmotic element in Dunaliella. The ability of Dunaliella to enhance photosynthetic activity at high salinity is remarkable because, in most plants and cyanobacteria, salt stress inhibits photosynthesis. The results demonstrated the power of MS BLAST searches for the identification of proteins in organisms whose genomes are not known and paved the way for dissecting molecular mechanisms of salinity tolerance in algae and higher plants.
Most plants can adapt to low or moderate salinities, but their growth is severely limited above 200 mm NaCl (Hasegawa et al., 2000). Salinity stress leads to a series of changes in basic biosynthetic functions, including photosynthesis and photorespiration, and amino acid and carbohydrate synthesis (Kawasaki et al., 2001; Ozturk et al., 2002; Seki et al., 2002). The response of plants to salt stress has previously been studied in model plant species with sequenced genomes, such as Arabidopsis (Consortium, 2000) and rice (Oryza sativa; Goff et al., 2002; Yu et al., 2002). Differential genomic screens carried out in Arabidopsis and rice have shown that plants respond to salt stress by up-regulation of a large number of genes involved in diverse physiological functions (Gong et al., 2001; Kawasaki et al., 2001; Kreps et al., 2002; Seki et al., 2002). However, since these species are not well adapted to high salinity, halophilic organisms may provide valuable models for identification of basic mechanisms of salinity tolerance (Glenn et al., 1999). A special example of adaptation to hypersaline conditions is the unicellular green algae Dunaliella, a dominant photosynthetic organism in many saline environments, which can adapt to practically the entire range of salinities. Dunaliella responds to salt stress by massive accumulation of glycerol (its internal osmotic element), enhanced elimination of Na+ ions, and accumulation of specific proteins (Pick, 2002). However, comprehensive analysis of salt up-regulated genes in Dunaliella was hindered by the paucity of accurate protein sequences. Sequence information for Dunaliella is limited to approximately 50 protein entries and 3,000 nucleotide entries in the National Center for Biotechnology Information (NCBI) database (DB; August, 2003), thus restricting the ability to identify proteins by a mass spectrometry (MS)-based proteomics approach using conventional DB-searching software.
Plant proteomics have encompassed a variety of species, including Arabidopsis (Mayfield et al., 2001), rice (Koller et al., 2002), maize (Zea mays; Chang et al., 2000), pea (Pisum sativum; Peltier et al., 2000), wheat (Triticum aestivum; Amiour et al., 2002), and poppy (Papaver somniferum; Decker et al., 2000). Proteomes are characterized most effectively by a combination of protein chromatography, proteolytic digestion, and peptide MS analysis (Aebersold and Goodlett, 2001; Mann et al., 2001). Genome sequences and corresponding protein sequence DBs provide a reference for the identification of proteins by MS by the correlation of masses of intact peptides (peptide mass fingerprinting, PMF) or their fragments (tandem mass spectrometry, MS/MS) with in silico processed sequences from DB entries. Conventional protein identification algorithms, such as Mascot (Perkins et al., 1999) and SEQUEST (Eng et al., 1994), are primarily capable of exact matching of analyzed peptides to DB sequences (Fenyo, 2000; Liska and Shevchenko, 2003b), thus hampering proteome characterization in many plant species with unsequenced genomes (van Wijk, 2001; Liska and Shevchenko, 2003a). We demonstrate here that this limitation can be overcome by advanced sequence-similarity DB-searching methods.
To characterize the proteome of the green alga Dunaliella salina, we applied two-dimensional (2-D) gel electrophoresis for protein separation, followed by MS and multiple DB-searching techniques (Liska and Shevchenko, 2003b). We used the conventional DB-searching algorithm, Mascot, and two sequence similarity DB-searching algorithms, MS BLAST and MultiTag, to probe protein and expressed sequence tag (EST) DBs. MS BLAST is a sequence-similarity searching tool, which utilizes redundant, degenerate, and partially inaccurate sequence candidates obtained by the interpretation of MS/MS spectra of peptides (Shevchenko et al., 2001). All sequence candidates obtained by the interpretation of all MS/MS spectra are merged into a single search query in an arbitrary order. Because of such queries, MS BLAST does not rely on E-values of reported alignments (Altschul et al., 1990). Instead, it uses an alternative scoring scheme, which is based on the precomputed threshold scores that are set conditionally on the number of retrieved high-scoring segment pairs and the total number of fragmented precursors (Habermann et al., 2004). MultiTag software (Sunyaev et al., 2003) utilizes short (2–4 amino acid residues) stretches of peptide sequences, combined with the masses of corresponding peptide fragments and the mass of intact peptides into a peptide sequence tag. The significance of hits is estimated by computing E-values of corresponding alignments, which include several error-tolerant matching peptide sequence tags (Mann and Wilm, 1994).
In an attempt to characterize the unique salinity tolerance of Dunaliella, we identified 61 proteins from three subcellular fractions: crude plasma membrane (cPm), chloroplast (chl)-soluble, and cytosol (cyt)-soluble that were up-regulated in 3 m NaCl. The induced proteins included enzymes of central metabolic pathways, such as photosynthesis, energy production, protein synthesis and turnover, and amino acid biosynthesis. Sequence-similarity protein identification techniques were essential for effective identification of more than one-half the proteins analyzed. We therefore expect the proteomics of many plants with unsequenced genomes to be more amenable to characterization than previously facilitated by conventional methods.
RESULTS
Biochemical Isolation and Fractionation of Dunaliella Proteins
D. salina cells that have been cultured continuously in either 0.5 m NaCl or in 3 m NaCl were utilized for differential protein analysis. In order to increase the resolution and dynamic range of protein detection, cells were fractionated into cPm, cyt, and chl fractions. Proteins were extracted following the protocols for each fraction and resolved on 2-D gels. Figure 1, A to C, shows a representative gel for each fraction at 0.5 and 3 m NaCl, where a sample of 250 μg protein was applied. Conventional staining with Coomassie Blue revealed approximately 800 distinct spots on each gel. The comparative analysis of 2-D gel images in all fractions suggested that, upon salinity stress in 3 m NaCl, only about 10% of detectable spots changed their intensity by more than 2-fold.
Figure 1.
Two-dimensional gel electrophoresis of different cellular fractions from D. salina: A, cPm; B, cyt soluble; C, chl soluble. Left, Proteins from 0.5 m NaCl cells; right, proteins from 3 m NaCl cells. Polypeptides (250 μg) were extracted and separated on 18-cm IPG strips and then applied on 11% SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250. Numbers indicate the selected spots and refer to the numbers in Table I; the circled spots are unidentified spots.
In the three separated fractions, about 100 proteins were reproducibly enriched by more than 2-fold in a high-salt environment. For comparison, analysis of proteins that was carried out on total cell extracts, by applying the same criteria, revealed only 30 spots (data not shown). Seventy-six spots from the combined three fractions, whose abundance increased upon salinity stress, were selected for identification by MS. Two additional noninduced spots were added as a reference (Fig. 1, spots A4 and A15).
Protein Identification by MS
From the three cellular fractions, PMF identified 9 out of 78 spots using reference protein DB sequences from Dunaliella sp. and Chlamydomonas sp., among others (Table I). The remaining 69 proteins from the three fractions were analyzed by nanoelectrospray MS/MS and Mascot protein DB searching, which identified 23 additional proteins (in one case two proteins were identified in one spot). An example of the MS and MS/MS spectra, obtained for one protein (spot C14), is presented in Figure 2. The complete set of MS/MS spectra from the analysis was further interpreted using the MS BLAST sequence-similarity protein identification approach. Amino acid sequences were predicted de novo from MS/MS spectra and assembled into modified BLAST queries for DB searching, as previously described (Shevchenko et al., 2001). MS BLAST identified 50 proteins, which included all of the proteins identified by Mascot (except one), plus 28 additional identifications (Table I).
Table I.
Identification of proteins induced by salinity stress
| Spot | Protein Identifications | FI | OMW | TMW | Species | Accession | PF | Ma-EST/Ac | Ma | MB | MT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Antioxidants | |||||||||||
| B30 | Fe-superoxide dismutase Pre | 2.9 | 27 | 27 | V. unguiculata | AAF28773 | 1 | ||||
| B31 | Thioredoxin peroxidase | 1.8 | 26 | 22 | T. elongatus | BAC09006 | 2 | 3 | |||
| C21 | Thioredoxin peroxidase | 2.8 | 25 | 23 | R. conorii | AAL02989 | 1 | ||||
| Chaperones | |||||||||||
| A22 | Chaperonin Pre | 4.1 | 70 | 63 | P. sativum | AAA66365 | 2/AV397884 | 4 | 8 | ||
| A24 | Heat shock Protein 70, hsp 70A2 | 16.0 | 100 | 70 | A. albimanus | AAC41543 | 2/BI874228 | 4 | 10 | ||
| C2 | Luminal binding protein S Pre | 3.6 | 98 | 74 | N. tabacum | CAA42660 | 2/AV643368 | 3 | 7 | ||
| C4 | Heat shock protein 70 Mit Pre, Put | 3.0 | 87 | 70 | O. sativa | AAO17017 | 2 | ||||
| Carbon Assimilation and Mobilization | |||||||||||
| A8 | Carbonic anhydrase | 20.0 | 47 | 64 | D. salina | AAC49378 | 4 | ||||
| A12 | Carbonic anhydrase | 14.0 | 35 | 64 | D. salina | AAC49378 | 3 | ||||
| A14 | Carbonic anhydrase | 30.0 | 60 | 64 | D. salina | AAC49378 | X | ||||
| A17 | Rubisco L Su Pre | 38.0 | 55 | 53 | C. moewusii | AAA84152 | X | ||||
| A4 | Rubisco L Su | 1.0 | 55 | 53 | C. reinhardtii | AAA84449 | 3/BI726309 | 6 | 9 | ||
| A15 | Rubisco L Su | 0.6 | 55 | 53 | Bacteria OTI-8 | BAA92486 | X | ||||
| B18 | Rubisco L Su | 4.3 | 60 | 52 | H. capensis | AAK96893 | X | ||||
| B13 | Rubisco activase Chl Pre | 1.6 | 75 | 45 | C. reinhardtii | AAA33091 | 2 | 4 | |||
| C7 | Rubisco activase Chl Pre | 2.0 | 75 | 45 | C. reinhardtii | AAA33091 | 4 | ||||
| A9 | Phosphoribulokinase Pre | 2.1 | 45 | 46 | S. oleracea | AAA34036 | 1/AV392278 | 6 | |||
| A20 | Phosphoribulokinase | 3.0 | 45 | 42 | C reinhardtii | AAA33090 | 1/BE453265 | 1 | 3 | ||
| A11 | NADP-Glyceraldehyde-3-P DH | 5.9 | 42 | 40 | Chlamydo. sp. | AB035312 | 8 | ||||
| B25 | Sedoheptulose-1,7-bisphosphatase | 3.6 | 42 | 39 | Chlamydo. sp. | BAA94305 | 6 | ||||
| B8 | Dihydroxyacetone kinase | 5.6 | 76 | 65 | S. pombe | AF059204 | 4 | ||||
| C3 | Dihydroxyacetone kinase-like | 4.0 | 88 | 64 | A. thaliana | BAB02871 | 8 | ||||
| B29 | Triose phosphate isomerase | 2.0 | 29 | 23 | B. belcheri | BAA22631 | 4 | ||||
| A1 | Glu-6-P-DH | 5.5 | 60 | 66 | D. bioculata | CAB52685 | X | ||||
| A3 | Glu-6-P-DH | 5.0 | 60 | 66 | D. bioculata | CAB52685 | X | ||||
| B10 | Phosphoglucomutase Chl Pre | 2.3 | 73 | 69 | S. tuberosum | AJ240053 | 2/BE128973 | 2 | 10 | ||
| B17 | 6-Phosphogluconate DH | 1.8 | 64 | 54 | M. Sativa | AAB41553 | 2/BF269268 | 4 | 8 | ||
| B15 | ADP-Glu pyrophosphorylase S Su | 2.1 | 66 | 55 | C. reinhardtii | AAF75832 | 5 | 10 | |||
| B16 | ADP-Glu pyrophosphorylase L Su | 3.4 | 65 | 57 | L. esculentum | AAC49943 | 5 | ||||
| B26 | Inorganic pyrophosphatase Pre | 7.0 | 34 | 31 | C. reinhardtii | CAC42762 | 1/BM498985 | 1 | 8 | ||
| C18 | Inorganic pyrophosphatase Pre | 2.7 | 39 | 31 | C. reinhardtii | CAC42762 | 6 | ||||
| Energy | |||||||||||
| B23 | Fd-NADP oxidoreductase, Put | 3.3 | 42 | 43 | A. thaliana | AAF19753 | 8 | ||||
| C15 | Fd-NADP oxidoreductase, Put | 2.3 | 49 | 43 | A. thaliana | AAF19753 | 2/BG647868 | 1 | 4 | ||
| C16 | Fd-NADP oxidoreductase, Put | 3.1 | 49 | 43 | A. thaliana | AAM65564 | 1/BG647868 | 1 | 3 | ||
| A2 | Duihydrolipoamide S-acetyltransferase | 4.0 | 60 | 44 | T. elongatus | BAC08851 | 8 | ||||
| A6 | Pyruvate DH E-1 α Su | 4.3 | 50 | 47 | A. thaliana | AAB86803 | 3 | ||||
| c13 | NADP-dependent malate DH, Chl | 2.4 | 58 | 47 | D. bioculata | CAC15546 | 2 | 3 | |||
| B19 | NADP-dependent malate DH, Chl | 3.8 | 50 | 47 | D. bioculata | CAC15546 | X | ||||
| B24 | Thiamin biosynthetic enzyme | 2.0 | 41 | 37 | G. max | BAA88227 | 1 | ||||
| C17 | Adenosine kinase | 3.1 | 48 | 38 | P. patens | CAA75628 | 1/BJ172248 | 1 | 1 | ||
| C5 | ATP synthase β; Su, Mit Pre | 2.0 | 86 | 62 | C. reinhardtii | CAA43808 | 1/BE642669 | 1 | 4 | ||
| C22 | ATP synthase δ Su, Chl Pre | 4.0 | 24 | 24 | C. reinhardtii | AAB51365 | 3 | ||||
| Pyrimidine and Amino Acid Biosynthesis | |||||||||||
| A7 | Glutamine synthetase | 20.0 | 50 | 42 | C. reinhardtii | AAB01817 | 5 | 7 | |||
| A20 | Glutamine synthetase | 3.0 | 45 | 41 | C. reinhardtii | AAB01818 | 2/AV623601 | 2 | 3 | ||
| B1 | Carbamoyl P synthetase L chain | 2.7 | 127 | 130 | A. thaliana | AAB67843 | 3 | 11 | |||
| B4 | Aspartate kinase-homoserine DH, Put | 3.3 | 95 | 100 | A. thaliana | BAC43372 | 3 | ||||
| B11 | Isopropylmalate synthase, Put | 3.8 | 72 | 74 | A. thaliana | AAF26002 | 2/AV631505 | 2 | 8 | ||
| B12 | Phosphoglycerate DH | 3.1 | 70 | 66 | A. thaliana | BAA20405 | 2/BF269268 | 4 | |||
| Protein Biosynthesis and Degradation | |||||||||||
| B2 | Zinc metalloprotease | 3.0 | 112 | 118 | A. thaliana | BAB02957 | 1/BE249333 | 2 | 5 | ||
| B3 | Zinc metalloprotease | 2.6 | 112 | 118 | A. thaliana | BAB02957 | 2/AV628512 | 4 | |||
| A10 | TGF-β receptor interacting homolog | 2.0 | 42 | 36 | A. thaliana | AAC49079 | 5 | ||||
| C9 | Mit Processing peptidase, Put | 2.1 | 72 | 59 | A. thaliana | AAF14827 | 7 | ||||
| A21 | 26S proteasome Reg particle triple-A | 3.0 | 58 | 50 | O. sativa | AB037154 | 4/AV620391 | 4 | 7 | ||
| C14 | Translation elongation factor Tu, Mit | 5.0 | 57 | 44 | R. americana | AAD11872 | 5 | ||||
| C1 | Translation elongation factor G, Mit | 4.0 | 103 | 78 | G. max | X71439 | 1/B1727515 | 2 | 9 | ||
| B22 | GDP-mannose pyrophosphorylase | 2.0 | 45 | 40 | A. thaliana | CAC35355 | 4 | ||||
| Cytoskeleton | |||||||||||
| C8 | β-Tubulin | 2.9 | 72 | 50 | C. incerta | AAB60936 | X | ||||
| C10 | αTubulin | 2.4 | 71 | 29 | Z. mays | S39969 | X | ||||
| Na+ Transport | |||||||||||
| B6 | NQR α Su | 5.4 | 79 | 51 | V. cholerae | AAF95439 | 1 | ||||
| B7 | NQR α Su | 3.5 | 77 | 51 | V. cholerae | AAF95439 | 1 | ||||
| Others | |||||||||||
| B5 | GTP-binding protein typA | 7.4 | 92 | 68 | A. thaliana | BAB08691 | 6 |
Spot, Numbers correspond to 2-D gels in Fig. 1; protein identification, proposed biochemical function based on MS analysis and DB searching; FI, fold induction by 3M NaCl; OMW, observed molecular weight in kD; TMW, theoretical molecular weight from DB entry; Species, origin of corresponding retrieved DB sequence; Accession, retrieved DB sequence; PF, peptide mass fingerprinting; X, positive identification; Ma-EST/Ac, number of peptides positively identified by Mascot EST DB searching/accession number; Ma, number of peptides positively identified by Mascot; MB, number of peptides matched by scripted or manually interpreted MS BLAST. Hits were statistically significant according to the MS BLAST scoring scheme; MT, number of peptides matched by MultiTag. The summarized numbers at the bottom of the table are the total number of identified proteins (IDs) by each method. Abbreviations: Put, putative; Mit, mitochondrial; Chl, chloroplast; L, large, S, small, Reg, regulatory, DH, dehydrogenase; P, phosphate, Fd, ferredoxin, Glu, glucose, Su, subunit; Pre, precursor. PMF IDs, 9; Mascot EST IDs, 20; Mascot Protein IDs, 23; MS BLAST(s) IDs, 50; MS BLAST(m) IDs, 2.
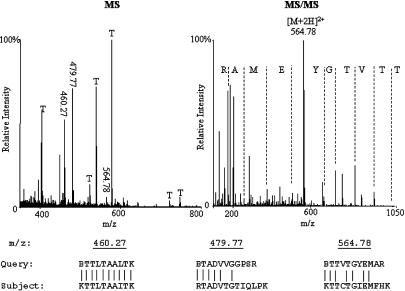
Figure 2.
MS of a protein digest and an MS/MS peptide spectrum. MS of a protein digest is shown on the left. Peaks labeled with T are trypsin autolysis peptides. An MS/MS of the doubly charged peptide precursor (m/z 564.78) is shown on the right, with an amino acid sequence produced by automated de novo interpretation. Altogether, 13 peptides were fragmented and 97 putative peptide sequences produced by automated interpretation of these MS/MS spectra were assembled into a single query and submitted for MS BLAST DB searching. Five peptides were matched to the protein sequence of EF-Tu from the heterotrophic flagellate Reclinomonas americana. Three high-scoring segment pairs—best-scoring alignments of the queried peptide sequences and corresponding peptides from the DB entry produced by a BLAST search engine—are presented in the section below. B in the peptide sequences stands for a generic trypsin cleavage site, either R or K.
Further EST DB searching of all MS/MS spectra with Mascot confirmed the protein identifications made in 20 of the cases, using primarily C. reinhardtii sequences (Table I). All unidentified spots were further analyzed by MS BLAST searching against EST DB. However, this did not contribute to the characterization of any of the unknown spots, although the method confirmed the identification of a few proteins previously identified (data not shown).
The MultiTag approach for sequence-similarity identification was used as a final attempt to identify the remaining proteins because of its demonstrated enhanced sensitivity over MS BLAST (Liska et al., 2004). This technique confirmed the identification made by Mascot and subsequently missed by MS BLAST, and contributed a single new identification, which relied upon a single peptide alignment.
Salt-Induced Proteins in Dunaliella
The overall 61 identified proteins that were up-regulated by high salt represent 45 individual proteins, according to their predicted function. This difference in number results either from cross-contamination between different fractions (six cases) or from the presence of several forms of proteins in the same fraction (eight cases).
The identified proteins, grouped according to their predicted function, are presented in Table I. The two largest categories contain enzymes of central metabolic pathways involved in carbon assimilation and mobilization or in metabolic energy production. The first group comprises five major Calvin cycle enzymes, including a special form of the Rubisco large subunit (RbcL), as well as Rubisco activase, which controls photosynthetic carbon fixation. It also contains three enzymes of the reductive pentose phosphate pathway, a starch biosynthetic enzyme, and three forms of plasma membrane carbonic anhydrase, which enhances CO2 uptake in Dunaliella at high salinity (Fisher et al., 1996). Up-regulation of Calvin cycle enzymes and of plasma membrane carbonic anhydrases might result from enhanced photosynthetic CO2 assimilation. In the independent experiment, we measured the rate of light-dependent carbon uptake at high and low salinity. Indeed, we found that carbon uptake activity was over 2-fold faster in cells grown in 3 m NaCl than in 0.5 m NaCl (Fig. 3).
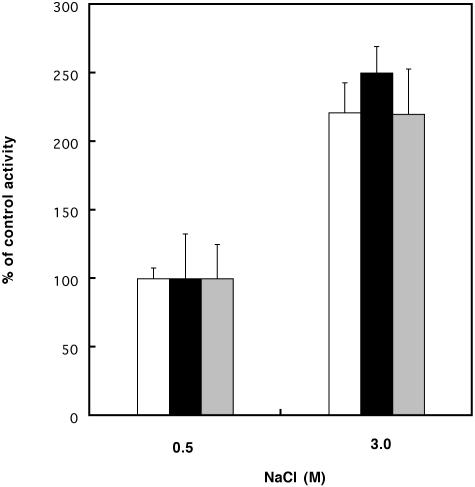
Figure 3.
Effect of salt concentration on photosynthetic CO2 fixation, oxygen evolution, and dark respiration activities. Dunaliella cells from 0.5 or 3 m NaCl cultures were washed and suspended in fresh growth media. Carbon fixation was measured from accumulation of 14C-bicarbonate (white). Rates of photosynthetic oxygen evolution (black) and dark respiration (gray) activities were measured with an oxygen electrode in the light and in the dark, respectively. Control (100%) activities represent the specific activities of 0.5 m NaCl cells. Control rates of carbon fixation, oxygen evolution, and dark respiration were 61.8 ± 4.8 μm C × mg chl−1 h−1, 96.8 ± 12.3 μm O2 evolved × mg chl−1 h−1, and 33.5 ± 5 μm O2 consumed × mg chl−1 h−1. The results represent averages of three measurements.
The second largest category includes enzymes involved in the generation of metabolic energy. It comprises enzymes in different metabolic pathways that function in generation and distribution of redox energy, such as the photosynthetic ferredoxin NADP oxidoreductase, pyruvate dehydrogenase (DH; A2, A6), which controls carbon flux into the respiratory pathway, reductive pentose phosphate pathway enzymes, and malate DH, which is involved in transfer of reducing equivalents across the chl and mitochondrial membranes through the malate/Asp shuttles in plant cells. These results suggest that high salt enhances energy metabolism in Dunaliella.
Another group of up-regulated proteins are key enzymes in biosynthesis of various amino acids: carbamoyl phosphate synthetase (Arg), Asp kinase-homoserine DH, (Thr, Met, and Ile), 2-isopropyl malate synthase (Leu), and 3-phosphoglycerate DH (Ser, Cys, and Gly). Related to this group is Gln synthetase (Gln synthesis), which is also a key enzyme in ammonia assimilation. Enhanced biosynthesis of amino acids may suggest dynamic changes in protein synthesis and/or turnover. This notion is further supported by up-regulation of various regulatory proteins involved in protein synthesis initiation (eukaryotic initiation factor [eIF]3 = TGF-β receptor-interacting protein), elongation factors (EF-Tu, EF-G), protein processing and glycosylation (GDP-Man pyrophosphorylase), and protein degradation (26S proteasome). A unique up-regulated protein is the NADH quinone reductase (NQR)-α subunit homolog, a component of a bacterial redox-driven Na+ extrusion system, which is associated with salinity tolerance in marine bacteria (Hayashi et al., 1995). Surprisingly, we identified this subunit in the cyt-soluble fraction, but not in the plasma membrane fraction. It is conceivable that the subunit dissociated from the membrane during purification. This subunit in bacteria has only one transmembrane domain and is easily dissociated from the bacterial complex during purification (Nakayama et al., 1998).
Two categories of general stress-related proteins in plants are up-regulated in Dunaliella at 3 m NaCl: antioxidants, involved in protection against oxidative stress, and chaperones that protect proteins against denaturation under stress conditions.
DISCUSSION
In this article we describe the first large-scale proteome analysis of salt up-regulated proteins in a lower plant whose genome remains largely uncharacterized. The analysis was limited mostly to major soluble proteins that are up-regulated by no less than 1.5-fold and did not account for minor proteins. We were able to identify about 80% of the selected proteins. The successful analysis was due to cellular fractionation, which enriched relatively minor up-regulated proteins, and the sequence-similarity search algorithm, MS BLAST.
Cellular Fractionation
Many of the identified proteins in the cPm fraction were soluble proteins derived from the chl or cyt. The contamination of plasma membrane preparations with soluble proteins has been observed in many previous analyses (Santoni et al., 2000) and probably results from adsorption of soluble proteins that are released during cell lysis. For example, the RbcL, which is released from the chl, was identified primarily in the plasma membrane fraction. Integral membrane proteins were not identified in the cPm fraction probably because they are underrepresented by isoelectric focusing (IEF; Santoni et al., 2000). Thylakoid membrane and integral plasma membrane proteins are currently being resolved and analyzed by different procedures. Considering these limitations, it may be expected that the overall number of salt up-regulated proteins in Dunaliella is much larger than revealed in this study.
Salinity Tolerance in Dunaliella
The identified salt-induced proteins in Dunaliella reveal an up-regulation of central metabolic networks that can shed light on the outstanding ability of this alga to survive at high salinity.
The ability of Dunaliella to maintain and enhance photosynthetic CO2 assimilation and energy production at high salt is remarkable when compared to the physiology of plants under salt stress; plants respond to salt stress by inhibition of photosynthesis and by stimulation of photorespiration, which result primarily from the large decrease in transpiration leading to CO2 limitation (Wingler et al., 2000). Salinity also limits the availability of CO2 for photosynthesis in aquatic ecosystems because of the decreased solubility of CO2 and shifts in pKs of carbonic acid and bicarbonate (Sass and Ben-Yaakov, 1977). Cyanobacteria and algae, including Dunaliella, overcome this limitation by up-regulation of plasma membrane carbonic anhydrases, which facilitate CO2 acquisition (Booth and Beardall, 1991; Fisher et al., 1996). We identified three carbonic anhydrases in the plasma membrane fraction (A8, A12, and A14). The 60-kD protein (spot A14) is a salt-resistant carbonic anhydrase that has been extensively characterized. The 47-kD protein (spot A8) was identified as a proteolytic product of the major 60-kD protein (Bageshwar et al., 2004). The 35-kD protein (spot A12) was found to be a distinct homolog of carbonic anhydrase protein (A. Zamir, personal communication).
In this study, we show that besides carbonic anhydrase, high salt up-regulates the expression of Rubisco, Rubisco activase, and other key enzymes in the Calvin cycle (Fig. 4), suggesting that the turnover of this CO2-assimilating pathway is stimulated at high salt. On the contrary, in plants, salt stress was reported to down-regulate transcript levels of Calvin cycle enzymes, consistent with the inhibition of photosynthetic CO2 fixation at high salt (Table II).
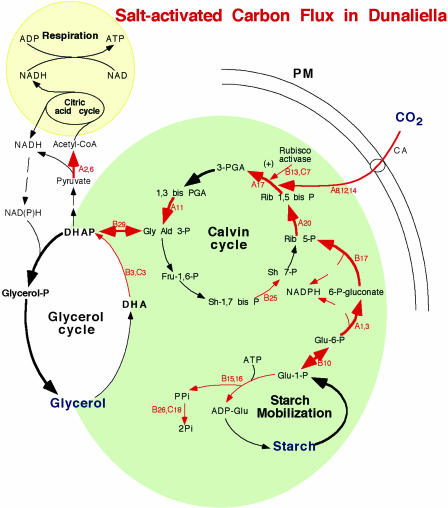
Figure 4.
Schematic presentation of the enhanced metabolic pathways under high salt in Dunaliella. Red arrows indicate the salt-induced enzymes identified in this work, thick arrows (black or red) show the direction of carbon flow leading to glycerol synthesis, and numbers indicate the identified enzymes according to Table I. PM, Plasma membrane; CA, carbonic anhydrase.
Table II.
Salt regulation of proteins/genes in plants in comparison to the identified proteins in Dunaliella
| Proteins | Salt or Drought Regulation in Plants | Reference |
|---|---|---|
| Carbonic anhydrase | − | Seki et al. (2002) |
| Rubisco | − | Ozturk et al. (2002); Seki et al. (2002) |
| Rubisco activase | − | Ozturk et al. (2002); Seki et al. (2002) |
| Phosphoribulokinase | − | Seki et al. (2002) |
| Sedoheptulose-1,7 bisphosphatase | − | Seki et al. (2002) |
| Glyceraldehyde 3-phosphate DH | − | Seki et al. (2002) |
| Glc 6-P DH | ± | Nemoto and Sasakuma (2000); Kreps et al. (2002) |
| 6-Phosphogluconate DH | + | Huang et al. (2003) |
| Triose phosphate isomerase | +D | Salekdeh et al. (2002) |
| Malate DH | ± | Ozturk et al. (2002); Seki et al. (2002) |
| D-3-phosphoglycerate DH | + | Ho and Saito (2001) |
| Gln synthetase | + | Silveira et al. (2003) |
| Adenosine kinase | + | Weretilnyk et al. (2001) |
| Translation initiation factors | + | Kawasaki et al. (2001),Seki et al. (2002) |
| Translation EF-Tu | +D | Salekdeh et al. (2002) |
| Heat shock proteins | + | Seki et al. (2002) |
| Fe-SOD chloroplastic | + | Gomez et al. (1999) |
| Peroxidases | + | Seki et al. (2002) |
| α-Tubulin | − | Kawasaki et al. (2001) |
| β-Tubulin | − | Seki et al. (2002) |
Left column, salt-up-regulated proteins in Dunaliella identified in this study. Middle column, +, up-regulation; −, down-regulation by salt stress or by drought stress (D) in plants.
The increased production of enzymes that generate ATP and redox energy suggests enhanced energy metabolism at high salt in Dunaliella. Consistent with this prediction, we found that photosynthetic oxygen evolution and dark respiration activities were stimulated by 150% and 120%, respectively, at high salt (Fig. 3). The increased ATP and redox energy production supplies the metabolic energy for enhanced CO2 assimilation and ion transport (Katz and Pick, 2001).
Why does Dunaliella enhance photosynthesis and energy utilization at high salt? We hypothesize that high salt enhances CO2 fixation, starch mobilization, and redox energy production to enable massive biosynthesis of glycerol (Fig. 4). The internal concentration of glycerol in Dunaliella, growing in 5 m NaCl medium, is close to 7 m, and constitutes the major carbon pool under these conditions (Avron, 1992). Specifically, glycerol is produced from enhanced CO2 assimilation in the Calvin cycle and/or from starch degradation to dihydroxyacetone phosphate, which is reduced by NADPH to yield glycerol. This interpretation is consistent with previous biochemical evidence for enhanced synthesis of glycerol in Dunaliella from starch and photosynthetic carbon assimilation (Avron, 1992). From four enzymes metabolizing glycerol through dihydroxyacetone phosphate, we identified only dihydroxyacetone kinase (Table I, C3 and B29), which converts glycerol into dihydroxyacetone phosphate (DHAP). In contrast, a proteomic analysis in yeast (Saccharomyces cerevisiae) revealed that all four enzymes, namely, glycerol-3-phosphate DH, glycerol-3-phosphate phosphatase, glycerol kinase, and dihydroxyacetone kinase, were up-regulated at high salinity (Norbeck and Blomberg, 1997). A possible reason for this discrepancy may be that in Dunaliella some of the glycerol-metabolizing enzymes are constitutive. Under optimal growth conditions (0.5–2 m NaCl), Dunaliella accumulates considerable amounts of glycerol and no significant change in the activity of the glycerol-metabolizing enzymes were observed upon salinity stress (Oren-Shamir, 1989).
The up-regulation of Glc-6-P DH and of 6-phosphogluconate DH, the two rate-limiting enzymes of the reductive pentose phosphate pathway (PPP), suggests that the PPP is enhanced at high salt in Dunaliella. This pathway is central in carbohydrate metabolism in plants and provides both reducing power and NADPH and precursors for biosynthesis of lipids, nucleotides, aromatic amino acids, and sugar derivatives. The increase of PPP activity in Dunaliella at high salt complements the up-regulation of the Calvin cycle and enhanced glycerol synthesis. The rapid consumption of glyceraldehyde-3-P DHAP in the synthesis of glycerol requires continuous replenishment of carbon intermediates, which can be provided as ribulose-5-P, the product of the PPP. It also provides NADPH, which can be utilized for reduction of DHAP. Salt stress was reported to increase the expression of Glc-6-P DH and 6-phosphogluconate DH, and to enhance the activity of the PPP in rice and wheat as well (Krishnaraj and Thorpe, 1996; Table II). These results suggest that the PPP may have a general role in diverting carbon metabolites under salt stress in plants and algae.
The up-regulation of key enzymes in amino acid biosynthesis, and of regulatory factors in protein initiation, elongation, and degradation, suggests that, in Dunaliella, high salt increases biosynthesis and turnover of proteins. However, this notion is rather ambiguous. For example, the large increase in the abundance of Gln synthetase and carbamoyl synthase may indicate either enhanced production of Gln and Arg as a source for biosynthesis of new proteins, or enhanced degradation of proteins. Elevation of Gln synthetase in plants usually reflects increased production of ammonia, whereas Arg is a storage form for nitrogen. Enhanced ammonia production in plants can result either from high photorespiration or from elevated protein degradation (Miflin and Habash, 2002). Since in Dunaliella we found no indication for enhanced photorespiration at high salt (stimulation of photosynthetic CO2 assimilation and oxygen evolution), we speculate that the increase in the abundance of Gln synthetase reflects ammonia production from enhanced protein degradation.
The significance of the two mitochondrial protein elongation factors, EF-G and EF-Tu, is also ambiguous, since these proteins may have dual functions. Complementary to their established role in protein biosynthesis in mitochondria and chl, bacterial homologs of both proteins have chaperone properties and were proposed to protect proteins against misfolding under stress (Caldas et al., 1998, 2000). Interestingly, salt stress was reported to up-regulate transcription of chl elongation factor, EF-Tu, in plants (Salekdeh et al., 2002).
Of particular interest is the dual-function eIF3 = TGF-β receptor-interacting protein (spot A10), which has been associated with stress responses and signal transduction. Homologs of eIF3 in mammals and in plants interact with plasma membrane receptors and, as such, are part of a signal transduction pathway in response to external stimuli (Chen et al., 1995; Jiang and Clouse, 2001). A fission yeast homolog of this protein, Sum1, is part of protein initiation complex 3 and was shown to be relocalized under salt or heat stress into distinct cyt domains (Dunand-Sauthier et al., 2002).
The mitochondrial processing peptidase, zinc metalloproteases, and GDP-Man pyrophosphorylase are involved in processing and glycosylation of proteins. The latter may be associated with glycosylation of major plasma membrane proteins, such as triplicated transferrin-like protein, that accumulate in Dunaliella at high salinity (Sadka et al., 1991; Fisher et al., 1997). The overall picture emerging from these results is of a dynamic remodeling of protein composition in different cellular compartments by synthesis and processing of novel proteins as well as by massive degradation of others.
The identification of a homolog of NQR-α is of special interest because it can provide a clue to clarify the exceptional ability of Dunaliella to eliminate Na+ ions at hypersaline solutions. Previously, we demonstrated a redox-driven Na+ extrusion system in Dunaliella (Katz and Pick, 2001).
The functional significance of increased production of cytoskeletal proteins (tubulin) under high salt is unclear, although it may reflect strengthening of the cell cytoskeleton.
Taken together, our findings suggest that the response of the halotolerant alga Dunaliella to salinity involves up-regulation of enzymes and metabolic pathways, some of which are specific to Dunaliella and others common to plants and related organisms.
Sequence-Similarity Protein Identification by MS
This study has demonstrated that sequence-similarity protein identification by MS can identify more than twice as many proteins as conventional approaches, thus greatly enhancing the proteome analysis in the alga D. salina. Of 78 protein spots analyzed, PMF identified 11% of the proteins, DB searching with MS/MS spectra and the conventional software, Mascot, identified 30% of the proteins, and the sequence-similarity methods, MS BLAST and MultiTag, together identified 67% of the proteins: in total, 78% of analyzed protein spots were identified from 2-D gels. Moreover, MS BLAST produced higher sequence coverage than the conventional software by aligning more peptides in an error-tolerant sequence-similarity manner, increasing the confidence of protein identifications by MS. MS BLAST recognized 266 peptides in total from MS/MS spectra (averaging 5.3 peptides/identification), whereas the conventional software was able to align only 60 peptides in total to DB entries (averaging 2.6 peptides/identification). In 19 cases, MS BLAST extended Mascot sequence coverage (i.e. from 1 to 8 peptides aligned); MS BLAST averaged 6 peptides per identification for this group. In 28 additional cases, MS BLAST made identifications where Mascot was unable to produce any significant alignments, averaging 4.4 peptide alignments per identification. EST DB searching with Mascot averaged 1.75 peptides/identification in 20 cases. False-positive functional assignments based on the sequence identity may be minimized by extending sequence coverage, but only to a certain degree. Rost has demonstrated, using bioinformatics, that proteins with high sequence similarity (70%–90%) can have diverse biochemical functions (Rost, 2002). Bona fide of the protein identifications reported here stems from the fact that multiple proteins were identified and recognized to fit into coherent metabolic pathways and groups (such as the Calvin cycle, reductive pentose phosphate pathway, antioxidation, etc.).
With sequence-similarity DB-searching methods and future related developments, the proteomes of plants with unsequenced genomes will be more amenable for characterization by high-throughput MS techniques. It will enable the identification of more conserved proteins in species that are distantly related to plants with sequenced genomes, as well as more diverse homologous proteins in species such as maize, wheat, and barley, using the closely related genomic sequences of rice. Using these methods for protein identification, immediate, rapid, and effective proteome analysis will be possible in plant biochemistry in many species, without having to wait for the completion of future genomic sequencing.
MATERIALS AND METHODS
Algal Material
Dunaliella salina, a green species, was obtained from the culture collection of Dr. W.H. Thomas (La Jolla, CA). Algae were cultured for several weeks in 0.5 m NaCl (control) or in 3 m NaCl (induced) media, as previously described (Katz and Avron, 1985).
Cellular Fractionation
Cellular fractionation was preformed essentially as previously described for plasma membrane preparation (Katz et al., 1986), with minor modifications (Zchut et al., 2003). Cells were harvested and washed with isotonic glycerol buffer, osmotically equivalent to the NaCl concentration in the growth medium, lysed by an osmotic shock (1:4 dilution), and followed by centrifugation at 5,000g for 15 min. The pellet contained chls and the supernatant contained cyt and cPm fractions. The supernatant was ultracentrifuged at 250,000g for 2 h. The ultracentrifugation supernatant was referred to as the cyt-soluble fraction. The pellet, which contained the cPm, was washed again with suspension buffer containing glycerol 0.5 m, Tris-MOPS 10 mm, pH 7.5, MgCl2 2 mm, KCl 10 mm, and ultracentrifuged as before. The ultracentrifugation pellet was resuspended in suspension buffer and referred to as cPm.
The chls were washed with chl buffer containing glycerol 0.5 m, Tris-MOPS 10 mm, pH 7.5, MgCl2 2 mm, and KCl 50 mm, followed by centrifugation at 3,000g for 15 min. The chls were lysed by two freezing-thawing cycles in buffer containing Tris-MOPS 10 mm, MgCl2 2 mm, and KCl 10 mm, followed by centrifugation at 10,000g for 30 min. The supernatant was referred to as the chl-soluble fraction, and the pellet as the chl membrane fraction.
Enzymatic Activities
For CO2 assimilation, D. salina cells cultured for 4 to 5 d in 0.5 or 3 m NaCl were washed and suspended in fresh growth media (at 3 × 106 cells mL−1) containing 10 mm 14C-NaHCO3 at 25°C. Cells were incubated for 30 min at saturating light or in dark. The reaction was terminated by three washes in fresh growth media at 4°C and cell pellets were treated with 5% TCA and incubated 2 h under reduced pressure to eliminate free 14CO2. The results presented are light minus dark values. Rates of dark oxygen uptake and of light-dependent oxygen evolution activities were measured with an oxygen electrode in cells suspended in fresh growth media at 2 × 106 cells mL−1 at 24°C.
Protein Isolation and 2-D Gels
For total cell protein extraction, 1×108 cells were harvested and washed in 90% glycerol + 10% NaCl medium, iso-osmotic to the growth medium. The pellet was suspended in 0.5 mL extraction buffer, containing 700 mm Suc, 100 mm KCl, 50 mm Tris-Cl, pH 8, 5 mm EDTA, 50 mm dithiothreitol (DTT), 0.5 mm phenylmethylsulfonyl fluoride, and plant protease inhibitor cocktail (Sigma P-9599; Sigma-Aldrich, St. Louis). The cells were frozen in liquid nitrogen, thawed, and sonicated twice, followed by centrifugation at 2,000g for 3 min (to get rid of cell debris). The supernatant was taken for phenol extraction.
For cPm extraction, 700 μg of protein from cPm fraction were suspended in 250 μL extraction buffer and incubated for 10 min at 4°C. Proteins from total cells and cPm were extracted by phenol as previously described (Usuda and Shimogawara, 1995). Proteins were extracted with 1.2:1 volumes of phenol to extraction buffer, mixed vigorously, and centrifuged to obtain phase separation; the phenolic upper phase was collected, and the proteins were precipitated by addition of 5 volumes 0.1 m ammonium acetate in methanol and incubated overnight at −20°C. After centrifugation at 10,000g for 20 min, the pellet was washed with ethanol and acetone and dried. Cyt- and chl-soluble fraction extraction proteins were precipitated by acetone, 1 volume of the soluble fraction was mixed with 4 volumes of cold acetone and incubated overnight at −20°C. The extraction was centrifuged at 20,000g for 20 min, and the pellet was air dried.
The pellets of all fractions were dissolved in 2-D sample buffer containing 7 m urea, 2 m thiourea, 4% CHAPS, 0.5% Triton, 5% glycerol, and 0.5% IPG buffer, and incubated for 1 h at room temperature. At this stage, a sample was taken for determination of protein concentration, using the modified Bradford-HCl assay (Ramagli and Rodrigues, 1985). Afterward, 20 mm DTT were added followed by centrifugation at 10,000g for 10 min. Samples of 360 μL containing 250 μg protein from each fraction were loaded on 18-cm IEF dry strips, pH 3 to 10 nonlinear (Amersham Biosciences AB, Uppsala), by cup loading for the total cell proteins, pH 3 to 10 NL with in-gel rehydration for the cPm, pH 4 to 7 with in-gel rehydration for the cyt- and chl-soluble fractions. The strips were subjected to IEF for 50,000 Vh using IPGphor (Amersham Biosciences). Focused gel strips were equilibrated in SDS equilibration buffer (50 mm Tris-Cl, pH 8.8, 30% glycerol, 2% SDS, 6 m urea), first with buffer containing 1% DTT (w/v) for 15 min, and afterward with buffer containing 4% iodoacetamide for 15 min. The strips were briefly washed with running buffer and loaded on top of a prepared SDS-PAGE Laemmli system, 11% acrylamide, and covered with 0.5% agarose. SDS-PAGE was run at 20°C, 70 V, 16 h using the Hoeffer Dalt system. The running buffer contained 50 mm Tris-base, 384 mm Gly, and 0.2% SDS. Gels were stained with 0.1% Coomassie Brilliant Blue R-250 in 50% methanol, 10% acetic acid (v/v). The stained gels were scanned using ImageScanner (Amersham Biosciences). The intensity of protein spots from the 3 and 0.5 m control gels were compared using ImageMaster 2-D software version 3.01 (Amersham Biosciences), and the Z3 software version 1.5 (Compugen, Tel Aviv). Comparative analyses were performed by analyzing images from three independent cultures, with at least two repetitions for each culture. The pixel intensity of each spot was normalized against the total pixel density arising from all the protein spots on the gel. Spots up-regulated more than 2-fold were selected for analysis by MS.
MS Analysis of Protein Spots
Individual protein spots were manually excised from 2-D gels and in-gel digested with trypsin as previously described (Shevchenko et al., 1996). Extracted protein digests were analyzed first by PMF on a Bruker Reflex IV matrix-assisted laser-desorption ionization time-of-flight (MALDI-TOF) MS in reflectron mode using AnchorChip 384/600 targets (Bruker Daltonics, Bremen, Germany) for preparing the probes (Thomas et al., 2004).
Proteins unidentified by PMF were analyzed by nanoelectrospray MS/MS on a modified MDS Sciex QSTAR Pulsar i quadruple time-of-flight (QqTOF) instrument using uncoated borosilicate glass capillaries (1.2 mm o.d. × 0.69 mm i.d.) from Harvard Apparatus (Holliston, MA; capillaries were drawn in-house on a Sutter P-97 puller).
DB Searching
PMF were used for DB searching by Mascot against the MSDB from NCBI (February 2003), with a mass tolerance of 150 ppm; no restrictions were imposed for protein molecular weight; species selection was set to Green Plants. Sets of MS/MS spectra from the analysis of unidentified proteins were first searched by Mascot against the above DB to identify proteins with peptides identical to those existing in silico, at a precursor mass tolerance of 0.1 D and fragment ion mass tolerance of 0.05 D, as above. Mascot queries were generated from MS/MS spectra using the processing script Mascot v.1.6b2 as an extension of Bioanalyst QS software from Applied Biosystems (Foster City, CA). Mascot EST DB searching used Other Green Plants EST_others (November 27, 2002). All MS/MS spectra were then analyzed by MS BLAST against the nonredundant nrdb protein DB at http://dove.embl-heidelberg.de/Blast2/msblast.html, as previously described (Shevchenko et al., 2001). Amino acid sequences were predicted with 0.1-D tolerance for precursor masses and a 0.05-D tolerance for fragment ions using Bioanalyst QS. Queries for MS BLAST DB searches were generated from MS/MS spectra using the ProBLAST v.1.0b11 data-processing script as an extension of Bioanalyst QS (Nimkar and Loo, 2002). In cases where scripted MS BLAST methods failed to identify a protein, an MS BLAST query was generated by manual interpretation of MS/MS spectra using Bioanalyst. If the analyzed protein remained unidentified after PMF, Mascot, and MS BLAST DB searching, with queries derived automatically and manually from MS/MS data, peptide sequence tags were constructed from MS/MS spectra by manual interpretation using Bioanalyst QS for DB searching and MultiTag analysis of search results (Sunyaev et al., 2003).
Acknowledgments
The authors acknowledge Dr. Henrik Thomas (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) for MALDI-TOF assistance for PMF, Yariv Spector for 2-D gel preparation assistance, Tal Varsano for preliminary development of sample preparation, and Professor Gad Galili for advice and fruitful discussions.
This work was supported by The Ministry of Commerce in Israel (MAGNET program), by The Avron-Minerva Center for Photosynthesis, and by Nature Beta Technologies, Eilat, Israel.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039438.
References
- Aebersold R, Goodlett DR (2001) Mass spectrometry in proteomics. Chem Rev 101: 269–295 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Amiour N, Merlino M, Leroy P, Branlard G (2002) Proteomic analysis of amphiphilic proteins of hexaploid wheat kernels. Proteomics 2: 632–641 [DOI] [PubMed] [Google Scholar]
- Avron M (1992) Osmoregulation. In M Avron, A Ben-Amotz, eds, Dunaliella: Physiology, Biochemistry, and Biotechnology. CRC Press, Boca Raton, FL
- Bageshwar UK, Premkumar L, Gokhman I, Savchenko T, Sussman J, Zamir A (2004) Natural protein engineering: a uniquely salt-tolerant, but not halophilic, α type carbonic anhydrase from algae proliferating in low- to hyper-saline enviroments. Protein Eng Des Sel 17: 191–200 [DOI] [PubMed] [Google Scholar]
- Booth WA, Beardall J (1991) Effects of salinity on inorganic carbon utilization and carbonic-anhydrase activity in the halotolerant alga Dunaliella salina (Chlorophyta). Phycologia 30: 220–225 [Google Scholar]
- Caldas T, Laalami S, Richarme G (2000) Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J Biol Chem 275: 855–860 [DOI] [PubMed] [Google Scholar]
- Caldas TD, El Yaagoubi A, Richarme G (1998) Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem 273: 11478–11482 [DOI] [PubMed] [Google Scholar]
- Chang WW, Huang L, Shen M, Webster C, Burlingame AL, Roberts JK (2000) Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiol 122: 295–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Miettinen PJ, Maruoka EM, Choy L, Derynck R (1995) A WD-domain protein that is associated with and phosphorylated by the type II TGF-beta receptor. Nature 377: 548–552 [DOI] [PubMed] [Google Scholar]
- Consortium S (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Decker G, Wanner G, Zenk MH, Lottspeich F (2000) Characterization of proteins in latex of the opium poppy (Papaver somniferum) using two-dimensional gel electrophoresis and microsequencing. Electrophoresis 21: 3500–3516 [DOI] [PubMed] [Google Scholar]
- Dunand-Sauthier I, Walker C, Wilkinson C, Gordon C, Crane R, Norbury C, Humphrey T (2002) Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol Biol Cell 13: 1626–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J, McCormack A, Yates J (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Fenyo D (2000) Identifying the proteome: software tools. Curr Opin Biotechnol 11: 391–395 [DOI] [PubMed] [Google Scholar]
- Fisher M, Gokhman I, Pick U, Zamir A (1996) A salt-resistant plasma membrane carbonic anhydrase is induced by salt in Dunaliella salina. J Biol Chem 271: 17718–17723 [DOI] [PubMed] [Google Scholar]
- Fisher M, Gokhman I, Pick U, Zamir A (1997) A structurally novel transferrin-like protein accumulates in the plasma membrane of the unicellular green alga Dunaliella salina grown in high salinities. J Biol Chem 272: 1565–1570 [DOI] [PubMed] [Google Scholar]
- Glenn EP, Brown JJ, Blumwald E (1999) Salt tolerance and crop potential of halophytes. CRC Crit Rev Plant Sci 18: 227–255 [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gomez JM, Hernandez JA, Jimenez A, del Rio LA, Sevilla F (1999) Differential response of antioxidative enzymes of chloroplasts and mitochondria to long-term NaCl stress of pea plants. Free Radic Res 31 (Suppl): S11–S18 [DOI] [PubMed] [Google Scholar]
- Gong Z, Koiwa H, Cushman MA, Ray A, Bufford D, Kore ES, Matsumoto TK, Zhu J, Cushman JC, Bressan RA, et al (2001) Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol 126: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann B, Oegema J, Sunyaev S, Shevchenko A (2004) The power and the limitations of cross-species protein identification by mass spectrometry-driven sequence similarity searches. Mol Cell Proteomics 3: 238–249 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Hirai K, Unemoto T (1995) Sequencing and the alignment of structural genes in the NQR operon encoding the Na+-translocating NADH-quinone reductase from ibrio alginolyticus. FEBS Lett 363: 75–77 [DOI] [PubMed] [Google Scholar]
- Ho CL, Saito K (2001) Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids 20: 243–259 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang HS, Wang JF, Yang JS (2003) Molecular cloning and characterization of rice 6-phosphogluconate dehydrogenase gene that is up-regulated by salt stress. Mol Biol Rep 30: 223–227 [DOI] [PubMed] [Google Scholar]
- Jiang J, Clouse SD (2001) Expression of a plant gene with sequence similarity to animal TGF-beta receptor interacting protein is regulated by brassinosteroids and required for normal plant development. Plant J 26: 35–45 [DOI] [PubMed] [Google Scholar]
- Katz A, Avron M (1985) Determination of intracellular osmotic volume and sodium concentration in Dunaliella. Plant Physiol 78: 817–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Kaback H, Avron M (1986) Na+/H+ antiport in isolated plasma membrane vesicles from the halotolerant alga Dunaliella salina. FEBS Lett 202: 141–144 [Google Scholar]
- Katz A, Pick U (2001) Plasma membrane electron transport coupled to Na+ extrusion in the halotolerant alga Dunaliella. Biochim Biophys Acta-Bioenergetics 1504: 423–431 [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller A, Washburn MP, Lange BM, Andon NL, Deciu C, Haynes PA, Hays L, Schieltz D, Ulaszek R, Wei J, et al (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99: 11969–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaraj S, Thorpe TA (1996) Salinity stress effects on [C-14-1]- and [C-14-6]-glucose metabolism of a salt-tolerant and salt-susceptible variety of wheat. Int J Plant Sci 157: 110–117 [Google Scholar]
- Liska AJ, Popov A, Sunyaev S, Shevchenko A, Habermann B, Bork P, Karenti E, Shevchenko A (2004) Homology-based proteomics by tandem mass spectrometry: application to the Xenopus microtubule-associated proteome. Proteomics 4: (in press) [DOI] [PubMed]
- Liska AJ, Shevchenko A (2003. a) Expanding the organismal scope of proteomics: cross-species protein identification by mass spectrometry and its implications. Proteomics 3: 19–28 [DOI] [PubMed] [Google Scholar]
- Liska AJ, Shevchenko A (2003. b) Combining mass spectrometry with database interrogation strategies in proteomics. TRAC Trends Anal Chem 22: 291–298 [Google Scholar]
- Mann M, Hendrickson RC, Pandey A (2001) Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem 70: 437–473 [DOI] [PubMed] [Google Scholar]
- Mann M, Wilm M (1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem 66: 4390–4399 [DOI] [PubMed] [Google Scholar]
- Mayfield JA, Fiebig A, Johnstone SE, Preuss D (2001) Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292: 2482–2485 [DOI] [PubMed] [Google Scholar]
- Miflin BJ, Habash DZ (2002) The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J Exp Bot 53: 979–987 [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Hayashi M, Unemoto T (1998) Identification of six subunits constituting Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. FEBS Lett 422: 240–242 [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Sasakuma T (2000) Specific expression of glucose-6-phosphate dehydrogenase (G6PDH) gene by salt stress in wheat (Triticum aestivum L.). Plant Sci 158: 53–60 [DOI] [PubMed] [Google Scholar]
- Nimkar S, Loo JA (2002) Application of a new algorithm for automated database searching of MS sequence data to identify proteins. In Proceedings of the 50th American Society of Mass Spectrometry and Allied Topics, Orlando FL, Abstract No. 334
- Norbeck J, Blomberg A (1997) Metabolic and regulatory changes associated with growth of Saccharomyces cerevisiae in 1.4 m NaCl—Evidence for osmotic induction of glycerol dissimilation via the dihydroxyacetone pathway. J Biol Chem 272: 5544–5554 [DOI] [PubMed] [Google Scholar]
- Oren-Shamir M (1989) Osmoregulatory metabolism in Dunaliella. Ph.D. thesis, Weizmann Institute of Science, Rehovot, Israel
- Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48: 551–573 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ (2000) Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12: 319–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pick U (2002) Adaptation of the halotolerant alga Dunaliella to high salinity. In A Lauchli, U Luthge, eds, Salinity: Enviroment, Plants, Molecules. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 97–112
- Ramagli LS, Rodrigues LV (1985) Quantitation of microgram amounts of protein in two dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis 6: 559–563 [Google Scholar]
- Rost B (2002) Enzyme function less conserved than anticipated. J Mol Biol 318: 595–608 [DOI] [PubMed] [Google Scholar]
- Sadka A, Himmmelhoch S, Zamir A (1991) A 150 kilodalton cell-surface protein is induced by salt in the halotolerant green-alga Dunaliella salina. Plant Physiol 95: 822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2: 1131–1145 [DOI] [PubMed] [Google Scholar]
- Santoni V, Kieffer S, Desclaux D, Masson F, Rabilloud T (2000) Membrane proteomics: use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophoresis 21: 3329–3344 [DOI] [PubMed] [Google Scholar]
- Santoni V, Molloy M, Rabilloud T (2000) Membrane proteins and proteomics: un amour impossible? Electrophoresis 21: 1054–1070 [DOI] [PubMed] [Google Scholar]
- Santoni V, Rouquie D, Doumas P, Mansion M, Boutry M, Degand H, Dupree P, Packman L, Sherrier J, Prime T, et al (1998) Use of a proteome strategy for tagging proteins present at the plasma membrane. Plant J 16: 633–641 [DOI] [PubMed] [Google Scholar]
- Sass E, Ben-Yaakov S (1977) Carbonate system in hypersaline solutions—Dead Sea brines. Mar Chem 5: 183–199 [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Sunyaev S, Loboda A, Bork P, Ens W, Standing KG (2001) Charting the proteomes of organisms with unsequenced genomes by MALDI- quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal Chem 73: 1917–1926 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Silveira JA, Viegas Rde A, da Rocha IM, Moreira AC, Moreira Rde A, Oliveira JT (2003) Proline accumulation and glutamine synthetase activity are increased by salt-induced proteolysis in cashew leaves. J Plant Physiol 160: 115–123 [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Liska AJ, Golod A, Shevchenko A, Shevchenko A (2003) MultiTag: multiple error-tolerant sequence tag search for the sequence-similarity identification of proteins by mass spectrometry. Anal Chem 75: 1307–1315 [DOI] [PubMed] [Google Scholar]
- Thomas H, Havlis J, Peychl J, Shevchenko A (2004) Dried-droplet probe preparation on AnchorChip trade mark targets for navigating the acquisition of matrix-assisted laser desorption/ionization time-of-flight spectra by fluorescence of matrix/analyte crystals. Rapid Commun Mass Spectrom 18: 923–930 [DOI] [PubMed] [Google Scholar]
- Usuda H, Shimogawara K (1995) Phosphate deficiency in maize. 6. Changes in the 2-dimensional electrophoretic patterns of soluble-proteins from 2nd-leaf blades associated with induced senescence. Plant Cell Physiol 36: 1149–1155 [Google Scholar]
- van Wijk KJ (2001) Challenges and prospects of plant proteomics. Plant Physiol 126: 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weretilnyk EA, Alexander KJ, Drebenstedt M, Snider JD, Summers PS, Moffatt BA (2001) Maintaining methylation activities during salt stress. The involvement of adenosine kinase. Plant Physiol 125: 856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC (2000) Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci 355: 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zchut S, Keren N, Ohad I, Pick U (2003) Cold-acclimation protects photosystem II against freezing damage in the halotolerant alga Dunaliella salina. J Plant Physiol 160: 185–192 [DOI] [PubMed] [Google Scholar]