Abstract
Fibroblast growth factor 16 (FGF-16) expression has previously been detected in mouse heart at mid-gestation in the endocardium and epicardium, suggesting a role in embryonic heart development. More specifically, exogenously applied FGF-16 has been shown to stimulate growth of embryonic myocardial cells in tissue explants. We have generated mice lacking FGF-16 by targeting the Fgf16 locus on the X chromosome. Elimination of Fgf16 expression resulted in embryonic death as early as day 11.5 (E11.5). External abnormalities, including hemorrhage in the heart and ventral body region as well as facial defects, began to appear in null embryos from E11.5. Morphological analysis of FGF-16 null hearts revealed cardiac defects including chamber dilation, thinning of the atrial and ventricular walls, and poor trabeculation, which were visible at E10.5 and more pronounced at E11.5. These findings indicate FGF-16 is required for embryonic heart development in mid-gestation through its positive effect on myocardial growth.
Keywords: Fibroblast growth factor, Heart, Myocardial defect, Mouse, Mid-gestation
Fibroblast growth factor (FGF) 16 (FGF-16) was originally characterized in the rat and shown to be expressed preferentially in the heart among adult tissues surveyed [1]. FGF-16 is a member of the FGF-9 subfamily (made up of FGF-9, FGF-16, and FGF-20), which has an amino acid sequence similarity with other FGFs of about 30% [2], while their similarity to one another ranges from 62% to 73% [1,3]. In fetal mouse heart studies, FGF-16 was shown to be expressed in the endocardium and epicardium at embryonic day (E) 10.5 and E12.5 in a unique but overlapping pattern with FGF-9 and FGF-20 [4]. Furthermore, introduction of FGF-16-coated beads into the myocardium at E10.5 stimulated proliferative potential, suggesting that FGF-16 released from the epicardium and endocardium and can have direct effects on embryonic cardiac myocyte growth and heart development [4]. Also, FGF-16 has been linked to the FGF signaling network that is involved in the expansion of anterior heart field progenitor cells [5]. Together, these studies suggest that removal of FGF-16 from the developing murine heart would result in compromised growth of the myocardium, the severity of which would depend on the ability of other FGFs to compensate for the loss of FGF-16. To test this hypothesis, we disrupted Fgf16 in mice by gene targeting and found its expression to be required in mid-gestation for normal heart development.
Materials and methods
Animals
All animals were housed and treated according to standards and guidelines set by the Canadian Council for Animal Care. All procedures used in this investigation conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996), and were approved by the Bannatyne Campus Protocol Management and Review Committee at the University of Manitoba.
Gene targeting and generation of Fgf16−/Y mice
Fgf16 genomic DNA was isolated from a 129Sv/J mouse genomic DNA library (Stratagene, La Jolla, CA) by PCR and used as a template to generate the Fgf16 targeting construct. A lacZ-neo cassette was flanked by a 3.7 kb 5′ homologous arm containing the flanking region of Fgf16 upstream of exon1 plus a 20-bp coding sequence in exon1 (including the ATG start codon) and a 3′ homologous arm of approximately 3 kb containing part of intron 1. Recombination thus resulted in the replacement of 254 bp of the 274 bp in the protein coding region of exon 1, as well as a portion of intron 1, with the lacZ-neo cassette. A KpnI/Acc65I restriction site was introduced at 3′end of lacZ-neo cassette to discriminate the wild-type and targeted alleles by DNA (Southern) blotting. The targeting construct was linearized with SstII before electroporation into RI embryonic stem (ES) cells. G418-resistant ES clones were screened for homologous recombination by Acc65I digestion, followed by DNA blotting analysis as described below in DNA analysis. Two independent homologous recombinant ES clones were microinjected into C57BL/6 J blastocysts and transferred into pseudopregnant recipients at the Transgenic Core Facilities of the University of California, San Diego. Male chimeras were bred with female Swiss Black mice to test for germline transmission of the agouti coat colour from 129-derived ES cells. Hemizygous male FGF-16 null mice (Fgf16−/Y) were generated and maintained by crossing female heterozygotes (Fgf16−/+) with wild-type Swiss Black breeders. All data used for this analysis were collected from offspring ranging from the third to seventh generations.
DNA analysis
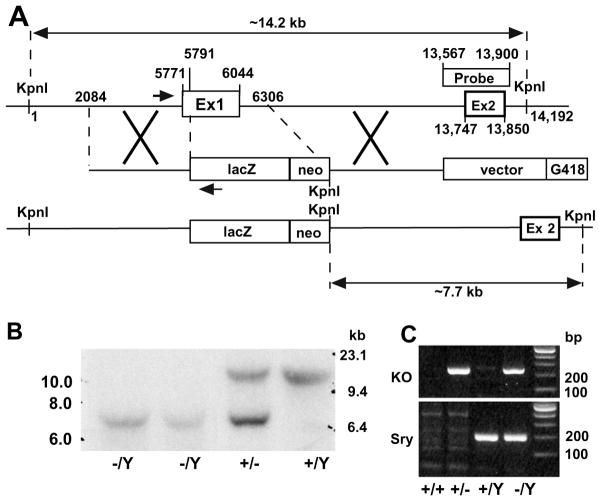
Genomic DNA was prepared from G418-resistant ES cell clones, whole embryos, yolk sac and tails by phenol–chloroform extraction. DNA blotting was done as described in [6] with modifications. Briefly, DNA was digested with Acc65I, electrophoresed on a 0.9% (w/v) agarose gel, following by transfer to nitrocellulose. A 334-bp fragment (shown in Fig. 1A) spanning Fgf16 exon 2 was generated by PCR using mouse genomic DNA and specific primers (forward, 5′-ggcatggaatgactgagcac-3′; reverse, 5′-caaacattggtggtcatagc-3′), and radiolabeled using 32P-dATP and 32P-dCTP by random priming (Prime-a-Gene Labeling System, Promega). DNA blots were hybridized to the radiolabeled probe at 42 °C for 24 h, washed 2 × 15 min at 65 °C, and visualized by autoradiography. The wild-type allele gave rise to a 14.2 kb band and the targeted allele produced a 7.7 kb band.
Fig. 1.
Generation of FGF-16 null mice by homologous recombination. (A) Schematic of Fgf16 targeting construct, showing the replacement of Fgf16 exon 1 (Ex 1) with the lacZ/neo cassette and introduction of a new KpnI site as shown. Maps of the relative genomic region of wild-type Fgf16 locus (top), the targeting locus (center), and the locus after recombination (bottom) are shown. Positions are numbered along the murine X-chromosome relative to first KpnI restriction site upstream of Fgf16 exon 1. The location of the probe used for DNA blot analysis in panel B is indicated, as are the positions of the PCR primers (shown by arrows) used in genotyping in panel C. (B) Genomic DNA from hemizygous (−/Y) Fgf16 null embryos (E12.5), as well as heterozygous female (+/−) and wild-type male (+/Y) littermates was digested with Acc65I (an isoschizomer of KpnI). The probe detected a 14.2 kb fragment in the wild-type (+/Y), a 7.7 kb fragment in null mice (−/Y), and both fragments in heterozygous females (+/−). (C) Genotyping of embryos by PCR. Primers to detect the recombinant (‘knockout’) allele amplified a 207-bp fragment from the targeted locus. Primers to detect the Y-linked Sry gene amplified a 211-bp fragment. The corresponding genotypes are shown below the image.
The genotype of each embryo used in this study was determined from genomic DNA (collected from either the yolk sac or the tail) by PCR using primers (positions shown by arrows in Fig. 1A) to detect the gene targeting construct (forward primer, 5′-gaagttctttggacgcgaac-3′; reverse primer, 5′-gttttcccagtcacgacgtt-3′) for 30 cycles at 58 °C annealing temperature. In addition, primers [7] were used to detect the presence of the Sry gene on the Y chromosome for sex determination (forward primer, 5′-gcacattgtggaggagaact-3′; reverse primer, 5′-cacaggctgtgtctctttag-3′) for 30 cycles at 55 °C annealing temperature. Products were visualized by ethidium bromide staining in an agarose gel.
Reverse-transcriptase (RT)-PCR
Gestation was timed based on the appearance of the vaginal copulation plug representing E0.5. Total RNA was isolated from tissues from the head (including the nasal area), pharyngeal arches, heart and tail of E9.5 and E10.5 embryos using the RNeasy Mini Plus RNA extraction kit (QIAGEN, Mississauga, ON). Complementary DNA was reverse transcribed from 1 μg of total RNA using the QuantiTect Reverse Transcription Kit (QIAGEN), and 2 μl of the RT product was amplified with primers that span exon 1 to exon 3 of Fgf16 to detect the cDNA with a product of 529 bp (forward, 5′-ctccttggactgggacctgc-3′; reverse, 5′-agtgagtgaatttctggtgtcg-3′, 36 cycles, 62 °C annealing temperature) or with primers to detect GAPDH as internal control (forward, 5′-tcaccaccatggagaaggc-3′; reverse, 5′-gctaagcagttggtggtgca-3′, 19 cycles, 60 °C annealing temperature).
Dissection and imaging
Embryos at various time points in gestation were dissected from maternal tissue, examined, and photographed.
Histology
Embryos were fixed in zinc fixative (2.84 mM calcium acetate, 22.8 mM zinc acetate, 36.7 mM ZnCl2 in 0.1 M pH 7.4 Tris–Hcl) at room temperature for 24 h, dehydrated through a graded series of ethanols, xylene and embedded in paraffin. Paraffin sections (6 μM) were dewaxed, rehydrated and stained with Hematoxylin and Eosin. Mutant embryos were somite-count matched to wild-type littermates.
Results
Targeted disruption of the Fgf16 gene in the mouse
The Fgf16 gene on the murine X chromosome was disrupted by replacing exon 1 with the LacZ gene by homologous recombination (Fig. 1 A). ES cells carrying the disrupted allele were identified by DNA blotting/hybridization and detection of a 7.7 kb KpnI/Acc65I fragment compared to the 14.2 kb fragment associated with the wild-type (data not shown). Successful germ-line transmission was also confirmed by DNA blotting in mid-gestation embryos (Fig. 1B). For subsequent genotyping of all embryos used in this analysis, genomic DNA from either the yolk sac (<E10.5) or a portion of the tail (≥E10.5) was analyzed by PCR to detect the recombined (‘knockout’) allele and the Y-linked Sry gene (Fig. 1C).
RT-PCR analysis was used to further assess the specificity of the Fgf16 gene disruption. Previously, Fgf16 RNA expression was detected by whole-mount in situ hybridization in the pharyngeal arches, otic vesicle and olfactory placode of E9.0–E9.5 wild-type embryos [8]as well as endocardial and epicardial cells in E10.5 embryos [4]. By dissecting the head (including developing otic and olfactory structures), pharyngeal arches and heart of embryos at E9.5 and E10.5, we were able to detect the presence of Fgf16 RNA in these structures by RT-PCR in wild-type embryos, and confirm that gene targeting had successfully removed this expression in Fgf16 null littermates (Fig. 2A and B).
Fig. 2.
RT-PCR analysis of Fgf16 expression in wild-type and Fgf16 null embryos. (A) RNA from the head (Hd, including the otic vesicle and olfactory placode), pharyngeal arches (Ph) and heart (Ht) of both wild-type (+/Y) and Fgf16 null (−/Y) E9.5 embryos were isolated and analyzed by RT-PCR using primers for FGF-16 and GAPDH as indicated. (B) Similar RT-PCR analysis in E10.5 embryos, with the addition of RNA from tails (Ta).
Loss of FGF-16 results in embryonic lethality with both craniofacial and cardiac defects
Heterozygous (female) mutant offspring (Fgf16+/−), maintained in a Swiss Black genetic background, were viable, fertile and were indistinguishable from their wild-type littermates. Embryonic lethality was suspected when no Fgf16 null pups were born in 30 litters analyzed by PCR genotyping (Table 1). Furthermore, the total number of male offspring was approximately half that of females (59 versus 137), supporting X-chromosome linked embryonic lethality. Examination of the offspring of timed-pregnant Fgf16+/− mothers revealed that Fgf16−/Y mice died between E10.5 and E12.5 (Table 1). Based on macroscopic analysis, Fgf16−/Y embryos appeared normal in early developmental stages, with no detectable external abnormality up to E10.5. External abnormalities were observed in some but not all null embryos at E11.5, and only growth retarded or reabsorbing Fgf16−/Y embryos were obtained at and after E12.5 (Fig. 3). The defects include the failure of the upper pharyngeal arch to fuse to the upper mandible, and, more importantly, hemorrhaging in and around the cardiac and the ventral body regions, which has been linked with cardiovascular defects [9].
Table 1.
Genotype analysisa of offspring from the crossing of female heterozygotes (Fgf16+/−) with wild-type Swiss Black male breeders (Fgf16+/Y)
| Stage | Number of litters | Mean litter size | F/M | +/+ | +/− | +/Y | −/Y |
|---|---|---|---|---|---|---|---|
| E10.5 | 16 | 10 | 76/84 | 39 | 37 | 39 | 45 |
| E11.5 | 6 | 10.8 | 31/34 | 11 | 20 | 14 | 20b |
| E12.5 | 4 | 8.5 | 17/17 | 9 | 8 | 10 | 7c |
| P1 | 30 | 6.5 | 137/59 | 66 | 71 | 59 | 0 |
Embryos and mice were genotyped by RT-PCR as in Fig. 1C. F/M, total females and males from each time point.
At E11.5, twenty (20) Fgf16−/Y embryos were identified by RT-PCR, 16 were living and four were not.
At E12.5, seven Fgf16−/Y embryos were identified but none were alive.
Fig. 3.

Gross morphology of embryonic development in Fgf16 null mice. Wild-type (WT, +/Y) and Fgf16 null (Nu, −/Y) embryos at E10.5, E11.5, and E12.5 are shown. (A) At E10.5, the null embryo is indistinguishable from its wild-type littermate. (B) For E11.5, right (top) and left (bottom) views of the same representative wild-type and null embryos are shown. Note that at this time point, some Fgf16 null embryos exhibited no differences from their age-matched wild-type counterparts. (C) At E12.5, the phenotype of all Fgf16 null embryos is represented by the figure. Affected embryos at E11.5 and all Fgf16 null embryos at E12.5 display hemorrhage in and around the cardiac and the ventral body regions, as well as marked craniofacial defects.
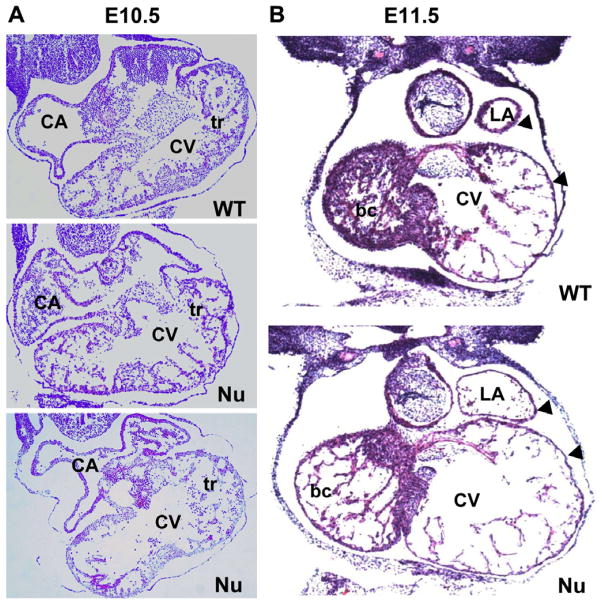
The histology of Fgf16−/Y embryos and somite-matched wild-type littermates was analyzed at various developmental stages. At E9.5, mutants showed a normal cardiac morphology compared to control mice (data not shown), but by E10.5 there was visible dilation of the common ventricle in Fgf16−/Y embryos. Compromised trabeculae were also apparent (Fig. 4A). At E11.5, in Fgf16−/Y embryos that were still viable at the time of dissection, the lumens of both atria and ventricles were enlarged compared to wild-type controls. Also, atrial and ventricular wall thickness was reduced in the null embryo compared to wild-type (arrows, Fig. 4B) and, as was the case at E10.5, Fgf16−/Y embryos at E11.5 had poor trabeculation. At this time point the trabecular insufficiency is more pronounced in the right part of the ventricle than the left, (Fig. 4B).
Fig. 4.
Histological assessment of heart defects in FGF-16 null mice. Mid-level cross-sections from E10.5 and E11.5 mouse embryos were stained by Hematoxylin and Eosin. (A) At E10.5, sections from two different null (Nu) embryos show an enlarged common ventricle (CV) and compromised trabeculation (tr) compared with a wild-type (WT) littermate. (B) At E11.5, both atrial and ventricular chamber enlargement are observed in null embryos (Nu) compared to WT. The changes to the trabeculation are now more pronounced, especially in the bulbus cordis (bc), which will become the future right ventricle. The arrow heads indicate thinning of the ventricular and atrial walls compared to WT. Other abbreviations: CA, common atrium; LA, left atrium.
Discussion
Previous reports have implicated that the production of FGF-16 by the epicardial and endocardial cells at E10.5 might be involved in the growth and development of the adjacent myocardium by increasing cardiac myocyte growth [4]. We have generated a mouse model bearing a null mutation of Fgf16 by homologous recombination, which resulted in compromised myocardial development as indicated by poor trabeculation, thinning of the myocardial wall as well as dilation of cardiac chambers in mid-gestation. Until now, evidence for the role of FGF-16 in cardiovascular development in vivo has been indirect, pertaining to the phenotype of mice lacking FGF-9 or FGF receptor (FGFR) 1 and FGFR2 [4,10]. The present study, however, is the first to demonstrate an in vivo requirement of FGF-16 expression for myocardial growth and development in mid-gestation.
Murine cardiac development begins with the specification of cardiac cell lineages and the subsequent formation of the linear heart tube, then rightward looping at E9.5. At E10.5, the cardiomyoblast population undergoes a robust expansion resulting in the emergence of trabeculae in the luminal layers of both ventricles [11]. This was proposed to be mediated by FGF-derived signals from the epicardium and endocardium [4]. In the same study, exogenously added FGF-16 was shown to have a proliferative effect on embryonic myocardial cells in tissue explants. In the work presented here, the removal of FGF-16 from the embryonic heart resulted in a marked reduction in the expansion of the myocardium, an effect that began at E10.5 in the common ventricle and persisted at E11.5 with more pronounced changes (Fig. 4). While causative relationships have yet to be established, the loss of FGF-16 correlates with decreased myocardial (and presumably myocyte) growth, and it is feasible that this lack of growth leads to compromised cardiac function evidenced by chamber dilation and wall thinning, and ultimately, embryonic death.
Using RT-PCR in wild-type embryos, we confirmed the presence of Fgf16 RNA in the hearts of E10.5 embryos, as had been shown by others using in situ hybridization [4]. Furthermore, the RT-PCR analysis revealed Fgf16 expression at E9.5 in the head region, which includes the otic vesicle, olfactory placode and pharyngeal arches. This is also consistent with the findings of others by in situ hybridization [8]. The expression of Fgf16 in the pharyngeal arches is particularly intriguing, given (1) the combined cardiovascular and craniofacial defects in Fgf16−/Y embryos at E11.5 and E12.5 (Fig. 3), and (2) the right versus left asymmetry of the decreased trabeculation in the ventricles of E11.5 Fgf16−/Y embryos (Fig. 4B). During embryogenesis, cells from the pharyngeal arches contribute to both mandibular and cardiovascular development [12,13]. While not lethal in utero, craniofacial defects are known to be linked to (often lethal) congenital cardiovascular defects in some human genetic disorders [14–16]. Interestingly, an Fgf8 mutant phenotype also displays defects in pharyngeal arch-derived structures, with remarkable resemblance to human 22q11 deletion syndrome [17,18].
The right–left asymmetry is interesting in light of the existence of two cell lineages (known as first and second) that contribute to cardiac structures during development [19]. Cells from the pharyngeal mesoderm are part of the second lineage, from which is derived a large portion of the right ventricle, the outflow tract and parts of both atria. A recent study indicated FGF-16 appears to be part of the FGF signaling network acting downstream of Wnt/β-catenin signaling that is required for expansion of anterior heart field progenitor cells, a subset of the second lineage [5]. Thus, a possible contribution of FGF-16 to normal heart development is the regulation of cardiac progenitor migration/expansion, particularly in the second lineage arising from pharyngeal mesoderm. Further study will be necessary to establish at a cellular level which lineages are affected by the loss of FGF-16 in this model.
Each of the three members of the FGF-9 subfamily (FGF-9, FGF-16, and FGF-20) was found to induce proliferation of myocardial cells in tissue explants [4]. Given this, combined with the homology and overlapping expression patterns of these genes in the developing heart, it was entirely feasible to find that the Fgf16−/Y mouse would have no immediately detectable phenotype. However, the defects we observed and ultimately the death of Fgf16−/Y embryos in mid-gestation point to a unique and specific role for FGF-16 apart from FGF-9 and FGF-20. This is further supported by the expression of Fgf16 in the pharyngeal arches (since there is no report of Fgf9 or Fgf20 expression in this region), and the similarity in phenotype to the Fgf8 mutant mouse, which involves pharyngeal arch-derived structures [17,18]. In conclusion, this mouse model identifies FGF-16 as an important growth factor required for mid-gestation heart development. The study of the cardiac defects in Fgf16−/Y embryos will be valuable to understanding the genetic and molecular basis of congenital heart defects.
Acknowledgments
We would like to thank Dr. N. Mesaeli for her generosity and technical support, as well as Drs. G. Hicks and H. Ding for providing access to microscopes. This work was supported by a grant from the Canadian Institutes of Health Research (MOP-62742). Dr. S.Y. Lu is the recipient of a Manitoba Institute of Child Health Postdoctoral Fellowship.
References
- 1.Miyake A, Konishi M, Martin FH, Hernday NA, Ozaki K, Yamamoto S, Mikami T, Arakawa T, Itoh N. Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochem Biophys Res Commun. 1998;243:148–152. doi: 10.1006/bbrc.1998.8073. [DOI] [PubMed] [Google Scholar]
- 2.Miyamoto M, Naruo K, Seko C, Matsumoto S, Kondo T, Kurokawa T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Mol Cell Biol. 1993;13:4251–4259. doi: 10.1128/mcb.13.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohmachi S, Watanabe Y, Mikami T, Kusu N, Ibi T, Akaike A, Itoh N. FGF-20, a novel neurotrophic factor, preferentially expressed in the substantia nigra pars compacta of rat brain. Biochem Biophys Res Commun. 2000;277:355–360. doi: 10.1006/bbrc.2000.3675. [DOI] [PubMed] [Google Scholar]
- 4.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Sheikh F, Hollander M, Cai C, Becker D, Chu PH, Evans S, Chen J. Embryonic atrial function is essential for mouse embryogenesis, cardiac morphogenesis and angiogenesis. Development. 2003;130:6111–6119. doi: 10.1242/dev.00831. [DOI] [PubMed] [Google Scholar]
- 7.Tsukamoto T, Inada K, Fukami H, Yamamoto M, Tanaka H, Kusakabe M, Bishop CE, Tatematsu M. Mouse strain susceptibility to diethylnitrosamine induced hepatocarcinogenesis is cell autonomous whereas sex-susceptibility is due to the micro-environment: analysis with C3H <–> BALB/c sexually chimeric mice. Jpn J Cancer Res. 2000;91:665–673. doi: 10.1111/j.1349-7006.2000.tb00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright TJ, Hatch EP, Karabagli H, Karabagli P, Schoenwolf GC, Mansour SL. Expression of mouse fibroblast growth factor and fibroblast growth factor receptor genes during early inner ear development. Dev Dyn. 2003;228:267–272. doi: 10.1002/dvdy.10362. [DOI] [PubMed] [Google Scholar]
- 9.Parlakian A, Tuil D, Hamard G, Tavernier G, Hentzen D, Concordet JP, Paulin D, Li Z, Daegelen D. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol Cell Biol. 2004;24:5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 12.Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 13.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 14.Guion-Almeida ML, Zechi-Ceide RM, Richieri-Costa A. Cleft lip/palate, abnormal ears, ectrodactyly, congenital heart defect, and growth retardation: definition of the acro-cardio-facial syndrome. Clin Dysmorphol. 2000;9:269–272. doi: 10.1097/00019605-200009040-00007. [DOI] [PubMed] [Google Scholar]
- 15.Giannotti A, Digilio MC, Mingarelli R, Dallapiccola B. An autosomal recessive syndrome of cleft palate, cardiac defect, genital anomalies, and ectrodactyly (CCGE) J Med Genet. 1995;32:72–74. doi: 10.1136/jmg.32.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zlotogora J, Ariel I, Ornoy A, Yagel S, Eidelman AI. Thomas syndrome: potter sequence with cleft lip/palate and cardiac anomalies. Am J Med Genet. 1996;62:224–226. doi: 10.1002/(SICI)1096-8628(19960329)62:3<224::AID-AJMG4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 17.Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abu-Issa R, Smyth G, Smoak I, Yamamura K, Meyers EN. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development. 2002;129:4613–4625. doi: 10.1242/dev.129.19.4613. [DOI] [PubMed] [Google Scholar]
- 19.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]