Summary
Aging is accompanied by loss of subcutaneous adipose tissue. This may be due to reduced differentiation capacity or deficiency in DNA damage repair (DDR) factors. Here we investigated the role of SNEVhPrp19/hPso4, which was implicated in DDR and senescence evasion, in adipogenic differentiation of human adipose stromal cells (hASCs). We showed that SNEV is induced during adipogenesis and localized both in the nucleus and in the cytoplasm. Knockdown of SNEV perturbed adipogenic differentiation and led to accumulation of DNA damage in hASCs upon oxidative stress. In addition, we demonstrated that SNEV is required for fat deposition in Caenorhabditis elegans. Consequently, we tested other DDR factors and found that WRN is also required for adipogenesis in both models. These results demonstrate that SNEV regulates adipogenesis in hASCs and indicate that DDR capacity in general might be a pre-requisite for this process.
Keywords: SNEV, Prp19, Pso4, adipogenesis, DNA damage repair, WRN, human adipose stromal cells, C. elegans
Graphical Abstract
Highlights
-
•
SNEV is required for adipogenic differentiation of human adipose stromal cells
-
•
SNEV modulates pro- and anti-adipogenic signaling pathways
-
•
SNEV regulates DNA repair capacity of human adipose stromal cells
-
•
SNEV modulates organismal fat deposition in Caenorhabditis elegans
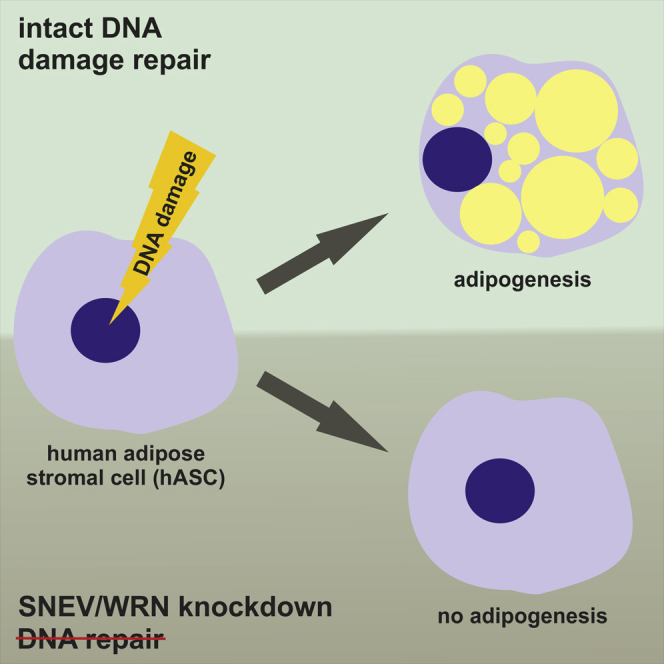
How does a DNA repair factor regulate adipogenic differentiation and organismal fat deposition? Grillari, Schosserer and colleagues have shown that the essential splicing and DNA repair factor, SNEVhPrp19/hPso4, modulates pro- and anti-adipogenic signaling pathways and ensures genome integrity during adipocyte differentiation, suggesting a novel failsafe mechanism to avoid accumulation of dysfunctional cells.
Introduction
Adipose tissue is formed at specific locations as a major energy storage compartment and is an important source of signaling activity. The distribution of adipose reservoirs within the body undergoes major changes during normal aging (Caso et al., 2013), while excess or dysfunctional fat tissue leads to reduced lifespan and accelerates the onset of age-related diseases (Ahima, 2009, Muzumdar et al., 2008). Moreover, loss of subcutaneous fat and increased visceral adiposity is observed in patients with segmental progeroid syndromes such as Werner syndrome (Mori et al., 2001), Cockayne syndrome, or trichothiodystrophy. These diseases, mirroring certain aspects of accelerated aging, are characterized by mutations in DNA damage repair (DDR) factors, leading to accumulation of DNA damage over time and hence potentially to reduced proliferation and differentiation or to senescence of pre-adipocytes (Tchkonia et al., 2010). Mouse models deficient in DNA repair also show adipose tissue degeneration (Karakasilioti et al., 2013). However, it remains unclear whether DNA repair factors themselves have an impact on adipogenic differentiation of human adipose stromal cells (hASCs).
WRN and SNEVhPrp19/hPso4 are members of a DDR protein complex (Zhang et al., 2005) and are involved in adipogenesis of mouse 3T3-L1 cells (Cho et al., 2007, Turaga et al., 2009). WRN is a helicase required for DNA recombination and repair and interacts with the SNEV complex during repair of interstrand crosslinks (Zhang et al., 2005). However, due to differences in the murine and human adipogenic differentiation processes (Mikkelsen et al., 2010), the impact of the human homologs on adipogenesis is still unclear.
SNEVhPrp19/hPso4, termed SNEV in the following, is highly conserved from yeast to humans and plays a role in several cellular pathways. It is an essential splicing factor (Grillari et al., 2005), possesses E3 ubiquitin ligase activity (Song et al., 2010), and interacts with the proteasome (Löscher et al., 2005). In addition, SNEV is involved in various types of DDR, such as DNA double-strand break repair (Mahajan and Mitchell, 2003) and homologous recombination (Abbas et al., 2014). It also interacts with two major DDR regulators: ataxia-telangiectasia mutated regulator phosphorylates SNEV after exposure to oxidative stress (Dellago et al., 2012), and SNEV contributes to the activation Rad3-related (ATR) regulator (Wan and Huang, 2014). SNEV is also linked to cellular senescence (Voglauer et al., 2006) and skin aging (Monteforte et al., 2016).
Here we show that SNEV indeed regulates adipogenesis in human cells and that these findings can be extended to other factors that counteract DNA damage during adipogenic differentiation of hASCs. This suggests that the ability to repair DNA might represent a checkpoint for adipogenesis and thereby provides a failsafe mechanism to reduce the risk of accumulating damaged and/or senescent cells with a pro-inflammatory phenotype in the adipose tissue.
Results
SNEV Expression Is Induced during Adipogenesis
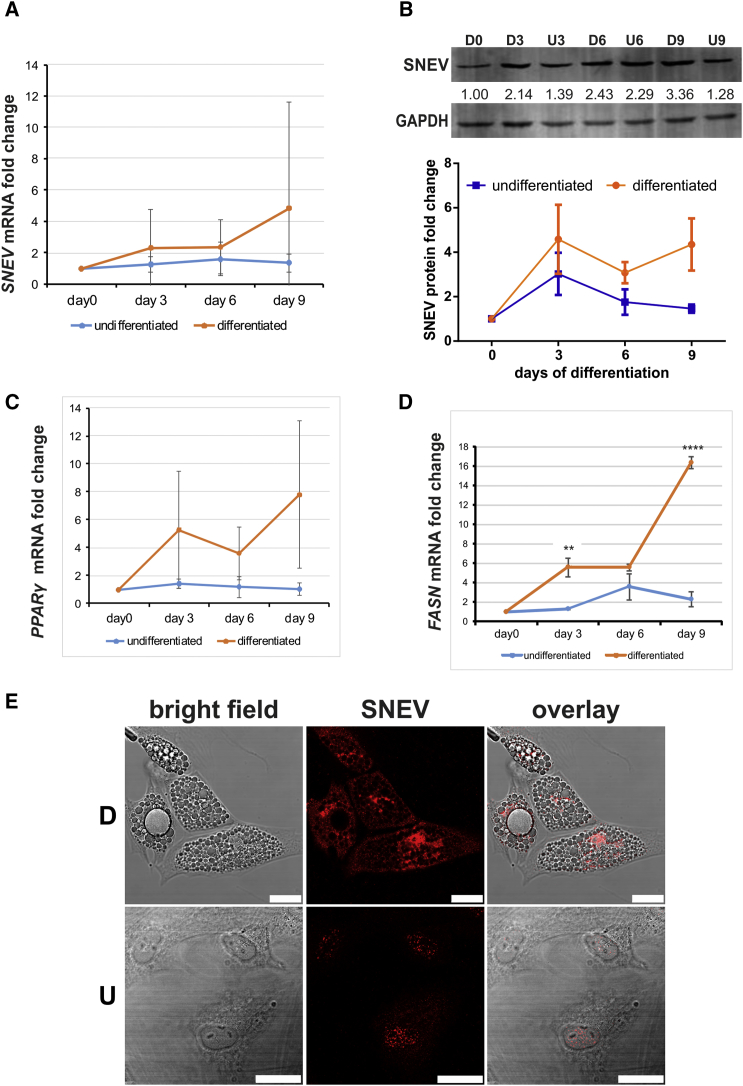
We analyzed SNEV expression on mRNA and protein level during adipogenic differentiation of hASCs at various time points. Adipogenic differentiation was confirmed by oil red O staining after 10 days. Indeed, SNEV mRNA (Figure 1A) as well as protein levels (Figure 1B) increased in a time-dependent manner over 9 days, in line with adipogenic markers PPARγ and FASN (Figures 1C and 1D). In addition, we observed changes in the cellular localization of SNEV from mainly nuclear in undifferentiated controls to also cytoplasmic in differentiated cells (Figure 1E).
Figure 1.
Expression of SNEV Is Induced during Adipogenesis
(A) SNEV mRNA expression in hASCs at different days post-induction of adipogenesis was quantified by RT-qPCR. Data points represent averages from three independent differentiation experiments (donors 803, 812, and 851). Error bars indicate SD. Two-way ANOVA p value of 0.045.
(B) Whole-cell lysate of differentiating (D) and undifferentiated (U) hASCs were submitted to western blotting and probed with anti-SNEV antibody. Anti-GAPDH was used to ensure equal loading. Numbers represent intensities of bands normalized to GAPDH. Upper panel, exemplary western blot. Lower panel, the graph shows average of band intensities normalized to GAPDH derived from three independent differentiation experiments (donors 803, 812, and 851). Error bars indicate SD. Two-way ANOVA p value of 0.032.
(C and D) Adipogenic differentiation was confirmed by qPCRs against adipogenic marker genes PPARγ and FASN. Data points represent the average from three independent differentiation experiments (donors 803, 812, and 851). Error bars indicate SD. Unpaired two-sided Student's t tests were performed to compare differentiated and undifferentiated samples at the same time points: ∗∗∗∗p < 0.0001, ∗∗p < 0.01.
(E) Differentiated (D) and undifferentiated (U) hASCs of donor 812 were stained with anti-SNEV antibody. Scale bar, 25 μm.
SNEV Regulates Adipogenic Differentiation of hASCs by Modulating PPARγ and Insulin Signaling
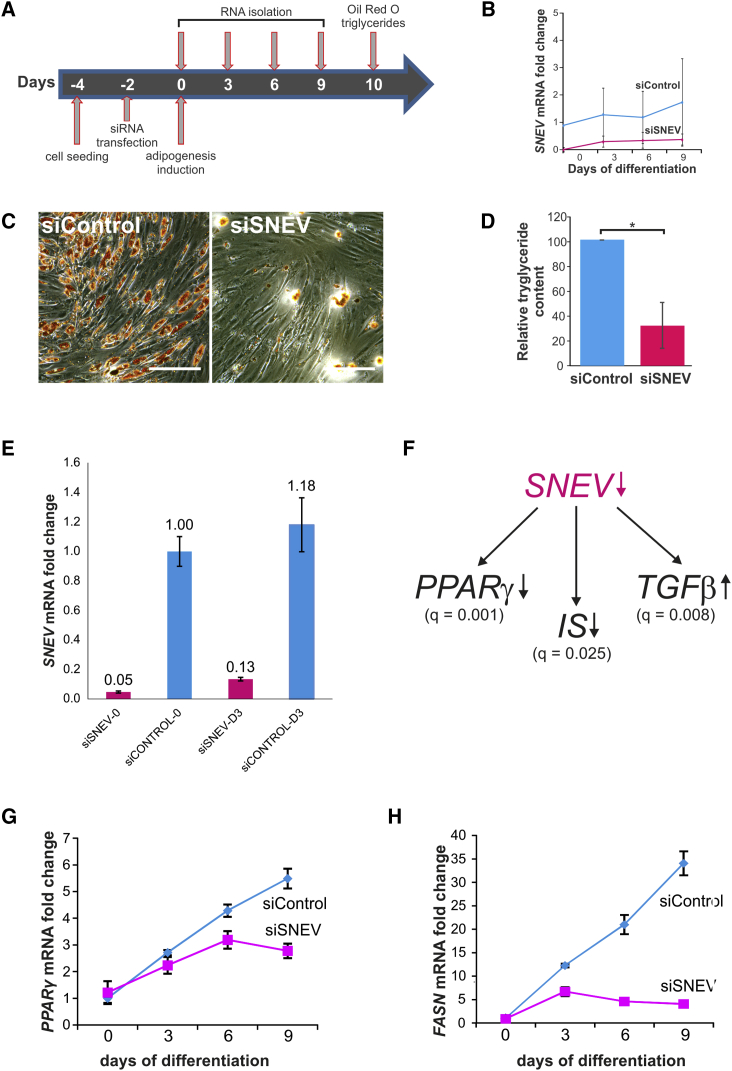
To determine whether SNEV is not only regulated, but also necessary for adipogenenic differentiation, hASCs were transfected with a small interfering RNA (siRNA) pool against SNEV (siSNEV) or a non-targeting control siRNA pool (siControl), and differentiation was induced 48 hr post-transfection (Figure 2A). siSNEV transfection resulted in a 90% knockdown of SNEV mRNA over the entire period of differentiation (Figure 2B). Indeed, this knockdown resulted in formation of fewer lipid droplets in comparison with control, as shown by oil red O staining (Figure 2C) and intracellular triglyceride content (Figure 2D).
Figure 2.
Knockdown of SNEV Inhibits Adipogenic Differentiation of hASCs by Downregulating Adipogenesis-Promoting Signaling Pathways
(A) Schematic representation of experimental design.
(B) Expression of SNEV mRNA during adipogenic differentiation after siSNEV or siControl transfection. Data points represent averages from four independent differentiation experiments (1× donor 803, 2× donor 812, and 1× donor 851). Error bars indicate SD. Two-way ANOVA p value < 0.001.
(C) Oil red O staining for triglyceride content in transfected hASCs after 10 days of differentiation. Scale bar, 100 μm.
(D) Intracellular triglyceride levels at day 10 of differentiation. Triglyceride content was normalized to total protein content and to the control. An unpaired two-sided Student's t tests without assuming homogeneity of variances was performed: ∗p < 0.05. Data points represent average from three independent differentiation experiments (donors 803, 812, and 851). Error bars indicate SD.
(E) RT-qPCR to determine knockdown efficiency in RNA samples subjected to microarrays. Numbers represent the day of harvest. The average of four technical replicates of cells from donor 812 is shown. Error bars indicate SD. Results with cells from donor 803 were similar.
(F) Microarray analysis of transcript expression after SNEV knockdown revealed a downregulation of the pro-adipogenic PPARγ and insulin signaling (IS) pathways and an upregulation of the anti-adipogenic transforming growth factor β (TGF-β) signaling pathway. q Values obtained from gene-set-enrichment analysis are depicted. The analysis was performed on two independent experiments with cells from two different donors (803 and 812).
(G and H) qPCR analysis of PPARγ (G) and FASN (H) confirming the microarray data. Unpaired two-sided Student's t tests were performed to compare control and siSNEV treated samples at the same time points; four technical replicates with cells from donor 851 are shown. Error bars indicate SD. See also Figures S1 and S2 and Table S1.
To investigate how SNEV might inhibit adipogenesis, we performed microarray analysis at day 3 of adipogenic differentiation in response to SNEV knockdown (Figure 2E). Thereby, 163 genes with at least 2-fold differential expression in siSNEV versus siControl were identified. Then, we performed gene-set-enrichment analysis and found that genes involved in the pro-adipogenic PPARγ (Figures 2F and S1A) and insulin signaling (Figures 2F and S1B) pathways were downregulated, whereas genes involved in the anti-adipogenic transforming growth factor β pathway (Figures 2F and S1C) were upregulated, among others (see also Table S1). We further confirmed the microarray data by qPCR of PPARγ (Figure 2G) and FASN (Figure 2H). Collectively, these data suggest that SNEV is necessary at an early step of adipogenesis, as it inhibits global changes of gene transcription that are usually induced by the differentiation process.
Our findings are corroborated by the observation that overexpression of SNEV in hASCs resulted in accelerated adipogenic differentiation (Figure S2). However, this finding strongly depended on the donor-specific differentiation propensity and on the construct used, hence on the precise degree of overexpression. Notably, SNEV overexpression had no inhibitory effect on adipogenesis in any case, while knockdown resulted in reduced lipid droplet accumulation in all cases. In addition, we did not observe obvious effects on osteogenic differentiation (Figures S1D–S1F), which is often seen to increase at the expense of adipogenic differentiation (James, 2013).
SNEV Is Required for Functional DDR in hASCs
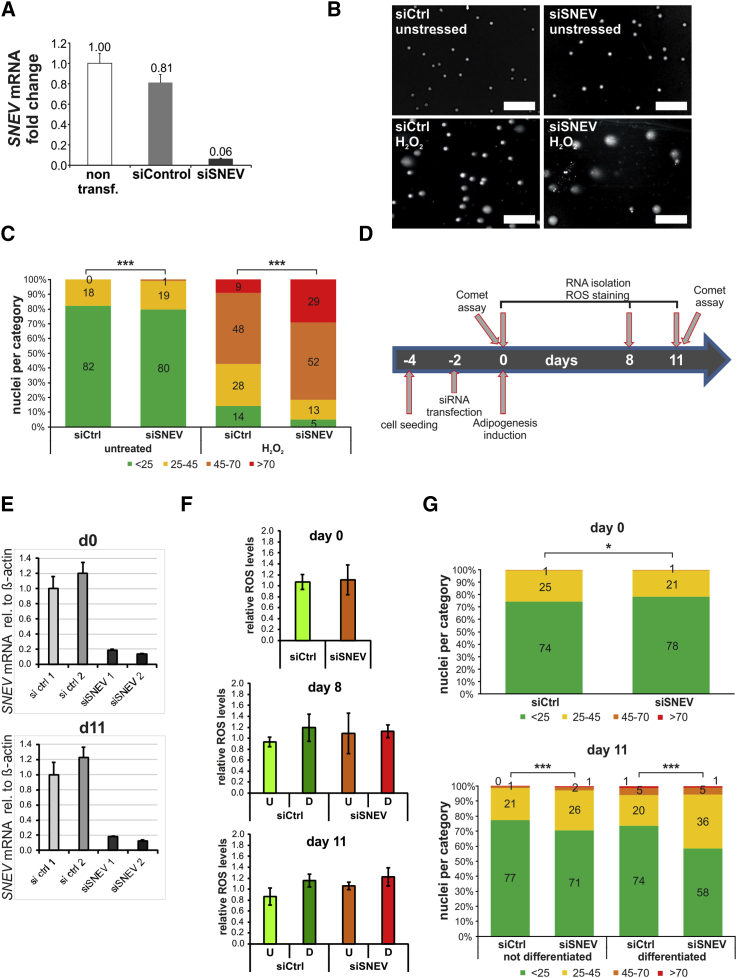
To prove that SNEV is involved also in DNA repair in ASCs, we knocked down SNEV (Figure 3A) and exposed ASCs to reactive oxygen species (ROS) which arise during adipogenesis. DNA damage was assessed by comet assay. Representative images of comet assays are shown in Figure 3B. The resulting comets were quantified by comparing the fluorescence intensity in the head versus the tail as a measure of nuclear DNA damage (Guo et al., 2013) and classified into four categories. Only low levels of DNA damage were detectable without H2O2 treatment in both siSNEV- and siControl-transfected cells. In contrast, upon H2O2 treatment, SNEV knockdown led to a marked increase of cells with high levels of DNA damage compared with siControl (Figure 3C). Manual classification into three categories according to the tail size gave highly similar results (Figure S3). Hence, SNEV is required for DNA repair after free radical damage in hASCs.
Figure 3.
SNEV Is Required for Repair of Oxidative DNA Damage during Adipogenic Differentiation of hASCs
hASCs were transfected with siSNEV or siControl, treated with 500 μM H2O2 for 90 and 60 min recovery, and submitted to comet assay.
(A) Transfection with siSNEV results in 90% reduction of SNEV mRNA expression. The average of four technical replicates of cells from donor 812 is shown. Error bars indicate SD.
(B) Representative images of comet assays. Scale bar, 100 μm.
(C) In hASCs transfected with siSNEV and treated with H2O2, the percentage of cells with high levels of DNA damage triples, while the percentage of cells with low DNA damage drops to one-third compared with control cells after H2O2 treatment. Numbers indicate the percentage of cells in the respective category. Pooled data from three biological replicates are shown (donors 803, 812, and 851). A minimum of 150 cells per condition and replicate were analyzed. Chi-square test was performed to compare results from control and SNEV knockdown: ∗∗∗p < 0.001.
(D) Schematic representation of experimental design. hASC were transfected with SNEV or control siRNAs and submitted to adipogenic differentiation. RNA samples to monitor knockdown and ROS measurements were taken on days 0, 8, and 11 of differentiation. Comet assays were performed on days 0 and 11.
(E) RT-qPCR shows stable knockdown over the course of differentiation.
(F) ROS formation was quantified by H2DCFDA staining. Mean values of three independent differentiation experiments are shown (2× donor 812 and 1× donor 803). Error bars indicate SD. Unpaired two-sided Student's t tests were performed to compare control and siSNEV treated samples and did not reveal statistical differences for any condition.
(G) Upper panel, 48 hr post-transduction, before adipogenic differentiation is induced, Comet assays reveal that DNA damage levels are slightly higher in siControl than siSNEV transfected ASCs. Lower panel, after adipogenic differentiation, SNEV knockdown leads to significant increase of DNA damage. Numbers indicate percentage of cells in the respective category. Pooled data from three independent differentiation experiments are shown (donors 803, 812, and 851). A minimum of 200 cells per condition and replicate were analyzed. Chi-square test was performed to compare results of control and SNEV knockdown: ∗∗∗p < 0.001, ∗p < 0.05. See also Figures S3 and S4.
To examine whether SNEV is necessary to prevent intrinsic DNA damage caused by ROS arising during adipogenic differentiation, we knocked down SNEV in hASCs (Figures 3D and 3E), induced adipogenic differentiation, and monitored formation of ROS and accumulation of DNA damage. On days 8 and 11, we observed elevated ROS levels in differentiating cells, independent of SNEV mRNA levels (Figure 3F). However, comet assays showed a significant increase of DNA damage in siSNEV versus control cells at day 11 of adipogenic differentiation (Figure 3G). Hence, SNEV does not interfere with ROS production during adipogenic differentiation of ASCs, but is required for efficient repair of the DNA damage inflicted by emerging ROS.
To test if the ability to modulate adipogenic differentiation is restricted to SNEV or a general property of DDR factors, we tested if genes mutated in segmental progeroid syndromes influence adipogenesis as well. First, we specifically visualized the expression of genes involved in DDR using existing transcriptomic data of an adipogenic differentiation time-course experiment (available at Gene Expression Omnibus, accession number GEO: GSE64845). From these, we selected WRN, CSA, and XPE for further analysis (Figures S4A–S4D). Mutations of these genes mirror aspects of accelerated aging and represent different DDR pathways, CSA being specific for transcription-coupled nucleotide excision repair (TC-NER), while XPE is involved in global genome- and TC-NER, as well as in homologous recombination. We knocked down WRN, CSA, and XPE in hASCs by siRNA transfection and induced adipogenesis. Knockdown was confirmed by qPCR (Figure S4E), and adipogenic differentiation was assessed by intracellular triglyceride accumulation at day 10 of differentiation. CSA and XPE knockdown reduced intracellular triglycerides slightly, but not statistically significantly, whereas WRN knockdown resulted in a significant reduction by 50% (Figure S4F).
Loss of SNEV and WRN Lead to Reduced Fat Deposition in C. elegans
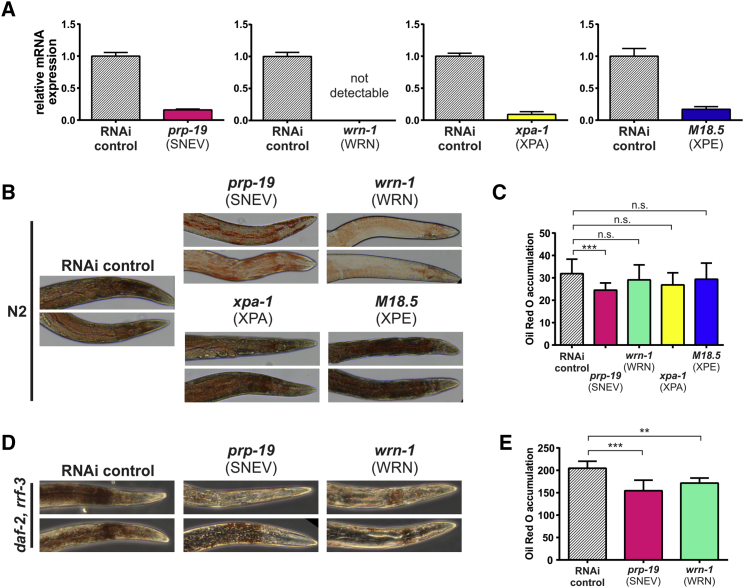
To test the functional conservation of the DDR factors SNEV, WRN, CSA, and XPE in adipogenesis, we assessed their role in fat deposition in C. elegans. For this purpose, we selected prp-19, wrn-1, M18.5, and xpa-1 as orthologs of the human DDR factors SNEV, WRN, XPE, and XPA. Since we did not detect major differences in developmental timing between RNAi-treated and control animals (data not shown), RNAi treatment was performed already upon hatching and led to a downregulation of the target mRNA by 80%–100% (Figure 4A). Young adult hermaphrodites (6 days after hatching) were subjected to oil red O staining to assess neutral lipid storage (Soukas et al., 2009). Indeed, prp-19 RNAi animals exhibited reduced fat mass compared with the RNAi control, while wrn-1, M18.5, and xpa-1 RNAi did not yield significant differences (Figures 4B and 4C).
Figure 4.
Loss of SNEV and WRN Reduces Fat Deposition in C. elegans
(A) Verification of RNAi efficacy upon depletion of prp-19, wrn-1, xpa-1, and M18.5 in wild-type C. elegans by qPCR. Knockdown efficiency was always between 80% and 100%. Error bars represent SEM of four technical replicates.
(B and C) Oil red O staining demonstrates significantly lower fat deposition upon prp-19 knockdown, compared with the empty vector (HT115) control. Representative images (B) and quantification (C) of three biological replicates with at least 20 worms per strain are shown. Error bars represent SEM. ∗∗∗p < 0.001. n.s., not significant.
(D and E) RNAi to prp-19 and wrn-1 in the high-fat and RNAi hypersensitive CF1841 strain, followed by oil red O staining confirms lower fat deposition compared with the empty vector control for both genes. Representative images (D) and quantification (E) of three replicates with at least 20 worms per strain. Error bars represent SEM. ∗∗p < 0.01, ∗∗∗p < 0.001.
We expected to further enhance the observed fat storage phenotype by using the CF1814 strain, which is mutated in rrf-3 and daf-2 and exhibits increased RNAi efficiency (Simmer et al., 2002), as well as elevated fat mass (Soukas et al., 2009). While the RNAi control group stained positive for oil red O throughout the whole body and especially surrounding intestine and pharynx, prp-19 and wrn-1 RNAi worms stained only weakly positive in close proximity to the pharynx (Figures 4D and 4E), now also detecting differences induced by WRN deficiency.
These findings indicate that loss of the conserved DDR factors prp-19 and wrn-1 reduces the accumulation of neutral lipids in C. elegans.
Discussion
SNEV regulates diverse cellular processes, such as mRNA splicing (Grillari et al., 2005), transcription (Chanarat et al., 2012), mitosis (Watrin et al., 2014), apoptosis (Lu et al., 2014), and multiple branches of DNA repair (reviewed in Mahajan, 2016). On the one hand, SNEV catalyzes the formation of polyubiquitin chains on replication protein A associated with single-stranded DNA arising at sites of DNA damage, which enhances ATR-ATRIP recruitment and consequently downstream DDR signaling (Wan and Huang, 2014). On the other hand, SNEV is recruited to RNA polymerase II via U2AF65, and this interaction stimulates co-transcriptional splicing in vitro (David et al., 2011). It is possible that SNEV senses the slowing of RNA polymerase II processivity when it encounters a lesion and acts as signal transducer to attract other DNA repair factors.
Albeit we cannot completely rule out off-target effects of the single siRNA pool we used in this study to deplete SNEV in hASCs, the requirement of SNEV for adipogenesis seems to be evolutionary well conserved, as we observed reduced fat deposition in the nematode C. elegans upon depletion of prp-19 by RNAi.
In addition to our data, several other lines of evidence point to a possible role for the DNA repair function of SNEV in adipogenesis. The loss of multiple stem cell functions with increasing age can be attributed to an accumulation of DNA damage (reviewed in Behrens et al., 2014), and hematopoietic stem cell self-renewal was already shown to be diminished after DNA damage (Wang et al., 2012), supporting the idea that the capacity of repairing DNA damage is a general checkpoint before differentiation.
But why does reduced DNA repair upon SNEV knockdown specifically block adipogenic, but not osteogenic differentiation? One hypothesis is based on the fact that ROS are specifically formed during adipogenic differentiation and are thought to reinforce pro-adipogenic signaling pathways (Kanda et al., 2011), while osteogenic differentiation is accompanied by a reduction of intracellular ROS (Atashi et al., 2015). Indeed, we observed accumulation of DNA damage in hASCs after SNEV knockdown, both after acute hydrogen peroxide treatment and after endogenous ROS accumulation resulting from adipogenic differentiation. Therefore, we suggest that only cells with sufficient DDR capacity might be allowed to enter adipogenic differentiation, as DDR is induced during early adipogenesis (Meulle et al., 2008). This lack of adipogenic differentiation together with adipose tissue degeneration induced by senescent adipocytes might contribute to the subcutaneous fat loss in segmental progeroid syndrome patients (Martin and Oshima, 2000) and mouse models of premature aging, such as ERCC1-, Csb-, or Xpa-deficient mice (Jaarsma et al., 2013, Kamenisch et al., 2010).
However, we cannot rule out the possibility that SNEV might play a more direct role during early adipogenic commitment of hASCs, e.g., via differential pre-mRNA splicing. Also, a scenario involving p21 regulation by SNEV is possible. After inducing adipogenesis in preadipocytes, cells undergo a transient increase in DNA synthesis, followed by an arrest in the G1 phase, which is characterized by an increase in p21 protein levels (Reichert and Eick, 1999). Interestingly, the SNEV/Cdc5L complex is recruited to the p21 gene and mRNA and is specifically required for protein expression of p21, but not of pro-apoptotic p53-targets (Chen et al., 2011).
To summarize, we suggest that availability of DDR might represent a checkpoint for cellular differentiation programs, which is of special importance for differentiation processes that involve high levels of ROS, or for long-lived cells such as adipocytes.
Experimental Procedures
Experimental details can be found in the Supplemental Experimental Procedures.
hASC Cultivation
Human subcutaneous adipose tissues were obtained from three different donors by liposuction (Table S2). Informed consent of the donors was obtained and therefore this study was performed according to the Declaration of Helsinki and approved by the local ethics commission (Ethikkommission des Landes Oberoesterreich). hASCs were isolated and cultivated as described by Wolbank et al. (2009) and characterized for surface markers (Table S3). Adipogenic and osteogenic differentiation, as well as oil red O and alizarin red staining were performed as described by Schosserer et al. (2015). Triglycerides were quantified by an Infinity Triglyceride Quantification Kit (Thermo Scientific) and normalized to total protein concentration as measured using the BCA Kit (Thermo Scientific).
siRNA against SNEV (ON-TARGETplus Human PRPF19 [27339] siRNA, SMARTpool) and control siRNA (ON-TARGETplus Non-targeting Pool) were purchased from Thermo Scientific. siRNAs against WRN, CSA, and XPE and a non-targeting control were purchased from Riboxx Pharmaceuticals.
A total of 14,000 hASCs was seeded into 1.9 cm2 plates. Forty-eight hours post-seeding, cells were transfected with siRNA using 50 nM of the respective siRNAs and DharmaFECT1 Transfection Reagent (Thermo Scientific).
Total RNA was extracted using TRIzol Reagent (Life Technologies). RNA concentration was on a NanoDrop ND-1000 spectrophotometer. cDNA was synthesized using 500 ng total RNA with the DyNAmo cDNA Synthesis Kit (Thermo Scientific). qPCR was performed using HOT FIREPol EvaGreen qPCR Supermix (Solis BioDyne). Primers for qPCR are listed in Table S4. mRNA expression was normalized to GAPDH.
SNEV cDNA was amplified by PCR and cloned into the retroviral plasmid pLenti6. The negative control vector contained only the blasticidin resistance gene. Retroviral particles were generated according to the manufacturer's protocol (Life Technologies). A total of 14,000/1.9 cm2 hASCs were seeded in growth medium and, after 48 hr, infected with retroviral particles at an MOI of 2 in DMEM (4.5 g/L glucose), supplemented with 4 mM L-glutamine, 10% fetal calf serum, 1 ng/mL basic fibroblast growth factor, and 8 μg/mL polybrene. The medium was replaced with the same medium without polybrene 24 hr post-transduction and with adipogenesis-inducing medium 48 hr post-transduction.
Microarray Analysis
Global gene expression analysis was performed by two-color microarrays for hASCs upon SNEV knockdown (donors 803 and 812 as independent biological replicates), as well as for human multipotent adipose-derived stem cells at various stages of adipocyte differentiation. Metadata (experimental parameters and detailed procedures), raw data files, and final (filtered and normalized) data are accessible via Gene Expression Omnibus (GEO: GSE64937 and GSE64845).
Western Blotting and Immunofluorescence
hASCs were harvested in radioimmunoprecipitation assay buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, and 1× protease and phosphatase inhibitor [Roche]), sonicated for 30 cycles (30 s on and 30 s off) at 4°C using a Bioruptor sonicator (Diagenode) and centrifuged for 30 min at 10,000 rpm. Protein concentration in the supernatant was determined using the BCA Kit (Thermo Scientific). Protein (30 μg) was mixed with 4× SDS loading dye (240 mM Tris-Cl [pH 6.8], 8% SDS, 40% glycerol, 0.05% bromophenol blue, and 5% β-mercaptoethanol), heated to 95°C for 10 min and submitted to SDS-PAGE and western blotting. SDS-PAGE and western blotting were carried out as described previously (Dellago et al., 2012).
At day 10, cells were fixed in paraformaldehyde and processed for immunofluorescence as described previously (Schosserer et al., 2015); antibody details can be found in the Supplemental Information.
Quantification of ROS
Adipogenic differentiation was induced 48 hr post-transfection as described above. On days 0, 8, and 11 of adipogenic differentiation, cells were harvested, resuspended in PBS containing 10 μM 2′,7'-dichlorodihydrofluorescein diacetate (H2DCFDA; Life Technologies) and incubated for 30 min at room temperature in the dark. After incubation, cells were put on ice and fluorescence was measured on a Gallios Flow Cytometer (Beckman Coulter).
Comet Assay
Undifferentiated hASCs were transfected with siSNEV and siControl as described above. Two days after transfection, cells were treated with 500 μM hydrogen peroxide (Sigma) in growth medium for 90 min. After 60 min recovery in growth medium, cells were harvested and processed for comet assays (Wojewódzka et al., 2002) and quantified as described previously (Guo et al., 2013).
C. elegans
C. elegans strains were cultured at 20°C under standard laboratory conditions on nematode growth medium agar as described previously (Brenner, 1974). Worms were synchronized by timed egg-lay on fresh RNAi plates and transferred to FUdR (Sigma)-containing plates upon adulthood. For knockdown, worms were fed double-stranded RNA expressed in bacteria as described previously (Timmons et al., 2001). RNAi constructs against prp-19, wrn-1, xpe-1, and M18.5 (xpa), derived from J. Ahringer's RNAi library, were obtained from Source BioScience. Oil red O staining was performed 6 days after hatching as described previously (Soukas et al., 2009). Stained worms were embedded in Mowiol and pictures were taken on a Leica DM IL LED inverted microscope with a 10× dry objective and staining was quantified as described previously (Yen et al., 2010). For qPCR, 30–40 worms were rinsed off plates and washed with S Basal, precipitated by gravity, and homogenized in Trizol using a pellet pestle. Samples were then processed as described for hASCs.
Statistical Analysis
Differences between datasets were tested for statistical significance using multiple-comparison adjusted Student's t tests, one-way ANOVA, or two-way ANOVA implemented in Prism QuickCalcs (GraphPad), and p < 0.05 was considered statistically significant. All error bars represent SDs of the mean if not indicated otherwise. For comet assays, counts from three donors were pooled, classified into categories, and analyzed by chi-square test.
Author Contributions
A.K., H.D., R.G.V., M.Scho., and J.G. designed the experiments; A.K., H.D., M.Scho., V.S., and L.T. performed the experiments with ASCs and analyzed the data; M.K. and M.Sche. designed and performed the microarrays and analyzed the data; S.W., F.H., A.H., and C.G. isolated and characterized the mesenchymal stem cells; C.M. and P.J.D. provided the lentiviral particles; M.Scho. performed the experiments with C. elegans and analyzed the data; A.K., H.D., M.K., M.Scho., and J.G. designed the figures and wrote the manuscript. All authors read and corrected the manuscript.
Acknowledgments
We are grateful to J. Binder, D. Pulferer, C. Heissenberger, and M. Mur for technical assistance. C. elegans strains were provided by the CGC, funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank the BOKU-VIBT Imaging Center for technical support with microscopy. This work was supported by the Austrian Science Fund (FWF, I2514 and P24498), by the Austrian Federal Ministry of Science, Research and Economy, and by the National Foundation for Research, Technology and Development; by the Higher Education Commission (HEC) of Pakistan as well as by the Christian Doppler Research Society. The financial support by the Austrian Federal Ministry of Economy, Family and Youth and the National Foundation for Research, Technology and Development is also gratefully acknowledged. J.G. and R.G.V. are founders and shareholders of Evercyte GmbH.
Published: December 29, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.12.001.
Contributor Information
Markus Schosserer, Email: markus.schosserer@boku.ac.at.
Johannes Grillari, Email: johannes.grillari@boku.ac.at.
Supplemental Information
References
- Abbas M., Shanmugam I., Bsaili M., Hromas R., Shaheen M. The role of the human psoralen 4 (hPso4) protein complex in replication stress and homologous recombination. J. Biol. Chem. 2014;289:14009–14019. doi: 10.1074/jbc.M113.520056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahima R.S. Connecting obesity, aging and diabetes. Nat. Med. 2009;15:996–997. doi: 10.1038/nm0909-996. [DOI] [PubMed] [Google Scholar]
- Atashi F., Modarressi A., Pepper M.S. The role of reactive oxygen species in mesenchymal stem cell adipogenic and osteogenic differentiation: a review. Stem Cells Dev. 2015;24:1150–1163. doi: 10.1089/scd.2014.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A., van Deursen J.M., Rudolph K.L., Schumacher B. Impact of genomic damage and ageing on stem cell function. Nat. Cell Biol. 2014;16:201–207. doi: 10.1038/ncb2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caso G., McNurlan M.A., Mileva I., Zemlyak A., Mynarcik D.C., Gelato M.C. Peripheral fat loss and decline in adipogenesis in older humans. Metabolism. 2013;62:337–340. doi: 10.1016/j.metabol.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanarat S., Burkert-Kautzsch C., Meinel D.M., Sträßer K. Prp19C and TREX: interacting to promote transcription elongation and mRNA export. Transcription. 2012;3:8–12. doi: 10.4161/trns.3.1.19078. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang L., Jones K.A. SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev. 2011;25:701–716. doi: 10.1101/gad.2002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.Y., Shin E.S., Park P.J., Shin D.W., Chang H.K., Kim D., Lee H.H., Lee J.H., Kim S.H., Song M.J. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J. Biol. Chem. 2007;282:2456–2465. doi: 10.1074/jbc.M608042200. [DOI] [PubMed] [Google Scholar]
- David C.J., Boyne A.R., Millhouse S.R., Manley J.L. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellago H., Khan A., Nussbacher M., Gstraunthaler A., Schosserer M., Mück C., Anrather D., Scheffold A., Ammerer G., Dürr P.J. ATM-dependent phosphorylation of SNEVhPrp19/hPso4 is involved in extending cellular life span and suppression of apoptosis. Aging (Albany NY) 2012;4:290–304. doi: 10.18632/aging.100452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillari J., Ajuh P., Stadler G., Löscher M., Voglauer R., Ernst W., Chusainow J., Eisenhaber F., Pokar M., Fortschegger K. SNEV is an evolutionarily conserved splicing factor whose oligomerization is necessary for spliceosome assembly. Nucleic Acids Res. 2005;33:6868–6883. doi: 10.1093/nar/gki986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Hanawalt P.C., Spivak G. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013;41:7700–7712. doi: 10.1093/nar/gkt524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaarsma D., van der Pluijm I., van der Horst G.T.J., Hoeijmakers J.H.J. Cockayne syndrome pathogenesis: lessons from mouse models. Mech. Ageing Dev. 2013;134:180–195. doi: 10.1016/j.mad.2013.04.003. [DOI] [PubMed] [Google Scholar]
- James A.W. Review of signaling pathways governing MSC osteogenic and adipogenic differentiation. Scientifica (Cairo) 2013;2013:684736. doi: 10.1155/2013/684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenisch Y., Fousteri M., Knoch J., von Thaler A.-K., Fehrenbacher B., Kato H., Becker T., Dollé M.E.T., Kuiper R., Majora M. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda Y., Hinata T., Kang S.W., Watanabe Y. Reactive oxygen species mediate adipocyte differentiation in mesenchymal stem cells. Life Sci. 2011;89:250–258. doi: 10.1016/j.lfs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Karakasilioti I., Kamileri I., Chatzinikolaou G., Kosteas T., Vergadi E., Robinson A.R., Tsamardinos I., Rozgaja T.A., Siakouli S., Tsatsanis C. DNA damage triggers a chronic autoinflammatory response, leading to fat depletion in NER progeria. Cell Metab. 2013;18:403–415. doi: 10.1016/j.cmet.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löscher M., Fortschegger K., Ritter G., Wostry M., Voglauer R., Schmid J.A., Watters S., Rivett A.J., Ajuh P., Lamond A.I. Interaction of U-box E3 ligase SNEV with PSMB4, the beta7 subunit of the 20 S proteasome. Biochem. J. 2005;388:593–603. doi: 10.1042/BJ20041517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Wang R., Cai C., Liang J., Xu L., Miao S., Wang L., Song W. Small kinetochore associated protein (SKAP) promotes UV-induced cell apoptosis through negatively regulating Pre-mRNA processing factor 19 (Prp19) PLoS One. 2014;9:e92712. doi: 10.1371/journal.pone.0092712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan K. hPso4/hPrp19: a critical component of DNA repair and DNA damage checkpoint complexes. Oncogene. 2016;35:2279–2286. doi: 10.1038/onc.2015.321. [DOI] [PubMed] [Google Scholar]
- Mahajan K.N., Mitchell B.S. Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc. Natl. Acad. Sci. USA. 2003;100:10746–10751. doi: 10.1073/pnas.1631060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.M., Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408:263–266. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- Meulle A., Salles B., Daviaud D., Valet P., Muller C. Positive regulation of DNA double strand break repair activity during differentiation of long life span cells: the example of adipogenesis. PLoS One. 2008;3:e3345. doi: 10.1371/journal.pone.0003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Xu Z., Zhang X., Wang L., Gimble J.M., Lander E.S., Rosen E.D. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteforte R., Beilhack G.F., Grausenburger R., Mayerhofer B., Bittner R., Grillari-Voglauer R., Sibilia M., Dellago H., Tschachler E., Gruber F., Grillari J. SNEV(Prp19/PSO4) deficiency increases PUVA-induced senescence in mouse skin. Exp. Dermatol. 2016;25:212–217. doi: 10.1111/exd.12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Murano S., Yokote K., Takemoto M., Asaumi S., Take A., Saito Y. Enhanced intra-abdominal visceral fat accumulation in patients with Werner’s syndrome. Int. J. Obes. Relat. Metab. Disord. 2001;25:292–295. doi: 10.1038/sj.ijo.0801529. [DOI] [PubMed] [Google Scholar]
- Muzumdar R., Allison D.B., Huffman D.M., Ma X., Atzmon G., Einstein F.H., Fishman S., Poduval A.D., McVei T., Keith S.W., Barzilai N. Visceral adipose tissue modulates mammalian longevity. Aging Cell. 2008;7:438–440. doi: 10.1111/j.1474-9726.2008.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M., Eick D. Analysis of cell cycle arrest in adipocyte differentiation. Oncogene. 1999;18:459–466. doi: 10.1038/sj.onc.1202308. [DOI] [PubMed] [Google Scholar]
- Schosserer M., Reynoso R., Wally V., Jug B., Kantner V., Weilner S., Buric I., Grillari J., Bauer J.W., Grillari-Voglauer R. Urine is a novel source of autologous mesenchymal stem cells for patients with epidermolysis bullosa. BMC Res. Notes. 2015;8:767. doi: 10.1186/s13104-015-1686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S.P., Nonet M.L., Fire A., Ahringer J., Plasterk R.H.A. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Song E.J., Werner S.L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J.W., Elledge S.J., Kirschner M.W., Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24:1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas A.A., Kane E.A., Carr C.E., Melo J.A., Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T., Morbeck D.E., Von Zglinicki T., Van Deursen J., Lustgarten J., Scrable H., Khosla S., Jensen M.D., Kirkland J.L. Fat tissue, aging, and cellular senescence. Aging Cell. 2010;9:667–684. doi: 10.1111/j.1474-9726.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D.L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Turaga R.V.N., Paquet E.R., Sild M., Vignard J., Garand C., Johnson F.B., Masson J.-Y., Lebel M. The Werner syndrome protein affects the expression of genes involved in adipogenesis and inflammation in addition to cell cycle and DNA damage responses. Cell Cycle. 2009;8:2080–2092. doi: 10.4161/cc.8.13.8925. [DOI] [PubMed] [Google Scholar]
- Voglauer R., Chang M.W.-F., Dampier B., Wieser M., Baumann K., Sterovsky T., Schreiber M., Katinger H., Grillari J. SNEV overexpression extends the life span of human endothelial cells. Exp. Cell Res. 2006;312:746–759. doi: 10.1016/j.yexcr.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Wan L., Huang J. The PSO4 complex associates with RPA and modulates the activation of ATR. J. Biol. Chem. 2014;289:6619–6626. doi: 10.1074/jbc.M113.543439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sun Q., Morita Y., Jiang H., Groß A., Lechel A., Hildner K., Guachalla L.M., Gompf A., Hartmann D. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Watrin E., Demidova M., Watrin T., Hu Z., Prigent C. Sororin pre-mRNA splicing is required for proper sister chromatid cohesion in human cells. EMBO Rep. 2014;15:1–8. doi: 10.15252/embr.201438640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojewódzka M., Buraczewska I., Kruszewski M. A modified neutral comet assay: elimination of lysis at high temperature and validation of the assay with anti-single-stranded DNA antibody. Mutat. Res. 2002;518:9–20. doi: 10.1016/s1383-5718(02)00070-0. [DOI] [PubMed] [Google Scholar]
- Wolbank S., Stadler G., Peterbauer A., Gillich A., Karbiener M., Streubel B., Wieser M., Katinger H., van Griensven M., Redl H. Telomerase immortalized human amnion- and adipose-derived mesenchymal stem cells: maintenance of differentiation and immunomodulatory characteristics. Tissue Eng. Part A. 2009;15:1843–1854. doi: 10.1089/ten.tea.2008.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K., Le T.T., Bansal A., Narasimhan S.D., Cheng J.-X., Tissenbaum H.A. A comparative study of fat storage quantitation in nematode Caenorhabditis elegans using label and label-free methods. PLoS One. 2010;5:e12810. doi: 10.1371/journal.pone.0012810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.X., Kaur R., Lu X.Y., Shen X., Li L., Legerski R.J. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem. 2005;280:40559–40567. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.