Summary
Inadequate silencing of exogenous genes represents a major obstacle to complete epigenetic reprogramming of porcine-induced pluripotent stem cells (piPSCs) by conventional pluripotency transcription factors (OSKM). We tested the hypothesis that epigenetic modification by active DNA or histone demethylation or by inhibition of histone deacetylase would enhance reprogramming and exogenous gene silencing in piPSCs. piPSCs induced by OSKM in combination with epigenetic factors, specifically Ten-Eleven Translocation (Tet1 or Tet3) or lysine (K)-specific demethylase 3A (Kdm3a), expressed higher levels of Rex1 and other genes representing naive state and exhibited more open chromatin status, compared with those of OSKM controls. Tet1 also improved differentiation capacity. Conversion with inhibitors of histone deacetylases (HDACi), NaB, TSA, or VPA, further increased Rex1 expression, while decreasing expression of exogenous genes. piPSCs induced by Tet1+OSKM followed by conversion with HDACi show high pluripotency. Together, epigenetic modifiers enhance generation of piPSCs and reduce their reliance on exogenous genes.
Keywords: iPSC, epigenetics, reprogramming, porcine
Highlights
-
•
Epigenetic modifiers facilitate induction and quality of porcine iPSCs
-
•
Tet1, Tet3, or Kdm3a increases naive pluripotency network in association with Rex1
-
•
Unlike cytoplasmic Rex1, nuclear expression of Rex1 is associated with high pluripotency
-
•
HDAC inhibitors further activate Rex1 and reduce reliance on the exogenous genes
In this article, Liu and colleagues show that epigenetic modifiers, Tet1, Tet3, or Kdm3a can significantly enhance induction and pluripotency of porcine iPSCs by conventional Yamanaka factors, as demonstrated by increased expression levels of Rex1 in the nuclei and its pluripotency network, and by teratoma formation test. Moreover, expression of exogenous genes is decreased by further treatment with HDAC inhibitor.
Introduction
Somatic cells can be reprogrammed to pluripotency through the ectopic expression of defined factors such as Oct4, Sox2, Klf4, and c-Myc (Takahashi et al., 2007, Takahashi and Yamanaka, 2006). The resultant induced pluripotent stem cells (iPSCs) provide unlimited cell sources in studying disease and in regenerative medicine (Yamanaka, 2012). Pigs show many similarities to humans in organ anatomy, physiology, and metabolism (Vodicka et al., 2005) and could be a suitable source of xenotransplantation and a model for the study of human diseases (Giraud et al., 2011). Derivation of pig iPSCs can complement research on human iPSCs (Montserrat et al., 2010). With fewer ethical concerns, robust transplantation experiments using pigs as a model could test the safety and effectiveness of iPSCs in pre-clinical translational medicine, such as retinal (Zhou et al., 2011) and myocardial therapy (Li et al., 2013).
Currently, the main obstacle in achieving fully reprogrammed porcine iPSCs (piPSCs) is inadequate silencing of exogenous genes (Esteban et al., 2009, Ezashi et al., 2009, Fujishiro et al., 2013, Montserrat et al., 2012, West et al., 2010), even after using non-integrating episomal plasmids (Du et al., 2015) or Sendai virus (Congras et al., 2016).
Epigenetic modification by active DNA demethylation or histone demethylation has been shown to facilitate iPSC induction in human and mouse (Huangfu et al., 2008a, Huangfu et al., 2008b). Tet1, a DNA methylcytosine dioxygenase, can facilitate iPSC induction by promoting Oct4 demethylation and re-activation, and even can replace Oct4 to initiate somatic cell reprogramming (Gao et al., 2013). H3K9me3 acts as a block to pluripotency, and Kdm3a/Jmjd1a as a histone H3K9 demethylase, or vitamin C that also can demethylate the histones, enhances reprogramming (Chen et al., 2013, Ma et al., 2008). Tet3 is another dioxygenase of Tet enzymes, and Tet3-mediated DNA hydroxylation is involved in epigenetic reprogramming of the zygotic paternal DNA (Gu et al., 2011); however, it has not been determined whether Tet3 can facilitate iPSC generation.
Recently, a new pluripotent state, called “F-class iPSCs” was found (Hussein et al., 2014, Tonge et al., 2014). The F-class iPSCs is at a Nanog-positive cell state that is stable, occurs frequently, and is dependent on high expression of reprogramming factors, and these cells do not form typical embryonic stem cell (ESC)-like colonies. The F-class cells express significantly reduced levels of many PluriNet genes (Muller et al., 2008), including Dnmt3b, Rex1 (Zfp42), and Tdgf1 (Cripto). Nevertheless, they also express many genes at ESC levels such as Sall4, endogenous Oct4, and Nanog. After treatment with histone deacetylase inhibitors (HDACi), sodium butyrate (NaB), or trichostatin A (TSA), these cells could be converted to transgene-independent ESC-like cells capable of contributing to chimeras and the germline (Tonge et al., 2014). We speculated that piPSCs might resemble F-class iPSCs to some extent given that the exogenous genes of piPSCs are not silenced, the edges of early piPSCs appear fuzzy, and piPSCs express relatively high levels of endogenous Oct4 and Nanog, but low level of Rex1 as one of important naive state marker genes (as seen below).
We tested whether epigenetic factors, including Tet3, Tet1, and Kdm3a, or small molecules that increase histone acetylation, could enhance epigenetic reprogramming and silencing of the exogenous genes in piPSCs.
Results
Epigenetic Regulatory Factors Activate Rex1
Rex1 is a naive pluripotent state marker (Nichols and Smith, 2009), and its expression has been positively linked to increased pluripotency in both mouse (Okita et al., 2007, Toyooka et al., 2008) and human ESCs and iPSCs (Brivanlou et al., 2003, Chan et al., 2009). Rex1 also is expressed in the inner cell mass of blastocyst and in trophectoderm cells or trophoblast-derived tissues during mouse and porcine embryo development (Liu et al., 2015, Rogers et al., 1991). Under certain conditions, piPSCs acquire features of naive pluripotency, characterized by expression of Rex1 and Stella (Rodriguez et al., 2012). However, pig epiblast stem cell lines (pEpiSC) do not express Rex1 (Alberio et al., 2010). We also found that piPSCs expressing Rex1 (Rex1+) showed higher expression levels of many genes associated with pluripotency, including E-cadherin, Utf1, Dppa2, Cripto, Eras, Cdc20, Nr6a1, Tert, and Terc, compared with piPSCs with minimal Rex1 (Rex1−) (Figure S1A). Moreover, Rex1+ piPSCs also expressed high levels of genes related to pluripotency regulation network in association with Rex1 (Wang et al., 2006), such as Nac1, Wdr18, Prmt1, Cdk1, and Arid3a (Figure S1B). Together, high expression levels of Rex1 can mark high pluripotency of piPSC lines.
To activate Rex1 and to promote the silence of exogenous genes of piPSCs, we overexpressed epigenetic regulatory factors, including Tet1, mouse Tet3 (mTet3) available, or Kdm3a, together with OSKM (4F) to generate piPSCs from porcine embryonic fibroblasts (PEF) as precursor cells. To achieve mouse ESC/iPSC-like colonies, we employed the conditions for mouse iPSC induction by adding human leukemia inhibitory factor but without basic fibroblast growth factor, and with NaB (Zhu et al., 2010), S-adenosylhomocysteine (Jeon et al., 2008), and BIX01294 (Shi et al., 2008). These small molecules are known to enhance induction and quality of mouse and human iPSCs.
To confirm the presence of the introduced epigenetic regulatory factors, cells were tested on day 9 of reprogramming for the expression of epigenetic factors. As expected, PEF infected by OSKM together with epigenetic factors expressed high levels of the corresponding factors (Figure S2A). Moreover, Rex1 was consistently elevated in the piPSC lines induced by 4F + Tet1, and variably activated in piPSC clones generated by addition of other epigenetic regulation factors (Figures S2B–S2E). Expression levels of Rex1 positively correlated with those of exogenous epigenetic regulatory factors (Figures S2B–S2E).
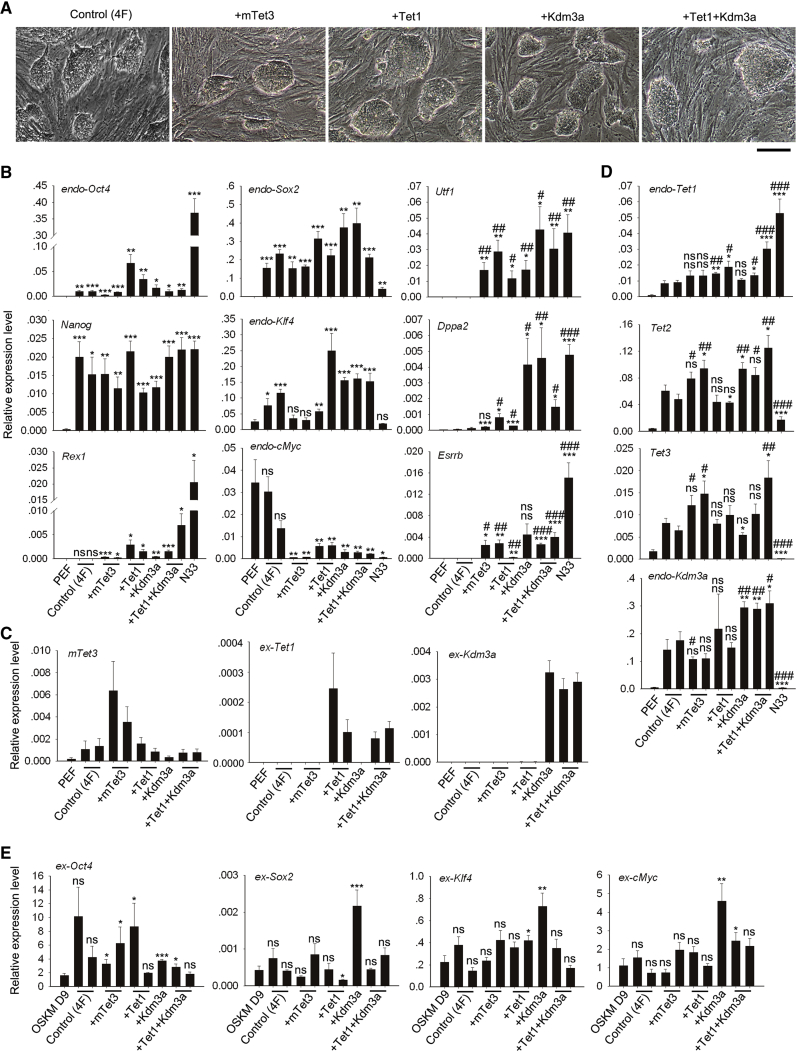
piPSCs were successfully generated from OSKM (4F, control), 4F + mTet3, 4F + Tet1, 4F + Kdm3a, and 4F + Tet1+Kdm3a. piPSC colonies derived by OSKM with epigenetic factors appeared as round and dome-shaped in contrast to the flattened shape formed by OSKM alone by day 15 (Figure S1C). piPSC clones induced by OSKM were loosened and their boundaries were fuzzy while piPSC clones induced by OSKM with epigenetic factors were compact with visible boundaries (Figure S1C). By randomly picking up a number of colonies, we obtained piPSC clones that resembled typical mouse ESCs in morphology, characterized by dome-shaped compact colonies with large nuclei and clear nucleoli in the cells, distinct from feeder fibroblasts (Figure 1A). Based on relatively high expression levels of Rex1, we further characterized the pluripotency of numerous piPSC lines, including two 4F + mTet3, two 4F + Tet1, one 4F + Kdm3a, and two 4F + Tet1+Kdm3a.
Figure 1.
Generation and Characterization of piPSCs Induced by OSKM or OSKM in Combination with Epigenetic Regulatory Factors
(A) Representative images showing piPSCs induced by various conditions. Scale bar, 100 μm.
(B) RT-qPCR analysis of endogenous pluripotent genes. Left and middle, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with PEF. Right, ∗/#p < 0.05, ∗∗/##p < 0.01, ∗∗∗/###p < 0.001, compared with control (4F) piPSC lines. Mouse ESCs (N33) served as positive control.
(C and D) RT-qPCR analysis of exogenous (C) and endogenous (D) epigenetic regulatory genes. p values as in (B).
(E) RT-qPCR analysis of exogenous genes. OSKM at day 9 (OSKM D9) served as positive control for exogenous genes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with OSKM D9. Data represent mean ± SEM from three independent experiments. ns, not significant.
See also Figures S1 and S2.
piPSCs Induced by Epigenetic Regulatory Factors Express High Levels of Genes Associated with Pluripotency
RT-qPCR analysis revealed that expression levels of endogenous Oct4 and Nanog in piPSCs were much higher than those of PEF (Figure 1B, left). Expression levels of endo-Oct4 were also higher in piPSC lines induced by OSKM with Tet1 (Figure 1B, left), consistent with the report that Tet1 can activate Oct4 (Gao et al., 2013), while endo-Oct4 expression levels in piPSC lines induced by other epigenetic factors were similar to those of OSKM controls (Figure 1B, left). Expression levels of Nanog did not differ among piPSC lines induced by OSKM with epigenetic factors (Figure 1B, left). In addition, endo-Sox2 and endo-Klf4 were also activated and expression of endo-cMyc reduced to some extent in piPSCs (Figure 1B, middle).
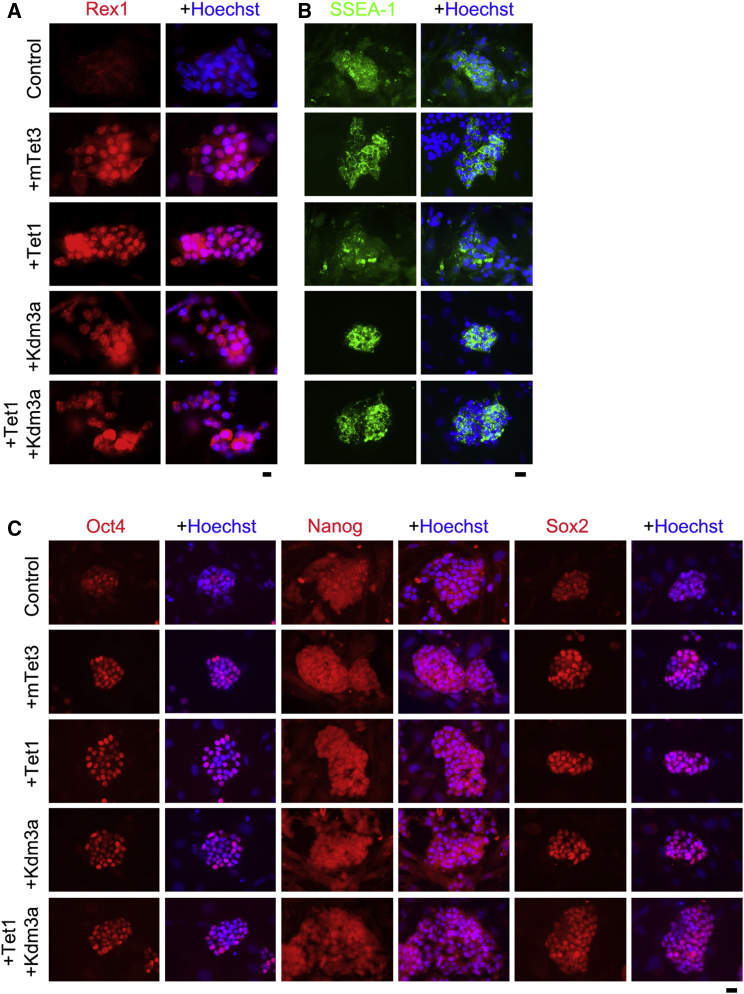
Furthermore, piPSC lines expressed higher levels of Rex1 induced by epigenetic factors, compared with 4F control (Figure 1B, left). Tet1 and Tet1+Kdm3a appeared to be more effective in activating Rex1 (Figure 1B, left). Notably, immunofluorescence microscopy showed trans-localization of Rex1 from the cytoplasm in piPSCs induced by 4F control to the nuclei in piPSCs derived by addition of epigenetic factors (Figure 2A). The nuclear localization of Rex1 may suggest its function in these piPSCs. Western blotting also revealed variable expression levels of Rex1 protein in piPSC lines induced by 4F control and addition of epigenetic factors (Figure S3A).
Figure 2.
Characterization of piPSCs by Immunofluorescence Microscopy
(A) Translocation of Rex1 from cytoplasm to nuclei in piPSCs induced by epigenetic factors.
(B) piPSCs induced by leukemia inhibitory factor without basic fibroblast growth factor express SSEA-1.
(C) Expression of pluripotent marker genes, Oct4, Nanog, and Sox2 in nuclei of piPSCs. Scale bars, 10 μm (A) or 20 μm (B) and (C). See also Figure S3.
In addition, piPSCs induced by OSKM with epigenetic factors expressed higher levels of other genes important for naive state, such as Utf1, Dppa2, and Esrrb (Valamehr et al., 2014) (Figure 1B, right), relative to piPSCs induced by OSKM alone. All three epigenetic factors were able to activate Utf1 and Dppa2, while mTet3, Tet1, and Tet1+Kdm3a significantly activated Esrrb, compared with 4F-piPSCs. Higher expression levels of Rex1, Utf1, Dppa2, and Esrrb in piPSCs generated by additional epigenetic modifiers suggest their pluripotency toward naive state.
The exogenous epigenetic factors were able to activate their corresponding endogenous genes. Expression levels of endo-Tet1 were much higher in piPSCs induced by OSKM in combination with Tet1 or Tet1+Kdm3a (Figure 1D). Expression levels of Tet2 were also higher in piPSCs induced by epigenetic factors except for two 4F + Tet1 piPSC lines (Figure 1D). Expression levels of Tet3 in 4F + mTet3 and 4F + Tet1+Kdm3a piPSC lines and of endo-Kdm3a in 4F + Kdm3a and 4F + Tet1+Kdm3a piPSC lines also were higher than those of controls (Figure 1D). Furthermore, these piPSCs expressed multiple pluripotent stem cell markers revealed by immunofluorescence, including Oct4, Nanog, and Sox2 in the nuclei and SSEA-1 on cell surface (Figures 2B and 2C).
By western blot, H3K9me3 protein levels declined in mTet3, Tet1, and Kdm3a-derived piPSCs, while H3K27me3 declined only in Tet1-derived piPSCs (Figure S3A). Unexpectedly, combination of Tet1 and Kdm3a did not reduce the protein levels of H3K9me3 and H3K27me3 (Figure S3A). These results suggest that different epigenetic factors further open chromatin to induce activation of pluripotency genes, although they may affect histone modifications differently.
Importantly, most piPSCs maintained normal karyotypes (Figure S3B) and can be propagated by single cells using TrypLE for more than 30 passages. Furthermore, the piPSCs derived by epigenetic factors showed transgene copy numbers at slightly higher but comparable levels with 4F control (Figure S3C). Thus, high expression levels of endogenous pluripotency genes in piPSCs induced by epigenetic factors are not likely due to differences in infection efficiency or transgene integration, but to the roles of these epigenetic factors in induction of piPSCs.
Various Expression Levels of Exogenous Genes in piPSCs
Exogenous mTet3 was exclusively expressed in piPSCs induced by 4F + mTet3, ex-Tet1 in piPSCs by 4F + Tet1 or 4F + Tet1+Kdm3a, and ex-Kdm3a in piPSCs by 4F + Kdm3a or 4F + Tet1+Kdm3a (Figure 1C). Exogenous transcription factor genes were either unchanged or upregulated in most of the piPSC lines regardless of addition of epigenetic factors, compared with those of OSKM at day 9 of induction (Figure 1E). Higher expression of ex-cMyc may compensate for the low expression levels of endo-cMyc. Together, exogenous genes were not effectively silenced in most of piPSCs induced by epigenetic factors, especially in Kdm3a-derived piPSCs (Figure 1E). Thus, while epigenetic factors can enhance reprogramming and activation of naive pluripotency genes in piPSCs, their effects on silencing exogenous genes are variable.
Pluripotency of piPSCs by Teratoma Test
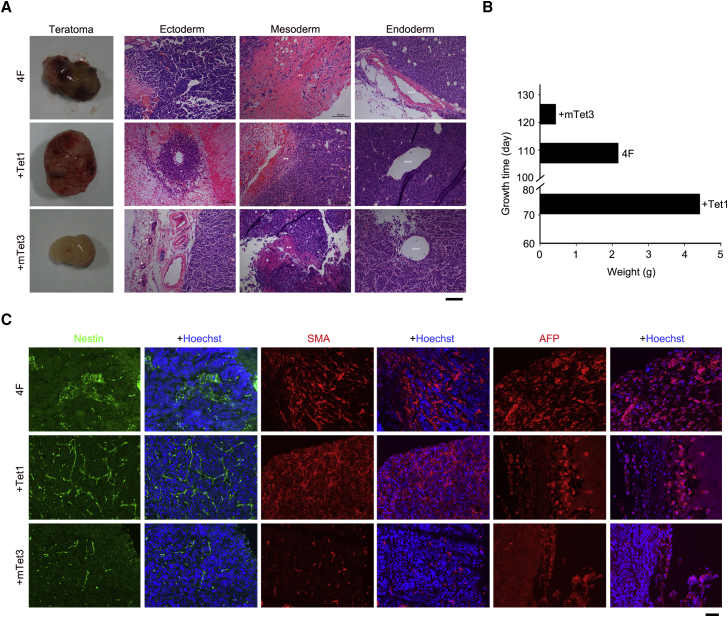
To examine the developmental potential of piPSCs generated by epigenetic regulation factors, we performed an in vivo differentiation test by teratoma formation (Lensch et al., 2007, Maherali and Hochedlinger, 2008). piPSCs induced by 4F, 4F + Tet1, and 4F + mTet3 formed teratomas following subcutaneous transplantation into non-obese diabetic/severe combined immunodeficiency mice, whereas 4F + Kdm3a and 4F + Tet1 + Kdm3a did not (Figure 3A). The 4F + Tet1-piPSCs formed teratomas faster and larger than did 4F- or 4F + mTet3-piPSCs (Figures 3A and 3B). In addition to the delayed formation, teratomas from 4F + mTet3-piPSCs were even smaller than those of OSKM (Figures 3A and 3B). The teratomas formed from these piPSCs showed the characteristic three embryonic germ layers, including neural epithelium (ectoderm), muscle (mesoderm), and gland epithelium (endoderm) (Figure 3A). Immunofluorescence microscopy validated that these teratomas expressed markers representative of three germ layers, including nestin (ectoderm), smooth muscle actin (mesoderm), and alpha 1-fetoprotein (endoderm) (Figure 3C), consistent with the results by immunohistochemistry (Figure S3D). Notably, piPSCs induced by Tet1 combined with OSKM exhibited enhanced differentiation capacity, while mTet3 seemed to reduce the differentiation potential of piPSCs (Figures 3A–3C).
Figure 3.
Differentiation Potential of piPSCs Induced by Addition of Epigenetic Factors
(A) Left, differentiation in vivo of piPSCs by teratoma formation test following injection into non-obese diabetic/severe combined immunodeficiency mice. Right, H&E staining of teratoma tissues derived from piPSCs. ∗, neural epithelium (ectoderm), ∗∗, muscle (mesoderm), ∗∗∗, gland epithelium (endoderm).
(B) Weight and time of teratomas formed from piPSCs.
(C) Immunofluorescence of the teratomas showing markers representative of three germ layers, nestin (ectoderm), smooth muscle actin (SMA, mesoderm), and alpha 1-fetoprotein (AFP, endoderm). Scale bars, 100 μm (A) or 50 μm (C). See also Figure S3.
Conversion of piPSCs by HDACi
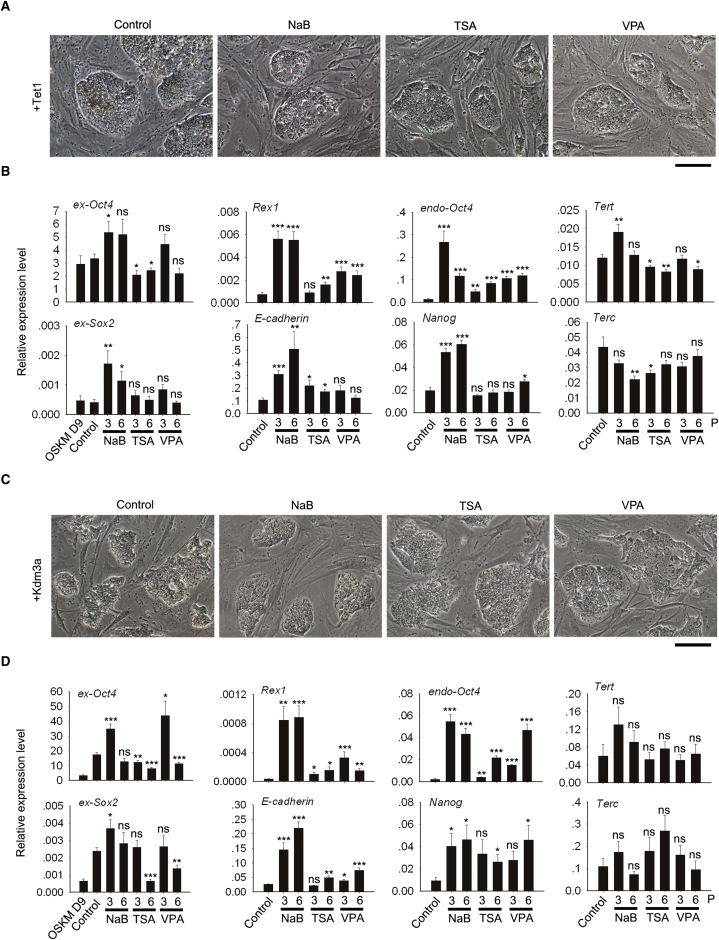
piPSCs failed from silencing of exogenous genes presumably are similar to the F-class iPSCs described recently, which, however, can be converted to naive iPSCs by HDACi (Tonge et al., 2014). To further reduce expression of exogenous genes and achieve the naive state, we attempted to convert two relatively high-quality piPSC lines, induced by epigenetic factors shown above (4F + Tet1 and 4F + Kdm3a), by adding HDACi, NaB, TSA, or valproic acid (VPA). Following culture in HDACi for three passages, piPSCs induced from 4F + Tet1 or 4F + Kdm3a maintained their original morphology (Figures 4A and 4C). With regard to the expression of exogenous genes, expression of ex-Oct4 was maintained in piPSCs induced by 4F + Tet1 after six passages by treatment with NaB or VPA but significantly declined after three and six passages by exposure to TSA (Figure 4B). Expression levels of ex-Sox2 remained unchanged after exposure to TSA or VPA (Figure 4B). piPSCs induced from 4F + Kdm3a showed lower expression of exogenous genes with increasing passages (Figure 4D). Expression levels of both ex-Oct4 and ex-Sox2 in 4F + Kdm3a were noticeably reduced after six passages in the presence of TSA or VPA, compared with controls (Figure 4D). Thus, exogenous genes were downregulated by HDACi.
Figure 4.
Conversion of piPSCs by HDACi
(A and C) Representative microscopy images showing morphology of piPSCs induced by 4F + Tet1 (A) or 4F + Kdm3a (C) after HDACi treatment for three passages, compared with control without HDACi. Scale bar, 100 μm.
(B and D) RT-qPCR analysis of selected exogenous genes and pluripotency genes of 4F + Tet1 (B) or 4F + Kdm3a (D). P, passaging number after exposure to HDACi. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant, compared with control. Data represent mean ± SEM from three independent experiments.
See also Figure S4.
HDACi Increases Expression of Pluripotency Genes in piPSCs
In piPSCs induced from 4F + Tet1, endo-Oct4 was upregulated but Tert expression minimally affected in the presence of HDACi. NaB significantly upregulated endo-Oct4, Nanog, Rex1, and E-cadherin (Figure 4B). Similarly, piPSCs induced from 4F + Kdm3a exhibited further increased expression levels of genes related to naive pluripotency following HDACi treatment. Both endo-Oct4 and endo-Nanog were upregulated (Figure 4D) and expression levels of Tert and Terc were maintained by HDACi (Figure 4D). Rex1 and E-cadherin also were upregulated by NaB (Figure 4D).
We also tested effects of 5-azacytidine and vitamin C on conversion of piPSCs. Consistent with a recent finding (Tonge et al., 2014), these two small molecules did not silence exogenous genes in piPSCs (Figure S4).
Discussion
We show that epigenetic regulatory factors can enhance reprogramming of piPSCs and addition of HDACi can further downregulate exogenous genes. Consistent with the notion that Rex1 is an indicator of the naive pluripotent state (Nichols and Smith, 2009), piPSCs induced by epigenetic factors, especially Tet1, show significantly activated Rex1 and other genes important for naive state, including Utf1, Esrrb, and Dppa2.
piPSCs are maintained dependent on exogenous genes, more like the “F-class” iPSCs (Hussein et al., 2014, Tonge et al., 2014). By exposure to HDACi, the two selected piPSC lines (4F + Tet1 and 4F + Kdm3a) show reduced expression of exogenous genes, especially in 4F + Kdm3a, and increased expression of endogenous genes associated with pluripotency. It is possible to convert F-class to naive-like state in some piPSCs. piPSCs in the presence of HDACi, including NaB, VPA, and suberoylanilide hydroxamic acid, express higher levels of Oct4, Nanog, Utf1, Rex1, Epcam, and Esrrb, compared with control (Petkov et al., 2016), confirmed in our study. VPA has been used for conversion of piPSCs from primed to naive-like state (Telugu et al., 2010). Moreover, we find that HDACi can moderately reduce expression of exogenous genes in piPSCs and increase expression of genes associated with pluripotency. Although exogenous genes are not effectively silenced, these approaches provide the basis for optimizing derivation and culture of piPSCs.
Despite the fact that epigenetic modifiers facilitate reprogramming and reduce reliance on exogenous genes, exogenous genes are still required to maintain pluripotency of piPSCs, likely suggesting that activation of endogenous genes is inadequate to maintain pluripotency of piPSCs, such that continued expression of exogenous genes complements the deficiency. It is also possible that some key pluripotency regulators in pigs may differ from the conventional reprogramming mixture. It is anticipated that methods that can fully activate endogenous pluripotent genes would allow piPSCs to self-renew without reliance on exogenous genes. Development of naive porcine ESCs and appropriate culture conditions become more critical for comparison of expression levels of endogenous pluripotent genes.
Experimental Procedures
Mice Used for Teratoma Tests
The care and use of mice for this research followed the guidelines and protocols for animal research approved by the Institutional Animal Care and Use Committee of Nankai University.
Generation of piPSCs
Retroviruses were produced and harvested following the protocol described previously (Ji et al., 2013).
For the induction of piPSCs, 1 × 105 PEF at passage 1–2 were plated in a 6-well dish prior to infection. Cells were infected with corresponding pMXs-based retroviral vectors (Oct4, Sox2, Klf4, c-Myc, mTet3, Tet1, and Kdm3a, 100 μL of concentrated virus suspension for each factor) for 12 hr twice with a 24-hr interval. Cells were induced to iPSCs in induction medium, and the medium changed daily. About 9 days after infection, 5 × 104 cells were passaged on to mitomycin C-inactivated mouse embryonic fibroblast (MEF) feeder cells in a 60-mm dish. ESC-like colonies were picked at days 18–20 following a standard protocol. See also Supplemental Experimental Procedures.
Cell Culture
The piPSCs were maintained on MEF treated with mitomycin C and cultured in piPSCs culture medium. piPSCs were passaged using TrypLE at a ratio of 1:10 every 3–4 days. See also Supplemental Experimental Procedures.
Author Contributions
L.L. designed the research; J.M. performed the main experiments; W.D. performed the H&E staining; Q.Z., H.W., K.L., H.F., and Q.Z. helped with the experiments; X.W. performed the RNA-seq experiments; L.L. and J.M. wrote the paper.
Acknowledgments
This work was supported by the China MOST National Major Basic Research Program (2011CBA01002, 2012CB911202), the National Natural Science Foundation of China (31271587), and PCSIRT (no. IRT13023). We thank Zhongcheng Zhou, Yudong Fu, and Yu Yin for assisting the experiments, and John Tsibris for critical reading of the manuscript.
Published: December 29, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.11.013.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE87361.
Supplemental Information
References
- Alberio R., Croxall N., Allegrucci C. Pig epiblast stem cells depend on activin/nodal signaling for pluripotency and self-renewal. Stem Cells Dev. 2010;19:1627–1636. doi: 10.1089/scd.2010.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brivanlou A.H., Gage F.H., Jaenisch R., Jessell T., Melton D., Rossant J. Stem cells. Setting standards for human embryonic stem cells. Science. 2003;300:913–916. doi: 10.1126/science.1082940. [DOI] [PubMed] [Google Scholar]
- Chan E.M., Ratanasirintrawoot S., Park I.H., Manos P.D., Loh Y.H., Huo H., Miller J.D., Hartung O., Rho J., Ince T.A. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Wu Y., Guo L. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Congras A., Barasc H., Canale-Tabet K., Plisson-Petit F., Delcros C., Feraud O., Oudrhiri N., Hadadi E., Griscelli F., Bennaceur-Griscelli A. Non integrative strategy decreases chromosome instability and improves endogenous pluripotency genes reactivation in porcine induced pluripotent-like stem cells. Sci. Rep. 2016;6:27059. doi: 10.1038/srep27059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Feng T., Yu D., Wu Y., Zou H., Ma S., Feng C., Huang Y., Ouyang H., Hu X. Barriers for deriving transgene-free pig iPS cells with episomal vectors. Stem Cells. 2015;33:3228–3238. doi: 10.1002/stem.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M.A., Xu J., Yang J., Peng M., Qin D., Li W., Jiang Z., Chen J., Deng K., Zhong M. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J. Biol. Chem. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T., Telugu B.P., Alexenko A.P., Sachdev S., Sinha S., Roberts R.M. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishiro S.H., Nakano K., Mizukami Y., Azami T., Arai Y., Matsunari H., Ishino R., Nishimura T., Watanabe M., Abe T. Generation of naive-like porcine-induced pluripotent stem cells capable of contributing to embryonic and fetal development. Stem Cells Dev. 2013;22:473–482. doi: 10.1089/scd.2012.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Chen J., Li K., Wu T., Huang B., Liu W., Kou X., Zhang Y., Huang H., Jiang Y. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Giraud S., Favreau F., Chatauret N., Thuillier R., Maiga S., Hauet T. Contribution of large pig for renal ischemia-reperfusion and transplantation studies: the preclinical model. J. Biomed. Biotechnol. 2011;2011:532127. doi: 10.1155/2011/532127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T.-P., Guo F., Yang H., Wu H.-P., Xu G.-F., Liu W., Xie Z.-G., Shi L., He X., Jin S.-g. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A.E., Melton D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Hussein S.M., Puri M.C., Tonge P.D., Benevento M., Corso A.J., Clancy J.L., Mosbergen R., Li M., Lee D.S., Cloonan N. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- Jeon B.G., Coppola G., Perrault S.D., Rho G.J., Betts D.H., King W.A. S-adenosylhomocysteine treatment of adult female fibroblasts alters X-chromosome inactivation and improves in vitro embryo development after somatic cell nuclear transfer. Reproduction. 2008;135:815–828. doi: 10.1530/REP-07-0442. [DOI] [PubMed] [Google Scholar]
- Ji G., Ruan W., Liu K., Wang F., Sakellariou D., Chen J., Yang Y., Okuka M., Han J., Liu Z. Telomere reprogramming and maintenance in porcine iPS cells. PLoS One. 2013;8:e74202. doi: 10.1371/journal.pone.0074202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lensch M.W., Schlaeger T.M., Zon L.I., Daley G.Q. Teratoma formation assays with human embryonic stem cells: a rationale for one type of human-animal chimera. Cell Stem Cell. 2007;1:253–258. doi: 10.1016/j.stem.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang F., Song G., Gu W., Chen M., Yang B., Li D., Wang D., Cao K. Intramyocardial injection of pig pluripotent stem cells improves left ventricular function and perfusion: a study in a porcine model of acute myocardial infarction. PLoS One. 2013;8:e66688. doi: 10.1371/journal.pone.0066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Bou G., Sun R., Guo S., Xue B., Wei R., Cooney A.J., Liu Z. Sox2 is the faithful marker for pluripotency in pig: evidence from embryonic studies. Dev. Dyn. 2015;244:619–627. doi: 10.1002/dvdy.24248. [DOI] [PubMed] [Google Scholar]
- Ma D.K., Chiang C.H., Ponnusamy K., Ming G.L., Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Montserrat N., Bahima E.G., Batlle L., Hafner S., Rodrigues A.M., Gonzalez F., Izpisua Belmonte J.C. Generation of pig iPS cells: a model for cell therapy. J. Cardiovasc. Transl. Res. 2010;4:121–130. doi: 10.1007/s12265-010-9233-3. [DOI] [PubMed] [Google Scholar]
- Montserrat N., de Onate L., Garreta E., Gonzalez F., Adamo A., Eguizabal C., Hafner S., Vassena R., Izpisua Belmonte J.C. Generation of feeder-free pig induced pluripotent stem cells without Pou5f1. Cell. Transplant. 2012;21:815–825. doi: 10.3727/096368911X601019. [DOI] [PubMed] [Google Scholar]
- Muller F.J., Laurent L.C., Kostka D., Ulitsky I., Williams R., Lu C., Park I.H., Rao M.S., Shamir R., Schwartz P.H. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Petkov S., Glage S., Nowak-Imialek M., Niemann H. Long-term culture of porcine induced pluripotent stem-like cells under feeder-free conditions in the presence of histone deacetylase inhibitors. Stem Cells Dev. 2016;25:386–394. doi: 10.1089/scd.2015.0317. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Allegrucci C., Alberio R. Modulation of pluripotency in the porcine embryo and iPS cells. PLoS One. 2012;7:e49079. doi: 10.1371/journal.pone.0049079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.B., Hosler B.A., Gudas L.J. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development. 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- Shi Y., Do J.T., Desponts C., Hahm H.S., Scholer H.R., Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Telugu B.P., Ezashi T., Roberts R.M. Porcine induced pluripotent stem cells analogous to naive and primed embryonic stem cells of the mouse. Int. J. Dev. Biol. 2010;54:1703–1711. doi: 10.1387/ijdb.103200bt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonge P.D., Corso A.J., Monetti C., Hussein S.M., Puri M.C., Michael I.P., Li M., Lee D.S., Mar J.C., Cloonan N. Divergent reprogramming routes lead to alternative stem-cell states. Nature. 2014;516:192–197. doi: 10.1038/nature14047. [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Valamehr B., Robinson M., Abujarour R., Rezner B., Vranceanu F., Le T., Medcalf A., Lee T.T., Fitch M., Robbins D. Platform for induction and maintenance of transgene-free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Rep. 2014;2:366–381. doi: 10.1016/j.stemcr.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodicka P., Smetana K., Jr., Dvorankova B., Emerick T., Xu Y.Z., Ourednik J., Ourednik V., Motlik J. The miniature pig as an animal model in biomedical research. Ann. N. Y. Acad. Sci. 2005;1049:161–171. doi: 10.1196/annals.1334.015. [DOI] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- West F.D., Terlouw S.L., Kwon D.J., Mumaw J.L., Dhara S.K., Hasneen K., Dobrinsky J.R., Stice S.L. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;19:1211–1220. doi: 10.1089/scd.2009.0458. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Zhou L., Wang W., Liu Y., Fernandez de Castro J., Ezashi T., Telugu B.P., Roberts R.M., Kaplan H.J., Dean D.C. Differentiation of induced pluripotent stem cells of swine into rod photoreceptors and their integration into the retina. Stem Cells. 2011;29:972–980. doi: 10.1002/stem.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.