Summary
Patients with glioblastoma die from local relapse despite surgery and high-dose radiotherapy. Resistance to radiotherapy is thought to be due to efficient DNA double-strand break (DSB) repair in stem-like cells able to survive DNA damage and repopulate the tumor. We used clinical samples and patient-derived glioblastoma stem cells (GSCs) to confirm that the DSB repair protein RAD51 is highly expressed in GSCs, which are reliant on RAD51-dependent DSB repair after radiation. RAD51 expression and RAD51 foci numbers fall when these cells move toward astrocytic differentiation. In GSCs, the small-molecule RAD51 inhibitors RI-1 and B02 prevent RAD51 focus formation, reduce DNA DSB repair, and cause significant radiosensitization. We further demonstrate that treatment with these agents combined with radiation promotes loss of stem cells defined by SOX2 expression. This indicates that RAD51-dependent repair represents an effective and specific target in GSCs.
Keywords: RAD51, glioblastoma, radiosensitization, glioma stem cell
Highlights
-
•
RAD51 is overexpressed in glioma stem cells
-
•
RAD51 expression levels fall when GSCs are differentiated
-
•
RAD51 inhibitors abrogate DNA repair leading to radiosensitization in GSCs
-
•
RAD51 inhibition + XR removes SOX2-expressing cells and abolishes clonogenicity
Short and colleagues show overexpression of RAD51 in a poorly differentiated stem-like population of glioma cells and that the homologous DNA double-strand break-repair pathway may represent a specific therapeutic target. RAD51-dependent repair is essential for recovery from radiation and temozolomide-induced DNA damage. Targeting this pathway may provide a tumor cell-specific effect in the context of chemo-radiotherapy.
Introduction
It has been widely postulated that a specific sub-population of glioblastoma (GBM) cells exhibit stem-like properties and that they underlie treatment resistance and recurrence due to their ability to survive DNA-damaging treatments and repopulate the tumor (Mannino and Chalmers, 2011). This population is dynamic and can be altered by specific growth conditions, including exposure to serum and bone morphogenic proteins (BMP), which render them non-tumorigenic (Piccirillo et al., 2006). These cells cannot be defined by a single marker, but the phenotype is enriched in Promonin1 (PROM1, known as CD133), SRY-box2 (SOX2) and Nestin (NES)-positive cells. The role of individual markers in contributing to the phenotype remains uncertain, but, for example, SOX2 has been functionally implicated in a rapidly proliferating, self-renewing population, and with maintenance of the undifferentiated state. Individual markers have rarely been shown to predict radioresistant sub-populations (Balbous et al., 2014, Berezovsky et al., 2014, Lemke et al., 2014). Upregulated DNA damage responses (DDRs) have been documented in glioblastoma stem cells (GSCs) including enhanced checkpoint signaling and recruitment of repair proteins (Bao et al., 2006, Cheng et al., 2011, Facchino et al., 2010, Zeppernick et al., 2008); however, the mechanisms underlying resistance to treatment are not fully understood. More importantly, it is not clear how specific resistance mechanisms align with the established phenotypic characteristics that drive recurrence or with marker positivity. Therefore, it remains unclear which repair pathways are the most relevant targets in GSCs.
Overexpression of the DNA repair protein, RAD51, the central protein involved in homologous repair (HR) of DNA double-strand breaks (DSBs), has been documented in glioma and numerous other cancers (Hannay et al., 2007, Maacke et al., 2000, Mehrara et al., 2007, Tennstedt et al., 2012, Welsh et al., 2009). Previously, we reported that targeting RAD51 using small interfering RNA-radiosensitized established glioma cell lines, and recent data confirm that targeting HR is more effective at radiosensitizing GSCs than inhibiting the major alternative DSB repair pathway, non-homologous end-joining (NHEJ) (Lim et al., 2014, Short et al., 2011). Inhibition of HR can be achieved through modulating expression, inhibiting nuclear translocation, or preventing DNA binding of RAD51, and small-molecule inhibitors have been developed, including B02 and RI-1. B02 impairs the RAD51-single-stranded DNA interaction at the primary site of RAD51 during nucleoprotein filament formation and at its secondary DNA binding site, where double-stranded DNA attaches during the search for homologous DNA (Huang et al., 2012). This agent displays synergy with the DNA crosslinking agent cisplatin, which requires HR for DNA repair (Huang and Mazin, 2014). RI-1 possesses a chloromaleimide moiety, which covalently binds to the thiol group in the cysteine at position 319 and occupies the interface between monomeric RAD51 proteins as well as an ATP binding loop. This alters RAD51-ATP interactions and subverts RAD51-RAD51 binding and polymerization, which is essential for filament elongation. RI-1 is synergistic with mitomycin C and both RI-1 and B02 are radiosensitizers (Budke et al., 2012a, Budke et al., 2012b, Huang and Mazin, 2014, Huang et al., 2012, Ward et al., 2015).
In this study, we tested the hypothesis that high RAD51 expression and RAD51 foci activation is specifically associated with GSCs and that small-molecule inhibitors are effective GSC radiosensitizers.
Results
RAD51 Is Highly Expressed in GSCs
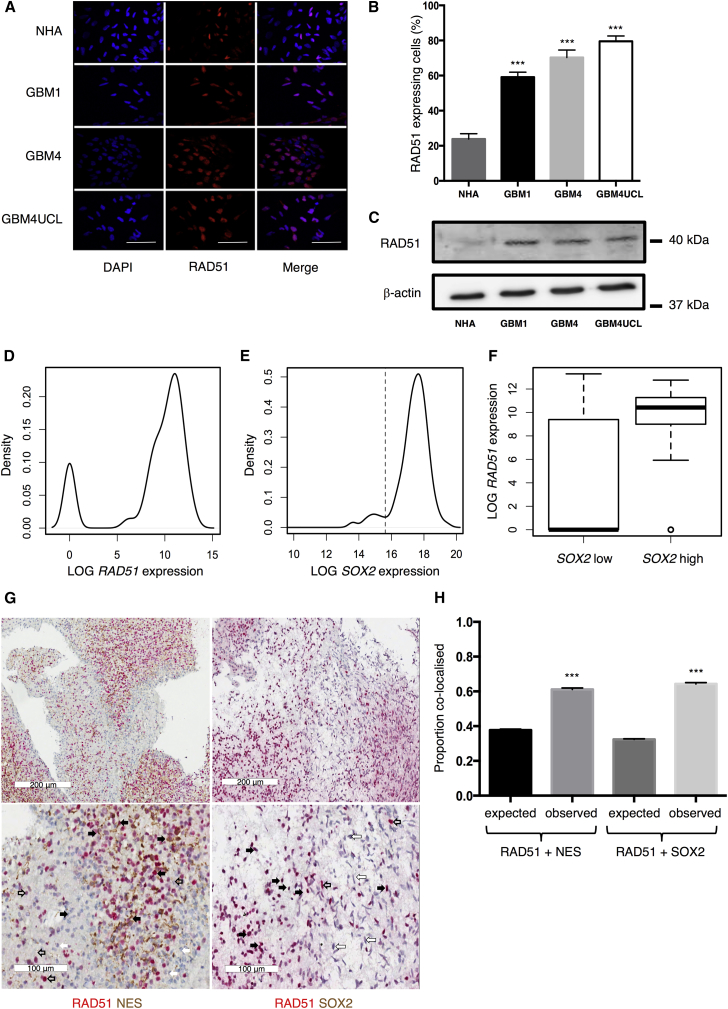
To confirm that RAD51 is a relevant target in GSCs, expression was examined in patient-derived GSCs and normal human astrocytes (NHAs). These GSCs are clonogenic cells propagated as cell lines from freshly resected glioblastoma tumors. Here, we use GBM1, GBM4, and GBM4UCL that express high levels of GSC markers NES and SOX2 and accurately recapitulate GBM when cultured in stem cell-permissive conditions, as described previously by ourselves and other authors using comparable protocols (Lee et al., 2006, Pollard et al., 2009, Wurdak et al., 2010). These cells maintain distinct morphologies and gene expression profiles during monolayer culture and form orthotopic tumors in mice with hallmarks of high-grade brain tumors. Figures 1A–1C show data confirming significantly greater RAD51 expression in all three GSCs compared with NHAs. Using immunofluorescence (IF) microscopy, 24% (±3%) of NHA cells were positive for RAD51, compared with 60% ± 3%, 72% ± 4%, and 84% ± 3% of GBM1, GBM4, and GBM4UCL cells, respectively (n = 6 independent experiments, p < 0.0001). Western blot confirms higher protein levels in GSCs than NHAs, with very low expression detectable in NHAs using this assay, which is less sensitive than IF.
Figure 1.
RAD51 Expression Is Elevated in Patient-Derived Glioma Cells
(A and B) Representative images of immunofluorescence (IF) staining for RAD51 in three GSCs in comparison with normal human astrocytes (NHAs) (A), quantified in (B) (n = 6 independent experiments with ≥100 cells counted per cell line).
(C) Western blots probed for RAD51 or β-actin in three GSCs and NHAs.
(D and E) Distributions of RAD51 and SOX2 expression (mRNA levels) in single GBM1 cells (n = 273 cells). The dotted line in (E) delineates cells with low and high SOX2 expression.
(F) RAD51 expression levels in SOX2-low (n = 30 cells) and SOX2-high (n = 243 cells) populations.
(G) Representative immunohistochemistry images from patient tumor samples stained for NES (brown) and RAD51 (red) (left hand panels) or SOX2 (brown) and RAD51 (red) (right hand panels). Examples of cells positive for both proteins are indicated with black arrows, cells negative for both with white arrows, cells positive for just NES or RAD51 indicated by open arrows. Statistical significance calculated by Student’s t test.
(H) Quantification of RAD51 and stem marker co-expression from ten tumor samples.
Error bars indicate SEM. Statistical significance, unless otherwise stated, calculated by one-way ANOVA. ∗∗∗p≤0.001. Scale bars, 100 μm.
RAD51 expression is cell-cycle regulated, being lowest in resting cells and highest in S and G2 phases (Johnson et al., 1992, Yamamoto et al., 1996). To establish the contribution of cell cycling to expression levels, we measured proliferation and S and G2 phase cell-cycle markers. The Ki67 proliferation marker revealed that our GSCs contain >40%–60% cycling cells, as do NHAs, suggesting that this is not the explanation for the higher RAD51 levels in GSCs (Figures S1A and S1B). As we described previously in established GBM cell lines, the RAD51-expressing cells in the GSC population are not limited to cells identified by Cyclin A staining (Short et al., 2011). A significant proportion are RAD51 positive/Cyclin A negative using IF (Figures S1C and S1D), indicating that RAD51 expression is not limited to G2/S phase in GSCs (p < 0.0001). By contrast, in NHAs the RAD51-positive/Cyclin-A-negative population is very small, suggesting the expected restriction to G2/S phase.
Since these data suggest significant RAD51 expression in a high proportion of GSCs, but do not define an association with any specific sub-population of cells, we investigated the distribution of RAD51 expression in the GSC population further using microfluidics-based single-cell qRT-PCR analysis. Our data show that RAD51 expression varies, with a distinctive bimodal distribution of low- and high-expressing cells (Figure 1D). In the same dataset, we defined the self-renewing fraction by high SOX2 expression, delineated by a minima at a log expression value of 15.6 (Figure 1E). When we tested the association between SOX2 positivity and RAD51 expression, we found it to be highly significant (p = 1.28 × 10−15), suggesting a correlation between RAD51 expression and the putative self-renewing fraction (Figure 1F). We confirmed these data using IF co-staining for SOX2 and RAD51 (Figure S1E) and also confirmed co-expression with NES (Figure S1F).
To confirm that RAD51 associates with a poorly differentiated, stem-like, self-renewing population in tumor material, ten samples from GBM resections were stained for RAD51, SOX2, and NES using immunohistochemistry (Figure 1G). We used χ2 tests to assess whether there was a greater than expected association with RAD51, considering that NES was detected in 37% of the tumor cells and SOX2 in 31%. These data show that 61% of RAD51 co-localized with NES, a significant difference from the expected value (χ2, p = 2.1 × 10−28). Similarly, 62% of RAD51 co-localized with SOX2 (χ2, p = 1.4 × 10−32) (Figure 1H). These data further confirm that stem cell marker positivity and high levels of RAD51 are significantly associated in GSCs.
RAD51 Expression Is Dependent on Differentiation Status of GSCs
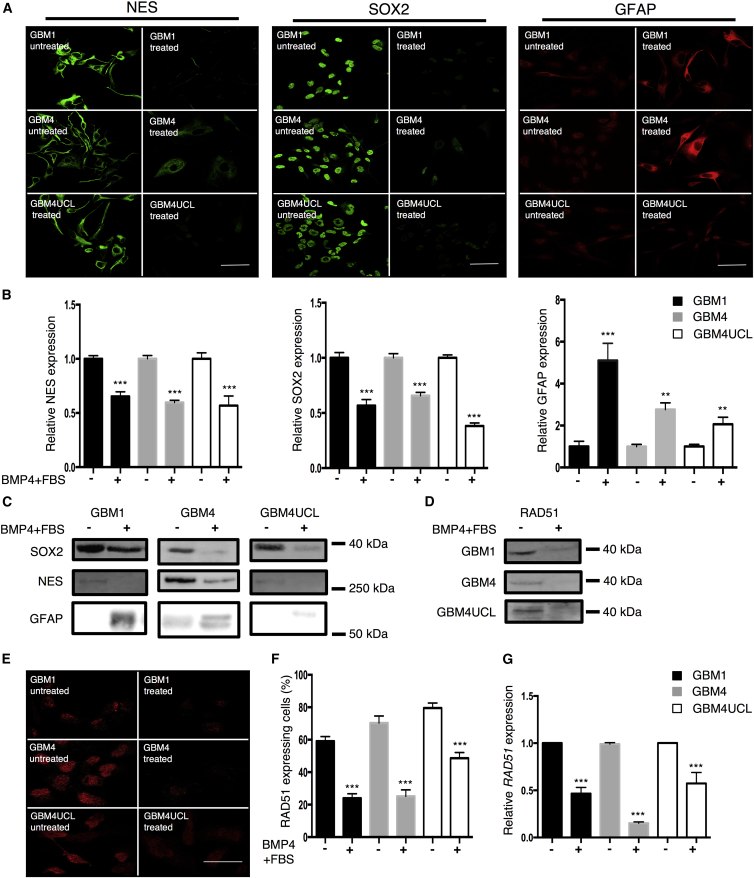
Because these data suggest that RAD51 may be specifically expressed in a self-renewing, SOX2-positive sub-population in GSCs, we hypothesized that RAD51 expression may change upon differentiation. To investigate this we used a forced differentiation paradigm (Piccirillo et al., 2006, Wurdak et al., 2010, Suva et al., 2014). We first confirmed that our GSCs responded to exposure to BMP and serum (fetal bovine serum [FBS]) with loss of stem cell markers. All three GSCs downregulated NES and SOX2 and upregulated glial fibrillary acid protein (GFAP) within 72 hr following treatment with BMP4 and serum, consistent with Wurdak et al. (2010) (Figures 2A–2C). We note that upregulation of GFAP was less marked in GBM4UCL, which is a recognized phenomenon in some cell lines (Restrepo et al., 2011). Loss of stem cell markers was associated with loss of clonogenicity (Figure S2A). We next investigated RAD51 expression in GSCs in response to differentiation cues. Western blots showed a marked reduction in RAD51 protein expression in all three GSCs following exposure to serum and BMP4 (Figure 2D). Furthermore, quantification of IF staining demonstrated a fall in the percentage of RAD51-expressing cells, defined by nuclear positivity, with reductions of 59%, 64%, and 39%, respectively (59% ± 3% to 24% ± 3% in GBM1, 70% ± 4% to 25% ± 4% in GBM4, and 80% ± 3% to 49% ± 3% in GBM4UCL, n = 6 independent experiments) (Figures 2E and 2F). Finally, qRT-PCR analysis revealed downregulation of RAD51 mRNA levels, demonstrating reductions of 53% ± 7%, 85% ± 1%, and 43% ± 12% for GBM1, GBM4, and GBM4UCL, respectively (n = 3 independent experiments, p < 0.01) (Figure 2G).
Figure 2.
Differentiation of GBM Stem Lines Causes a Reduction in RAD51 Expression
(A) Immunofluorescence microscopy in three GSCs (GBM1, GBM4, and GBM4UCL) grown in NB medium (untreated) or NB-BMP4-FBS (treated) and stained for SOX2, NES, and GFAP.
(B) Mean fluorescence intensity of whole slides from (A) (n = 3 independent experiments with ≥100 cells counted per condition). NB medium (−), NB-BMP4-FBS (+).
(C) Western blots probed for SOX2, NES, or GFAP.
(D) Western blots assessing expression of RAD51.
(E) GSCs grown in NB medium (untreated) or NB-BMP4-FBS (treated), stained for RAD51 and visualized by IF microscopy.
(F) Quantification of RAD51 expression from (E) (n = 6 independent experiments with ≥100 cells counted per cell line).
(G) qRT-PCR of RAD51 mRNA in GSC cultured in NB medium (−) or NB-BMP4-FBS (+) (n = 3 independent experiments).
Error bars indicate SEM. Statistical significance was calculated using one-way ANOVA with ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 20 μm.
Although differentiation is expected to lead to cell-cycle exit, it has recently been shown that BMP-directed differentiation does not result in an irreversible G0 state, due to failed silencing of key cell-cycle and mitosis regulators (Caren et al., 2015, Dirks, 2008, Wurdak et al., 2010). Interestingly, in two of our GSC models (GBM1 and GBM4UCL), the changes in RAD51 expression occurred before we could detect a significant reduction in Ki67 (Figures S2B and S2C). Although differentiation led to a significant reduction in Cyclin A positivity, suggesting a shift out of G2/S phase, there was also a marked reduction in the proportion of cells that are RAD51 positive/Cyclin A negative (Figures S2D and S2E). Taken together, these data suggest that downregulation of RAD51 may occur as part of an early change in a transcriptional network during differentiation that is not entirely dependent on cell-cycle exit.
GSC Preferentially Activate RAD51 after Radiation Damage
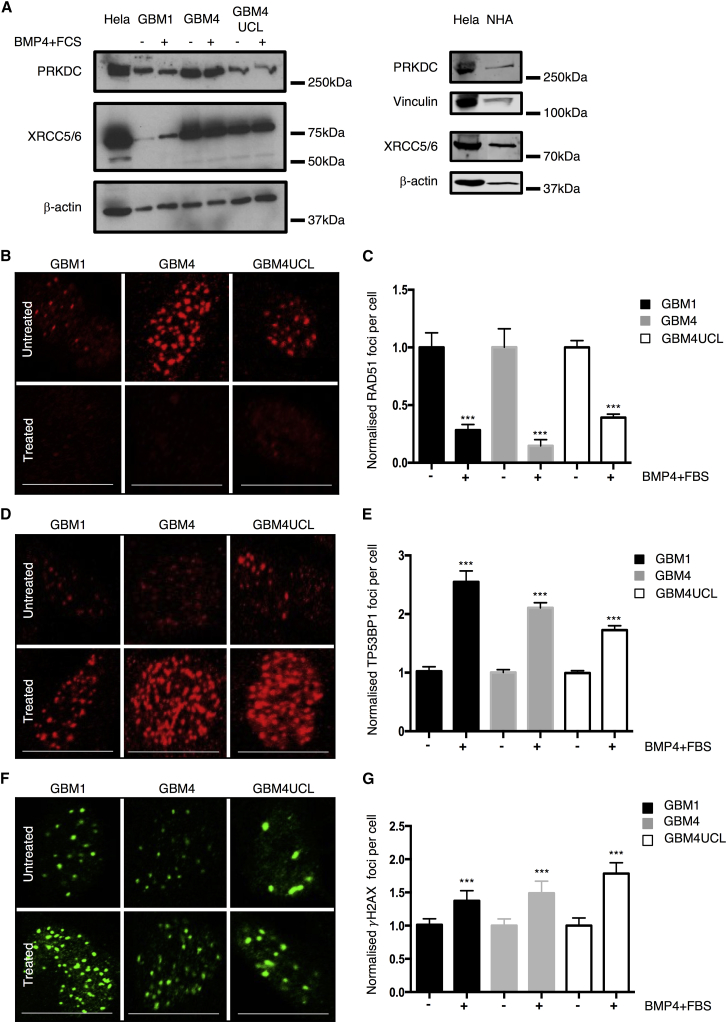
To investigate whether changes in RAD51 expression that occur upon differentiation are associated with a general downregulation of DSB repair protein activity, we measured levels of other proteins including PRKDC (DNA-PKcs), XRCC5 (Ku80), and XRCC6 (Ku70). No significant changes were observed in expression of any of these proteins in GSCs in response to differentiation conditions, and all are expressed in NHAs (Figure 3A).
Figure 3.
Differentiation Alters the Ability of GBM Stem Lines to Repair DNA Damage Caused by Ionizing Radiation
(A) Western blots probed for PRKDC or XRCC5/XRCC6 (Ku80/Ku70) in GSC grown in NB medium or NB-BMP4-FBS. β-Actin or Vinculin was a loading control. HeLa included as positive control and NHA for comparison.
(B) Representative images of RAD51 foci in the nuclei of GSCs grown in either NB medium or NB-BMP4-FBS, 4 hr after treatment with 3 Gy radiation.
(C) Quantification of RAD51 foci from (B) (n = 3 independent experiments with ≥100 cells counted per condition).
(D) Representative images of GSC nuclei stained for TP53BP1 foci 30 min post-treatment with 3 Gy radiation.
(E) Quantification of TP53BP1 foci from (E) (n = 6 independent experiments with ≥100 cells counted per condition).
(F) Representative images of GSC nuclei stained for γH2AX 24 hr following 3 Gy radiation.
(G) Quantification of γH2AX foci from (F) (n = 3 independent experiments with ≥100 cells counted per condition).
Error bars indicate SEM. Statistical significance was calculated using one-way ANOVA with ∗∗∗p < 0.001. Scale bars, 20 μm.
To confirm that RAD51 was functional in these cells, we next examined foci formation in the presence or absence of BMP4/serum 4 hr after 3 Gy X-ray. The resulting data showed a robust increase in RAD51 foci numbers in GSCs after XR that is attenuated when GSCs are exposed to FBS and BMP4 for 72 hr (Figures 3B and 3C). More detailed analysis of RAD51 foci kinetics in GBM1 confirmed lower foci numbers in differentiated cells over a repair time course of 24 hr (Figure S3A). Since this suggests that differentiation is associated with both a change in RAD51 expression and reduced activation at repair foci, we next measured TP53BP1 foci formation, which marks DSB for repair by NHEJ, primarily by blocking end resection (Callen et al., 2013, Chapman et al., 2012, Chapman et al., 2013, Xie et al., 2007). Thirty minutes after irradiation, we observed higher numbers of TP53BP1 foci per cell in all three GSCs exposed to serum and BMP4, compared with cells in standard growth conditions (Figures 3D and 3E). These data were also confirmed by examining foci over a 24 hr repair time course in GBM1 cells (Figure S3B). A comparison of DSB repair kinetics using γH2AX foci demonstrated that GSC in stem cell-permissive or differentiating conditions had slower repair kinetics than NHAs, with more residual unrepaired DSB at 24 hr (Figure S3C). A detailed comparison at 24 hr post-irradiation in all three GSCs demonstrated that the serum/BMP4-treated cells have more residual unrepaired DSB (Figures 3F and 3G), despite there being no differences in initial foci numbers (Figure S3C), implying less efficient repair.
These data suggest that differentiation does not cause a global change in levels of DNA DSB repair proteins, but rather that GSCs in stem cell conditions preferentially upregulate RAD51 and activate RAD51 foci at sites of damage, whereas in differentiating conditions, GSCs maintain lower RAD51 protein levels, show less activation of RAD51 foci at sites of damage, and preferentially activate TP53BP1 foci.
RAD51 Inhibitors Radiosensitize GSCs
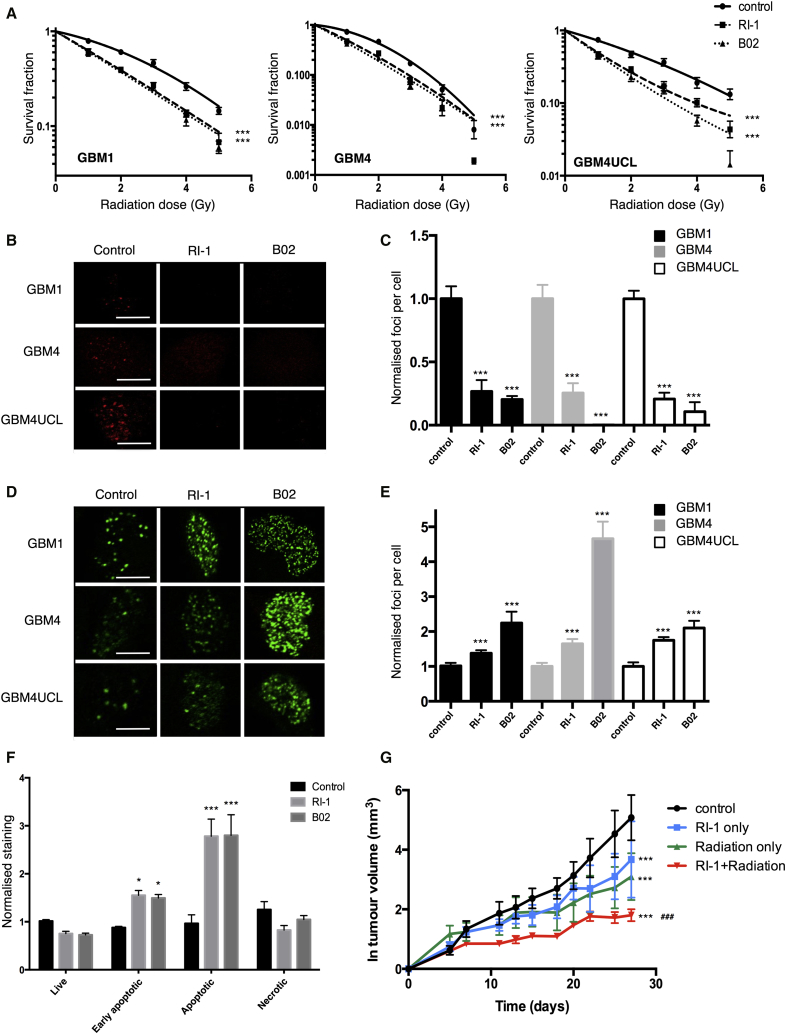
Because these data confirmed that RAD51 may be a particularly relevant target in GSCs, the effect of combining small-molecule RAD51 inhibitors with radiation was investigated.
Figure 4A shows the clonogenic survival data for the three GSCs treated with RI-1 (1.5 μM), B02 (1.2 μM), or DMSO and radiation doses between 1 and 5 Gy. The mean inactivation dose (MID, dose causing 50% cell death) and dose-modifying factors (DMF, ratio of untreated to treated MID, where values >1 indicate radiosensitization) were calculated by fitting the data to the linear-quadratic model (Hall and Giaccia, 2006). In GBM1, an MID of 2.69 Gy in cells exposed to XR only was reduced to 1.94 and 1.87 Gy after treatment with RI-1 or B02, respectively (DMFs of 1.39, 1.44). Similar effects were observed in GBM4 cells (DMF of 1.43, 1.55) and GBM4UCL (DMF of 1.54 and 1.70). Comparison of survival curves by two-way ANOVA demonstrated that, for each cell line, treatment with either inhibitor resulted in a statistically significant difference in survival compared with untreated cells (p < 0.001 in each case). These data confirm radiosensitization in GSCs using either of the small-molecule RAD51 inhibitors (RI-1 or B02).
Figure 4.
RAD51 Inhibitors Radiosensitize Cells Expressing High RAD51 In Vitro and In Vivo
(A) Clonogenic survival of GSCs treated with DMSO, RI-1 (1.5 μM), or B02 (1.2 μM) in combination with radiation (doses between 0 and 5 Gy). Data were fitted using the linear-quadratic model and statistical significance determined by two-way ANOVA (n = 3 independent experiments).
(B) IF microscopy staining for RAD51 in three GSCs showing the effect of RI-1 and B02 (30 μM) on number of RAD51 foci 4 hr after treatment with 3 Gy radiation.
(C) Quantification of RAD51 foci in (B) (n = 3 independent experiments with ≥100 cells counted per treatment).
(D) Effect of RAD51 inhibitors (30 μM RI-1 or B02) or BMP4 (10 ng/mL) + FBS (10%) on γH2AX foci (as visualized by IF microscopy) in GSCs 24 hr after treatment with 3 Gy radiation.
(E) Quantification of γH2AX foci from (D) (n = 3 independent experiments with ≥100 cells counted per treatment).
(F) Annexin V/PI staining of GBM1 cells treated with B02 or RI-1 (7.5 μM) 24 hr prior to irradiation (2 Gy) followed by incubation at 37°C for 5 days prior to staining. Statistical significance in (C), (D), and (F) was calculated using one-way ANOVA (n = 3 independent experiments).
(G) Log-transformed tumor growth data from untreated mice, mice treated with RI-1 (100 μL of 20 μM RI-1), or mice treated with radiation (15 Gy in 3 × 5 Gy) on alternate days alone, or mice treated with a combination of RI-1 and radiation (n = 10 animals per treatment group).
Statistical significance was determined by one-way ANOVA. For all panels, error bars indicate SEM, ∗p< 0.05, ∗∗∗p < 0.001, ###p < 0.001 for combination compared with individual treatments. Scale bars, 20 μm.
To confirm that RI-1 and B02 were directly affecting DNA repair as predicted by their known mechanisms of action, RAD51 and γH2AX foci were assessed in irradiated and RAD51 inhibitor-treated cells. Treatment of GSCs with either agent reduced RAD51 foci formation post-irradiation (4 hr, 3 Gy) by >75% in all three cell lines (Figures 4B and 4C) and consistently increased numbers of γH2AX foci at 24 hr, with no effect on foci induction at early time points (30 min), indicating a reduced DSB repair capacity (Figures 4D, 4E, S4A, and S4B). By contrast in NHAs, the RAD51 inhibitors did not affect the number of γH2AX foci at 24 hr (Figures S4C and S4D), suggesting no effect on repair capacity.
To confirm that these inhibitors caused additional cytotoxicity when used in combination with radiation, apoptosis was measured 5 days after treatment using Annexin V/propidium iodide (PI) labeling. The results showed an increase in both early apoptotic (Annexin V positive) and apoptotic cells (Annexin V and PI positive) following treatment of GBM1 with either RAD51 inhibitor (Figure 4F). A 1.8-fold increase in apoptotic cells was observed with either RI-1 or B02 (p < 0.0001). Early apoptotic cells were increased 1.67- and 1.61-fold with RI-1 and B02, respectively (p < 0.05). Similar results were obtained in GBM4 and GBM4UCL cells (Figures S4E and S4F).
Taken together, these data suggest that GSCs are radiosensitized by small-molecule RAD51 inhibitors, consistent with a significant reliance on RAD51-dependent repair events after irradiation.
Since neither of the RAD51 inhibitors are suitable for treating intracranial tumors, the established U87 glioma cell line was implanted as a subcutaneous xenograft and used as a model to test the effects of radiation ± RI-1 in vivo. Although these cells do not express high levels of conventional stem cell markers, they do express high levels of RAD51 required for this approach to radiosensitization (Short et al., 2011). The effects of RI-1 and B02 on U87 were first confirmed in vitro, demonstrating that the inhibitors reduced the number of RAD51 foci and radiosensitized the cells (Figure S5). Xenografts were treated in four groups: control, radiotherapy only, RI-1 only, and RI-1 combined with radiotherapy. Radiotherapy was delivered in 3 × 5 Gy fractions on alternate days with RI-1 or vehicle delivered intratumorally 3 hr before irradiation. Tumor growth was recorded over time, normalized to volumes at day 0 and log-transformed to allow for fitting of data using linear regression (Figure 4G). The linear regression of the increase in tumor volume was used to determine the tumor growth rate, from which doubling time (DT) was calculated (Demidenko, 2010). The DT was 3.85 days for vehicle-only-treated tumors, 5.61 days for RI-1, 7.16 days for radiotherapy only, and 11.1 days for the combination of RI-1 and radiotherapy. All treatments caused a significant increase in DT when compared with control, and the combination of both treatments significantly enhanced growth retardation (p < 0.0001) compared with either treatment individually. These data confirm a radiosensitizing effect of pharmacologic inhibition of RAD51 in vivo.
Combination Treatment with Radiation and a RAD51 Inhibitor Results in Loss of the SOX2+ Clonogenic Population
Since we have shown that inhibiting RAD51 is an effective means of radiosensitizing GSCs and that there is a significant association between RAD51 expression and SOX2 expression, we further hypothesized that targeting RAD51 repair may specifically remove SOX2 and RAD51 double-positive cells, potentially removing a radioresistant, self-renewing cellular fraction. We therefore examined the expression of both genes in single GSCs before and after exposure to radiation (2 Gy), in the presence or absence of the RAD51 inhibitor RI-1 (1.5 μM). We measured expression of RAD51 and SOX2 mRNA 1 and 3 weeks post-treatment, including the time point at which apoptosis occurs in response to treatment (1 week) and when most cell death has occurred, leaving a population that has survived treatment (3 weeks).
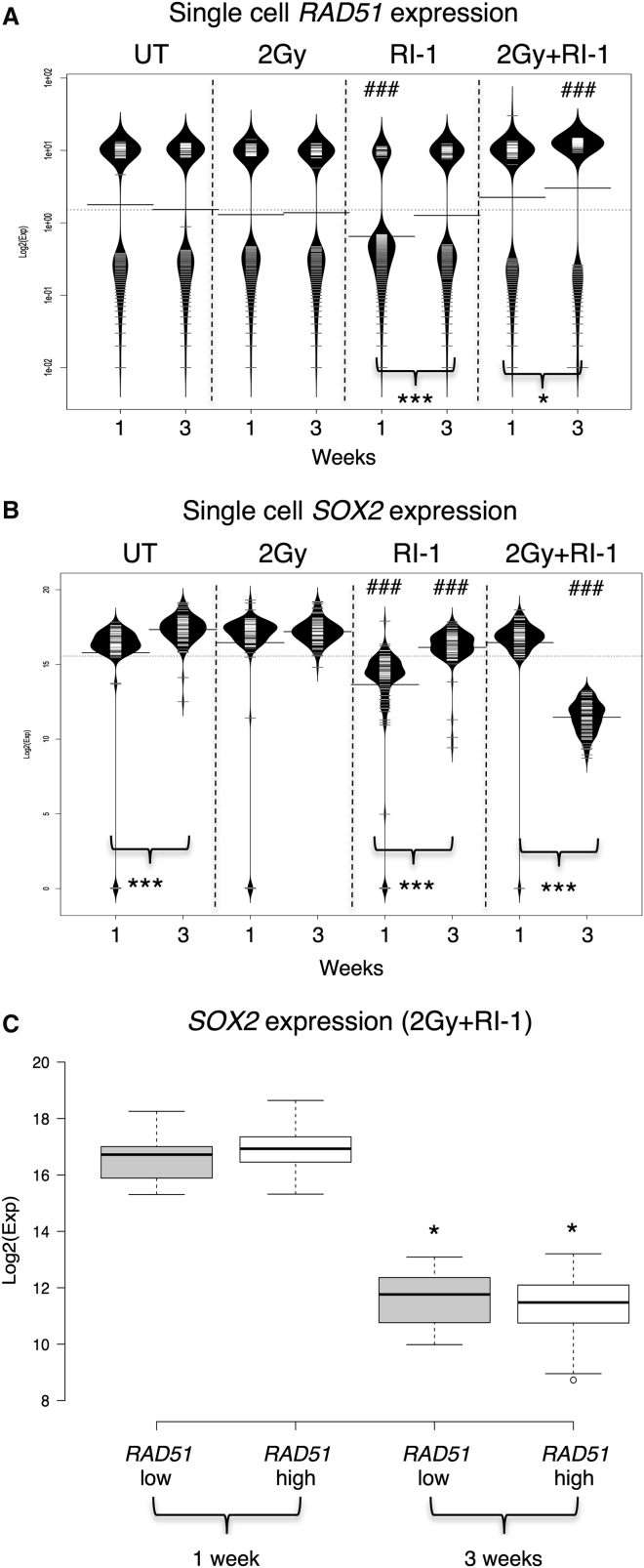
The distributions of RAD51 single-cell expression data are shown as bean plots for each treatment condition in Figure 5A, and demonstrate a major sub-population with high expression and a second population with lower, more variable expression in all conditions, as demonstrated previously (Figure 1D). In contrast, all cells express high levels of SOX2 (Figure 5B). In untreated cells, there is no change in the distribution of RAD51 expression at either 1 or 3 weeks (Figure 5A, first column), but a slight increase in SOX2 is observed at 3 weeks, which may be explained by overgrowth by SOX2+ cells in permissive culture conditions (Figure 5B, first column). Treatment with a single dose (2 Gy) of radiation does not affect the expression of RAD51 or SOX2 (Figures 5A and 5B, second columns), suggesting that this population is stable over the course of this experiment and not affected by XR. In contrast, exposure to the RAD51 inhibitor RI-1 alone causes a marked shift in the distribution for both RAD51 and SOX2 after 1 week (Figures 5A and 5B; third columns). There is a fall in the proportion of cells with high RAD51 expression and reduction in median expression levels (from 6.1 ± 0.5 in untreated cells to 2.7 ± 0.4 in RI-1-treated cells; p < 0.0001, n = 92 cells). Similarly, SOX2 expression is reduced across the entire population with the median level falling from 15.8 ± 0.3 in untreated cells to 13.7 ± 0.3 (p < 0.0001, n = 92–96 cells). However, this effect is transient, and by 3 weeks after treatment the distribution of RAD51 expression is not significantly different from untreated controls (median expression 5.7 ± 0.7 in untreated cells and 4.9 ± 0.5 in RI-1-treated cells; p > 0.05, n = 92 cells). SOX2 expression has also recovered significantly at 3 weeks, although it remains lower in cells exposed to RI-1 compared with untreated controls (median levels 17.3 ± 0.1 [untreated] and 16.1 ± 0.1 [RI-1]; p < 0.0001, n = 92–96 cells). These data are consistent with early toxicity and apoptosis of cells with high RAD51 expression, followed by replacement of this population.
Figure 5.
Combined Treatment with RAD51 Inhibitor and Radiation Results in Loss of the SOX2-Positive Clonogenic Population
(A) Bean plots illustrating expression of RAD51 mRNA (Log2(Exp)) in single cells cultured in a clonogenic assay, with and without radiation (2 Gy) exposure and/or RI-1 (1.5 μM), harvested at 1 or 3 weeks. Black lines show the median expression, each white line represents a single cell, and the width of the bean indicates the density of single cells with specific expression levels. UT, untreated.
(B) Bean plots illustrating expression of SOX2 mRNA (Log2(Exp)) in single cells cultured in a clonogenic assay, with and without radiation (2 Gy) exposure and/or RI-1 (1.5 μM), harvested at 1 or 3 weeks.
(C) Boxplot to demonstrate levels of SOX2 mRNA expression in RAD51 low- and high-expressing GBM1 cells at 1 and 3 weeks after treatment with radiation and RI-1.
Error bars indicate SEM. For comparisons between time points, statistical significance was calculated using the Student’s t test for each treatment (∗p < 0.05, ∗∗∗p < 0.001). For comparisons between treated and untreated cells, statistical significance was calculated using one-way ANOVA (###p < 0.001). For each treatment, 96 cells were captured and cDNA amplified on a single 96.96 Dynamic Array IFC. Number of cells for which data was obtained are mentioned in the text.
Remarkably, combining RI-1 treatment with radiation has a dramatically different effect than RAD51 inhibition alone. After 1 week, the distribution of both RAD51 and SOX2 expression is no different to untreated cells (Figures 5A and 5B, fourth columns). This is in marked contrast to the downregulation seen on exposure to RI-1 alone, indicating that radiation has abrogated the effects of the RAD51 inhibitor at this time point. However, by 3 weeks post-treatment, a very different population emerges with significantly reduced SOX2 expression (median level 11.4 ± 0.1, compared with 17.3 ± 0.1 in untreated cells; p < 0.0001, n = 92 cells). Within this population, the bimodal distribution of RAD51 is maintained, but expression levels have increased (median expression is 9 ± 0.6 versus 5.7 ± 0.7 in untreated cells at 3 weeks, 5.7 ± 0.7; p < 0.0001, n = 92 cells). We examined SOX2 expression specifically in cells expressing either low or high levels of RAD51 and confirmed that, 3 weeks after combined radiation and RI-1 treatment, SOX2 is significantly lower in both populations (Figure 5C). These cells are non-clonogenic (plating efficiency [PE] 0, compared with control PE 0.36, SD 0.09), indicating a redistribution of population dynamics toward cells with low self-renewing capacity.
Overall, these data show the potential for recovery in GSCs that express RAD51 and SOX2 after XR treatment, since after 2 Gy both of these populations persisted, confirming their contribution to the surviving population. Our data suggest that although RI-1 significantly reduced the RAD51-expressing population of cells 1 week after exposure, combining RI-1 with XR annulled this effect and, by 3 weeks, the RAD51-expressing population re-emerged in both treatment conditions. However, they also suggest a significant effect of combined treatment with XR and RI-1 on the SOX2-expressing population, which does not re-emerge after combination treatment. These data demonstrate the potential for abolishing clonogenicity by combining RI-1 with radiation.
Discussion
Identifying targets in the DDR pathway as a means to sensitize cancers to DNA-damaging cytotoxic treatments including radiotherapy has become of increasing interest with the availability of new inhibitors and the demonstration of their effectiveness in specific contexts. A notable example is the use of PARP inhibitors in BRCA-deficient tumors, based on the synthetic lethality paradigm (Audeh et al., 2010, Helleday, 2011, Tutt et al., 2010). In glioma, attempts to improve the outcome by adding agents to increase radiation sensitivity have been the subject of much research, but none of these approaches have been successful. The stem cell model has added a new perspective to these investigations, since GSCs are believed to be the relevant target population. However, specific repair targets have not been linked with this cell population. In this study, we demonstrated that RAD51 and the HR pathway represent a specific DNA repair target in GSC.
We first demonstrated that GSCs express high levels of RAD51 and that this is associated with robust foci formation after irradiation. This is surprising since in most circumstances HR is responsible for a small fraction of DSB repair. However, the complex regulation of the balance between repair pathways is still not fully understood and hyper-recombination with significant utilization of HR documented in tumor cell lines may be associated with loss of normal TP53 function, which can increase HR by promoting BRCA1 binding at DSBs (Dong et al., 2015).
We further show that RAD51 expression is associated with markers of stemness both in vitro and in tumor material, and that a forced differentiation paradigm using BMP4/FBS reduces this expression within 72 hr. It has recently become clear that this manipulation is not equivalent to terminal differentiation and that associated changes in DNA methylation, chromatin structure, and transcription occur with variable and often delayed kinetics. Interestingly a failure to repress SOX2-driven transcription programs is suggested to be an important influence on the ability of these cells to re-enter the cell cycle (Caren et al., 2015). The transcriptional programs that underlie these changes, including those that may explain the close correlation between SOX2 and RAD51 expression are the subject of ongoing research. Interestingly, recent data suggest that SOX2 and RAD51 may be regulated by overlapping transcription factors, including FOXM1 (Lee et al., 2015, Zhang et al., 2012).
The data presented here also demonstrate that inhibiting RAD51 is an effective means of sensitizing GSCs, as would be predicted from evidence that RAD51 is very active in contributing to DNA repair in these cells. This is consistent with previous data specifically examining the role of HR in GSCs (Lim et al., 2012, Lim et al., 2014) but may also explain the important contribution of cell-cycle checkpoint upregulation to resistance in these cells since homology-directed repair has a long half-time (Jeggo et al., 2011, Qin et al., 2014).
When we examined our GSCs following treatment with a RAD51 inhibitor and radiation, we found that, contrary to our expectations, a RAD51-expressing population reappeared after treatment and that exposure to XR seemed to enhance this. Fascinatingly, however, combination treatment was effective in removing the SOX2-expressing population and rendered surviving cells non-clonogenic. The exact mechanisms that explain sensitivity to XR combined with RAD51 inhibition in these cells and the association between RAD51 expression and surviving, non-clonogenic cells after XR requires further investigation. Nevertheless, our study provides a strong rationale for targeting RAD51-mediated repair to specifically radiosensitize GSCs.
Experimental Procedures
Cell Lines and Reagents
GBM1 and GBM4 (a gift from H. Wurdak) were maintained as described previously (Wurdak et al., 2010). They were cultured in Neural Basal medium consisting of Neurobasal Medium (Life Technologies), N2 and B27 supplements (Life Technologies, 0.5× each), recombinant basic fibroblast growth factor (bFGF), and epidermal growth factor (EGF) (Peprotech and R&D Systems, respectively, 40 ng/mL). GBM4UCL cells (a gift from S. Brandner, UCL Institute of Neurology) were cultured in DMEM/F12 (1:1 mixture), with 15 mM HEPES and 2 mM L-glutamine (Sigma), 1× B27, and 20 ng/mL each of recombinant EGF and bFGF. GSCs were cultured on laminin-coated (2 μg/mL; Sigma-Aldrich) surfaces. NHAs were purchased from Lonza and grown in AGM astrocyte growth medium (Lonza). U87-MG (ECACC) cells were grown in DMEM supplemented with 2 mM L-glutamine (Sigma) and 10% FBS (Biosera). BMP4 was obtained from Life Technologies. Protease inhibitor cocktail and trypsin were purchased from Sigma-Aldrich. The RAD51 inhibitors B02 and RI-1 were purchased from Sigma-Aldrich and Merck, respectively, and stored as 10 mM stock solutions in DMSO.
Immunofluorescence Microscopy
Cells were cultured on laminin-coated coverslips, fixed in 4% paraformaldehyde (PFA), and blocked with blocking buffer (10% FBS, 0.2% Triton X-100, and 0.1% sodium azide in PBS) for 2 hr at room temperature (RT). Cells were incubated with primary antibody overnight at 4°C in a humidified chamber. The antibodies used were anti-RAD51 (1:1,000) and anti-γH2AX (1:800) (PC130, JBW130, respectively; Merck), anti-SOX2 (1:50) and anti-nestin (1:1,000) (MAB2018, MAB1259, respectively; R&D Systems), and anti-TP53BP1 (ab36823, 1:1,000; Abcam). After washing (3 × 5 min in PBS), cells were incubated with secondary antibodies (donkey anti-rabbit-Cy3, 711165152, Jackson ImmunoResearch; and goat anti-mouse-Alexa 488, A11029, Life Technologies, 1:1,000) for 1 hr at RT. The cells were then washed in PBS with DAPI (0.1 μg/mL; Biotium) and mounted on glass slides with Fluoromount-G (SouthernBiotech). Cells were viewed using a Nikon A1 confocal microscope and images analyzed in Fiji (Schindelin et al., 2012). For the detection of RAD51, background fluorescence (determined from secondary AB-only controls) was subtracted and any cell with detectable fluorescence in the nucleus was scored positive. Foci were counted manually with a minimum of 50 cells scored per condition.
Western Blotting
Whole-cell extracts were prepared in radioimmunoprecipitation assay buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor cocktail), fractionated by SDS-PAGE, and transferred to a nitrocellulose membrane according to the manufacturer's instructions (Invitrogen). Membranes were blocked with LI-COR blocking buffer (LI-COR Biosciences) and incubated with antibodies against RAD51 (D4B10, 1:1,000; Cell Signaling Technology), SOX2 (1:1,000) and nestin (1:1,000) (MAB2018, MAB1259, respectively; R&D Systems), GFAP (Z0334, 1:100; Dako), β-actin (ab8227, 1:20,000), PRKDC (ab32566, 1:5,000), XRCC5/XRCC6 (Ku80/Ku70) (ab53126, 1:100,000), and vinculin (ab129002, 1:20,000) (all from Abcam). Antibody binding was visualized by: (1) incubation with Alexa Fluor 680-conjugated secondary anti-mouse/rabbit antibodies (1:5,000; Life Technologies) and imaged on an Odyssey Infrared Imaging System (LI-COR Biosciences) or (2) using a SuperSignal West Pico Complete Mouse/Rabbit IgG Detection Kit (Thermo Fisher Scientific) according to the manufacturer's instructions, imaging with X-ray film.
Quantitative Real-Time PCR
Total cellular RNA was extracted from cells 48 hr after seeding using RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. Reverse transcription of approximately 400–800 ng of total RNA was carried out to obtain cDNA as described previously (Burchill et al., 1994). For quantitative real-time RT-PCR analysis, cDNA was added to 20 μL of PCR mix to produce a 1× TaqMan Universal PCR Master Mix (containing AmpliTaq Gold DNA Polymerase; Life Technologies) with a 1× RAD51 or β-actin TaqMan Gene Expression Assay (Life Technologies) in RNase-free H2O. Samples were analyzed using an Applied Biosystems 7500 Real-Time PCR instrument. Ct values for candidate genes were normalized to the mean Ct values for β-actin for both test and control samples. The resulting test values were normalized to control samples.
Single-Cell Experiments
GBM1 cells were seeded into 6-well plates (500–1,500 cells/well) and untreated or treated with 2 Gy radiation, RI-1 (1.5 μM), or RI-1 and 2 Gy radiation, and then incubated for 1–3 weeks at 37°C. Cells were harvested by trypsinization and resuspended in culture medium at 2.5 × 105 cells/mL. Cells were diluted in C1 cell suspension reagent at a ratio of 3:2, respectively. Single cells were captured onto a medium (10–17 μM) C1 Single-Cell Auto Prep IFC for PreAmp according to the manufacturer's instructions using the Fluidigm C1 Single-Cell Auto Prep System (Fluidigm). Preamplification was conducted using pooled TaqMan primers (180 nM) for the targets of interest (Life Technologies). Amplified products were diluted with C1 DNA dilution reagent (25 μL) and stored at −20°C until use. Bulk tube controls were prepared from the remaining GBM1 cells according to the manufacturer's instructions. Single-cell cDNA was amplified for the genes of interest on a 96.96 Dynamic Array IFC using the Fluidigm Biomark system. Gene expression was analyzed using Fluidigm analysis software. Log2(Exp) expression values were calculated by subtracting the individual Ct values from 30. A Ct value of 30 was used as the limit of detection for the assays. BoxPlotR software was used to generate bean plots of the Log2(exp) values. R was used to generate density and boxplots. SINGuLAR Analysis Toolset was used to calculate significance between treatment groups.
Immunohistochemistry
Tumor samples were collected from consenting patients undergoing surgery at the Leeds Teaching Hospitals NHS Trust in accordance with local ethics. Samples were fixed for 24 hr in 4% PFA at 4°C and transferred to 70% ethanol before embedding in paraffin. Tumor sections (5 μm) were mounted onto Superfrost Plus slides (Thermo Fisher Scientific). Dewaxing and antigen retrieval was performed using MenaPath slide wash and access EDTA (pH 8.5–8.7) in a pressure cooker. Slides were incubated with 3% hydrogen peroxide for 20 min to block endogenous peroxide. Slides were mounted in Sequenza racks, washed with Tris-buffered saline (TBS) and blocked with casein-blocking solution (Vector) for 30 min at RT. Antibodies to RAD51 (E19100 Spring Bioscience, 1:75), SOX2, and NES (MAB2018; 1:800, MAB1259; 1:10,000, respectively; R&D Systems) were incubated with the slides overnight at 4°C. After washing with TBS, the cells were incubated with an X-Cell Plus multiplex HRP-ALP Kit (MenaPath) for 30 min. Bound antibodies were visualized with Vulcan Fast Red chromogen or diaminobenzidine (MenaPath) for rabbit and mouse antibodies, respectively, before counterstaining with hematoxylin, dehydration, and mounting with coverslips. Slides were scanned on an Aperio ScanScope AT slide scanner (Leica), tumor regions were manually annotated, and 100 sub-regions for analysis randomly assigned using RandomSpot (Wright et al., 2015). Image patches were extracted using the coordinates in the annotated XML files. Extracted images were split into separate staining channels using color deconvolution, which were individually thresholded into foreground and background components using Otsu's bimodal histogram method. The two resulting masks were then used to determine the amount of staining present for each stain. Using the spatial distribution of both masks, the intersection of each channel was calculated in order to identify the level of co-localization of stains. The presence of stains was reported as a percentage of pixels within the staining channel masks in relation to the total number of pixels in the image.
Clonogenic Survival Assay
Clonogenic survival assays were carried out as described previously (Franken et al., 2006). In brief, cells were seeded into 24-well plates at 125 cells/well in medium containing the appropriate treatment and incubated for 2 hr at 37°C before irradiation. The cells were returned to the incubator for 3 weeks. Cells were fixed with 4% PFA for 15 min and stained with methylene blue (1%, w/v, 50%, v/v, ethanol) for 30 min. After washing in water the colonies were counted using a Gallenkamp colony counter. Data were analyzed in Prism using the LQ model of radiation survival described by the equation S = exp(−αd − βd2), where S is the survival fraction, d is the XR dose, and α and β are constants (Douglas and Fowler, 1976).
Annexin V/PI Assay
Cell killing was determined using the BD Biosciences FITC Annexin V Apoptosis Detection Kit II according to the manufacturer's instructions. Cells were seeded at 5 × 104 cells/well in a 24-well plate and incubated at 37°C. After 24 hr, the cells were treated with RI-1 or B02 at 7.5 μM for a further 24 hr and then irradiated with 2 Gy. The cells were returned to the incubator for 5 days then harvested and stained using an FITC Annexin V Apoptosis Detection Kit II and analyzed on an Applied Biosystems Attune acoustic focusing cytometer. Data were analyzed using Attune software. Unstained cells were considered live, Annexin V-positive cells were designated to be early apoptotic, PI-only-positive cells necrotic, and double-positive cells apoptotic.
In Vivo Studies
Animals were purchased from the breeding facility, University of Leeds. U87-MG cells (1 × 106), in 50 μL of PBS, were injected subcutaneously into the right flank of 7–9-week-old female BALB/c nude mice. Once tumors were palpable (approximately 5 mm diameter), animals were randomly assigned into experimental groups. Treatment consisted of three cycles over 5 days. RAD51 inhibitor RI-1 was administered at a concentration of 20 μM in a solution of 30% DMSO, 35% PEG 400, and 35% PBS. On each treatment day, animals received intratumoral injections of 100 μL of PBS/DMSO/PEG 400 solution (control) or 20 μM RI-1. Three hours after injection, animals were either mock irradiated (anesthetized but not irradiated) or received image-guided radiotherapy delivered by SARRP (XStrahl). For each treatment, a dose of 5 Gy was targeted to the tumor using a 10 × 10 mm collimator and two opposing beams. After treatment, animal welfare was monitored daily, and tumors were measured three times a week using calipers. Animals were killed once tumors attained a mean diameter of 12.5 mm. Tumor volumes were calculated using (a2b)/2, where a and b represent the smallest and largest dimensions of the tumor, respectively. Tumor growth rate was determined as described by Demidenko (2010). Plotting the natural log of tumor volume against time gives a straight line described by the equation lnV = α + βt, where V is the tumor volume, α is the y intercept (tumor starting volume), β is the growth rate, and t is the time. Tumor DT was calculated using DT = ln2/β (Demidenko, 2010, Mehrara et al., 2007). All experiments were performed in accordance with the Animals (Scientific Procedures) Act 1986, under license number 70/7340.
Author Contributions
H.O.K., T.B., H.L.P., H.W., and S.C.S. conceived and designed experiments. H.O.K., T.B., H.L.P., T.E., and A.P performed experiments. H.O.K., T.B., H.L.P., A.W., L.F.S., A.P., H.W., and S.C.S analysed and interpreted data. H.O.K., T.B., H.L.P., T.A.W., K.P., H.W., and S.C.S. prepared figures and wrote the manuscript. All authors contributed critical evaluation of the manuscript.
Acknowledgments
This work was funded by CRUK (grant A10065). A.P. and H.W. thank Worldwide Cancer Research for support (grant 13-0146).
Published: January 10, 2017
Footnotes
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.12.005.
Supplemental Information
References
- Audeh M.W., Carmichael J., Penson R.T., Friedlander M., Powell B., Bell-McGuinn K.M., Scott C., Weitzel J.N., Oaknin A., Loman N. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Balbous A., Cortes U., Guilloteau K., Villalva C., Flamant S., Gaillard A., Milin S., Wager M., Sorel N., Guilhot J. A mesenchymal glioma stem cell profile is related to clinical outcome. Oncogenesis. 2014;3:e91. doi: 10.1038/oncsis.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Berezovsky A.D., Poisson L.M., Cherba D., Webb C.P., Transou A.D., Lemke N.W., Hong X., Hasselbach L.A., Irtenkauf S.M., Mikkelsen T. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia. 2014;16:193–206. doi: 10.1016/j.neo.2014.03.006. 206.e19–e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B.B., Logan H.L.H., Kalin J.H.J., Zelivianskaia A.S.A., McGuire W.W.C., Miller L.L.L., Stark J.M.J., Kozikowski A.P.A., Bishop D.K.D., Connell P.P.P. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 2012;40:7347–7357. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B., Kalin J.H., Pawlowski M., Zelivianskaia A.S., Wu M., Kozikowski A.P., Connell P.P. An optimized RAD51 inhibitor that disrupts homologous recombination without requiring Michael acceptor reactivity. J. Med. Chem. 2012;56:254–263. doi: 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill S.A., Bradbury F.M., Smith B., Lewis I.J., Selby P. Neuroblastoma cell detection by reverse transcriptase-polymerase chain reaction (RT-PCR) for tyrosine hydroxylase mRNA. Int. J. Cancer. 1994;57:671–675. doi: 10.1002/ijc.2910570510. [DOI] [PubMed] [Google Scholar]
- Callen E., Di Virgilio M., Kruhlak M.J., Nieto-Soler M., Wong N., Chen H.T., Faryabi R.B., Polato F., Santos M., Starnes L.M. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caren H., Stricker S.H., Bulstrode H., Gagrica S., Johnstone E., Bartlett T.E., Feber A., Wilson G., Teschendorff A.E., Bertone P. Glioblastoma stem cells respond to differentiation cues but fail to undergo commitment and terminal cell-cycle arrest. Stem Cell Rep. 2015;5:829–842. doi: 10.1016/j.stemcr.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Sossick A.J., Boulton S.J., Jackson S.P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell. Sci. 2012;125:3529–3534. doi: 10.1242/jcs.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Wu Q., Huang Z., Guryanova O.A., Huang Q., Shou W., Rich J.N., Bao S. L1CAM regulates DNA damage checkpoint response of glioblastoma stem cells through NBS1. EMBO J. 2011;30:800–813. doi: 10.1038/emboj.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko E. Three endpoints of in vivo tumour radiobiology and their statistical estimation. Int. J. Radiat. Biol. 2010;86:164–173. doi: 10.3109/09553000903419304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks P.B. Brain tumor stem cells: bringing order to the chaos of brain cancer. J. Clin. Oncol. 2008;26:2916–2924. doi: 10.1200/JCO.2008.17.6792. [DOI] [PubMed] [Google Scholar]
- Dong C., Zhang F.M., Luo Y., Wang H., Zhao X.P., Guo G.S., Powell S.N., Feng Z.H. p53 suppresses hyper-recombination by modulating BRCA1 function. DNA Repair. 2015;33:60–69. doi: 10.1016/j.dnarep.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas B.G., Fowler J.F. The effect of multiple small doses of X rays on skin reactions in the mouse and a basic interpretation. Radiat. Res. 1976;66:401. [PubMed] [Google Scholar]
- Facchino S., Abdouh M., Chatoo W., Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J. Neurosci. 2010;30:10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken N.A.P., Rodermond H.M., Stap J., Haveman J., van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- Hall E.J., Giaccia A.J. Lippincott Williams & Wilkins; 2006. Radiobiology for the Radiologist. [Google Scholar]
- Hannay J.A.F., Liu J., Zhu Q.-S., Bolshakov S.V., Li L., Pisters P.W.T., Lazar A.J.F., Yu D., Pollock R.E., Lev D. RAD51 overexpression contributes to chemoresistance in human soft tissue sarcoma cells: a role for p53/activator protein 2 transcriptional regulation. Mol. Cancer Ther. 2007;6:1650–1660. doi: 10.1158/1535-7163.MCT-06-0636. [DOI] [PubMed] [Google Scholar]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Mazin A.V. A small molecule inhibitor of human RAD51 potentiates breast cancer cell killing by therapeutic agents in mouse xenografts. PLoS One. 2014;9:e100993. doi: 10.1371/journal.pone.0100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.F., Mazina O.M.O., Zentner I.J.I., Cocklin S.S., Mazin A.V.A. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. J. Med. Chem. 2012;55:3011–3020. doi: 10.1021/jm201173g. [DOI] [PubMed] [Google Scholar]
- Jeggo P.A., Geuting V., Lobrich M. The role of homologous recombination in radiation-induced double-strand break repair. Radiother. Oncol. 2011;101:7–12. doi: 10.1016/j.radonc.2011.06.019. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Henderson S.T., Petes T.D., Prakash S., Bankmann M., Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 1992;12:3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kotiliarova S., Kotiliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim K.H., Kim D.G., Cho H.J., Kim Y., Rheey J., Shin K., Seo Y.J., Choi Y.S., Lee J.I. FoxM1 promotes stemness and radio-resistance of glioblastoma by regulating the master stem cell regulator Sox2. PLoS One. 2015;10:e0137703. doi: 10.1371/journal.pone.0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke D., Weiler M., Blaes J., Wiestler B., Jestaedt L., Klein A.-C., Löw S., Eisele G., Radlwimmer B., Capper D. Primary glioblastoma cultures: can profiling of stem cell markers predict radiotherapy sensitivity? J. Neurochem. 2014;131:251–264. doi: 10.1111/jnc.12802. [DOI] [PubMed] [Google Scholar]
- Lim Y.C., Roberts T.L., Day B.W., Harding A., Kozlov S., Kijas A.W., Ensbey K.S., Walker D.G., Lavin M.F. A role for homologous recombination and abnormal cell-cycle progression in radioresistance of glioma-initiating cells. Mol. Cancer Ther. 2012;11:1863–1872. doi: 10.1158/1535-7163.MCT-11-1044. [DOI] [PubMed] [Google Scholar]
- Lim Y.C., Roberts T.L., Day B.W., Stringer B.W., Kozlov S., Fazry S., Bruce Z.C., Ensbey K.S., Walker D.G., Boyd A.W. Increased sensitivity to ionizing radiation by targeting the homologous recombination pathway in glioma initiating cells. Mol. Oncol. 2014;8:1603–1615. doi: 10.1016/j.molonc.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maacke H., Jost K., Opitz S., Miska S., Yuan Y., Hasselbach L., Luttges J., Kalthoff H., Sturzbecher H.W. DNA repair and recombination factor RAD51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000;19:2791–2795. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- Mannino M., Chalmers A.J. Radioresistance of glioma stem cells: intrinsic characteristic or property of the “microenvironment-stem cell unit?”. Mol. Oncol. 2011;5:374–386. doi: 10.1016/j.molonc.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrara E., Forssell-Aronsson E., Ahlman H., Bernhardt P. Specific growth rate versus doubling time for quantitative characterization of tumor growth rate. Cancer Res. 2007;67:3970–3975. doi: 10.1158/0008-5472.CAN-06-3822. [DOI] [PubMed] [Google Scholar]
- Piccirillo S.G., Reynolds B.A., Zanetti N., Lamorte G., Binda E., Broggi G., Brem H., Olivi A., Dimeco F., Vescovi A.L. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Pollard S.M., Yoshikawa K., Clarke I.D., Danovi D., Stricker S., Russell R., Bayani J., Head R., Lee M., Bernstein M. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Qin Q., Cheng H.Y., Lu J., Zhan L.L., Zheng J.C., Cai J., Yang X., Xu L.P., Zhu H.C., Zhang C. Small-molecule survivin inhibitor YM155 enhances radiosensitization in esophageal squamous cell carcinoma by the abrogation of G(2) checkpoint and suppression of homologous recombination repair. J. Hematol. Oncol. 2014;7:62. doi: 10.1186/s13045-014-0062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A., Smith C.A., Agnihotri S., Shekarforoush M., Kongkham P.N., Seol H.J., Northcott P., Rutka J.T. Epigenetic regulation of glial fibrillary acidic protein by DNA methylation in human malignant gliomas. Neuro Oncol. 2011;13:42–50. doi: 10.1093/neuonc/noq145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short S.C., Giampieri S., Worku M., Alcaide-German M., Sioftanos G., Bourne S., Lio K.I., Shaked-Rabi M., Martindale C. RAD51 inhibition is an effective means of targeting DNA repair in glioma models and CD133+ tumor-derived cells. Neuro Oncol. 2011;13:487–499. doi: 10.1093/neuonc/nor010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva M.L., Rheinbay E., Gillespie S.M., Patel A.P., Wakimoto H., Rabkin S.D., Riggi N., Chi A.S., Cahill D.P., Nahed B.V. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennstedt P., Fresow R., Simon R., Marx A., Terracciano L., Petersen C., Sauter G., Dikomey E., Borgmann K. RAD51 overexpression is a negative prognostic marker for colorectal adenocarcinoma. Int. J. Cancer. 2012;132:2118–2126. doi: 10.1002/ijc.27907. [DOI] [PubMed] [Google Scholar]
- Tutt A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N., Friedlander M., Arun B., Loman N., Schmutzler R.K. The future is “PARP” – phase-II-activity of PARP inhibitor among patients with BRCA-deficient breast cancer. Breast Care. 2010;5:364–365. [Google Scholar]
- Ward A., Khanna K.K., Wiegmans A.P. Targeting homologous recombination, new pre-clinical and clinical therapeutic combinations inhibiting RAD51. Cancer Treat. Rev. 2015;41:35–45. doi: 10.1016/j.ctrv.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Welsh J.W., Ellsworth R.K., Kumar R., Fjerstad K., Martinez J., Nagel R.B., Eschbacher J., Stea B. RAD51 protein expression and survival in patients with glioblastoma multiforme. Int. J. Radiat. Oncol. Biol. Phys. 2009;74:1251–1255. doi: 10.1016/j.ijrobp.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Wright A., Grabsch H., Treanor D. RandomSpot: a web-based tool for systematic random sampling of virtual slides. J. Pathol. Inform. 2015;6:8. doi: 10.4103/2153-3539.151906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdak H., Zhu S., Romero A., Lorger M., Watson J., Chiang C.-Y., Zhang J., Natu V.S., Lairson L.L., Walker J.R. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell. 2010;6:37–47. doi: 10.1016/j.stem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F.W., Chen J. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Taki T., Yagi H., Habu T., Yoshida K., Yoshimura Y., Yamamoto K., Matsushiro A., Nishimune Y., Morita T. Cell cycle-dependent expression of the mouse RAD51 gene in proliferating cells. Mol. Gen. Genet. 1996;251:1–12. doi: 10.1007/BF02174338. [DOI] [PubMed] [Google Scholar]
- Zeppernick F., Ahmadi R., Campos B., Dictus C., Helmke B.M., Becker N., Lichter P., Unterberg A., Radlwimmer B., Herold-Mende C.C. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin. Cancer Res. 2008;14:123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- Zhang N., Wu X., Yang L., Xiao F., Zhang H., Zhou A., Huang Z., Huang S. FoxM1 inhibition sensitizes resistant glioblastoma cells to temozolomide by downregulating the expression of DNA-repair gene RAD51. Clin. Cancer Res. 2012;18:5961–5971. doi: 10.1158/1078-0432.CCR-12-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.