Abstract
Synechocystis PCC 6803 contains two types of glutathione peroxidase-like proteins (GPX-1 and GPX-2) that utilize NADPH but not reduced glutathione and unsaturated fatty acid hydroperoxides or alkyl hydroperoxides. The steady-state transcript level of gpx-1 gradually increased under oxidative stress conditions imposed by high light intensity, high salinity, or application of methylviologen or t-butyl hydroperoxide in the wild-type and GPX-2 knock-out mutant (gpx-2Δ) cells. To examine the ability of GPX-1, GPX-2, and thioredoxin peroxidase to scavenge lipid hydroperoxide in vivo, we measured the photosynthetic evolution of O2 and the level of lipid peroxidation in the wild-type and each type of mutant cell after the application of t-butyl hydroperoxide or H2O2. The data reported here indicate that GPX-1 and GPX-2 are essential for the removal of lipid hydroperoxides under normal and stress conditions, leading to the protection of membrane integrity.
Oxidative damage poses a great risk to the survival of cells because it can undermine cellular structures. Aerobic organisms have evolved a variety of enzymatic and nonenzymatic mechanisms to cope with the deleterious effects of active oxygen species (AOS). In photosynthetic organisms ascorbate peroxidase is the major H2O2 scavenging enzyme, while in animals glutathione peroxidase (GPX) is the major enzyme (Ursini et al., 1995; Shigeoka et al., 2002). GPX can also reduce alkyl and lipid hydroperoxides to protect the cells from lipid peroxidation.
Several cDNAs encoding proteins similar to animal GPX have been identified in higher plants (Criqui et al., 1992; Holland et al., 1993; Churin et al., 1999; Li and Sherman, 2000), Chlamydomonas (Yokota et al., 1988), and yeast (Saccharomyces cerevisiae; Inoue et al., 1999). GPX-like proteins of plants exhibit the highest identity to the mammalian selenium-dependent phospholipid hydroperoxide GPX. However, these plant genes carry a codon for Cys residues instead of selenocysteine residue in mammalian GPXs, resulting in lower activity by three orders of magnitude compared to the activity of the homologous animal GPX (Maiorino et al., 1995). These facts thus lead to a limited understanding of the potential physiological role of GPX-like proteins in photosynthetic organisms.
Synechocystis PCC 6803 contains catalase-peroxidase and thioredoxin peroxidase (TPX) as the scavenging system for H2O2 and/or alkyl hydroperoxides (Jakopitsch et al., 1999; Yamamoto et al., 1999). Recently, we characterized two genes encoding GPX-like proteins (GPX-1 and GPX-2) from Synechocystis PCC 6803 and demonstrated that the activities of both recombinant enzymes could be detected using NADPH but not reduced glutathione, and unsaturated fatty acid hydroperoxides or alkyl hydroperoxides (Gaber et al., 2001).
In this study, to clarify the physiological role of both enzymes in vivo, we studied the effect of some stress conditions on the transcript and protein levels of GPX-1 and GPX-2 in wild-type and the respective gene-disrupted mutant cells. The expression pattern observed indicated that each gpx gene has a different response to stress conditions.
RESULTS
Targeted Disruption of the Genes for GPX-1 and GPX-2 in Synechocystis PCC 6803
We performed PCR analysis with DNA of wild-type, gpx-1Δ, or gpx-2Δ mutant cells that had been transformed with the vector pT7-gpx-1/kanr (Kanamycin resistance cartridge gene) or pT7-gpx-2/kanr, which had been constructed for the transformations. PCR with chromosomal DNA of wild-type cells as a template amplified a 0.5-kb DNA fragment for gpx-1 and a 0.46-kb DNA fragment for gpx-2, while PCR with DNA from cells with disrupted gpx-1 or gpx-2 yielded a fragment of 1.76 kb (data not shown). These results indicated that both genes in all mutant cells had been disrupted by the insertion of the kanr gene.
We investigated the effect of disruption of each gene on the cell viability of Synechocystis PCC 6803 cells. Under illumination at 30 μE m−2 s−1, the growth and chlorophyll levels of both types of mutant cells were the same as those of the wild-type cells. There were no significant differences in the rate of NaHCO3-dependent O2 evolution between wild-type and mutant cells at 90, 250, 600, and 1,000 μE m−2 s−1 or 0.5, 1, and 2 mm NaHCO3 at 27°C (data not shown). We also found no effect on the activities of other antioxidant enzymes in each mutant.
Next, we attempted to produce double-mutant cells (gpx-1Δ and gpx-2Δ) using the kanr gene and the chloramphenicol resistance cartridge gene (chmr). On the first segregation of gpx-2Δ cells transformed with pgpx-1/chmr, PCR amplification of gpx-1 from chromosomal DNA yielded both the gpx-1 gene and disrupted gene (pgpx-1/chmr). At the following segregation, the cells could not grow in Allen's medium (Allen, 1968) supplemented with chloramphenicol and Kanamycin under various light intensities (10, 30, 100, and 240 μE m−2 s−1). The same results were obtained in the gpx-1Δ cells transformed with pgpx-2/chmr. Thus, these results suggest that Synechocystis PCC 6803 needs at least one of the gpx genes for survival.
Supply of NADPH for Both GPX Isoenzymes
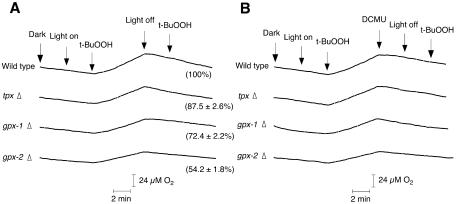
The H2O2-dependent evolution of O2 by Synechocystis PCC 6803 occurred only in the light (Miyake et al., 1991). When GPX-1 and GPX-2 scavenge peroxides using NADPH, the evolution of O2 from PSII should occur in the presence of t-butyl hydroperoxide (t-BuOOH) in the light. The addition of 10 mm glycolaldehyde, which inhibits CO2 fixation in cyanobacteria (Miller and Canvin, 1989), caused a rapid inhibition of steady-state O2 evolution. Then, the addition of t-BuOOH to wild-type cells and each type of mutant cells induced the t-BuOOH-dependent evolution of O2 in the light, but the evolution of O2 stopped in the dark (Fig. 1A). Further addition of t-BuOOH did not cause the evolution of O2 in the dark. The addition of 10 μm 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) caused the t-BuOOH-dependent evolution of O2 to completely stop in the light (Fig. 1B). The rate of O2 evolution of gpx-1Δ or gpx-2Δ mutants was lower than that of wild-type cells and was related to the level of GPX-1 and GPX-2 in each mutant cell. In the tpxΔ mutant, the t-BuOOH-dependent evolution of O2 could be detected and was higher than those of the gpx-1Δ or gpx-2Δ mutants.
Figure 1.
t-BuOOH-dependent evolution of oxygen in wild-type, gpx-1Δ, gpx-2Δ, and tpxΔ mutant cells in the absence (A) or presence (B) of DCMU. Glycolaldehyde was added at 10 mm to the suspension of cells to inhibit CO2 fixation. Light (1,000 μE m−2 s−1) was turned on and off, and t-BuOOH (70 μm) and DCMU (10 μm) were added to the suspension of cells at the times indicated by labeled arrows. The number in parentheses indicates the rate of t-BuOOH-dependent evolution of oxygen; the rate at 100% corresponds to 43.9 μmol O2 evolved mg chlorophyll−1 h−1. The data are the mean value ± sd of three independent experiments.
Changes of the Levels of Transcript and Protein of GPX-1 and GPX-2 under Stress Conditions
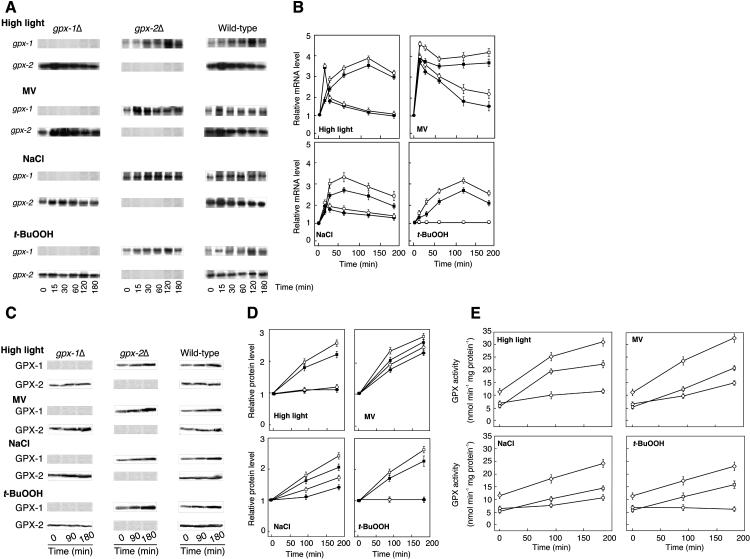
The transcript levels of gpx-2 in the wild-type and GPX-1 knock-out mutant (gpx-1Δ) cells increased after 15 min in response to high light at 350 μE m−2 s−1 followed by its decline until it reached almost the same value as that in the untreated cells (Fig. 2, A and B), which was in agreement with the results reported by Hihara et al. (2001). In contrast, there was a significant up-regulation of the gpx-1 mRNA level, which was steadily increased by approximately 3.5-fold until 2 h. Next, wild-type and each type of mutant cells were treated with 1.0 μm methylviologen (MV), 200 mm NaCl, or 0.2 mm t- BuOOH. All of these types of environmental stress increased the level of gpx-1 transcript (Fig. 2, A and B). MV and salt stress conditions increased the expression level of gpx-2 by approximately 2-fold and 1.5-fold, respectively. The level of gpx-2 transcript did not change in response to treatment with t-BuOOH.
Figure 2.
The effect of various stress conditions on transcript and protein levels and activities of GPX-1 and GPX-2. A, Northern-blot analysis. Detailed conditions for experiments are described in “Materials and Methods.” B, Relative transcript levels of GPX-1 and GPX-2 in the wild-type and each type of mutant cells. The mRNA level of each sample was expressed as the mean value ± sd of three independent experiments. The value of each transcript level at zero time was set to 1. C, Immunoblot analysis. D, Relative levels of GPX-1 and GPX-2 proteins. The protein ratio in each sample was densitometrically quantified and presented as the mean value of three independent experiments. E, Enzyme activities. ▪, GPX-1 in wild-type cells; •, GPX-2 in wild-type cells; □, GPX-1 in gpx-2Δ mutant; ○, GPX-2 in gpx-1Δ mutant; ⋄, GPX-1 plus GPX-2 in wild-type cells.
Furthermore, we investigated the changes of GPX-1 and GPX-2 proteins in response to these stress conditions (Fig. 2, C and D). The change in the level of GPX-1 protein was in agreement with the change in the transcript level under all types of environmental stress. The protein level of GPX-2 increased approximately 3-fold and 1.6-fold at 3 h with the MV and NaCl treatments, respectively. Under high light conditions, the protein level of GPX-2 increased approximately 1.2-fold. However, no significant change in the protein level of GPX-2 was observed upon treatment with t-BuOOH (Fig. 2, C and D).
The activities of GPX-1 and GPX-2 increased in response to MV and NaCl treatment in parallel with the transcript abundance (Fig. 2E). The activity of GPX-1 was increased by treatment with high light and t-BuOOH, whereas the activity of GPX-2 was not changed by treatment with high light or t-BuOOH.
Effect of Oxidative Stress Treatments on Oxygen Evolution in gpx-1Δ, gpx-2Δ, tpxΔ, and Wild-Type Cells
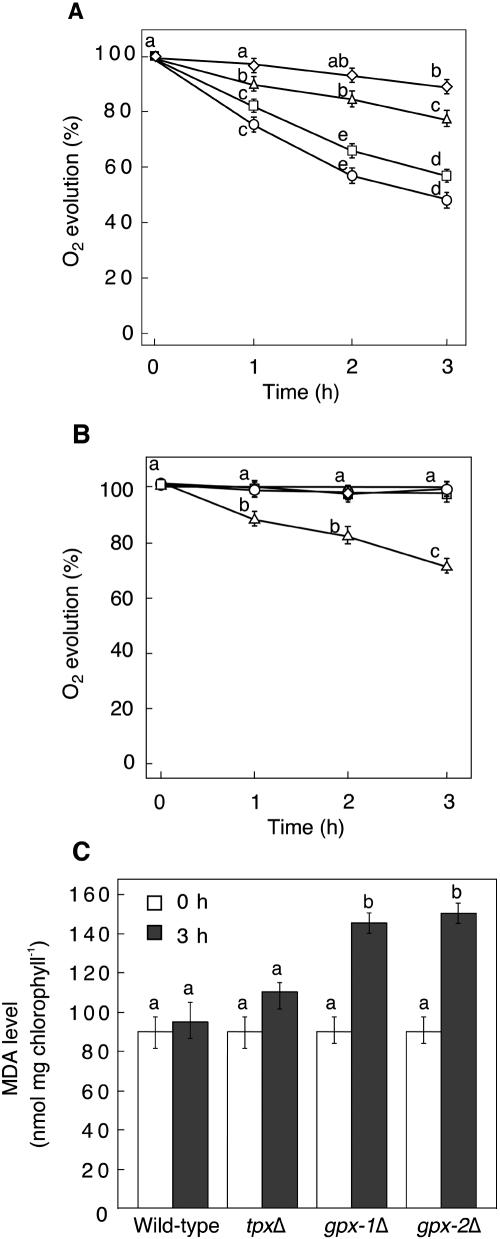
The photosynthetic evolution rates of O2 in gpx-1Δ and gpx-2Δ mutants were markedly decreased by incubation with 0.2 mm t-BuOOH, while the rate in tpxΔ mutant cells was only slightly decreased compared with that in wild-type cells (Fig. 3A). In response to incubation with 0.2 mm H2O2, the O2 evolution in wild-type and gpx-1Δ and gpx-2Δ mutant cells was not changed. However, under the same conditions, the rate of O2 evolution in tpxΔ mutant cells was significantly decreased (Fig. 3B).
Figure 3.
The effect of treatment with t-BuOOH or H2O2 on the viability of the wild-type, gpx-1Δ, gpx-2Δ, and tpxΔ mutant cells. A, Changes in O2 evolution rate during incubation in the presence of 0.2 mm t-BuOOH. B, Changes in O2 evolution rate during incubation in the presence of 0.2 mm H2O2. The rate at 100% corresponded to 250 μmol O2 evolved mg chlorophyll−1 h−1. ⋄, Wild type; ▵, tpxΔ mutant; □, gpx-2Δ mutant; ○, gpx-1Δ mutant cells. C, Lipid hydroperoxide content in the wild-type and mutant cells after treatment with t-BuOOH for 3 h. The data are the mean value ± sd of three independent experiments. Different letters indicate that the mean values are significantly different from those of the wild-type cells (P < 0.05).
The changes in lipid hydroperoxide induced by addition of 0.2 mm t-BuOOH were measured by determining the malondialdehyde (MDA). The production of MDA was increased approximately 1.5-fold in both types of mutant cells (Fig. 3C). In contrast, there was no significant change in the MDA contents of wild-type or tpxΔ mutant cells under the same conditions.
Effect of Disruption of the tpx Gene on the Expression of GPX Isoenzymes
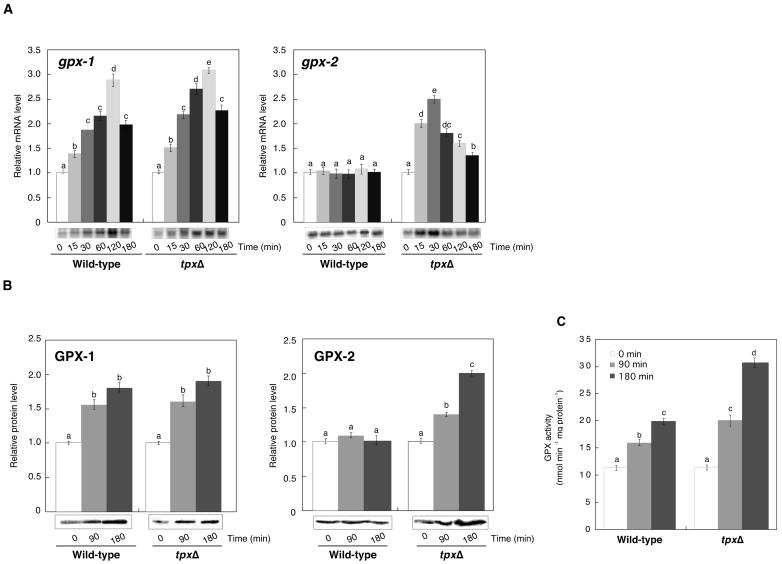
Transcripts, proteins, and enzyme activities of GPX-1 and GPX-2 were determined in wild-type and tpxΔ mutant cells after the treatment with 0.2 mm t-BuOOH. The transcript levels of gpx-2 were increased 15 min after the treatment in the tpxΔ mutant but not in wild-type cells (Fig. 4A). The level of GPX-2 protein increased approximately 2-fold in the tpxΔ mutant, though there was no significant increase in wild-type cells (Fig. 4B). The transcript and protein of GPX-1 in tpxΔ mutant cells increased rapidly in agreement with those in wild-type cells (Fig. 4, A and B). The combined GPX activities in the tpxΔ mutant were approximately 1.5-fold higher than those in wild-type cells (Fig. 4C).
Figure 4.
Effect of tpx null mutation on the transcription, protein levels, and activities of GPX-1 and GPX-2 during treatment with t-BuOOH. A, Relative transcript levels of GPX-1 and GPX-2 in the tpxΔ mutant and wild-type cells. B, Relative levels of GPX-1 and GPX-2 proteins. C, NADPH-dependent GPX activities after treatment with 0.2 mm t-BuOOH for 3 h. The data are the mean value ± sd of three independent experiments. Different letters indicate that the mean values are significantly different from those of the cells at zero time (P < 0.05).
DISCUSSION
The Reduction of t-BuOOH by GPX-1 and GPX-2 via NADPH with Electrons from PSI
We extended our observation that GPX-like proteins (GPX-1 and GPX-2) in Synechocystis PCC 6803 have the ability to reduce fatty acid hydroperoxides or alkyl hydroperoxides using NADPH (Gaber et al., 2001). These results raise the question of how NADPH is supplied. It has been reported that the peroxidases use the electrons generated during the photosynthetic electron transport and these activities are not observed in the presence of DCMU or in the dark (Miyake et al., 1991). As shown in Figure 1, the t-BuOOH-dependent evolution of O2 was coupled to the photosynthetic electron transport system in Synechocystis PCC 6803, indicating that GPX-1 and GPX-2 reduce t-BuOOH using electrons from PSI.
The Induction of GPX-1 and GPX-2 Expressions in Response to Several Stress Conditions
It has been reported that the steady-state levels of gpx mRNA and/or GPX protein in photosynthetic organisms, including eukaryotic algae, increase in response to high light, high osmolarity, t-BuOOH, MV, H2O2, or high salinity (Sugimoto and Sakamoto, 1997; Roeckel-Drevet et al., 1998; Leisinger et al., 1999). Escherichia coli cells expressing the cDNA for a putative Citrus GPX were more tolerant to MV treatment than the wild-type cells; moreover, the gpx-related gene from Citrus sinensis was strongly induced by salinity stress (Holland et al., 1994). The transcript levels of the gpx-1 and gpx-2 genes increased after the exposure of wild-type and mutant cells to high light (Fig. 2, A and B). Treatment of wild-type and gpx-1Δ and gpx-2Δ mutant cells with MV drastically increased the transcript levels and protein levels of both GPX-1 and GPX-2 (Fig. 2, A–D). Salinity stress also led to increases in the levels of the transcript and protein of GPX-1 and GPX-2 in wild-type and mutant cells. Thus, GPX-1 and GPX-2 seem to contribute to the scavenging of lipid hydroperoxide generated by oxidative stress caused by high light, MV treatment, and high salinity.
Alkyl hydroperoxides like t-BuOOH readily react with transition-metal reductants or catalysts to form radicals and cause oxidative damage over long distances (Asada, 1994). Thus, defense mechanisms by which the peroxide intermediates are degraded are essential for the cells. Both GPX-1 and GPX-2 can utilize alkyl hydroperoxide (Gaber et al., 2001). However, the levels of transcript and protein of GPX-2 were not changed in response to treatment with t-BuOOH (Fig. 2, A–D), suggesting that GPX-1 and GPX-2 may have some different functions, at least in the case of t-BuOOH, and GPX-1 may be more important than GPX-2 for protection against this oxidant. The expression level of the mRNA of gpx-2 was increased within 15 min and then decreased under oxidative stress conditions imposed by high light, salinity, or treatment with MV (Fig. 2, A and B), so possible alternative roles for GPX-2 must be taken into consideration. It has been speculated that some member of the GPX family such as mammalian selenium-dependent phospholipid hydroperoxide GPX might be involved in signal transduction rather than in the detoxification of hydroperoxides (Ursini et al., 1997).
The Contribution of GPX-1 and GPX-2 to the Scavenging of Lipid Hydroperoxide
Synechocystis PCC 6803 contains catalase-peroxidase, TPX, and GPX isoenzymes as the scavenging system for AOS. Among them, it has been reported that TPX functions in the reduction of both H2O2 and alkyl hydroperoxides (Yamamoto et al., 1999). The cell viability of gpx-1Δ and gpx-2Δ mutants markedly decreased compared with that of the tpxΔ mutant during the incubation of cells with t-BuOOH (Fig. 3A). In contrast, the tpxΔ mutant but not gpxΔ mutants was more hypersensitive to H2O2 than t-BuOOH (Fig. 3B). These results indicate that TPX in Synechocystis PCC 6803 predominantly scavenges H2O2 rather than alkyl hydroperoxide. Furthermore, we demonstrated that expression of the transcript and protein of GPX-2 was increased in the tpxΔ mutant, resulting in the increase of GPX activities (Fig. 4, A–C). Accordingly, it seems likely that when Synechocystis PCC 6803 lost the activity of TPX, the de novo synthesis of GPX-2 proteins was induced as a backup system for TPX.
Lipid hydroperoxides are generated by the enzymatic catalysis of lipooxygenase or the chemical reaction of AOS (Gueta-Dahan et al., 1997). The rate of lipid peroxidation was clearly increased in both types of gpxΔ mutant cells compared with wild-type and tpxΔ mutant cells (Fig. 3C). These results clearly indicate that GPX isoenzymes act to scavenge lipid hydroperoxides for the survival of Synechocystis PCC 6803 cells under normal and stress conditions. This view is supported by the fact that we could not produce double-mutant cells of gpx-1 and gpx-2 genes.
In summary, the results reported here demonstrate that disruption of the gpx-1 or gpx-2 genes is not lethal for the survival of Synechocystis PCC 6803 cells under normal conditions. However, under stressful conditions, both enzymes are essential for protecting/stabilizing the photosynthetic apparatus and perhaps other aspects of the metabolic machinery of the cell from oxidative damage.
MATERIALS AND METHODS
Materials
Reduced glutathione, H2O2, linolenic acid, and t-BuOOH were obtained from Sigma (St. Louis). Hydroperoxides of unsaturated fatty acids were prepared by the method described previously (Gaber et al., 2001). Chemicals were of the highest purity grade commercially available.
Culture Conditions
The wild-type strain of Synechocystis PCC 6803 and its mutant cells were grown photoautotrophically at 27°C in Allen's medium at 30 μE m−2 s−1 under fluorescent lamps. Log-phase cells of Synechocystis PCC 6803 (A730 = 0.6–1.0) were subjected to stress treatments.
Assay of GPX
The cell extracts were prepared as described previously, and the GPX activity was assayed spectrophotometrically (Gaber et al., 2001). Protein was determined by the method of Bradford (1976) with bovine serum albumin as a standard.
Targeted Disruption of the ORFs gpx-1 and gpx-2
The 0.6-kb DNA fragments containing the regions encoding gpx-1 and gpx-2 were amplified by PCR with chromosomal DNA from Synechocystis PCC 6803 and the following primers for the N termini of gpx-1 and gpx-2: 5′-GCTAAATCATATGACTGCCC-3′ and 5′-CTTAACACATATGCCATTAC-3′, respectively, and the following primers for the C termini of gpx-1 and gpx-2: 5′-AGAAAATTACAACAATTTCT-3′ and 5′-ATAGCACACAATGTTTGTGC-3′, respectively. The amplified DNA fragments were cloned into pT7Blue-T (Novagen, Madison, WI), and their nucleotide sequences were confirmed by the dideoxy chain primer method with an automated DNA sequencer (model 310; Perkin Elmer/Applied Biosystems, Chiba, Japan). The kanr (1.2 kb) was excised from pUC4K (Amersham Biosciences, Uppsala; Taylor and Rose, 1988) by digestion with HincII and inserted into the ClaI site of gpx-1 in pT7Blue-T and into the HpaI site of gpx-2 in pT7Blue-T. The resultant plasmids were designated pgpx-1/kanr and pgpx-2/kanr. Then, Synechocystis PCC 6803 cells were transformed with these plasmids to generate gpx-1Δ and gpx-2Δ mutants (Golden et al., 1987). Transformed cells were selected on agar-solidified Allen medium supplemented with 50 μg mL−1 kanamycin, and complete segregation of gpx-1 or gpx-2 in the mutant cells was confirmed by PCR with appropriate primers.
For the generation of double-mutant cells (gpx-1Δ and gpx-2Δ), gpx-2Δ mutant cells were transformed with plasmid pgpx-1/chmr, which was derived from pT7/gpx-1 by interrupting the gpx-1 gene at the ClaI site with chmr (1.5 kb) which was isolated from plasmid pUC 303 (Amersham Biosciences). On the other hand, gpx-1Δ mutant cells were transformed with plasmid pgpx-2/chmr, which was derived from pT7/gpx-2 by interrupting the gpx-2 gene at the HpaI site with chmr.
Determination of Oxygen Evolution Rate
A Clark-type O2 electrode (Hansatech, Norfolk, UK) placed in a water circulation chamber at 27°C was used to measure the photosynthetic rates of cyanobacterial cells. For measurement of cell viability after the treatment with stress compounds, cell samples (2 mL) were directly transferred to an O2 electrode chamber and 2 mm NaHCO3. O2 evolution rates were measured at 1,000 μE m−2 s−1 after 0, 15, 30, 60, 120, or 180 min.
The t-BuOOH-dependent evolution of O2 by the cells was monitored at 27°C at 1,000 μE m−2 s−1. Glycolaldehyde was added at 10 mm to the suspension of cells to inhibit the CO2 fixation via the Calvin cycle (Miller and Canvin, 1989). After the evolution of O2 had stopped in the light, 70 μm t-BuOOH was added to the suspension of cells. The rate of O2 evolution was calculated in terms of μmol of O2 evolved mg chlorophyll−1 h−1. The chlorophyll content was determined by the method of Lichtenthaler (1987).
Analysis of Lipid Peroxidation
Lipid hydroperoxide contents were determined by measuring MDA using the 2-thiobarbituric acid assay as described previously (Roxas et al., 1997). MDA was estimated by measuring A535 and using a molar absorption coefficient of 1.56 × 105 mm−1 cm−1 (Gueta-Dahan et al., 1997).
Northern-Blot Analysis
Total RNA (20 μg) was isolated from the cells (Los et al., 1997) and used for northern-blot analysis with gpx-1 and gpx-2 DNA probes (Yoshimura et al., 2000). The mRNA was quantified using a Mac BAS 1000 image scanner (Fuji Photo Film, Tokyo).
SDS-PAGE and Immunoblotting
Cell extracts were homogenized with SDS-loading buffer (150 mm Tris-HCl, pH 6.8, 4% [w/v] SDS, and 10% [v/v] 2-mercaptoethanol). The homogenates were boiled for 5 min and centrifuged at 10,000g for 5 min at 4°C. The supernatants (40 μg) were analyzed by 15% (w/v) SDS-PAGE (Laemmli, 1970), and then immunoblot analysis was performed for GPX-1 and GPX-2 proteins using polyclonal antibodies raised against each protein (Yoshimura et al., 2000; Gaber et al., 2001).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers BAA18344 for gpx-1 and BAA17881 for gpx-2.
Acknowledgments
We thank Dr. Norio Murata and Dr. Hiroshi Yamamoto, National Institute for Basic Biology, for the generous gift of the tpxΔ mutant.
This work was supported by the Japan Society for the Promotion of Science Research for the Future Program (grant no. JSPS–RFTF 00L01604), by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (grant no. 15380078), and by the Ministry of Agriculture, Forestry, and Fisheries, Japan (grant to S.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044842.
References
- Allen MM (1968) Simple conditions for the growth of unicellular blue-green algae on plates. J Phycol 4: 1–4 [DOI] [PubMed] [Google Scholar]
- Asada K (1994) Production and action of active oxygen species in photosynthetic tissues. In C Foyer, PM Mullineaux, eds, Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants. CRC Press, Boca Raton, FL, pp 77–104
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 717: 1448–1454 [DOI] [PubMed] [Google Scholar]
- Churin Y, Schilling S, Börner T (1999) A gene family encoding glutathione peroxidase homologues in Hordeum vulgare (barley). FEBS Lett 459: 33–38 [DOI] [PubMed] [Google Scholar]
- Criqui MC, Jamet E, Parmentier Y, Marbach J, Durr A, Fleck J (1992) Isolation and characterization of a plant cDNA showing homology to animal glutathione peroxidases. Plant Mol Biol 18: 623–627 [DOI] [PubMed] [Google Scholar]
- Gaber A, Tamoi M, Takeda T, Nakano Y, Shigeoka S (2001) NADPH-dependent glutathione peroxidase-like proteins (Gpx-1, Gpx-2) reduce unsaturated fatty acid hydroperoxides in Synechocystis PCC 6803. FEBS Lett 499: 32–36 [DOI] [PubMed] [Google Scholar]
- Golden SS, Brusslan J, Haselkorn R (1987) Genetic engineering of the cyanobacterial chromosome. Methods Enzymol 153: 215–231 [DOI] [PubMed] [Google Scholar]
- Gueta-Dahan Y, Yaniv Z, Zilinskas BA, Ben-Hayyim G (1997) Salt and oxidative stress: similar and specific responses and their relation to salt tolerance in Citrus. Planta 203: 460–469 [DOI] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland D, Ben-Hayyim G, Faltin Z, Camoin L, Strosberg AD, Eshdat Y (1993) Molecular characterization of salt stress-associated protein in citrus: protein and cDNA sequence homology to mammalian glutathione peroxidases. Plant Mol Biol 21: 923–927 [DOI] [PubMed] [Google Scholar]
- Holland D, Faltin Z, Perl A, Ben-Hayyim G, Eshdat Y (1994) A novel plant glutathione peroxidase-like protein provides tolerance to oxygen radicals generated by paraquat in Escherichia coli. FEBS Lett 337: 52–55 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Matsuda T, Sugiyama KI, Izawa S, Kimura A (1999) Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J Biol Chem 274: 27002–27009 [DOI] [PubMed] [Google Scholar]
- Jakopitsch C, Rüker F, Regelsberger G, Dockal M, Peschek GA, Obinger C (1999) Catalase-peroxidase from the cyanobacterium Synechocystis PCC 6803: cloning, overexpression in Escherichia coli, and kinetic characterization. Biol Chem 380: 1087–1096 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Leisinger U, Rüfenacht K, Zehnder AJB, Eggen RIL (1999) Structure of a glutathione peroxidase homologous gene involved in the oxidative stress response in Chlamydomonas reinhardtii. Plant Sci 149: 139–149 [Google Scholar]
- Li H, Sherman LA (2000) A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 182: 4268–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–380 [Google Scholar]
- Los DA, Ray M, Murata N (1997) Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis PCC 6803. Mol Microbiol 25: 1167–1175 [DOI] [PubMed] [Google Scholar]
- Maiorino M, Aumann KD, Brigelius-Flohé R, Doria D, van den Heuvel J, McCarthy J, Roveri A, Ursin F, Flohé L (1995) Probing the presumed catalytic triad of selenium-containing peroxidases by mutational analysis of phospholipid hydroperoxide glutathione peroxidase (PHGPx). Biol Chem Hoppe Seyler 376: 651–660 [DOI] [PubMed] [Google Scholar]
- Miller AG, Canvin DT (1989) Glycolaldehyde inhibits CO2 fixation in the cyanobacterium Synechococcus UTEX 625 without inhibiting the accumulation of inorganic carbon or the associated quenching of chlorophyll a fluorescence. Plant Physiol 91: 1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake C, Michihata F, Asada K (1991) Scavenging of hydrogen peroxide in prokaryotic and eukaryotic algae: acquisition of ascorbate peroxidase during the evolution of cyanobacteria. Plant Cell Physiol 32: 33–43 [Google Scholar]
- Roeckel-Drevet P, Gagne G, de Labrouhe D, Dufaure J, Nicolas P, Drevet J (1998) Molecular characterization, organ distribution and stress-mediated induction of two glutathione peroxidase-encoding mRNAs in sunflower (Helianthus annuus). Physiol Plant 103: 385–394 [Google Scholar]
- Roxas VP, Smith RK, Allen ER, Allen RD (1997) Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat Biotechnol 15: 988–991 [DOI] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53: 1305–1319 [PubMed] [Google Scholar]
- Sugimoto M, Sakamoto W (1997) Putative phospholipids hydroperoxide glutathione peroxidase gene from Arabidopsis thaliana induced by oxidative stress. Genes Genet Syst 72: 311–316 [DOI] [PubMed] [Google Scholar]
- Taylor LA, Rose RE (1988) A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res 16: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F, Mariorino M, Brigelius-Flohé R, Aumann KD, Roveri A, Schomburg D, Flohé L (1995) Diversity of glutathione peroxidase. Methods Enzymol 252: 38–53 [DOI] [PubMed] [Google Scholar]
- Ursini F, Maiorino M, Roveri A (1997) Phospholipid hydroperoxide glutathione peroxidase (PHGPx): more than an antioxidant enzyme? Biomed Environ Sci 10: 327–332 [PubMed] [Google Scholar]
- Yamamoto H, Miyake C, Dietz KJ, Tomizawa KI, Murata N, Yokota A (1999) Thioredoxin peroxidase in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 447: 269–273 [DOI] [PubMed] [Google Scholar]
- Yokota A, Shigeoka S, Onishi T, Kitaoka S (1988) Selenium as inducer of glutathione peroxidase in low-CO2-grown Chlamydomonas reinhardtii. Plant Physiol 86: 649–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Yabuta Y, Ishikawa T, Shigeoka S (2000) Expression of spinach ascorbate peroxidase isoenzymes in response to oxidative stress. Plant Physiol 123: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]