Abstract
Recent experiments indicate that nitric oxide (NO) plays a pivotal role in disease resistance and several other physiological processes in plants. However, most of the current information about the function of NO in plants is based on pharmacological studies, and additional approaches are therefore required to ascertain the role of NO as an important signaling molecule in plants. We have expressed a bacterial nitric oxide dioxygenase (NOD) in Arabidopsis plants and/or avirulent Pseudomonas syringae pv tomato to study incompatible plant-pathogen interactions impaired in NO signaling. NOD expression in transgenic Arabidopsis resulted in decreased NO levels in planta and attenuated a pathogen-induced NO burst. Moreover, NOD expression in plant cells had very similar effects on plant defenses compared to NOD expression in avirulent Pseudomonas. The defense responses most affected by NO reduction during the incompatible interaction were decreased H2O2 levels during the oxidative burst and a blockage of Phe ammonia lyase expression, the key enzyme in the general phenylpropanoid pathway. Expression of the NOD furthermore blocked UV light-induced Phe ammonia lyase and chalcone synthase gene expression, indicating a general signaling function of NO in the activation of the phenylpropanoid pathway. NO possibly functions in incompatible plant-pathogen interactions by inhibiting the plant antioxidative machinery, and thereby ensuring locally prolonged H2O2 levels. Additionally, albeit to a lesser extent, we observed decreases in salicylic acid production, a diminished development of hypersensitive cell death, and a delay in pathogenesis-related protein 1 expression during these NO-deficient plant-pathogen interactions. Therefore, this genetic approach confirms that NO is an important regulatory component in the signaling network of plant defense responses.
Plants have evolved several mechanisms to defend themselves from bacterial or fungal invasion. The rapid recognition of pathogenic microbes is based on the interaction of products from a pathogen-derived avirulence gene and a plant-derived resistance gene and represents a prerequisite to specific resistance in incompatible plant-pathogen interactions (Flor, 1956). The multicomponent defense responses associated with specific resistance include a burst of reactive oxygen intermediates (ROI; Lamb and Dixon, 1997), transcriptional activation of defense genes encoding phenylpropanoid pathway enzymes, lytic and antimicrobial pathogenesis-related (PR) proteins (Lamb et al., 1989), increase of intracellular levels of salicylic acid (SA; Malamy et al., 1990; Métraux et al., 1990), and development of the hypersensitive response (HR). The HR results in the rapid appearance of a dry, necrotic lesion at the infection site that is clearly delimited from surrounding healthy tissue and is thought to contribute to the limitation of pathogen spread (Keen, 1990).
One of the earliest events following pathogen recognition is a burst of oxidative metabolism leading to the generation of superoxide (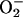 ) and subsequent accumulation of H2O2 (Lamb and Dixon, 1997). These ROI are directly protective and drive the oxidative cross-linking of cell wall structural proteins (Brisson et al., 1994). The H2O2 originating from the oxidative burst induces some plant genes involved in cellular protection and defense such as gluthathione S-transferase (GST) and is necessary for the initiation of host cell death following the HR (Levine et al., 1994).
) and subsequent accumulation of H2O2 (Lamb and Dixon, 1997). These ROI are directly protective and drive the oxidative cross-linking of cell wall structural proteins (Brisson et al., 1994). The H2O2 originating from the oxidative burst induces some plant genes involved in cellular protection and defense such as gluthathione S-transferase (GST) and is necessary for the initiation of host cell death following the HR (Levine et al., 1994).
Recent pharmacological experiments indicate that nitric oxide (NO), which acts as a signal in the immune, nervous, and vascular system in vertebrates (Schmidt and Walter, 1994), also plays an important role in plant disease resistance. Generation of NO by chemical NO donors augments the induction of hypersensitive cell death by H2O2 in soybean (Glycine max) suspension cultures (Delledonne et al., 1998, 2001). Likewise, inhibitors of NO synthesis as well as NO scavengers are able to block the HR induced by avirulent Pseudomonas syringae in soybean cell cultures and in Arabidopsis plants. Compared to ROI, NO induces a complementary set of plant defense genes, including two key enzymes of the phenylpropanoid pathway, namely Phe ammonia lyase (PAL) and chalcone synthase (CHS). Furthermore, NO-treated tobacco (Nicotiana tabacum) cells were shown to induce the pathogenesis-related protein 1 (PR-1) together with an accumulation of SA (Durner et al., 1998), a key molecule for the expression of systemic acquired resistance (Gaffney et al., 1993). Moreover, the molecular components of NO signaling in plants appear to be similar to those in animals, regarding the involvement of NO producing NO synthases (NOS; Chandok et al., 2003; Guo et al., 2003) and cGMP as a second messenger (Clark et al., 2000).
As an increasing number of recent reports suggests, a regulatory function of NO in plants seems to be essential in other physiological processes, including guard cell abscisic acid signaling (Desikan et al., 2002; Garcia-Mata and Lamattina, 2003), regulation of iron homeostasis (Graziano et al., 2002; Murgia et al., 2002), execution of programmed cell death in barley (Hordeum vulgare) aleurone layers (Beligni et al., 2002), root organogenesis (Pagnussat et al., 2002), and wound signaling (Orozco-Cardenas and Ryan, 2002). However, despite the recent identification of a pathogen-inducible NOS (Chandok et al., 2003), assumptions of NO function in plants emerging from all these studies are almost exclusively based on pharmacological studies, i.e. either exogenous application of NO donors, NO scavengers, and inhibitors of mammalian NOS or detection of NO by essentially indirect methods using fluorescent dyes or photometric indicator molecules (Delledonne et al., 1998; Foissner et al., 2000). If indeed the application of pharmacological compounds reflects a physiological NO situation without exerting nonspecific side effects is far from clear, and additional experimental approaches are therefore desirable.
We report here a novel genetic approach to manipulate NO levels in planta, which has been used to gain a better understanding of the function of NO in the signaling network underlying incompatible plant-pathogen interactions. We first generated transgenic Arabidopsis plants overexpressing the Escherichia coli hmp gene encoding NO dioxygenase (NOD), a flavohemoglobin capable of converting NO to nitrate by use of NAD(P)H and O2 (Vasudevan et al., 1991; Gardner et al., 1998; Poole and Hughes, 2000). In this way, we attempted to directly reduce the levels of NO in plant cells. We then compared the defense responses of NO-deficient plants and wild-type Arabidopsis following challenge with avirulent P. syringae pv tomato (Pst) bacteria. Additionally, we employed avirulent Pst expressing the hmpX gene from Erwinia chrysanthemi [Pst(avr-hmpX)] (Favey et al., 1995) that encodes a highly similar NOD to lower NO levels specifically at the side of pathogen infection. We then challenged wild-type Arabidopsis and hmp-expressing Arabidopsis plants with Pst(avr-hmpX) to study the effect of NO removal at both the plant and the pathogen side.
RESULTS
Arabidopsis Plants Expressing a Bacterial NOD
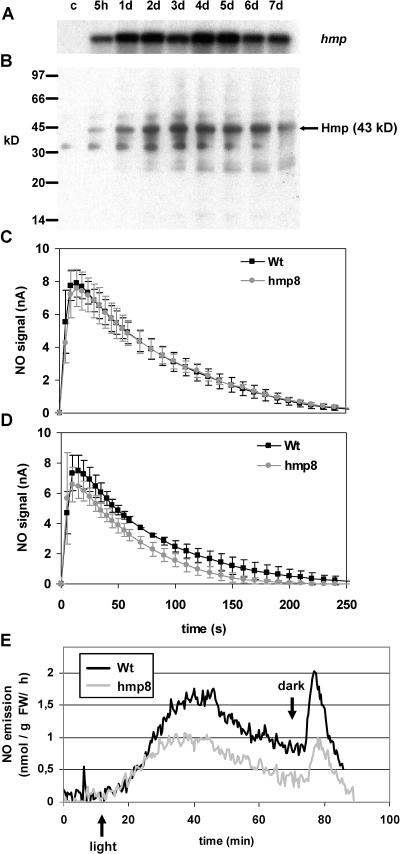
For the production of Arabidopsis ecotype Col-0 plants expressing a functional NOD, the hmp coding sequence from E. coli (Vasudevan et al., 1991) was cloned into the dexamethasone (DEX)-based pTA7001-inducible vector system (Aoyama and Chua, 1997). After Agrobacterium-mediated transformation of wild-type Col-0 plants with the vector construct, homozygous T3 plants segregating for a single T-DNA insertion were used for further experiments. Upon treatment with 3 μm DEX, the various lines expressed the hmp transgene at different levels (data not shown). We selected one of the lines that expressed hmp to a higher degree, designated hmp8, for more detailed analysis. In hmp8 plants, hmp transcripts accumulated at 5 h after spraying rosette leaves with DEX (Fig. 1A). Transcriptional levels increased 1 d after DEX treatment and remained nearly constant for at least 1 week. Western-blot analysis with antibodies raised against E. coli Hmp revealed the production of a full-size Hmp (43 kD) protein in planta (Fig. 1B). The kinetics of Hmp protein expression was similar to transcriptional hmp induction.
Figure 1.
Expression of E. coli hmp in transgenic Arabidopsis as a functional NOD. A, Northern-blot analysis illustrating time-dependent accumulation of hmp transcripts after treatment of hmp8 plants with 3 μm DEX (c, no DEX). B, Western blot demonstrating the appearance of correctly sized Hmp protein in leaf extracts; time course as in A. C and D, Electrochemically measured degradation kinetics of 10 μm NO in leaf extracts of wild-type and hmp8 transgenic Arabidopsis. Error bars represent the sds of five independent measurements. C, Hmp8 plants without DEX-induced transgene expression. D, Wild-type and hmp8 plants 1 d after DEX treatment. NO concentrations of less than 1 μm were reached for wild-type, noninduced hmp8, and DEX-induced hmp8 plants at 197 s, 202 s, and 131 s, respectively. E, NO emission from intact wild-type and hmp8 plants 1 d after DEX treatment. Plants were first incubated in the dark for 10 min and then illuminated for 60 min to cause nitrate reductase-dependent NO emission. The light was switched off again, which gave rise to a characteristic light-off peak (see “Results”). Experiments were repeated three times with similar results.
The functional effects of Hmp in transgenic plants were first investigated by studying the capability of isolated leaf protein extracts to degrade NO. Whereas wild-type plants and noninduced hmp8 plants showed highly similar degradation kinetics for NO (Fig. 1C), plant extracts from the DEX-induced hmp8 line significantly accelerated the degradation of NO (Fig. 1D).
In leaves, NO can be produced from nitrite by nitrate reductase, and this NO production is measurable as emission by chemiluminescence (Rockel et al., 2002). A temporary rise of NO emission resulting from increased nitrate reductase activity was detected when dark-adapted plants were transferred to light. When the light source was switched off again, a light-off peak caused by transient nitrite accumulation resulted (Kaiser et al., 2002). Both the light-induced NO emission and the NO light-off peak were significantly lower in DEX-treated hmp8 plants compared to the wild type (Fig. 1E).
P. syringae Expressing a Bacterial NOD
To expand our genetic approach to study NO deficiency in incompatible plant-pathogen interactions, we transformed Pst (avrB) with the hmpX gene from E. chrysanthemi. Like Hmp from E. coli, E. chrysanthemi HmpX represents a NOD (M. Delledonne and R. Poole, unpublished data). The expression of HmpX in the bacterium should decrease the concentration of the diffusible molecule NO specifically at the site of pathogen infection. In this way, we were able to study plant-pathogen interactions in which NO is simultaneously removed at the infection site from both the plant and the pathogen side.
Plant Defense Responses under NO-Deficient Conditions
Pst carrying the avrB avirulence gene is recognized by Arabidopsis ecotype Col-0 carrying the Rpm1 resistance gene (Bisgrove et al., 1994). The recognition results in the oxidative burst, a production of ROI like H2O2 and 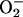 at the site of infection, induction of defense and cellular protectant genes, development of the HR, and a limitation of pathogen growth in comparison to isogenic virulent Pst.
at the site of infection, induction of defense and cellular protectant genes, development of the HR, and a limitation of pathogen growth in comparison to isogenic virulent Pst.
NO Production
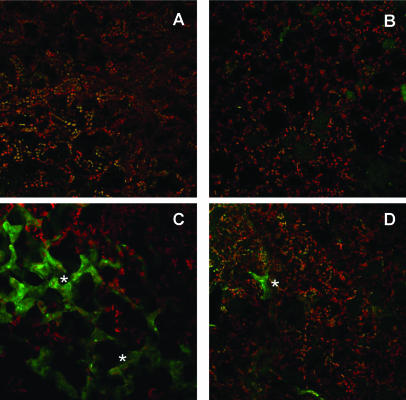
To investigate whether NO is produced during the infection of Col-0 plants with avirulent bacteria, we infiltrated DEX-treated wild-type leaves with the NO-sensitive, cell-permeable fluorescent dye 4,5-diaminofluorescein diacetate (DAF2-DA; Foissner et al., 2000) 3 h after challenge with 2 × 106 colony-forming units (cfu) mL−1 Pst (avrB). Inside the area of pathogen infiltration, various brightly green fluorescing cell groups were discernible 1 h after DAF2-DA treatment (Fig. 2C, white stars), whereas MgCl2-infiltrated control leaves only showed a weak fluorescence when treated with the fluorophore for the same period of time (Fig. 2B). Pathogen infiltration without fluorophore treatment did not cause any fluorescence (Fig. 2A). As compared to wild-type leaves, DEX-induced hmp8 plants challenged with Pst (avrB) and subsequently treated with DAF2-DA showed a markedly reduced fluorescence at the site of pathogen infiltration (Fig. 2D). A similar reduction of DAF2-DA fluorescence was observed when wild-type leaves were challenged in the presence of 100 μm CPTIO, a NO scavenging compound (data not shown). These results suggest that NO is produced during the earlier stages of the Arabidopsis-Pst (avrB) interaction and that this NO burst is attenuated in NOD-expressing hmp8 plants.
Figure 2.
Pathogen-induced DAF2-DA fluorescence as a measure for NO production in DEX-treated wild-type and hmp8 plants. Leaves were pretreated with Pst (avrB) or MgCl2 for 3 h and subsequently infiltrated with 10 μm DAF2-DA or control buffer (10 mm Tris/KCl, pH 7.2). Infiltrated leaf areas were analyzed 1 h later by confocal laser scanning microscopy. DAF2-DA fluorescence (green) was recorded using a channel with a 505- to 530-nm band-pass filter, and autofluorescence of chloroplast (red) was captured with a channel equipped with a 560-nm long-pass filter. A, Treatment of a wild-type Arabidopsis leaf with Pst (avrB) and control buffer. B, Wild-type Arabidopsis-MgCl2 and DAF2-DA. C, Wild-type Arabidopsis-Pst (avrB) and DAF2-DA. D, Hmp8-Pst (avrB) and DAF2-DA. Seven independent samples were recorded for each condition, and representative leaf areas are shown.
Oxidative Burst
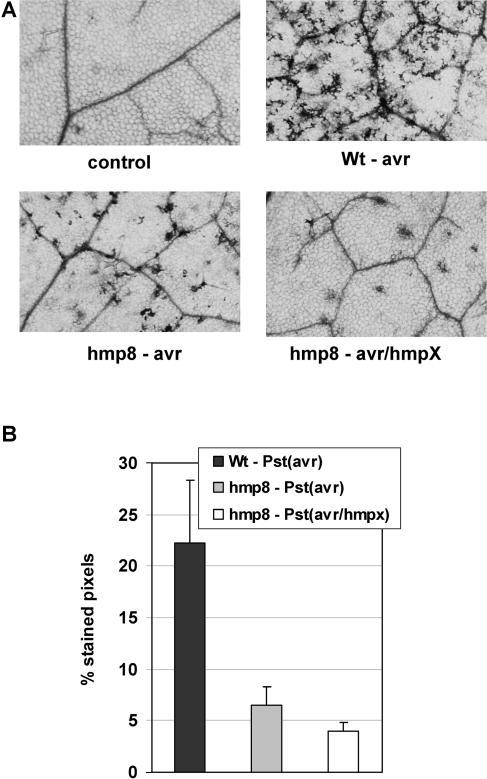
To assess the accumulation of ROI in response to infection with an avirulent pathogen, DEX-treated leaves from wild-type and hmp8 plants were infiltrated with 2 × 106 cfu mL−1 Pst (avrB) and stained with diaminobenzidine (DAB), a histochemical reagent that forms a reddish-brown precipitate upon contact with H2O2 (Thordal-Christensen et al., 1997). Wild-type leaves showed a strong production of H2O2 at the infection site 4 h after pathogen challenge (Fig. 3). By contrast, leaves from hmp8 plants showed significantly lower levels of H2O2 compared to the wild type when challenged with Pst (avrB). Moreover, when DEX-treated leaves from hmp plants where challenged with Pst (avrB/hmpX) bacteria, the suppression of H2O2 levels during the oxidative burst was even more pronounced.
Figure 3.
DAB staining of Arabidopsis leaves to assess H2O2 accumulation during the oxidative burst in DEX-treated wild-type and hmp8 transgenic plants. Solutions of Pst (avrB) were infiltrated into Arabidopsis leaves, and DAB staining was initiated 4 h after infection. A, Staining patterns of representative, MgCl2-infiltrated wild-type or hmp8 leaves (control), Pst (avrB)-infected wild-type Arabidopsis leaves (Wt-avr), Pst (avrB)-infected hmp8 plants (hmp8-avr), and Pst (avrB/hmpX)-infected hmp8 plants (hmp8-avr/hmpX) 4 h after the respective treatment (100-fold magnification). B, Quantification of DAB staining in Pst (avrB)-infected wild-type and hmp8 leaves. The percentage of stained pixels inside the infiltration area was assessed as described in “Materials and Methods.” Values are shown as the mean ± sd of at least five leaves from different plants. Experiments were repeated three times with similar results.
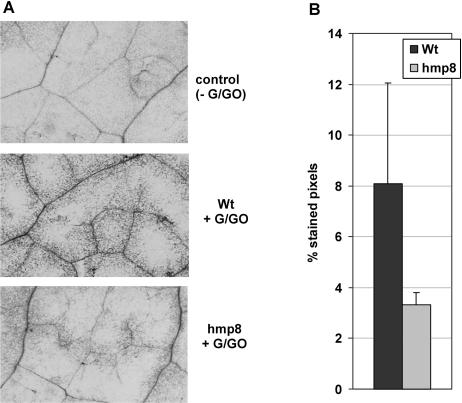
This observation prompted us to test whether the reduced H2O2 levels were a consequence of less H2O2 production or, once produced, an effect of increased H2O2 degradation. We infiltrated equal amounts of the H2O2-generating system Glc/Glc oxidase into leaves of wild-type and hmp8 plants and performed DAB staining 1 h after infiltration. Again, wild-type leaves showed stronger staining patterns with respect to induced hmp8 leaves (Fig. 4), suggesting that the action of Hmp increased the ability of the plants to degrade H2O2.
Figure 4.
DAB staining of Arabidopsis leaves to assess their capability to degrade H2O2 in DEX-treated wild-type and hmp transgenic plants. Solutions of 2.5 mm Glc/2.5 units mL−1 Glc oxidase were infiltrated into leaves and DAB staining was performed 1 h after infiltration. A, Staining patterns of representative leaves inside the infiltrated area (100-fold magnification). B, Percentage of stained pixels inside the infiltration zone. Values are shown as the mean ± sd of at least five leaves from different plants. Experiments were repeated three times with similar results.
Defense Gene Expression
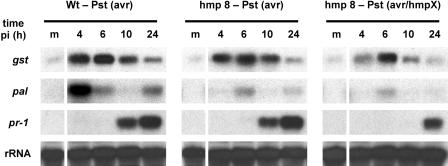
We next examined whether the expression of three typical defense-related genes, GST, PAL, and PR-1, was affected in the hmp8 line (Fig. 5). GST functions in cellular protection, and gst transcripts are induced during the oxidative burst (Levine et al., 1994; Delledonne et al., 2001). Gst transcripts accumulated 4 to 10 h after DEX-treated wild-type plants were challenged with Pst (avrB). Similar induction kinetics were observed when DEX-induced hmp8 plants were challenged with Pst (avrB) or Pst (avrB/hmpX). Only in the latter case was the amount of gene induction slightly diminished, presumably reflecting the extremely low H2O2 levels during the oxidative burst.
Figure 5.
Expression of defense and cellular protectant genes in wild-type and hmp8 transgenic plants after Pst (avrB) challenge. Three parallel leaf samples were collected at the indicated times after infection for RNA extraction and northern-blot analysis. MgCl2 (m)-infiltrated leaves were collected 4 h after infection. Experiments were repeated three times with similar results.
PAL catalyzes the first step in phenylpropanoid biosynthesis and possibly initiates the synthesis of lignin, antibiotics, and SA. A strong induction of pal transcripts occurred 4 h after pathogen infection in wild-type plants challenged with Pst (avrB) (Fig. 5). This strong induction of pal was highly suppressed in NO-deficient interactions, i.e. when DEX-induced hmp8 plants where challenged either with Pst (avrB) or Pst (avrB/hmpX).
Transcriptional induction of the antimicrobial PR-1 protein occurred 10 h after challenge of both wild-type and hmp8 plants with Pst (avrB) (Fig. 5). PR-1 induction was delayed, albeit not fully suppressed, when hmp8 plants were challenged with Pst (avrB/hmpX).
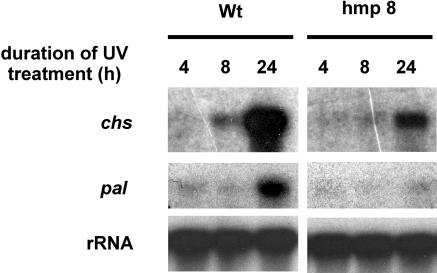
The finding that pathogen-induced pal expression is blocked under NO-deficient conditions prompted us to test whether NO could act as a general signal for the activation of the phenylpropanoid pathway. Another stimulus activating this pathway is UV light (Chappell and Hahlbrock, 1984), and we examined the UV-induced expression of PAL and CHS, the key enzyme for flavonoid biosynthesis, in DEX-treated wild-type and hmp8 plants (Fig. 6). Whereas wild-type plants strongly expressed chs and pal after 24 h of UV treatment, this UV-induced gene expression was markedly attenuated in hmp8 plants, indicating that NO is required for phenylpropanoid pathway activation by different environmental stimuli.
Figure 6.
Expression of PAL and CHS in DEX pretreated wild-type and hmp8 plants after UV exposure. Three parallel leaf samples were collected at the indicated times after the beginning of UV treatment. Experiments were repeated three times with similar results.
Development of Hypersensitive Cell Death
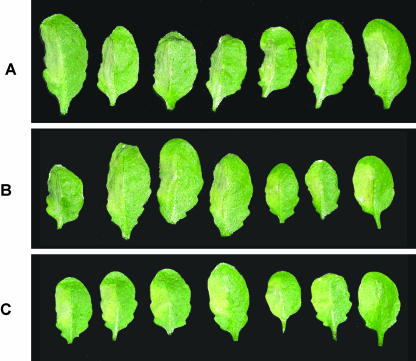
When leaves from wild-type plants were challenged with Pst (avrB) at concentrations of 5 × 106 cfu mL−1, a dry, colorless lesion limited to the site of pathogen infiltration developed within 2 d in at least 5 out of 7 leaves (Fig. 7A). These macroscopic symptoms characteristic of hypersensitive cell death developed to the same extent when noninduced hmp8 plants were used (data not shown). However, when infected with Pst (avrB), DEX-treated hmp8 plants showed a more chlorotic lesion that was reminiscent of the symptoms caused by isogenic virulent Pst in about 50% of leaves (Fig. 7B). Infection of induced hmp8 plants with Pst (avrB/hmpX) further increased this percentage. In this case, virtually every challenged leaf developed chlorotic symptoms (Fig. 7C).
Figure 7.
Macroscopic HR symptoms 2 d after infiltration of DEX-pretreated wild-type and hmp8 plants with avirulent Pseudomonas. Bacteria were infiltrated into the left side of leaves. Seven parallels are shown for each condition. A, Wild-type plants-Pst (avrB). B, hmp8 plants-Pst (avrB). C, hmp8 plants-Pst (avrB/hmpX).
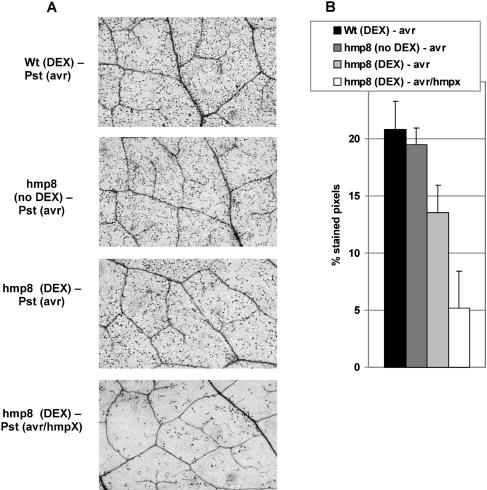
To characterize the HR development in more detail, we performed Trypan blue staining of infected leaves 24 h after bacterial inoculation (Fig. 8). Blue-stained dead cells or patches of dead cells appeared to a similar extent at the sites of Pst (avrB) infiltration in DEX-treated wild-type leaves and nontreated hmp8 leaves. In DEX-treated hmp8 leaves, the density of dead cells was reduced when challenged with Pst (avrB), and Pst (avrB/hmpX) challenge led to a dramatic reduction of HR lesions.
Figure 8.
Microscopic cell death after pathogen challenge of DEX-pretreated wild-type and hmp8 plants. Trypan blue staining was performed 24 h after infection. A, Staining patterns of representative leaves inside the infiltrated area (100-fold magnification). B, Percentage of stained pixels inside the infiltration area. Values are shown as the mean ± sd of at least six leaves from different plants. Experiments were repeated three times with similar results.
SA Levels and Bacterial Growth
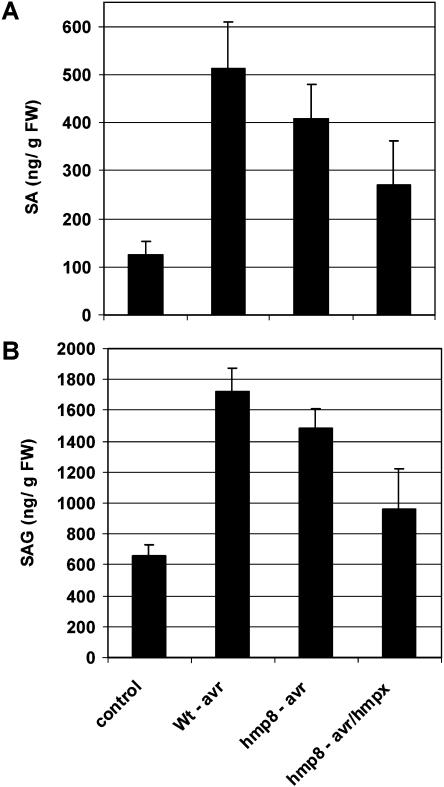
SA plays a central role in the activation of plant defense responses, and a relationship between NO and SA signaling pathways has been discussed (Klessig et al., 2000). To determine the effects of NO on SA levels in the Pst (avrB)-Col-0 interaction, we determined the levels of SA and SA glucoside (SAG) in our system 8 h after pathogen infection (Fig. 9). DEX-treated wild-type and hmp8 plants infiltrated with 10 mm MgCl2 had SA levels of about 120 ng g−1 leaf fresh weight and SAG contents of about 600 ng g−1. Upon Pst (avrB) challenge, SA levels increased in wild-type plants about 4-fold to 500 ng g−1, and SAG levels reached values of 1,700 ng g−1, representing a 3-fold increase. In DEX-treated hmp8 plants, the SA content in Pst (avrB)-challenged leaves showed a small, albeit statistically not significant, decrease to about 80% of the wild-type value (Fig. 9A). A similar trend was observed for the glucoside (Fig. 9B). Again, the most striking effect was detected when induced hmp8 plants were infiltrated with Pst (avrB/hmpX). Here, SA and SAG were reduced to about 50% of the wild type-Pst (avrB) values, reflecting a pathogen-induced increase of only about 2-fold (SA) and 1.5-fold (SAG), respectively.
Figure 9.
SA contents of wild-type and hmp8 transgenic plants after challenge with avirulent Pst (± hmpX). Leaf samples were collected 8 h postinfection. Leaves were pretreated with DEX for 16 h. Bars indicated mean values of three independent measurements. Control, MgCl2-infiltrated plants. A, Free SA. B, SAG.
To test whether these changes of defense responses in our genetically different pathosystems affected bacterial growth in planta, we determined the number of colony-forming bacteria in the apoplast 2 d after leaf inoculation (Table I). Compared to wild-type plants, bacterial growth was slightly, but statistically insignificantly, enhanced in DEX-treated hmp8 plants when challenged with Pst (avrB). A more pronounced growth enhancement was detected when DEX-treated hmp8 plants were challenged with Pst (avrB/hmpX). However, this enhancement did not reach the extent of growth found in the compatible interaction of wild-type plants and the isogenic virulent Pst strain (Table I).
Table I.
Bacterial growth of different Pst in Arabidopsis leaves of wild-type Arabidopsis and hmp8 transgenic plants
| Time | Wild Type vir | Wild Type avrB | hmp8 avrB | hmp8 avrB/hmpX |
|---|---|---|---|---|
| 1 h | 2.33 (±1.14) | 2.2 (±0.67) | 2.00 (±0.76) | 1.57 (±0.77) |
| 2 d | 2,700 (±361) | 152 (±47) | 170 (±53) | 267 (±76) |
Leaves were pretreated with DEX 16 h before pathogen inoculation, infiltrated with 106 cfu mL−1 Pst, and harvested 1 h and 2 d after inoculation. Values ×104 represent means of cfu per cm2 (±sd), each from five sets of three leaf discs. Experiments were repeated twice with similar results. vir, Virulent; avrB, avirulent; avrB/hmpX, avirulent expressing hmpX.
DISCUSSION
Pharmacological methodologies in different laboratories using mainly mammalian NOS inhibitors, NO scavengers, and NO-releasing systems have implicated a pivotal role for NO in plant disease resistance (Delledonne et al., 1998; Durner et al., 1998). Moreover, the recent report that activity suppression of a pathogen-inducible NOS in tomato (Lycopersicon esculentum) increases susceptibility to P. syringae demonstrates the involvement of a NO-generating enzyme in plant defense (Chandok et al., 2004). This plant pathogen-inducible NOS represents a variant form of the P protein of the Gly decarboxylase complex that shares some biochemical characteristics with animal NOS, such as sensitivity to inhibitors (Chandok et al., 2003), indicating the validity of the data obtained with the use of pharmacological approaches. However, it cannot be fully excluded that pharmacological compounds not only interfere with the metabolic pathway of interest but also have nonspecific effects. The widely used NADPH oxidase inhibitor diphenylene iodonium, for instance, has been shown to be a potent inhibitor of mammalian NOS and other flavoproteins (Stuehr et al., 1991; Bolwell, 1999). To generalize and broaden the knowledge of NO function in plant defense, we employed a genetic approach to interfere with NO signaling by expressing a NOD in transgenic Arabidopsis plants as well as in avirulent P. syringae.
Transgenic Arabidopsis plants were produced that express the Hmp protein from E. coli, and whole-plant NO emission was evaluated. The emission was shown to be markedly reduced in Hmp-expressing plants, and leaf extracts from transgenic plants degraded NO significantly faster than extracts from control plants. These findings demonstrate that Hmp is a functional NOD in planta (Fig. 1). Using the NO-sensitive fluorescence indicator DAF2-DA, we furthermore showed that NO is produced during the incompatible interaction of Arabidopsis and Pst (avrB) and that this NO burst is attenuated in NOD-expressing plants (Fig. 2). We also produced avirulent Pseudomonas expressing HmpX, a similar NOD from E. chrysanthemi. Biochemical experiments suggest that HmpX is located in the periplasm and represents a functional NOD in transformed Pseudomonas (R. Poole and M. Delledonne, unpublished data).
With these genetic tools, we examined the hypersensitive disease resistance response when NO accumulation was attenuated by the action of NODs in two different surroundings. We could generally state that the removal of NO from the plant resulted in strikingly similar tendencies compared to NO removal from the pathogen side. When comparing the interaction of hmp8 plants with Pst (avrB) on the one hand and the interaction of wild-type plants with Pst (avrB/hmpX) on the other hand, we found very similar tendencies (data not shown). Moreover, when combining the two genetically modified systems, i.e. the interaction of hmp8 plants with Pst (arvB/hmpX), we observed additive effects in all examined defense responses.
We first detected significantly lower H2O2 levels during the oxidative burst in NO-deficient interactions, and removal of NO from both the plant and the pathogen side had an additive effect (Fig. 3). Less H2O2 staining in the presence of NOD was also obvious when plants were infiltrated with Glc/Glc oxidase, a H2O2-generating system (Fig. 4), indicating that the in planta capability to degrade H2O2 was increased by the action of the NOD. This effect might be due to the NO-degrading function of the NOD or possibly by direct degradation of H2O2 by NOD. Because the rate of H2O2 degradation was identical in leaf extracts from wild-type and hmp8 plants (data not shown), we conclude that direct H2O2 degradation through NOD does not take place. Rather, a factor differing in hmp8 and wild-type plants but not in the corresponding extracts might account for the different observation in intact plants and extracts, respectively. This factor might be the concentration of NO, which is delivered by the intact plant continuously but not necessarily by extracts. Following this interpretation, the higher in planta H2O2 degradation capability of NO-deficient Hmp plants suggests an inhibitory effect of NO toward H2O2-degrading enzymes. In fact, the predominant H2O2 scavenging enzymes are catalase and ascorbate peroxidase. Mammalian catalase is reversibly inhibited by NO (Brown, 1995), and it has been shown in vitro that NO inhibits both tobacco catalase and ascorbate peroxidase (Clark et al., 2000). In accordance to previous reports (Delledonne et al., 1998; Foissner et al., 2000), our data reveal that a local burst of NO coincides with the oxidative burst at the site of pathogen infection. Therefore, it is conceivable that this NO burst locally contributes to maintain more sustained and higher H2O2 levels that can then either act directly as an antimicrobial oxidant or indirectly by triggering various defense responses (Lamb and Dixon, 1997).
The most striking differences in defense gene expression concerned the induction of pal transcripts, which was significantly attenuated in hmp plants and in the presence of avirulent Pseudomonas expressing HmpX (Fig. 5). This observation confirms pharmacology-based findings demonstrating a reduction of pal expression by NOS inhibitors in soybean cells and pal induction by NO donors and recombinant NOS in soybean and tobacco, respectively (Delledonne et al., 1998; Durner et al., 1998). In addition, the UV-induced expression of pal and chs, the first committed enzyme in anthocyanin biosynthesis, was strongly repressed in hmp plants (Fig. 6). Based on this experimental evidence, NO appears to play a pivotal regulatory role in the signaling processes leading to expression of phenylpropanoid pathway genes.
PAL is the key enzyme for the general phenylpropanoid pathway, and possible outcomes are lignin, anthocyanin, and/or SA biosynthesis. However, despite the strong repression of pal, SA levels only showed a 20% reduction in the presence of Hmp in plants or HmpX in bacteria (Fig. 9). This supports the findings that in Arabidopsis, SA is produced by alternative routes, e.g. by the isochorismate pathway (Wildermuth et al., 2001). The observed induction of pal in the Pseudomonas-Arabidopsis pathosystem might then feed alternative processes like lignification or the production of other phenylpropanoid compounds to a significant extent. If SA is the causative agent of PR-1 induction (Yalpani et al., 1991; Uknes et al., 1992), the rather weak attenuation of SA induction might explain the fact that in these cases the expression of PR-1 is not affected (Fig. 5). The levels of SA might still be above a threshold value necessary for full pathogen-induced PR-1 expression. In the double experiment in which NO is reduced from both the pathogen and the plant, however, SA was reduced to 50% of the usual value and PR-1 expression was clearly delayed, although not fully suppressed. It is worth noting that the expression of PR-1 is also up-regulated in plants challenged with virulent pathogens but slower than in the corresponding incompatible interaction (de Torres et al., 2003). Compared to pharmacological experiments with tobacco and soybean cells (Delledonne et al., 1998; Durner et al., 1998), our results demonstrate a similar tendency of NO involvement in SA and PR-1 production and a strong regulatory role of NO toward the synthesis of phenylpropanoid pathway enzymes like PAL.
When NO is scavenged by Hmp or HmpX alone, dry lesion development is delayed but not eliminated (Fig. 7), and the appearance of microscopic HR lesions is only moderately suppressed (Fig. 8). However, when NO is scavenged by the simultaneous action of Hmp and HmpX, the macroscopic dry HR lesions are yellowish with less pronounced symptoms, and the microscopic HR lesions are significantly reduced (Fig. 8). Therefore, HR lesion development is clearly affected by a reduced NO content in Arabidopsis, and the correlation with SA levels suggests a mediatory role of SA in these processes. This is in accordance with findings that SA is required for induction of the HR in response to bacterial pathogens in soybean (Tenhaken and Rubel, 1997) and that SA is needed for cell death initiation in Arabidopsis lsd mutants (Weymann et al., 1995). The execution of hypersensitive cell death in soybean cells challenged with avirulent P. syringae is strongly diminished by NO scavenging compounds and NOS inhibitors (Delledonne et al., 1998). Furthermore, a poised production of ROI and NO is necessary to trigger the HR, and NO together with H2O2, but not 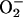 , are indicated as the essential players in this process (Delledonne et al., 2001). One possible mechanism for this cooperation could be that NO ensures maintenance of high, persistent H2O2 levels that are necessary to trigger the HR. The threshold H2O2 levels for HR development in soybean cells is about 6 μm (Levine et al., 1994), and in our experiments, H2O2 levels may not have significantly fallen below a comparable threshold value in the cases of Hmp or HmpX action alone. In the double experiment, however, when H2O2 levels showed the strongest decrease (Fig. 3), the H2O2 threshold might not have been exceeded and, consequently, a significant reduction in HR development could be observed. A similar reasoning could explain why a significant induction of gst transcription still took place under NO deficiency, even in the double experiment (Fig. 5). For gst expression, H2O2 threshold levels have been shown to be below the HR value, which are around 2 μm in soybean cells (Levine et al., 1994).
, are indicated as the essential players in this process (Delledonne et al., 2001). One possible mechanism for this cooperation could be that NO ensures maintenance of high, persistent H2O2 levels that are necessary to trigger the HR. The threshold H2O2 levels for HR development in soybean cells is about 6 μm (Levine et al., 1994), and in our experiments, H2O2 levels may not have significantly fallen below a comparable threshold value in the cases of Hmp or HmpX action alone. In the double experiment, however, when H2O2 levels showed the strongest decrease (Fig. 3), the H2O2 threshold might not have been exceeded and, consequently, a significant reduction in HR development could be observed. A similar reasoning could explain why a significant induction of gst transcription still took place under NO deficiency, even in the double experiment (Fig. 5). For gst expression, H2O2 threshold levels have been shown to be below the HR value, which are around 2 μm in soybean cells (Levine et al., 1994).
Blockage of the accumulation of NO by NO scavengers or mammalian NOS inhibitors has previously been shown to enhance bacterial growth of avirulent Pseudomonas in Arabidopsis leaves, although to a lower extent in comparison to the growth of virulent strains (Delledonne et al., 1998). In this work, NO scavenging by NOD led to a similar, albeit weaker, tendency of bacterial growth enhancement of avirulent Pst (Table I). Obviously, the removal of NO in these and other recent experiments is not sufficient for a strong growth enhancement of avirulent pathogens, despite the apparent effects on progression of the described defense responses. This behavior is in some respects reminiscent of the dnd (defense, no death) class of Arabidopsis mutants that show the ability to limit pathogen growth in a gene-for-gene resistance without developing an HR (Yu et al., 1998). Dnd1 encodes a cyclic nucleotide-gated ion channel (Clough et al., 2000). In mammals, cyclic nucleotides like cGMP or cADP-Rib are second messengers closely associated with NO signaling (Galione and White, 1994; Schmidt and Walter, 1994), and this has been suggested for plants as well (Pfeiffer et al., 1994; Durner et al., 1998; Clarke et al., 2000). It will be of interest to investigate if generation of NO leads to activation of Dnd1, which contributes to the development of symptoms characteristic of the HR.
The complete scavenging of a highly diffusible and reactive molecule like NO is difficult to achieve for a single protein, and the (physiologically active) reaction of NO with other cellular molecules might to a certain extent still compete with the NOD reaction. This is also reflected by the fact that Hmp-overexpressing plants still emitted about one-half the amount of gaseous NO than wild-type plants (Fig. 1E). Therefore, not all cellular NO-mediated effects might have been fully suppressed by this transgenic approach. Compared to the very similar salicylate hydroxylase (NahG) strategy applied by Gaffney et al. (1993), which addressed the role of the far less reactive signaling compound SA in disease resistance, these data have some limitations due to the physicochemical properties of NO. In general, the possibility that protein overexpression in plants leads to side effects not associated with the physiological process under investigation cannot be fully excluded. For instance, it was demonstrated recently that effects on plant defense responses observed by overexpression of NahG might not necessarily result from a lack of SA accumulation but could partly be a consequence of the presence of the SA degradation product catechol (van Wees and Glazebrook, 2003). Because Hmp converts NO, O2, and NAD(P)H to nitrate in equistoichiometric amounts, it is conceivable that physiological side effects resulting from oxygen and NAD(P)H consumption or nitrate accumulation exist in hmp-overexpressing plants. Such side effects, however, should be minimal in hmp plants considering the comparatively low NO levels produced in plants (Fig. 1E) and taking into account the use of an inducible vector system, which restricted the action of Hmp to a small experimental window. The parallel tendencies observed in two different genetic backgrounds, i.e. the action of Hmp in Arabidopsis and HmpX in Pseudomonas, further support that the described effects on plant defenses were a direct consequence of NO degradation rather than a result of indirect effects caused by NOD on plant metabolism. Besides its NOD activity, however, it was shown that Hmp-overexpressing E. coli strains are capable of generating ROI (Poole and Hughes, 2000). We can rule out that this metabolic activity took place in transgenic hmp plants during the course of our pathogen and UV experiments due to the findings that transcriptional up-regulation of gst, a sensitive marker for ROI production (Levine et al., 1994), and a positive DAB reaction could not be detected in uninfected but DEX-induced hmp plants (Figs. 3 and 5). The latter statement, however, could not be maintained when hmp plants were exposed to DEX for longer periods of time. About 4 d after transgene induction, we detected elevated levels of ROI in leaves of hmp plants by DAB staining and the expression of gst and pr-1 in the absence of pathogens (data not shown). Obviously, a new physiological situation in hmp plants appeared when plants perpetually accumulated Hmp protein, either caused by the constant removal of NO or by an emerging ROI-generating activity. Interestingly, in a recent transgenic approach, the constitutive overexpression of an alfalfa (Medicago sativa) hemoglobin with putative NO scavenging properties in tobacco similarly resulted in increased basal ROI levels, besides elevated pathogen-induced SA levels and reduced disease symptoms after P. syringae infection (Seregélyes et al., 2003). These findings indicate that a temporally controlled expression rather than constitutive or prolonged overexpression of certain transgenes like (flavo) hemoglobins might be crucial in overexpression studies. The use of the inducible vector system permitted us to use a window of 3 to 4 d after transgene expression to perform pathogen and physiological experiments without undesirable side effects like ROI production.
MATERIALS AND METHODS
Generation of Hmp-Overexpressing Arabidopsis
To generate transgenic Arabidopsis overexpressing the Escherichia coli hmp gene, pTA7001, a dexamethasone-inducible expression system, was used (Aoyama and Chua, 1997). The hmp coding sequence was generated by PCR with the primers 5′-CGGCTCGAGATGCTTGACGCTCAAACCATC-3′ and 5′-GGACTAGTACGCGGCAATTTAAACCGCGTC-3′ using a full-length hmp clone as a template, which was kindly provided by A.M. Gardner (University of Cincinnati). The PCR product was subcloned into pGEMT-Easy (Promega, Madison, WI), sequenced, and introduced into pTA7001 by the use of 5′-XhoI and 3′-SpeI restriction sites. The construct was transformed into Agrobacterium tumefaciens (strain GV3101) and the latter used for plant transformation of Arabidopsis ecotype Col-0 by the floral dip method (Clough and Bent, 1998). After transformation, seeds were harvested from T0 plants and surface sterilized, and positive transformants were selected on phytagar plates supplemented with Murashige minimal organics medium (Life Technologies, Paisley, UK) containing 15 μg L−1 hygromycin.
Homozygous T3 plants from single insert lines were used for all experiments, and plants were grown at 22°C under a 9-h-light/15-h-dark cycle. For transgene induction, hmp plants were sprayed with a solution of 3 μm DEX in 0.01% Tween 20. Control experiments were performed with wild-type Col-0 plants treated with 3 μm DEX in 0.01% Tween 20 and/or hmp transgenic plants solely sprayed with 0.01% Tween 20. Pathogen infiltrations followed 16 h after DEX/Tween 20 treatment.
Growth of Plant-Pathogens and Infection
Pseudomonas syringae pv tomato carrying the avirulence gene avrB were transformed with a pRK415 broad host vector (Keen et al., 1988) carrying the complete coding sequence of hmpX (Favey et al., 1995) under control of the Lac promoter, which is constitutively active in Pseudomonas. Detailed description of the construction of pRK415-hmpX and maintenance in Pseudomonas is not provided (M. Boccara, C. Mills, J. Zeier, C. Anzi, C. Lamb, R. Poole, and M. Delledonne, unpublished data).
Pst strains were grown overnight at 28°C in King's B medium containing the appropriate antibiotics (concentrations: rifampicin 50 μg L−1, kanamycin 50 μg L−1, tetracycline 15 μg L−1). Bacteria were pelleted, washed three times with 10 mm MgCl2, resuspended, and diluted in 10 mm MgCl2 to the desired concentration (generally 2 × 106 cfu mL−1, for symptom development 5 × 106 cfu mL−1, for bacterial growth 106 cfu mL−1). The bacterial solutions were infiltrated from the abaxial side into one-half of a sample leaf using a 1-mL syringe without a needle. Control (mock) inoculations were performed with 10 mm MgCl2. Macroscopic symptoms were documented 2 d after infection. Bacterial growth was assessed by homogenizing discs originating from infiltrated areas of three different leaves in 1 mL of 10 mm MgCl2, plating appropriate dilutions on King's B medium containing Rifampicin, and quantifying colony numbers after 2 to 3 d.
UV Treatment of Arabidopsis Plants
Five-week-old Arabidopsis wild-type and hmp8 plants were pretreated with DEX for 16 h and placed into a growth chamber equipped with UV-A light-emitting black light tubes (Phillips TL 8 W/08; Eindhoven, The Netherlands).
Histochemical Staining and Quantification of H2O2 Levels and Microscopic HR Lesions
DAB and Trypan blue staining were performed as described by Thordal-Christensen et al. (1997) and Koch and Slusarenko (1990), respectively. Exogenous H2O2 was generated by infiltrating 10 μL of 2.5 mm d-Glc and 2.5 units mL−1 Aspergillus niger Glc oxidase (Calbiochem, San Diego) in 20 mm Na phosphate buffer, pH 6.5, into Arabidopsis leaves.
For quantification of the number of stained pixels inside the infected leaf area, the histogram function of Adobe Photoshop 6.0 (Adobe Systems, Mountain View, CA) was used. Microscopic photographs were reduced to grayscale mode, and all pixels inside the infiltration zone with a gray tone value <125 were quantified. To account for background staining, the corresponding value for an area of equal size inside the noninfected opposite side of the leaf was subtracted from the latter value and the result divided by the total amounts of considered pixels to yield the relative number of stained pixels in percentage.
RNA and Protein Analysis
Total RNA was isolated from Arabidopsis leaves using Trizol reagent (Life Technologies) following the manufacturer's instructions. RNA-blot hybridization (Levine et al., 1994) was performed with probes of the Arabidopsis gst, pal, and PR-1 genes: ATGST (GenBank accession no. U70672), ATPAL1A (X62747), and ATHRPRP1A (M90508). Equal loading was verified by gel staining with ethidium bromide and by hybridization with an rDNA probe.
For protein extraction, three leaves were homogenized with 1 mL of extraction buffer (15 mm HEPES, 40 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, pH 7.6). The mixture was centrifuged for 30 min at 19,000g and 4°C. The supernatant constituted the protein extract. Protein samples were subjected to SDS-PAGE on 10% (w/v) polyacrylamide (Sambrook et al., 1989) and electroblotted to a polyvinylidene difluoride membrane (Hybond-P; Amersham Pharmacia, Little Chalfont, UK). Probing and detection of western blots were performed as described in the ECL western-blotting detection kit (Amersham). The primary antibody raised against E. coli Hmp was kindly provided by Robert Poole (Sheffield, UK) and used at a dilution of 1:3,000. A goat anti-rabbit IgG horseradish peroxidase conjugate (Sigma, St. Louis) was used as the secondary antibody with a dilution of 1:10,000.
Determination of SA Levels
Measurements of SA and SAG essentially followed the protocol of Raskin et al. (1989). Briefly, 0.15 g of frozen leave tissue was homogenized in 1 mL of 90% methanol and extracted for 10 min at 40°C. The mixture was centrifuged for 5 min at 14,000g, and the pellet was extracted for another 10 min at 40°C with 100% methanol. Supernatants from both extractions were combined and dried under a gentle stream of N2 at 40°C. The residue was resuspended in 1.5 mL of 0.1 m HCl, and 100 ng of o-anisic acid was added as an internal standard. After centrifugation for 10 min at 14,000g, the aqueous solution was extracted three times with 2 mL of cyclopentane/ethylacetate (1:1). The extracts were combined and the solvent removed under N2 at 40°C. The residue was dissolved in 50 μL of methanol and passed through a solid-phase extraction column filled with ODS-H optimal packing material (Capital HPLC, Broxburn, UK) using 6 mL of methanol. The eluate was dried under N2 at 40°C, dissolved in 100 μL of methanol, and used for HPLC analysis. For detection of SAG, the aqueous, acidic phase from the first extraction step was heated to 100°C for 30 min to convert the glucoside to free SA. The above protocol was repeated starting with addition of the internal standard.
HPLC analysis was performed using an ODS-H optimal column (10 × 2.1 mm, Capital HPLC) on a Shimadzu (Columbia, MD) LC-5A chromatograph. For separation, a linear gradient from 95% of H2O/BuOH/HOAc (98.3/1.2/0.5) to 90% acetonitrile/BuOH/HOAc (98.3/1.2/0.5) in 20 min and flow rate of 0.7 mL min−1 was applied. For detection, a Waters (Milford, MA) 474 scanning fluorescence detector with an excitation wavelength of 300 nm was used. The emission wavelength was switched at 7 min elution time from 365 nm to 405 nm to ensure highest sensitivities for o-anisic and SA, respectively.
NO Degradation Assay
The kinetics of NO degradation were measured electrochemically using an Iso-NO meter (World Precision Instruments, Sarasota, FL). A saturated, 2 mm NO solution was prepared by bubbling 10 mL of NO gas through 5 mL of HEPES buffer (see above). Protein extracts were prepared as described above using six fully grown Arabidopsis leaves and 1 mL of HEPES-extraction buffer (without dithiothreitol). For NO degradation measurements, 1 mL of plant extract was supplemented with 10 μL of 10 mm NADH and the temperature of the solution kept at 24°C in a water bath. An Iso-NO electrode was calibrated according to the manufacturer's instructions and submerged into the protein solution inside a gas-tight vial. Under stirring, 5 μL of NO solution was added, and the time-dependent changes of the NO signal were recorded.
NO Emission Measurements
Rosette leaves were cut from root parts of Arabidopsis plants and immediately floated on deionized water. The leaves from three different plants were placed in a transparent lid container with 2 L of air volume. A constant flow of measuring gas (NO-free air conducted through a custom-made charcoal column) of 1.5 L/min was pulled through the container and subsequently through the chemiluminescence detector (CLD 770 AL ppt; Eco-Physics, Dürnten, Switzerland) by a vacuum pump connected to an ozone destroyer. Light was provided by a 400 W HQi-lamp (Schreder, Winterbach, Germany) above the container. The quantum flux density could be adjusted at 100 μmol m−2 s−1 photosynthetic active radiation by a polyester sieve (pore size is 210 μm) on the lid of the container. Air temperature in the container was usually about 23°C in the dark and 23°C to 25°C under light conditions.
NO Detection using DAF2-DA Fluorescence
Arabidopsis leaves were treated with 2 × 106 cfu mL−1 Pst (avrB) or 10 mm MgCl2 as described above, and 3 h later, 10 μm DAF2-DA (Sigma) dissolved in 10 mm Tris/KCl, pH 7.2, was infiltrated into the pretreated leaf areas. One hour after DAF2-DA infiltration, leaf areas were analyzed microscopically using a Zeiss Axioskop 2 fluorescence microscope equipped with a confocal laser scanner (LSM 5 PASCAL; Zeiss, Oberkochen, Germany). Leaves were excited with an argon laser (488 nm). DAF2-DA fluorescence was recorded using a channel with a 505- to 530-nm band-pass filter, and autofluorescence of chloroplast was captured with a channel equipped with a 560-nm long-pass filter.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers X58872, X75893, U70672, X62747, and M90508.
Supplementary Material
Acknowledgments
We thank Robert Poole (Sheffield, UK) and Anne M. Gardner (Cincinnati) for kindly providing hmp antibodies and the full-length hmp clone, respectively. Werner M. Kaiser (Würzburg, Germany) is gratefully acknowledged for providing the opportunity to perform NO-emission experiments. J.Z. is a Fellow of the Alexander-von-Humboldt Foundation. The corresponding fellowship supervision by Robert Chow (Edinburgh, UK) is also gratefully acknowledged.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042499.
References
- Aoyama T, Chua N-H (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol 129: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW (1994) A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6: 927–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP (1999) Role of active oxygen species and NO in plant defence responses. Curr Opin Plant Biol 2: 287–294 [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C (1994) Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 6: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GC (1995) Reversible binding and inhibition of catalase by nitric oxide. Eur J Biochem 232: 188–191 [DOI] [PubMed] [Google Scholar]
- Chandok MR, Ekengren SK, Martin GB, Klessig DF (2004) Suppression of pathogen-inducible NO synthase (iNOS) activity in tomato increases susceptibility to Pseudomonas syringae. Proc Natl Acad Sci USA 101: 8239–8244 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chandok MR, Ytterberg AJ, van Wijk KJ, Klessig DF (2003) The pathogen-inducible nitric oxide synthase (iNOS) in plants is a variant of the P protein of the glycine decarboxylase complex. Cell 113: 469–482 [DOI] [PubMed] [Google Scholar]
- Chappell J, Hahlbrock K (1984) Transcription of plant defence genes in response to UV light or fungal elicitor. Nature 311: 76–78 [Google Scholar]
- Clarke A, Desikan R, Hurst RD, Hancock JT, Neill SJ (2000) NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J 24: 667–678 [DOI] [PubMed] [Google Scholar]
- Clark D, Durner J, Navarre DA, Klessig DF (2000) Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol Plant Microbe Interact 13: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Lippok B, Smith Jr RK, Yu I-c, Bent AF (2000) The Arabidopsis dnd1 “defence, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Griffiths R, Hancock JT, Neill SJ (2002) A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 16319–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres M, Sanchez P, Fernandez-Delmond I, Grant M (2003) Expression profiling of the host response to bacterial infection: the transition from basal to induced defence responses in RPM1-mediated resistance. Plant J 33: 665–676 [DOI] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF (1998) Defence gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favey S, Labesse G, Vouille V, Boccara M (1995) Flavohaemoglobin HmpX: a new pathogenicity determinant in Erwinia chrysanthemi strain 3937. Microbiology 141: 863–871 [DOI] [PubMed] [Google Scholar]
- Flor HH (1956) The complementary genetic systems in flax and flux rust. Adv Genet 8: 29–54 [Google Scholar]
- Foissner I, Wendehenne D, Langebartels C, Durner J (2000) In vivo imaging of an elicitor-induced nitric oxide burst in tobacco. Plant J 23: 1–10 [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Galione A, White A (1994) Ca2+ release induced by cyclic ADP-ribose. Trends Cell Biol 4: 431–436 [DOI] [PubMed] [Google Scholar]
- Garcia-Mata C, Lamattina L (2003) Abscisic acid, nitric oxide and stomatal closure—is nitrate reductase one of the missing links? Trends Plant Sci 8: 20–26 [DOI] [PubMed] [Google Scholar]
- Gardner PR, Gardner AM, Martin LA, Salzman AL (1998) Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci USA 95: 10378–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano M, Benigni MV, Lamattina L (2002) Nitric oxide improves internal iron availability in plants. Plant Physiol 130: 1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F-Q, Okamoto M, Crawford NM (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- Kaiser WM, Weiner H, Kandlbinder A, Tsai CB, Rockel P, Sonoda M, Planchet E (2002) Modulation of nitrate reductase: some new insights, an unusual case and a potentially important side reaction. J Exp Bot 53: 875–882 [DOI] [PubMed] [Google Scholar]
- Keen NT (1990) Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet 24: 447–463 [DOI] [PubMed] [Google Scholar]
- Keen NT, Tamaki S, Kobayashi D, Trollinger D (1988) Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70: 191–197 [DOI] [PubMed] [Google Scholar]
- Klessig DF, Durner J, Zhou JM, Kumar D, Navarre R, Zhang S, Shah J, Wendehenne D, Du H, Trifa Y, et al (2000) NO and salicylic acid signalling in plant defence. Proc Natl Acad Sci USA 97: 8849–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E, Slusarenko AJ (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lamb CJ, Lawton MA, Dron M, Dixon RA (1989) Signals and transduction mechanisms for activation of plant defences against microbial attack. Cell 56: 215–224 [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon RA, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid. A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002–1004 [DOI] [PubMed] [Google Scholar]
- Métraux J-P, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Raschdorf K, Schmid E, Blum W, Inverardi B (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250: 1004–1006 [DOI] [PubMed] [Google Scholar]
- Murgia I, Delledonne M, Soave C (2002) Nitric oxide mediates iron-induced ferritin accumulation in Arabidopsis. Plant J 30: 521–528 [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas ML, Ryan CA (2002) Nitric oxide negatively modulates wound signalling in tomato plants. Plant Physiol 130: 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Simontacchi M, Puntarulo S, Lamattina L (2002) Nitric oxide is required for root organogenesis. Plant Physiol 129: 954–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer S, Janistyn B, Jessner G, Pichorner H, Ebertmann R (1994) Gaseous nitric oxide stimulates guanosine-3′5′-cyclic monophosphate (cGMP) formation in spruce needles. Phytochemistry 36: 259–262 [Google Scholar]
- Poole RK, Hughes MN (2000) New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol 36: 775–783 [DOI] [PubMed] [Google Scholar]
- Raskin I, Turner IM, Melander WR (1989) Regulation of heat production in the inflorescences of an Arum lily by endogenous salicylic acid. Proc Natl Acad Sci USA 86: 2214–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53: 103–110 [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schmidt HHHW, Walter U (1994) NO at work. Cell 78: 919–924 [DOI] [PubMed] [Google Scholar]
- Seregélyes C, Barna B, Hennig J, Konopka D, Pasternak TP, Lukács N, Fehér A, Gábor V, Horváth GV, Dudits D (2003) Phytoglobins can interfere with nitric oxide functions during plant growth and pathogenic responses: a transgenic approach. Plant Sci 165: 541–550 [Google Scholar]
- Stuehr DJ, Fasehun OA, Kwon NS, Gross SS, Gonzalez JA, Levi R, Nathan CF (1991) Inhibition of macrophage and endothelial cell nitric oxide synthase by diphenyleneiodonium and its analogs. FASEB J 5: 98–103 [DOI] [PubMed] [Google Scholar]
- Tenhaken R, Rubel C (1997) Salicylic acid is needed in hypersensitive cell death in soybean but does not act as a catalase inhibitor. Plant Physiol 115: 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei YD, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194 [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SC, Glazebrook J (2003) Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J 33: 733–742 [DOI] [PubMed] [Google Scholar]
- Vasudevan SG, Armarego WL, Shaw DC, Lilley PE, Dixon NE, Poole RK (1991) Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol Gen Genet 226: 49–58 [DOI] [PubMed] [Google Scholar]
- Weymann K, Hunt M, Uknes S, Neuenschwander U, Lawton K, Steiner H-Y, Ryals J (1995) Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7: 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yalpani N, Silverman P, Wilson TM, Kleier DA, Raskin I (1991) Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3: 809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I-c, Parker J, Bent AF (1998) Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95: 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.