Abstract
Plants naturally cycle amino acids across root cell plasma membranes, and any net efflux is termed exudation. The dominant ecological view is that microorganisms and roots passively compete for amino acids in the soil solution, yet the innate capacity of roots to recover amino acids present in ecologically relevant concentrations is unknown. We find that, in the absence of culturable microorganisms, the influx rates of 16 amino acids (each supplied at 2.5 μm) exceed efflux rates by 5% to 545% in roots of alfalfa (Medicago sativa), Medicago truncatula, maize (Zea mays), and wheat (Triticum aestivum). Several microbial products, which are produced by common soil microorganisms such as Pseudomonas bacteria and Fusarium fungi, significantly enhanced the net efflux (i.e. exudation) of amino acids from roots of these four plant species. In alfalfa, treating roots with 200 μm phenazine, 2,4-diacetylphloroglucinol, or zearalenone increased total net efflux of 16 amino acids 200% to 2,600% in 3 h. Data from 15N tests suggest that 2,4-diacetylphloroglucinol blocks amino acid uptake, whereas zearalenone enhances efflux. Thus, amino acid exudation under normal conditions is a phenomenon that probably reflects both active manipulation and passive uptake by microorganisms, as well as diffusion and adsorption to soil, all of which help overcome the innate capacity of plant roots to reabsorb amino acids. The importance of identifying potential enhancers of root exudation lies in understanding that such compounds may represent regulatory linkages between the larger soil food web and the internal carbon metabolism of the plant.
Plant roots normally are surrounded by a collection of functionally and nutritionally interconnected species known as the soil food web (Brussaard et al., 1997). Root-colonizing bacteria and fungi, which form a resource base for this heterotrophic complex, use root exudates as a carbon source while simultaneously supplying mineralized nitrogen to the plant as they are consumed by nematodes and protozoa (Griffiths, 1994; Ferris et al., 1998). Experiments show that supplying such micropredators to roots colonized by microorganisms increases biomass of both the plant and soil organisms (Ingham et al., 1985; Setälä and Huhta, 1991; Jentschke et al., 1995; Scheu et al., 1999). One can reasonably hypothesize that such strong reciprocal benefits to both the autotrophic plant and the heterotrophic food web could have selected for regulatory linkages between these interdependent entities that influence exudation. Analogous linkages could be important for symbiotic rhizobial bacteria and mycorrhizal fungi, which also function outside the root cell plasma membrane using root exudates as a carbon source (Day et al., 2001; Hahn and Mendgen, 2001; Trevaskis et al., 2002). Defining how microorganisms affect root exudation, therefore, may increase our understanding of all these plant-microbe interactions.
Root exudation of soluble compounds, specifically amino acids, is actually a net release composed of both influx and efflux components (Jones and Darrah, 1994). The passive efflux of amino acids is driven primarily by large differences in concentration between the inside (e.g. 10 mm) and outside (e.g. 0.1–10 μm) of root cells, while influx is fueled by outward pumping proton-ATPases that create and maintain an electrochemical potential difference across the plasma membrane to support uptake into roots by proton-coupled amino acid transporters (Farrar et al., 2003). The extent to which plants compete effectively for amino acid exudates with root-colonizing microorganisms, therefore, depends on the innate capacity of root cells to recover released compounds. The realistic capacity, however, is as yet undefined because quantitative relationships between amino acid influx and efflux rates in axenic roots have not been measured at ecologically relevant concentrations. In maize (Zea mays), 120 μm Gly has an influx rate approximately 60% greater than the efflux rate (Jones and Darrah, 1994), but Gly concentrations in tundra soils with high organic matter actually range from 4 to 14 μm (Kielland, 1994). In fact, when the external concentration of [3H]Lys was decreased from 100 to 1 μm, uptake rates in barley roots declined 96% (Soldal and Nissen, 1978). By extension, the influx rate for Gly in the low micromolar range probably is much reduced from that measured at 120 μm and could potentially be less than the efflux rate.
Answers to two questions will help define how plant roots interact with the soil food web and may secondarily contribute to understanding rhizobial and mycorrhizal symbioses. First, do axenic roots efflux amino acids faster than they are absorbed at ecologically relevant external concentrations? If so, then simple, opportunistic uptake already described (Owens and Jones, 2001) can explain how soil microorganisms compete with roots for these compounds. Alternatively, if axenic roots absorb low micromolar concentrations of amino acids from an external solution faster than they are released, then the traditional concept of amino acid exudation (Rovira, 1956) requires clarification. Specifically, can microorganisms enhance net amino acid efflux from roots with any common metabolites? A wide variety of potential disruptors exist, including aromatic and fatty acid products released from most microbial populations, as well as more specialized compounds, such as 2,4-diacetylphloroglucinol (DAPG) from Pseudomonas bacteria (Cook et al., 1995) and zearalenone (Z) from Fusarium fungi (Jimenez et al., 1996), which have been studied as mycotoxins but apparently not tested for their effects on root exudation. We addressed these issues using a sensitive method for detecting amino acids (Cohen and Michaud, 1993) and report relevant influx and efflux rates under various experimental conditions in the presence and absence of several microbial products.
RESULTS
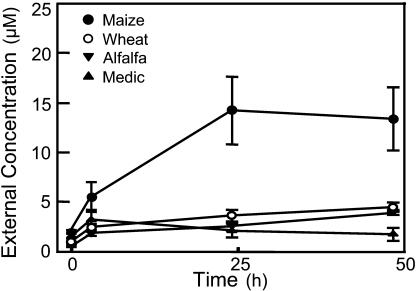
Initial trials showed that after 24 h in hydroponic nutrient solutions at pH 6.5 with 1.0 mm nitrate, axenic seedlings established a steady-state external concentration of 16 amino acids (Fig. 1). One striking result from these measurements was the similarity of concentrations produced by the C3-species alfalfa (Medicago sativa), medic (Medicago truncatula), and wheat (Triticum aestivum) compared to the 4- to 5-fold higher concentration around roots of maize, a C4-species. Amino acid concentrations around alfalfa roots remained steady until at least 96 h (data not shown). Such data support the traditional concept that roots release amino acids, but the relatively constant values measured after 24 h could reflect either a balance between efflux and influx rates or a pool of amino acids that is no longer influenced by the root.
Figure 1.
Stabilization of amino acid concentrations in solution surrounding roots of axenic plants. Intact seedlings of B73 maize, Anza wheat, Moapa 69 alfalfa, and A17 medic were transferred to fresh solution at t0 and total concentration of 16 amino acids was measured at the times indicated.
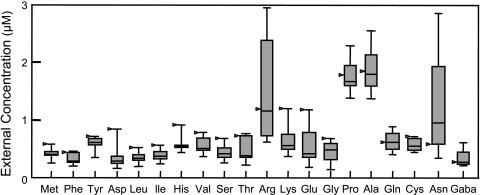
Within M. Sativa, host genetic traits had limited effects on the steady-state concentration of amino acids in the root hydroponic solution. Tests with nine diverse alfalfa germplasm pools representing both unselected natural populations and the highly selected agronomic cultivar Moapa 69 showed a relatively narrow range of concentrations for most amino acids in root exudates after 96 h (Fig. 2). These experiments, which were done without nitrate at pH 6.5, may represent the spectrum of amino acids available to symbiotic Sinorhizobium meliloti bacteria that infect roots under these conditions. Other experiments with Moapa 69 alfalfa showed little effect of swirling the root solution before sampling, altering pH (6.5 versus 8.0), or adding nitrate (0 versus 1 mm) on net efflux of amino acids from roots (data not shown). Likewise, no difference in the net efflux of amino acids from Moapa 69 alfalfa seedlings was detected at the end of 12-h dark and light periods (data not shown). Based on these results, we concluded that amino acid exudation from alfalfa roots is a conservative trait, which is not easily influenced by short-term changes in several important environmental parameters.
Figure 2.
Variation in amino acids released by roots of diverse alfalfa germplasms. Compounds in axenic root solutions from plants of nine alfalfa germplasm pools (see “Materials and Methods”) were analyzed after 48 h. “Box-and-whisker” plots show four quartiles surrounding each median value. The agronomic cultivar Moapa 69 (black arrow) produced slightly higher levels of many, but not all, amino acids than less selected populations.
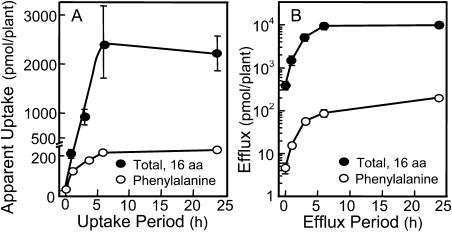
HPLC measurements easily detected apparent uptake (AU) of amino acids present at 2.5 μm in the root solution (Fig. 3A). The amount of efflux (E) into root solution containing no amino acids was determined (Fig. 3B) as a prelude to calculating influx (I). The AU and E values generally saturated after 6 h, so actual rates were calculated over the initial 3-h period when reasonably linear changes were large enough to permit reliable measurement. Rapid seedling growth required that data in this 24-h experiment be expressed on a per plant basis, as opposed to rates reported per gram root fresh weight in 3-h experiments. Rates calculated over the first 3-h period in Figure 3 were similar to those measured in subsequent experiments, assuming comparable fresh weights at 3 h. Tests in alfalfa with carbonyl cyanide m-chlorophenylhydrazone (CCCP) concentrations ranging from 0.1 to 100 μm established that 10 μm CCCP was the minimum concentration required to maximize efflux of all amino acids measured (data not shown), consistent with common usage of this compound for blocking amino acid uptake (Jones and Darrah, 1994).
Figure 3.
AU and E of amino acids by alfalfa seedling roots. HPLC analyses quantified the total of 16 amino acids for (A) AU from an initial concentration of 2.5 μm for each and (B) efflux into an amino acid-free mineral solution containing 10 μm CCCP, a protonophore that blocks influx. Data for Phe, one amino acid included in the total, also are shown.
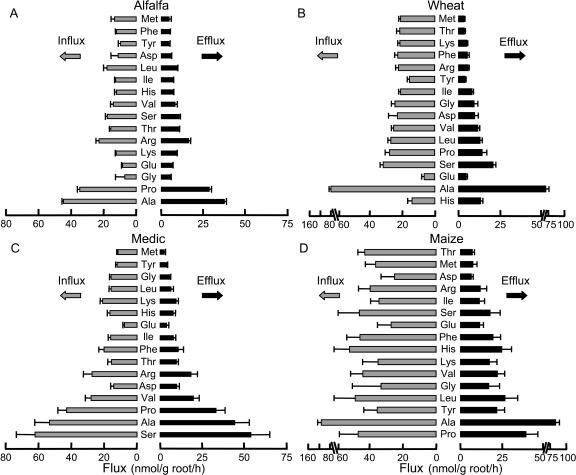
Based on these results, the values for I and E over an initial 3-h period in subsequent experiments were determined for 16 amino acids in four plant species (Fig. 4). Calculations of average fluxes (mean ± se, nmol/g root fresh weight/h) across all 16 amino acids for alfalfa (I = 16.3 ± 2.4; E = 10.3 ± 2.2), medic (I = 23.8 ± 3.9; E = 15.5 ± 3.9), wheat (I = 25.4 ± 4.1; E = 12.7 ± 4.1), and maize (I = 44.5 ± 5.2; E = 22.0 ± 4.7) showed that both influx and efflux were higher in maize than the other three species. For ecological purposes, amino acid exudation patterns may be analyzed most productively using the I/E ratios for individual amino acids. When I/E ≥ 1.0, the plant is theoretically capable of recovering all of the amino acid released. If, however, I/E < 1.0, then the plant almost certainly is losing the amino acid to the soil food web. In the absence of culturable microorganisms, all four species had I/E > 1.0 for each of the 16 amino acids examined here (Table I). Ordering individual amino acids by their I/E rank value (Table I; Fig. 4) predicts that Met will be available outside roots in relatively low concentrations while Ala will be present at higher levels. This prediction generally was supported for alfalfa (Fig. 2) as well as the other three species (data not shown).
Figure 4.
Amino acid fluxes in axenic plant roots. Mean efflux and influx rates +se of 16 amino acids were determined for (A) Moapa 69 alfalfa, (B) Anza wheat, (C) A17 medic, and (D) B73 maize. Amino acids are ranked by I/E ratios from Table I.
Table I.
Influx/Efflux (I/E) ratios for 16 amino acids near roots of four plant species
| Alfalfa
|
Medic
|
Wheat
|
Maize
|
||||
|---|---|---|---|---|---|---|---|
| aa | I/E | aa | I/E | aa | I/E | aa | I/E |
| Met | 3.00 | Met | 4.65 | Met | 6.45 | Thr | 5.93 |
| Phe | 2.55 | Tyr | 3.22 | Thr | 6.33 | Met | 4.78 |
| Tyr | 2.23 | Gly | 2.85 | Lys | 4.25 | Asp | 3.73 |
| Asp | 2.09 | Leu | 2.39 | Phe | 4.25 | Arg | 3.31 |
| Leu | 1.97 | Lys | 2.21 | Arg | 3.97 | lle | 2.97 |
| Ile | 1.94 | His | 2.15 | Tyr | 3.91 | Ser | 2.57 |
| His | 1.86 | Glu | 2.05 | lle | 2.57 | Glu | 2.31 |
| Val | 1.79 | lle | 2.05 | Gly | 2.55 | Phe | 2.31 |
| Ser | 1.71 | Phe | 1.86 | Asp | 2.35 | His | 2.07 |
| Thr | 1.58 | Thr | 1.59 | Val | 2.16 | Lys | 2.00 |
| Arg | 1.45 | Arg | 1.50 | Leu | 2.10 | Val | 1.96 |
| Lys | 1.40 | Asp | 1.45 | Pro | 1.93 | Gly | 1.94 |
| Glu | 1.39 | Val | 1.41 | Ser | 1.51 | Leu | 1.80 |
| Gly | 1.36 | Pro | 1.28 | Glu | 1.35 | Tyr | 1.60 |
| Pro | 1.25 | Ala | 1.20 | Ala | 1.17 | Ala | 1.37 |
| Ala | 1.23 | Ser | 1.14 | His | 1.05 | Pro | 1.17 |
Influx and efflux rates, determined as described in “Materials and Methods,” were obtained from axenic plants growing with 1.0 mm nitrate at pH 6.5. aa, Amino acid.
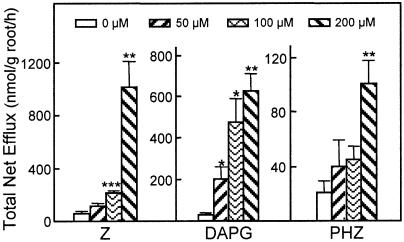
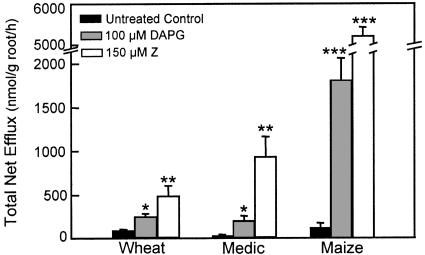
Several microbial products increased net efflux of the 16 amino acids studied here by substantial amounts. No unique effect on any individual amino acid was detected, so results are reported as the sum of all 16 amino acids. Dose response tests with alfalfa established that net efflux was increased by micromolar concentrations of Z, DAPG, and phenazine (PHZ; Fig. 5). Normalized across numerous alfalfa experiments, 200 μm treatments increased the 3-h efflux total by 200% for PHZ, 1,600% for DAPG, and 2,600% for Z. Alfalfa seedlings treated with PHZ or PHZ-1-carboxylate at 200 μm produced nearly identical total net effluxes of the 16 amino acids (data not shown). Tests with 150 μm Z and 100 μm DAPG on the other three plant species established that these compounds were active on all plant species examined (Fig. 6) Another aromatic microbial product, lumichrome, which increases root respiration and plant growth (Phillips et al., 1999), had no detectable effect over the concentration range of 5 nm to 200 μm. Myristic and palmitic acids, known to have protonophoric effects on mitochondria (Wojtczak et al., 1998), also had no effect on net amino acid efflux at 200 μm.
Figure 5.
Effects of microbial products on net efflux of amino acids in hydroponic solutions surrounding axenic alfalfa roots. Total efflux +se of 16 amino acids was determined over a 3-h period for plants treated with Z, DAPG, or PHZ. Means identified with *, **, or *** differed significantly from the untreated control by P ≤ 0.05, 0.01, or 0.001, respectively.
Figure 6.
Effects of two microbial products on net efflux of amino acids from roots of three plant species. Roots of Anza wheat, A17 medic, and B73 maize were treated with 100 μm DAPG or 150 μm Z, and the total release of 16 amino acids was measured over 3 h. Means identified with *, **, or *** differed significantly from the untreated control by P ≤ 0.05, 0.01, or 0.001, respectively. Note breaks in the y axis.
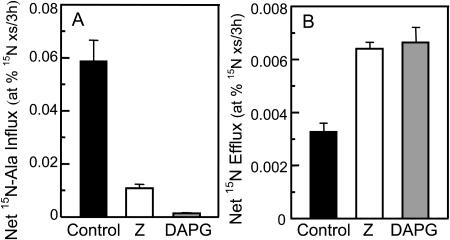
Experiments with 15N established that the two most powerful microbial factors, Z and DAPG, increased net efflux through different mechanisms (Fig. 7). When 2.5 μm 15N-Ala was supplied outside the root of unlabeled alfalfa seedlings, 100 μm DAPG blocked 98% of Ala uptake measured in the untreated control (P < 0.001), whereas roots exposed to 150 μm Z contained an amount of 15N intermediate between the untreated control and the DAPG-treated roots (Fig. 7A). In contrast, using alfalfa seedlings labeled with 15N, both Z and DAPG doubled net efflux of nitrogenous compounds (P < 0.01; Fig. 7B). Taken together, these results are consistent with the concept that DAPG blocks amino acid uptake and Z promotes efflux of nitrogenous compounds, including amino acids, from roots. The 15N-efflux tests employed a highly selected agronomic population UC2705 because a seed-producing population was already available in the field for labeling with (15NH4)2SO4. The limited effect of genetic traits in alfalfa on the total net efflux of amino acids (Fig. 2) suggests that this population has amino acid exudation characteristics similar to Moapa 69.
Figure 7.
Effects of two microbial products on net influx and net efflux of 15N-containing compounds. Axenic alfalfa roots treated with 150 μm Z or 100 μm DAPG were tested for effects on influx of 15N-Ala (A) and, separately, for effects on efflux of 15N-containing compounds from seedlings labeled with (15NH4)2SO4 in the previous generation (B). Data are expressed as mean changes (+ se) over a 3-h period in atom % 15N excess, relative to a reference of 0.36679 atom % 15N.
DISCUSSION
Amino acid efflux and influx rates, assessed here in roots of four plant species, document both the general uniformity of these transport systems (Fig. 4; Table I) and a shared vulnerability to disruption by certain microbial compounds that increase net efflux of amino acids (Figs. 5 and 6). Together these facts clarify how plants minimize amino acid losses while root-colonizing microorganisms simultaneously can enhance the availability of amino acids in the root zone. Two different mechanisms of disruption found here apparently involve blocking amino acid uptake by the bacterial reagent DAPG and promoting efflux by the fungal compound Z (Fig. 7). This diversity of mechanisms for compounds from dissimilar microorganisms, together with the differing responses of plant species to these compounds (Fig. 6), suggests these phenomena are potentially important factors that may alter structure, and possibly productivity, of microbial and plant communities. Our data from intact plants do not allow any speculation on the contribution of different root components, such as border cells or the elongation zone.
Results reported here update quantitative details on the physiology of root exudation. Previous work with maize showed that influx of 120 μm Gly exceeds normal efflux (Jones and Darrah, 1994). Our finding that influx of 16 amino acids exceeds efflux by 5% to 545% (Table I) quantifies that relationship across four species in the absence of any culturable microorganisms using appropriately low (2.5 μm) amino acid concentrations (Kielland, 1994; Figs. 1 and 2) that could be measured with a newer fluorescence technique (Cohen and Michaud, 1993). Evidence here that amino acid influx rates exceed efflux (Fig. 4; Table I) suggests these compounds should not be present in root exudates, in contrast to published reports (Rovira, 1956; Fan et al., 2001) and our own data (Figs. 1 and 2). This apparent paradox presumably is resolved in axenic hydroponic cultures by diffusional forces, which allow some molecules to escape immediate uptake after they are released from the root. In soil, physical and chemical adsorptive forces could prevent amino acid reabsorption by the plant (Jones and Darrah, 1994) and ensure that microorganisms obtain significant amounts either by competitive uptake (Owens and Jones, 2001) or by active disruption of fluxes (Figs. 5 and 6). The somewhat higher apparent equilibrium concentration maintained by maize roots (Fig. 1) may be associated with elevated rates of efflux compared with the other species (Fig. 4).
The observation that specific microbial products increase net efflux of amino acids from roots (Figs. 5, 6, and 7B) establishes a previously unrecognized principle: Some nonpathogenic microorganisms can potentially increase their access to plant carbon resources. Pseudomonas bacteria, long known as major colonists on healthy plant roots and suppressors of fungal pathogens (Cook and Rovira, 1976), play that role primarily through the production of antifungal compounds, such as PHZ-1-carboxylate and DAPG (Cook et al., 1995; Weller et al., 2002). PHZ is also a common product of these bacteria (Taraz et al., 1990). Z is an antifungal compound produced by various species of the soil fungus Fusarium (Jimenez et al., 1996). If these compounds increase the availability of amino acids in root exudates in addition to impairing the growth of competing microorganisms, then clearly this combination of benefits could promote growth of the organisms producing them. By extension, anything that micropredators, such as protozoa, nematodes, or collembola, do physically or chemically to simulate the synthesis or release of these compounds in microorganisms could enhance foundation resources for the entire food web. Thus, this identification of active chemical compounds establishes the existence of a potentially important molecular link between the heterotrophic soil food web and the autotrophic plant.
15N studies suggest that DAPG and Z affect amino acid flux by different mechanisms (Fig. 7). These experiments compared effects of the compounds on two contrasting fluxes of 15N in alfalfa roots: (1) net uptake of 15N-Ala (Fig. 7A), and (2) net efflux of diverse 15N-compounds, including amino acids, from seedling roots labeled in the previous generation with (15NH4)2SO4 (Fig. 7B). DAPG decreased 15N-Ala uptake 98%, whereas plants treated with Z accumulated 19% as much 15N from Ala as untreated controls (Fig. 7A). Because neither DAPG nor Z preferentially affected the net efflux of specific individual amino acids (extracted from summed data in Figs. 5 and 6), we sought a general explanation for their action. The simplest interpretation is that inhibition of Ala uptake, and presumably other amino acids, by DAPG is responsible for accumulation of amino acids after their release from the root (Figs. 5 and 6). Z clearly allowed some 15N-Ala uptake (Fig. 7A), but it also promoted a large increase in net efflux of amino acids (Figs. 5 and 6). If Z has only one effect on transport processes, then these results could be explained by a general enhancement of efflux, which recycled 15N-Ala quickly back across the root cell plasma membrane. The increase in net efflux of 15N compounds from roots treated with either DAPG or Z (Fig. 7B) is consistent with the proposed effects of these compounds on different portions of the efflux-influx cycle; amino acids accumulate outside the root in both cases but for different reasons. Molecular explanations of how these microbial compounds alter amino acid transport await results from ongoing tests for specific effects on transporters, proton-pumping ATPases, signaling, and membrane integrity.
The ecological relevance of active concentrations studied here is supported by data reported for DAPG (Bonsall et al., 1997). In that study direct extraction of rhizosphere soil adhering to wheat roots recovered as much as 2.1 μg DAPG/g of soil plus root. While calculating relevant concentrations of DAPG at the plasma membrane from such data is imprecise, the 100 μm concentration active in this study (Figs. 5 and 6) is certainly feasible. For example, 2.1 μg DAPG in 1 g of a typical cultivated soil with a bulk density of 1.45 g/cm3 and 45% pore space calculates to 32 μm DAPG with complete saturation and as high as 128 μm DAPG at the permanent wilting point (Brady, 1974). Patchy distributions of bacterial colonies along roots could increase the local concentration of DAPG considerably above these levels.
Microbial compounds tested here have not been reported in N2-fixing rhizobial bacteria or endomycorrhizal mycorrhizal fungi, and it seems likely these symbionts would benefit more from a regulated modification, rather than a general disruption, of transport processes in the plant plasma membrane. Data reported here are useful because they document the amino acid mixtures that could be available to microsymbionts associated with root cells in four plant species (Fig. 4). The fact that rhizobia send large amounts of N2-derived nitrogen through the plasma membrane as Ala (Waters et al., 1998) is consistent with the high rates of Ala influx measured here (Fig. 4). Rhizobia may obtain other amino acids used in the symbiosis (Lodwig et al., 2003) by passive absorption of those that diffuse normally through the plasma membrane. Endomycorrhizal symbioses also depend on an orderly exchange of plant carbon and fungal phosphate through the plasma membrane (Hahn and Mendgen, 2001), and these fungi, like rhizobia, may obtain sufficient amino acids by passive absorption.
Current ecological theory argues that soil communities use feedback mechanisms and indirect interactions to influence plant growth (Moore et al., 2003). Data reported here (Figs. 5 and 6) offer specific molecular examples of how microbial products can directly elicit changes in plant processes. Other results show that additional microbial compounds at much lower concentrations induce both proteomic (Mathesius et al., 2003) and physiological (Phillips et al., 1999; Joseph and Phillips, 2003) changes in plants. Defining how such short-term changes are integrated into longer term plant responses will clarify any role these molecular interactions may play in structuring plant communities and possibly driving ecosystem functions such as plant biodiversity, productivity, and variability (Van der Heijden et al., 1998; Bever, 2003).
MATERIALS AND METHODS
Experimental Materials
Alfalfa (Medicago sativa) germplasm pools were collected and supplied for testing by the U.S. Department of Agriculture as plant introduction (PI) materials that represent primarily unselected, landrace populations from the indicated geographical regions: PI162787, Argentina; PI170532, Turkey; PI205329, Peru; PI213005, India; PI217648, Iraq; PI399551, Romania; PI435232, Egypt; and PI478545, Peru. In addition, the highly selected, agronomic cultivar Moapa 69 was used as indicated. For wheat (Triticum aestivum), the agronomic variety Anza was employed. Two other plants favored as genetic models also were examined: Medicago truncatula Gaertner genotype A17 from Jemalong (termed medic in this study) and maize (Zea mays) line B73. An additional agronomic alfalfa line, UC2705, was used to produce 15N-labeled seeds.
Plant Cultures
Axenic plant populations were produced as described below. Repeated water rinses were crucial for removing antibiotics to prevent growth impairment. All sterile water rinses thus consisted of at least five rapid rinses with 100 mL, a 1- to 2-h soaking on a shaker at 50 rpm, and an additional series of at least five rapid rinses with 100 mL. Without this extensive rinsing regime, seedlings sometimes were chlorotic and unsuited for further study.
Alfalfa
Unscarified seeds were treated 10 min with concentrated H2SO4, rinsed with water, soaked 30 min in 5.25% NaClO (w/v), and rinsed with water. Then seeds were soaked overnight at 4°C in a solution containing ampicillin (100 μg/mL), kanamycin (100 μg/mL), and rifampicin (10 μg/mL) and rinsed with water. Seeds (120/container) were germinated in the standard growth chambers in petri plates on a sterile cheesecloth bed supported by wire mesh over 25 mL of unaerated Fåhraeus solution (Fåhraeus, 1957) with or without 1.0 mm KNO3 and adjusted to pH 6.5 or 8.0.
Medic
Unscarified seeds were treated 8 min in concentrated H2SO4 and rinsed with water, soaked 3 min in 0.25% NaClO (w/v)/0.1% Tween 20 and rinsed with water, then soaked 30 min in 1% Chloramine T/0.1% Tween 20 and rinsed with water. After soaking seeds overnight at 4°C in the alfalfa antibiotic solution and rinsing with water, seeds were germinated 3 to 5 d in the dark at 25°C on petri plates containing Fåhraeus solution with 1.0 mm KNO3, adjusted to pH 6.5, and solidified using 8 g/L phytagel (Sigma, St. Louis). Seedlings were then transferred to inverted 16-mm test-tube caps containing 7 mL of unaerated Fåhraeus solution, 1.0 mm KNO3, pH 6.5. Each replicate cap contained 5 to 20 plants and was supported with other replicates inside Magenta jars (77 × 77 × 97 mm) in the standard growth chamber.
Wheat
Seeds were treated 30 min with 5.25% NaClO (w/v) and rinsed with water, soaked 20 min in 1% Chloramine T/0.1% Tween 20 and rinsed with water, then soaked overnight at 4°C in the alfalfa antibiotic solution and rinsed with water. Seeds were germinated 4 d in Magenta jars on wire mesh racks in contact with unaerated Fåhraeus solution, 1.0 mm KNO3, pH 6.5, in the standard growth chamber.
Maize
Seeds were rinsed briefly in 70% ethanol and then soaked 20 min in 1% NaClO (w/v)/0.1% Tween 20 and rinsed with water, soaked 20 min in 1% Chloramine T/0.1% Tween 20 and rinsed with water, then soaked overnight at 4°C in the alfalfa antibiotic solution and rinsed with water. Seeds were germinated on phytagel plates as with medic for 6 d, then transferred onto wire mesh racks as for wheat in the standard growth chambers.
Experimental Conditions
Standard growth chamber conditions employed a photosynthetically active (400–700 nm) photon flux density of 100 μmol photons/m2 s, a 12-h day/12-h night cycle, and 25°C/20°C. Before experimental treatments were imposed, plants were equilibrated, unless specified, for the following periods: alfalfa, 3 d; medic, 2 d; wheat, 4 d; and maize, 2 d. Seedlings produced by these methods had typical root fresh weights (mg/plant) of alfalfa, 6; medic, 17; wheat, 30; and maize, 77 at the conclusion of the experiments.
Microbiological Sterility
At the beginning and conclusion of each experiment, sterility of hydroponic root solutions was assessed by testing for culturable microorganisms on TY agar (Beringer, 1974). Only samples showing no microbial growth were analyzed further. In addition, macerates of all roots studied, except those reported in Figures 2 and 7B, also were verified as being free of culturable microorganisms by the same test procedure. This second, more rigorous criterion for axenic roots proved generally superfluous because only three of more than 500 tests showed bacteria present in root macerates but absent in the hydroponic solution.
Chemical Treatments
All treatments were added to the hydroponic Fåhraeus solution, which contained 1.0 mm KNO3 unless specified otherwise. AU was measured using a mixture of HPLC-grade, L isomers of 16 amino acids supplied at 2.5 μm for each amino acid. This solution, Amino Acid Standard H (Pierce, Rockford, IL), contained Ala, Arg, Asp, Cys, Glu, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr, Tyr, and Val. The protonophore CCCP (Sigma) was supplied at 10 μm unless specified otherwise. DAPG (Toronto Research Chemicals, Toronto), Lumichrome (Aldrich, Milwaukee, WI), myristic acid (Sigma), palmitic acid (Sigma), PHZ (Sigma), PHZ-1-carboxylic acid (Chembridge, San Diego), and Z (Sigma) were used at various, specified concentrations.
Experiments were initiated by draining the hydroponic medium and rinsing roots twice with sterile Fåhraeus solution, containing 1.0 mm KNO3, unless noted. After replacing the solution for a third time, 0.5-mL samples were removed for the initial time point. All samples were collected with 1.5-mL plastic pipette tips taking care to minimize root disturbance.
Amino Acid Analyses
Amino acids in root solutions were measured by HPLC analysis of fluorescent derivatives produced by reaction with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (Cohen and Michaud, 1993), using commercially available reagents and an AccQ•Tag column (Waters, Milford, MA). With a Waters 2475 fluorescence detector, changes of 1 to 10 pmol were measured reproducibly for all amino acids. To amplify the signal, 500-μL samples from root solutions were freeze dried and then derivatized in 100 μL before analyzing a 40 μL aliquot.
15N Experiments
Two types of 15N experiments were conducted. Influx tests measured the effects of microbial compounds on Ala uptake in Moapa 69 alfalfa using l-Ala-15N (99 atom % excess, C/D/N Isotopes, Pointe-Claire, Canada), which was supplied at 2.5 μm in the standard hydroponic solution. Roots, harvested 1 min and 3 h after supplying 15N-Ala, were rinsed 1 min in 1 mm CaSO4, frozen, and ground in Eppendorf tubes with a mechanized pestle. Extracts were freeze dried and analyzed for 15N content. Efflux tests were conducted with 15N-labeled seeds produced from a field population of alfalfa, UC2705, which had been fertilized with (15NH4)2SO4. Seeds were sterilized and germinated in the standard hydroponic seedling protocol to test whether microbial compounds altered efflux of 15N-containing compounds, including amino acids. Solution surrounding roots was collected at t0 and again after treating plants for 3 h with test compounds, freeze dried, and measured for changes in 15N content. 15N enrichments were measured by ratio mass spectrometry at the University of California, Davis Stable Isotope Facility. Data are reported as changes in 15N atom percent excess, relative to a constant reference value of 0.36679 atom % 15N, over the 3-h period to account for both experimental treatment effects and initial differences in 15N enrichment (Hauck and Bremner, 1976).
Experimental Design
Every experiment contained three to six replicates for each treatment, and all experiments were repeated at least once. Because each replicate contained 8 (maize) to 120 (alfalfa) seedlings, data were drawn from large numbers of individual plants: alfalfa (>25,000), medic (>1,000), wheat (>1,800), and maize (>600). Statistical analyses were conducted using Statistix 7 (Analytical Software, Tallahassee, FL).
Amino Acid Influx and Efflux Calculations
All rates were calculated from linear changes during the initial 3 h of treatment. AU was measured as the disappearance of amino acids from the initial 2.5 μm solution. E was determined separately in the absence of added amino acids by blocking normal reabsorption of amino acids with 10 μm CCCP. I was calculated from the relationship
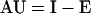 |
with all parameters defined as positive for the purposes of this study. Because CCCP has no effect on plant cell plasma membrane permeability (Porter et al., 1985) and functions as a protonophore to decrease influx (DiTomaso et al., 1992), external accumulation of amino acids in the presence of CCCP is interpreted as one measure of efflux (Jones and Darrah, 1994).
Acknowledgments
We thank D. Harris for 15N analyses and W.D. Bauer for helpful discussions.
This work was supported by the National Science Foundation Division of Environmental Biology (grant no. DEB–0120169), by the Binational Agricultural Research and Development Fund (award no. US–3353–02), and by the University of California Institute for Mexico and the United States-Consejo Nacional de Ciencia y Tecnológia.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044222.
References
- Beringer JE (1974) R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84: 188–198 [DOI] [PubMed] [Google Scholar]
- Bever JD (2003) Soil community feedback and the coexistence of competitors: conceptual frameworks and empirical tests. New Phytol 157: 465–473 [DOI] [PubMed] [Google Scholar]
- Bonsall RF, Weller DM, Thomashow LS (1997) Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl Environ Microbiol 63: 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady NC (1974) The Nature and Property of Soils, Ed 8. Macmillan, New York
- Brussaard L, Behan-Pelletier VM, Bignell DE, Brown VK, Didden W, Folgarait P, Fragoso C, Freckman DW, Gupta V, Hattori T, et al (1997) Biodiversity and ecosystem functioning in soil. Ambio 26: 563–570 [Google Scholar]
- Cohen SA, Michaud DP (1993) Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Anal Biochem 211: 279–287 [DOI] [PubMed] [Google Scholar]
- Cook RJ, Rovira AD (1976) The role of bacteria in the biological control of Gaeumannomyces graminis by suppressive soils. Soil Biol Biochem 8: 269–273 [Google Scholar]
- Cook RJ, Thomashow LS, Weller DM, Fujimoto D, Mazzola M, Bangera G, Kim D (1995) Molecular mechanisms of defense by rhizobacteria against root disease. Proc Natl Acad Sci USA 92: 4197–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DA, Poole PS, Tyerman SD, Rosendahl L (2001) Ammonia and amino acid transport across symbiotic membranes in nitrogen-fixing legume nodules. Cell Mol Life Sci 58: 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTomaso JM, Hart JJ, Linscott DL, Kochian LV (1992) Effect of inorganic cations and metabolic inhibitors on putrescine transport in roots of intact maize seedlings. Plant Physiol 99: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fåhraeus G (1957) The infection of clover root hairs by nodule bacteria, studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Fan TW-M, Lane AN, Shenker M, Bartley JP, Crowley D, Higashi RM (2001) Comprehensive chemical profiling of gramineous plant root exudates using high-resolution NMR and MS. Phytochemistry 57: 209–221 [DOI] [PubMed] [Google Scholar]
- Farrar J, Hawes M, Jones D, Lindow S (2003) How roots control the flux of carbon to the rhizosphere. Ecology 84: 827–837 [Google Scholar]
- Ferris H, Venette RC, vanderMeulen HR, Lau SS (1998) Nitrogen mineralization by bacterial-feeding nematodes: verification and measurement. Plant and Soil 203: 159–171 [Google Scholar]
- Griffiths BS (1994) Soil nutrient flow. In JF Darbyshire, ed, Soil Protozoa. CAB International, Wallingford, UK, pp 65–91
- Hahn M, Mendgen K (2001) Signal and nutrient exchange at biotrophic plant-fungus interfaces. Curr Opin Plant Biol 4: 322–327 [DOI] [PubMed] [Google Scholar]
- Hauck RD, Bremner JM (1976) Use of tracers for soil and fertilizer nitrogen research. Adv Agron 28: 219–266 [Google Scholar]
- Ingham RE, Trofymow JA, Ingham ER, Coleman DC (1985) Interactions of bacteria, fungi and their nematode grazers: effects on nutrient cycling and plant growth. Ecol Monogr 55: 119–140 [Google Scholar]
- Jentschke G, Bonkowski M, Godbold DL, Scheu S (1995) Soil protozoa and forest tree growth: non-nutritional effects and interaction with mycorrhizae. Biol Fertil Soils 20: 263–269 [Google Scholar]
- Jimenez M, Manez M, Hernandez E (1996) Influence of water activity and temperature on the production of zearalenone in corn by three Fusarium species. Int J Food Microbiol 29: 417–421 [DOI] [PubMed] [Google Scholar]
- Jones DL, Darrah PR (1994) Amino-acid influx at the soil-root interface of Zea mays L. and its implications in the rhizosphere. Plant Soil 163: 1–12 [Google Scholar]
- Joseph CM, Phillips DA (2003) Metabolites from soil bacteria affect plant water relations. Plant Physiol Biochem 41: 189–192 [Google Scholar]
- Kielland K (1994) Amino acid absorption by arctic plants: implications for plant nutrition and nitrogen cycling. Ecology 75: 2373–2383 [Google Scholar]
- Lodwig EM, Hosie AHF, Bordes A, Findlay K, Allaway D, Karunakaran R, Downie JA, Poole PS (2003) Amino-acid cycling drives nitrogen fixation in the legume—Rhizobium symbiosis. Nature 422: 722–726 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Mulders S, Gao MS, Teplitski M, Caetano-Anolles G, Rolfe BG, Bauer WD (2003) Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc Natl Acad Sci USA 100: 1444–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JC, McCann K, Setälä H, De Ruiter PC (2003) Top-down is bottom-up: Does predation in the rhizosphere regulate aboveground dynamics? Ecology 84: 846–857 [Google Scholar]
- Owens AG, Jones DL (2001) Competition for amino acids between wheat roots and rhizosphere microorganisms and the role of amino acids in plant N acquisition. Soil Biol Biochem 33: 651–657 [Google Scholar]
- Phillips DA, Joseph CM, Yang GP, Martínez-Romero E, Sanborn JR, Volpin H (1999) Identification of lumichrome as a Sinorhizobium enhancer of alfalfa root respiration and shoot growth. Proc Natl Acad Sci USA 96: 12275–12280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter GA, Knievel DP, Shannon JC (1985) Sugar efflux from maize (Zea mays L.) pedicel tissue. Plant Physiol 77: 524–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira AD (1956) Plant root excretions in relation to the rhizosphere effect. Plant Soil 7: 178–194 [Google Scholar]
- Scheu S, Theenhaus A, Jones TH (1999) Links between the detritivore and the herbivore system: effects of earthworms and Collembola on plant growth and aphid development. Oecologia 119: 541–551 [DOI] [PubMed] [Google Scholar]
- Setälä H, Huhta V (1991) Soil fauna increase Betula pendula growth: laboratory experiments with coniferous forest floor. Ecology 72: 665–671 [Google Scholar]
- Soldal T, Nissen P (1978) Multiphasic uptake of amino acids by barley roots. Physiol Plant 43: 181–188 [Google Scholar]
- Taraz K, Schaffner EM, Budzikiewicz H, Korth H, Pulverer G (1990) 2,3,9-Trihydoxyphenazin-1-carbonsäure: ein unter Berylliumeinwirkung gebildetes neues Phenazinderivat aus Pseudomonas fluorescens. Z Naturforsch 45b: 552–556 [Google Scholar]
- Trevaskis B, Colebatch G, Desbrosses G, Wandrey M, Wienkoop S, Saalbach G, Udvardi MK (2002) Differentiation of plant cells during symbiotic nitrogen fixation. Comp Funct Genom 3: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolfengel R, Boller T, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396: 69–72 [Google Scholar]
- Waters JK, Hughes BL, Purcell LC, Gerhardt KO, Mawhinney TP, Emerich DW (1998) Alanine, not ammonia, is excreted from N2-fixing soybean nodule bacteroids. Proc Natl Acad Sci USA 95: 12038–12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller DM, Raaijmakers JM, Gardener BBM, Thomashow LS (2002) Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol 40: 309–348 [DOI] [PubMed] [Google Scholar]
- Wojtczak L, Wieckowski MR, Schonfeld P (1998) Protonophoric activity of fatty acid analogs and derivatives in the inner mitochondrial membrane: a further argument for the fatty acid cycling model. Arch Biochem Biophys 357: 76–84 [DOI] [PubMed] [Google Scholar]