Abstract
Steady state population pharmacokinetics of a non-commercial immediate release metformin (hydrochloride) drug product were characterized in 28 severely obese children with insulin resistance. The concentration-time profiles with double peaks were well described by a one-compartment model with two absorption sites. Mean population apparent clearance (CL/F) was 68.1 L/hr and mean apparent volume of distribution (V/F) was 28.8 L. Body weight was a covariate of CL/F and V/F. Estimated glomerular filtration rate was a significant covariate of CL/F (p<0.001). SLC22A1genotype did not significantly affect metformin pharmacokinetics. The response to 6 months of metformin treatment (HbA1c, HOMA IR, fasting insulin, and glucose changes) was not different between SLC22A1 wild type subjects and carriers of presumably low activity SLC22A1 alleles. However, SLC22A1 variant carriers had smaller reductions in percentage of total trunk fat after metformin therapy, although the percentage reduction in trunk fat was small. The median % change in trunk fat was −2.20 % (−9.00 % – 0.900 %) and −1.20 % (−2.40 % – 7.30 %) for the SLC22A1 wild-type subjects and variant carriers, respectively. Future study is needed to evaluate the effects of SLC22A1 polymorphisms on metformin-mediated weight reduction in obese children.
Keywords: Pharmacokinetics, Obesity, Pediatric, Pharmacogenomics, Metformin, Weight Loss
Introduction
Childhood obesity is a serious worldwide issue and is one of the major health challenges in the 21st century.1, 2 Since the 1960s, prevalence rates have quadrupled in many countries.3 The importance of childhood obesity as a predisposing condition for impaired glucose and lipid homeostasis, cardiovascular diseases, and mortality in adulthood has been increasingly recognized.4–7 Yet, there are no approved weight-loss medications for children under 12 years of age.
The biguanide metformin has been considered a promising compound for amelioration of adolescent and childhood obesity.8–12 Metformin almost exclusively exists in cationic form at physiological pH; its passage through cell membranes is mediated via different transporters including the organic cation transporter member 1 (OCT1, encoded by SLC22A1), multidrug and toxic compound extrusion proteins (MATEs), and plasma membrane monoamine transporter (PMAT). Given the fact that OCT1 is predominantly expressed on the basolateral side of hepatocytes,13 OCT1 is believed to be the most important transporter for the pharmacological action of metformin. Shu et al. demonstrated that OCT1 serves a critical function in the glucoregulatory therapeutic response to metformin, and the glucose lowering effect of metformin was ablated in OCT1 knockout mice due to reduced hepatic metformin uptake.14 OCT1 also appeared to mediate metformin’s inhibitory effects on adipocyte differentiation in visceral and subcutaneous adipocytes.15
The interpatient variability of metformin pharmacokinetics (PK) and pharmacodynamics (PD) is substantial.16–18 Mean metformin renal clearance (CLR) from different studies has been reported to range from 280 – 636 mL/min,19 with an 80-fold variability in steady state trough concentration in patients with type 2 diabetes.18 Several factors contribute to the individual variations in PK, including age, renal function, and genetic polymorphisms of metformin transporters.20 The variability in PK likely affects metformin pharmacological effects; a population PK/PD model has been established between metformin concentration and the glucose-lowering effects of metformin.21 Recently, a single nucleotide polymorphism (SNP) near the Ataxia Telangiectasia Mutated (ATM) gene was identified to be accountable for a small fraction (2.5 %) of the variance in metformin response.22
Despite the evidence suggesting that metformin treatment is associated with considerable variability in terms of weight reduction, to our knowledge there are no data relating metformin pharmacogenetics to metformin-induced weight loss in obese children.23 In addition, information of metformin PK in children is limited, and the influence of pharmacogenetics on metformin PK is not well established.
Previously, we conducted a randomized study to investigate the effects of metformin on body weight and composition in obese insulin-resistant children.12 We collected and analyzed PK samples in 30 subjects to determine the PK of metformin in obese children. We also determined the SCL22A1 genotype of these subjects to evaluate the contribution of this genotype to the variability in PK/PD response in this population.
Methods
Study Sample
Obese, insulin-resistant, but otherwise healthy children, between the ages of 6 and 12y were recruited for a 6-month placebo-controlled, randomized trial of metformin12 by advertisements in local newspapers and by referral from physicians. Obesity was defined as BMI at or above the 95th percentile for US children of the same age and sex. Included patients were pre-pubertal or had at most early pubertal development (breast Tanner stages I, II, or III for girls; testes size ≤ 8 mL for boys) and were hyperinsulinemic (fasting insulin concentration ≥107 pmol/L [15 mIU/mL]).12 Patients were excluded if they had significant renal, cardiac, pulmonary or hepatic disease, evidence for impaired fasting glucose (≥100 mg/dL) or were diabetic (fasting plasma glucose ≥ 126 mg/dL or HgbA1C ≥6.5%), weight loss of 2% bodyweight within the past 6 months, presence of other endocrinologic disorders leading to obesity, recent use (within six months) of anorexiant medications, or medical treatment for hypertension or dyslipidemia. For this secondary analysis, only results from subjects treated with metformin who gave permission for genomic DNA collection and analysis are reported. The study was approved by the Institutional Review Board of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), NIH (clinicaltrials.gov NCT00005669). Written assent and consent were obtained from children and their parents.
PK sampling was performed at steady state (i.e. at the end of the 6-month treatment period. Six mL of blood was collected in K2EDTA containing Vacutainer tubes immediately before metformin administration, and then at 0.25, 0.5, 1, 1.5, 2, 2.5, 3, 4, 6, 8 and 12 hours following the dose. Immediately after collection from the test subjects, the blood samples were stored at 4°C. For the first 10 PK samples, plasma was separated from red blood cells by centrifugation at 3200 r.p.m for 10 minutes at 4°C within 3 hours of collection, transferred to cryo vials, and stored at −80°C until analysis. The last two PK samples were processed within 14 hours of collection and stored as described above. The PK samples were collected over a period of eight years and analyzed nine years after collecting samples from the first subject.
A medication dosing history was documented for 48 hours prior to the PK assessment. With the exception of one subject who received metformin 500 mg bid, all children received metformin 1000 mg bid. The immediate-release formulation used in this study was manufactured and packaged in 250 mg capsules by the NIH Clinical Center Pharmaceutical Development Section.
Analytical Determination of Metformin Concentrations
Metformin and buformin internal standard were separated and quantified using a newly developed ultra-performance liquid chromatography (UPLC) method. The UPLC system consisted of a Waters Acquity Ultra Performance Liquid Chromatography liquid handling system with an integrated column heater/cooler, and an Acquity photodiode array detector set at λ=276 nm (Waters Corp., Milford, MA, USA). The UPLC system and the assay parameters were controlled using the Empower (Version 5.0) chromatography manager software. The analytical column used was an Acquity BEH Shield RP18, 1.7 μm, 2.1 × 50 mm reverse-phase analytical column, and was preceded by an Acquity BEH Shield RP18, 1.7 μm, 2.1 × 5 mm guard column (Waters Corp., Milford, MA, USA). The samples were extracted and eluted isocratically at 0.50 mL/min. with a controlled column temperature at T=25°C for 2.5 minutes and a 0.6 min injection delay using a mobile phase consisting of (34:66, v/v) acetonitrile and 2mM Sodium Dodecyl Sulfate (SDS) buffer.
Metformin and buformin internal standard were isolated from human plasma by a newly developed off-line solid-phase extraction (SPE) method using Oasis WCX 1cc/30mg cartridges (Waters). Briefly, in a 13×100 mm test tube mix 300 μL plasma sample with 20 μL of buformin internal standard (10.0 μg/mL) solution followed by two 300 μL aliquots of 50mM ammonium acetate (pH 7) buffer. Using a RapidTrace solid-phase extraction module (Biotage LLC) the SPE cartridges were conditioned with 1.0 mL methanol followed by 1.0 ml water Milli-Q, 1.0 mL of 50 mM ammonium acetate (pH 7) buffer, and the samples were subsequently loaded onto the cartridges. The SPE cartridges were next washed with 1.0 mL of 50 mM ammonium acetate (pH 7) buffer followed by 1.0 mL [40:60] methanol/water, and the analytes were subsequently eluted with 1.0 mL of 2% formic acid in methanol. The eluate was evaporated to dryness using a TurboVap (Biotage LLC) for 30 minutes at 40°C under a stream of nitrogen. The samples were immediately reconstituted with 200 μL of 50% acetonitrile, vortexed for 35 seconds, and transferred to an injection vial. An aliquot of 1.0 μL was injected into the Acquity UPLC system, and eluted isocratically at 0.50 mL/min over 2.5 minutes with a 0.6 mL injection delay.
Calibration curves for metformin were linear from 0.010 μg/mL – 10.0 μg/mL with R2 > 0.998. Percent errors, as a measure of accuracy, were <15% and the inter- and intra-assay coefficients of variation for MTF were 5.20 – 7.39% and 4.35 – 9.63%, respectively, at three different drug concentrations. The limit of quantitation for metformin was 0.010 μg/mL and the limit of detection was 0.005 μg/mL. During the validation, stability of the drug in plasma was performed at 25°C, 4°C, and −80°C. Repeated freezing and thawing (three cycles) of plasma was evaluated at −80°C. The overall recovery of metformin and buformin was >90%.
Population PK Analysis of Metformin
Plasma metformin concentration-time profiles were analysed by nonlinear mixed-effects modelling using NONMEM® software (version 7.2, ICON Development Solutions, Ellicott City, MD, USA). Double precision, first-order conditional estimation with interaction and subroutines ADVAN5 TRANS1 were used.
Plasma concentrations of metformin were natural log-transformed before the analysis. Visual inspection of the concentration-time plots demonstrated a monexponential decay with double peaks in most of the subjects, consistent with a one-compartment PK model with two absorption sites. The model selection was based on goodness-of-fit criteria, including diagnostic plots, minimum objective function value (MOFV) after accounting for the number of fitted parameters, Akaike information criterion (AIC; equal to the MOFV plus twice the number of parameters), precision, and the physiological plausibility of the estimates. The inter-subject variability in the PK parameters was modelled assuming an exponential model. Residual variability was modelled using an additive model.
The one-compartment model (Figure 1) used in the present study was parameterized in terms of CL/F and V/F. The between-subject variability (BSV) of the model parameters was described using a lognormal variance model. An allometric exponential covariate model was used to account for effect of body weight on both CL/F and V/F, with a weight exponent of 0.75 and 1 for CL/F and V/F, respectively.24, 25
Figure 1. Diagram of the one-compartment open model with two parallel sites of absorption used in the pharmacokinetic analysis of metformin.
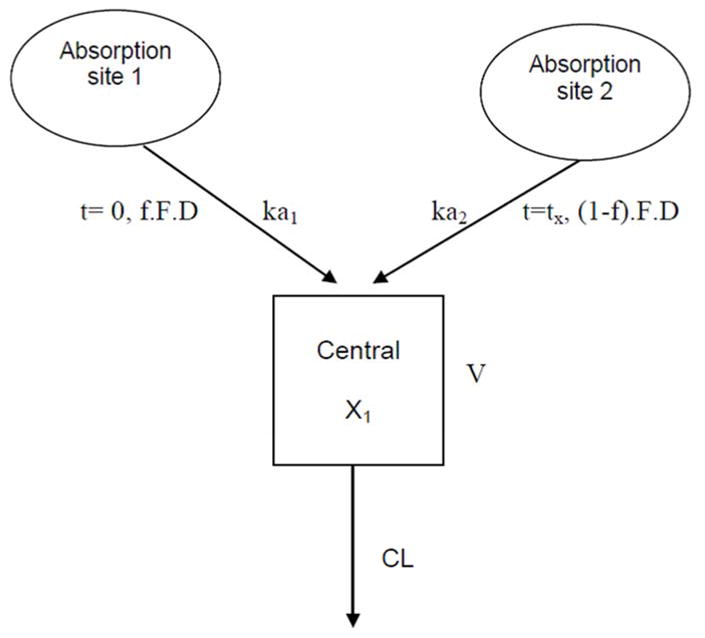
X1, amount of metformin in the central compartment; D, dose; F, drug bioavailability; f, fraction of metformin absorbed at the first absorption site; ka1, absorption rate constant at the first absorption site; ka2, absorption rate constant at the second absorption site; tx, time for the second fraction of the drug to begin to be absorbed; V, volume of distribution of the central compartment; CL, clearance of metformin from the central compartment
| (1) |
| (2) |
where; ηCL/Fi is the difference between individual (CL/Fi) and typical value or population mean (TVCL/F) clearance on log scale, ηV/Fi is the difference between individual (V/Fi) and typical value or population mean (TVV/F) volume of distribution on log scale, 0.75 and 1 are the allometric exponents for the effect of individual body weight (WTi) on CL/F and V/F, respectively. The parameters ηCL/Fi and ηV/Fi were both assumed to follow a normal distribution independent of each other, with mean of zero and variances of ω2CL/F and ω2V/F respectively.
To identify covariates that significantly affect the PK of metformin, covariates were examined graphically via plots of eta versus covariate values. The candidate covariates that were identified during graphical screening were then tested in NONMEM 7.2, one at a time in a univariate analysis. The effect of continuous covariates (such as age, body weight, height, body mass index, cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, triglycerides, serum creatinine, and estimated glomerular filtration rate [eGFR] as calculated by the revised Schwartz equation26) on the PK parameters was modelled according to a power model scaled to the population median covariate value. The effect of categorical covariates including sex, race, ethnicity, and genotype group on the PK parameters were modelled using a proportional relationship.
The statistical significance of the covariates was assessed using a likelihood ratio test corresponding to a decrease in MOFV of < 3.84 (with one degree of freedom) in comparisons between two hierarchical models. These covariates were then included in the basic population model to form the full model. A stepwise backward deletion of covariates from the full model was performed to determine the covariates to be retained in the final model at a significance level of p < 0.001. This corresponds to an increase in MOFV >10.8 (one degree of freedom) when comparing between the two hierarchical models. This was repeated and continued until all remaining covariates were significant in the final model.
The predictability of the final model was evaluated using a visual predictive check. One thousand datasets each for metformin were simulated from the final model. Within the 2.5th and 97.5th percentiles of the simulated concentration, a 95% prediction interval was constructed and plotted along with the observed data and the median of the simulated concentration. A bootstrap procedure was performed to determine the stability and robustness of the final model. One thousand bootstrap re-samples of the original dataset were created, and these were evaluated using the final model.
DNA Purification and Genotyping
Genotyping was performed by direct sequencing of nested PCR products. Genomic DNA was extracted from the plasma of the PK samples using Epigentek Fitamp Plasma/Serum DNA isolation kit (Epigentek, NY, USA) according to the manufacturer’s protocol. Target gene areas were amplified by nested PCR, using 2 sets of primers. Oligonucleotide primers were designed to span all 11 exons of the SLC22A1 gene (GenBank accession number NM_003057) with approximately 20 bp of flanking intronic sequence of each adjacent intron. Most of the outer primer designs are described in Kang et al.27 The inner primers and outer primers for exons 1, 2, and 9 were designed using open source program Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi).
We slightly modified the PCR method reported by Kang et al. The reaction volume was 50 μL containing 1X PCR buffer, 300 ng genomic DNA, 0.2 mM dNTPs (Invitrogen), 0.2 μM concentration of each primer, and 1 U of Taq polymerase (Invitrogen). PCR was performed using GeneAmp PCR 9700 (Applied Biosystems, Foster City, CA). The cycling conditions were as follows: initial denaturation at 94°C for 5 or 8 minutes, denaturation at 95°C for 1 min, annealing at 55 to 59 °C for 1 min, and extension at 72°C for 1 min for 45 cycles. The final termination of elongation step was 5 min at 72°C. The products of the primary PCR were then amplified again with the same method. All amplicons were visualized by agarose gel electrophoresis and ethidium bromide staining. The amplification products were purified and had both strands directly sequenced by Macrogen INC (Rockville, MD, USA) using an ABI3730 XL DNA analyzer. Sequence results were aligned and assembled into contigs using Sequencher® version 5.0 (Gene Codes Corporation, Ann Arbor, MI USA). The primers and conditions for genotyping are summarized in Table 1.
Table 1.
Summary of polymorphisms in the SLC22A1 gene detected in obese children
| NCBI SNP ID | Exon | Positiona | Allele | Nucleotide Change (5′ to 3′) | Amino Acid substitution | N | ||
|---|---|---|---|---|---|---|---|---|
| wt/wt | wt/v | v/v | ||||||
| rs1867351 | 1 | 156 | T>C | agagT/Ccctg | Ser52Ser | 18 | 6 | 4 |
| rs12208357 | 1 | 181 | C>T | ccagC/Tgctg | Arg61Cys | 27 | 1 | |
| Novel | 1 | 283 | T>C | ggacT/Cggaa | Trp95Cys | 27 | 1 | |
| Novel | 1 | 343 | C>T | gagcC/Tacct | His115Tyr | 27 | 1 | |
| rs683369 | 2 | 480 | C>G | tcttC/Gtttg | Leu160Phe | 22 | 5 | 1 |
| rs3413415 | 3 | 558 | C>T | tcaaC/Tgcgg | Asn186Asn | 27 | 1 | |
| rs2282143 | 6 | 1022 | C>T | acgcC/Tgcgc | Pro341Leu | 24 | 1 | |
| rs34205214 | 6 | 1025 | G>A | ccgcG/Acctg | Arg342His | 23 | 1 | |
| rs34130495 | 7 | 1201 | G>A | cgtgG>Agccg | Gly401Ser | 27 | 1 | |
| rs628031 | 7 | 1222 | A>G | ggccA/Gtgtc | Met408Val | 5 | 10 | 13 |
| rs34888879 | 7 | 1239 | G>A | tggcG/Agggg | Ala413Ala | 27 | 1 | |
| rs72552763 | 7 | 1260 | GAT>del | tcatGAT/-tttt | Met420del | 20 | 7 | 1 |
| rs35270274 | 9 | 1463 | G>T | ctgaG/Tggag | Arg488Met | 23 | 1 | |
| rs41267797 | 10 | 1503 | G>A | cggtG/Attgg | Val501Val | 24 | 4 | |
| rs78899680 | 10 | 1555 | G>T | ggggG/Ttcgc | Val519Phe | 27 | 1 | |
A of the translation initiation codon ATG is numbered 1
wt, wild type; v, variant
Statistical Analyses
To evaluate whether the variations in SLC22A1 gene would influence the response to metformin, subjects were classified into two groups based on their SLC22A1 genotypes: variant and reference groups. Subjects who had any of the six polymorphisms (His115Tyr, Arg61Cys, Trp95Cys, Gly401Ser, Met420del, or Val 519 Phe) were classified into the variant group (n=10), whereas those in the reference group had wild type allele at these positions in the SLC22A1 gene (n = 18). The variants Arg61Cys, Gly401Ser, Met420del were chosen because they were known to have reduced uptake and activity of metformin in cellular assays.28 The other 3 variants were selected because they were determined to be “possibly damaging” using the PolyPhen-2 analyses.
The differences in PD variables between males and females and between the reference and variant genotype groups were analyzed using Mann Whitney U test using Statistica v.12 (StatSoft, Inc. Tulsa, OK). The variables explored included total and trunk fat determined by dual-energy X-ray absorptiometry (DEXA), abdominal adipose tissue subcompartments measured by magnetic resonance imaging (MRI), fasting glucose and insulin, HA1c, and indexes for insulin sensitivity and resistance.12 The level of statistical significance was set at P<0.05. Because all analyses were exploratory, no adjustments were made for multiple comparisons.
Results
Study Population
Serial blood PK samples at steady state were available for 30 obese pre-pubertal or early pubertal children whose BMI was >95th percentile and who had fasting hyperinsulinemia and were randomized to take metformin 1000mg twice daily in a double-blind, placebo-controlled, randomized trial. At baseline and after 6-months of treatment, metabolic measures and dual-energy X-ray absorptiometry (4500A, Hologic Inc., Bedford MA, software version 11.2) to assess regional adiposity were obtained as previously described.12 The median (range) of fasting inulin at study enrollment was 23.2 (15.0 – 71.3) μU/mL. Seventeen of the subjects were Caucasians, 12 were African Americans, and one was Asian; none were Native Americans. The median age at the time of PK collection was 11.2 (7.7 – 13.5) years, weight was 77 (50.5 – 118) kg, BMI was 33.5 (24.2 – 43.6), and baseline eGFR was 116 (93.0 – 180) mL/min/1.73 m2.
SCL22A1 Genotype
SLC22A1 genotype was determined for 28 of the 30 subjects with PK samples. DNA sample was not available for one subject, and there was difficulty in genotyping the sample for another subject. Fifteen SNPs were detected in 10 subjects: 11 were nonsynonymous SNPs and 4 were synonymous SNPs. Two of the nonsynonymous SNPs, c.283 T>C (95 Trp>Cys) and c.343 C>T (115His>Tyr) in exon 1, were newly discovered (Table 1) and were predicted to be possibly damaging mutations by Polymorphism Phenotyping v.2 (PolyPhen-2) software.29 Five subjects were heterozygous for Met420del, and 1 subject was heterozygous for Val519P. One child was a His115Tyr homozygote. One subject was heterozygous for the Arg61Cys and Met420del alleles, and another was heterozygous for Gly410Ser and Met420del. The last subject was homozygous for Met420del and heterozygous for Trp95Cys.
Of note, the Pro341Leu polymorphism was not determined in three subjects and the Arg342His and Arg488Met polymorphisms were not determined in four subjects because of insufficient DNA samples. Since these SNPs were not used in genotype grouping as described below, the lack of information for these SNPs in the four subjects did not affect our analysis results.
Steady State Population PK of Metformin
Among the 30 subjects with PK samples, metformin concentration data from two subjects were excluded from the analysis because of missing or incomplete data required for analysis. As a result, a total of 328 concentration data from 28 subjects were used in the dataset. The metformin PK was studied at steady state; concentrations at time zero were similar to the concentrations at 12 hours post dose. The median (range) concentration at time zero was 0.34 (0.03 – 1.74) mg/L and the median concentration at 12 hours post dose was 0.32 (0.14 – 1.77) mg/L. The PK characteristics of metformin in pediatric subjects were best described using a one-compartment model with two first-order absorption rate constants from two different absorption sites in the gastrointestinal tract (Figure 1). This structural model was used because double absorption peaks after oral absorption were identified in 20 out of 28 individual PK profiles. Using this model, a univariate analysis of clinical characteristics and laboratory values was performed for all individuals. The univariate analysis identified several possible covariates affecting the PK of metformin, as summarized in Table 2. However, the results of a multivariate analysis indicated that the values of apparent clearance of metformin (CL/F) were related to the eGFR of the subject (p< 0.001). The median post-hoc estimate of individual CL/F was 68.4 (40.4 – 149) L/hr.
Table 2.
Significant covariates for the univariate and multivariate analyses
| Significant covariates | ΔMOFV | p-value |
|---|---|---|
| Univariate analysis | ||
| Effect of GLU on V/F | −4.674 | <0.05 |
| Effect of eGFR on CL/F | −6.658 | <0.01 |
| Effect of LAMF on ka1 | −6.587 | <0.01 |
| Multivariate analysis | ||
| Effect of eGFR on CL/F | 52.145 | <0.001 |
MFOV, minimum objective function; GLU, glucose; V/F, apparent volume of distribution; eGFR, estimated glomerular filtration rate; CL/F, apparent total clearance of metformin; LAMF, left arm fat; ka1, absorption rate constant at the first absorption site
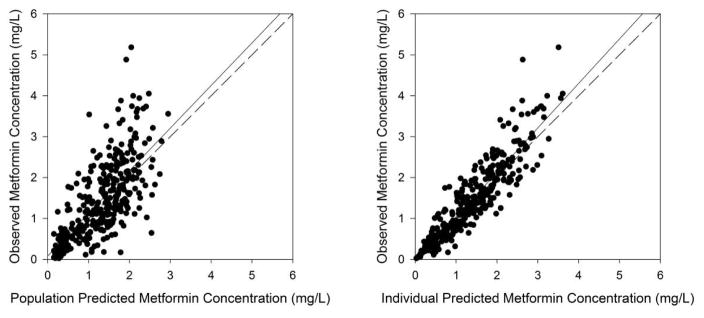
Figure 2 shows the basic goodness-of-fit plots from the final model. It shows clustering of both population and individual predicted values around the line of unity in the goodness-of-fit plots. The plots of conditional weighted residuals (CWRES) versus population predictions and time after dose does not show any trends and most of the CWRES were randomly distributed within ± 2 units. The parameters of the final model, including bootstrap medians and 95% confidence intervals for the PK parameters, are presented in Table 3. Figure 3 presents the results of the visual predictive check for metformin.
Figure 2. The goodness-of-fit plots of the final model.
The population-predicted (left) and individual-predicted (right) concentrations for metformin vs observed concentrations. The solid lines represent the regression lines; the dotted lines represent the lines of identity (slope=1, intercept=0).
Table 3.
Parameter estimates for the population pharmacokinetics model of metformin and results of the bootstrap validation of the final model
| Parameter (units) | Estimate (%RSE) | CV% | Bootstrap median (95% CI)* |
|---|---|---|---|
| Fixed effects | |||
| V/F (L) | 28.8 (17.5) | 63.7 | 29.2 (1.00, 61.9) |
| CL/F (L/hr) | 68.1 (5.48) | 28.4 | 68.3 (59.6, 79.3) |
| ka1 (hr−1) | 0.335 (17.1) | 32.1 | 0.292 (0.124, 0.689) |
| ka2 (hr−1) | 0.139 (13.6) | 56.9 | 0.141 (0.00139, 47.8) |
| tx (hr) | 2.28 (3.48) | 2.20 (0.0318, 2.85) | |
| f | 0.446 (13.4) | 0.468 (0.280, 0.985) | |
| Effect of eGFR on CL/F | 1.10 (34.5) | 1.08 (0.008, 1.84) | |
| Random effects | |||
| Inter-subject variability (ω2) | |||
| V/F | 0.406 (37.7) | 0.498 (0.120, 12.8) | |
| CL/F | 0.0804 (38.3) | 0.0811 (0.0306, 0.218) | |
| V/F~CL/F | 0.129 (48.7) | 0.158 (0.0269, 1.11) | |
| ka1 | 0.103 (38.3) | 0.0804 (1×10−6, 0.227) | |
| ka2 | 0.324 (39.2) | 0.327 (1×10−6, 224) | |
| Residual error | |||
| Std. deviation (mg/L) | 1.58 (16.9) | 1.14 (1.09, 1.24) | |
Results of 1000 bootstrap re-samples of the final model.
RSE, relative standard error; CV, coefficient of variation; CI, confidence interval
V/F, apparent volume of distribution; CL/F, apparent total clearance of metformin; ka1, absorption rate constant at the first absorption site; ka2, absorption rate constant at the second absorption site; tx, time for the second fraction of the drug to begin to be absorbed; f, fraction of metformin absorbed at the first absorption site; eGFR, estimated glomerular filtration rate
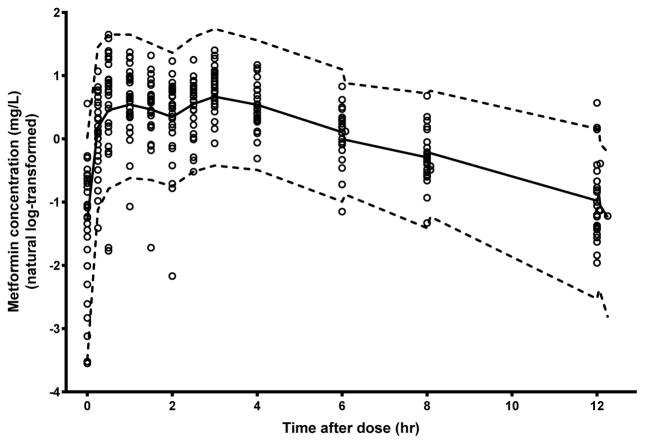
Figure 3. Visual predictive check of metformin observed data compared with the 97.5th, 50th (median) and 2.5th percentiles for 1000 simulated data sets.
Comparison of median (solid line) and 2.5–97.5th percentile interval (dotted lines). Observed data are plotted using open circles.
Effects of Genetic Polymorphisms on Metformin PD
The two genotype groups responded similarly to metformin in variables related to glucose homeostasis (Table 4). There were no significant differences between the two genotype groups in the changes from baseline to six months of metformin therapy for hemoglobin A1c (HbA1c), homeostasis model assessment for insulin resistance (HOMA IR), fasting insulin, or glucose concentrations. However, the percentage (%) of trunk fat reduction was significantly less in the variant group than in the reference group (p=0.035, Table 4). The median % change in trunk fat by DEXA was −2.20 % (−9.00 % – 0.900 %) and −1.20 % (−2.40 % – 7.30 %) for the reference group and the variant group, respectively. The reduction in trunk fat mass (p=0.053) and total % fat (p=0.053) also tended to be higher in the reference group (Table 4). There were no differences in abdominal adipose tissue compartments examined by MRI between the two genotype groups (Table 4).
Table 4.
Changes from baseline in body fat composition, glucose metabolism, and index of insulin resistance after 6 months of metformin therapy between the SLC22A1 variant and reference groups
| Median (Range) | |||
|---|---|---|---|
|
| |||
| Parameter | Reference Group (n = 18) | SLC22A1 Variant Group (n = 10) | P value |
|
| |||
| Total Fat by DEXA (g) | −429 (−9502, 2246) | 580 (−2649, 7824) | NS |
|
| |||
| Trunk Fat by DEXA (g) | −523 (−4952, −1550) | 532.500 (−844, 3351) | 0.053 |
|
| |||
| Total Fat by DEXA (%) | −1.95 (−7.00, 0.500) | −0.700 (−2.00, 8.80) | 0.053 |
|
| |||
| Trunk Fat by DEXA (%) | −2.20 (−9.00, 0.900) | −1.20 (−2.40, 7.30) | 0.035 |
|
| |||
| Abdominal adipose tissue by MRI (cc) | |||
| L2-3 subcutaneous | 8.70 (−146 – 93.8) | −12.6 (−169 – 140) | NS |
| L4-5 subcutaneous | −12.1 (−247 – 58.9) | −14.7 (−1334 – 92.5) | NS |
| L2-3 intra-abdominal | −8.60 (63.3 – 166) | −14.6 (−26.0 – 20.3) | NS |
| L4-5 intra-abdominal | −2.95 (−73.2 – 248) | −6.57 (−35.2 – 23.0) | NS |
|
| |||
| Fasting Insulin (mIU/mL) | −2.40 (−17.8, 21.5) | 0.000 (−14.0, 15.1) | NS |
|
| |||
| Fasting Glucose (mg/dL) | −5.50 (−12.0, 5.00) | −2.50 (−8.00, 5.00) | NS |
|
| |||
| HbA1c (%) | −0.200 (−0.900, 0.300) | 0.000 (−0.400, 0.300) | NS |
|
| |||
| HOMA IR index | −0.687 (−3.98, 5.36) | −0.118 (−3.18, 3.40) | NS |
|
| |||
| QUICKI | 0.010 (−0.029, 0.072) | 0.001 (−0.030, 0.040) | NS |
DEXA, dual-energy X-ray absorptiometry; MRI, magnetic resonance imaging; HOMA IR, homeostasis model assessment for insulin resistance; HbA1c, glycated hemoglobin; QUICKI, quantitative insulin sensitivity check index; NS, not significant
Discussion
This report is the first describing the population PK of metformin in severely obese but nondiabetic children with insulin resistance. Initial analysis of the concentration-time profiles revealed the presence of double peaks, which has been reported with several orally administered drugs and is associated with such factors as gastrointestinal transit time, absorption window effect, irregular gastric emptying, and enterohepatic cycling.30, 31 We used a one-compartment model with two absorption sites, one of which has a delayed onset, to describe this double peak phenomenon. The model fit reasonably well to the data, and a mean onset of 2.28 hours at the delayed absorption site was estimated. This time delay was consistent with the time of food intake on the PK study day, when the subjects had lunch approximately 2 hours after the initiation of a hyperglycemic clamp. We suspect that the second peak may be related to a combination of hyperglycemia and food effect; the former has been shown to slow gastric emptying and modulate gastric motility in healthy subjects.32, 33 Nonetheless, metformin is a Biopharmaceutics Classification System (BCS) Class III drug, for which the double-peak phenomenon is not uncommon, and the attribution of administering multiple capsules to the double-peak phenomenon cannot be ruled out. Previously, metformin double peaks have been observed at 1 and 3 hours upon administration of an immediate-release tablet of metformin in Chinese healthy male volunteers.34
While the prevalence of type 2 diabetes and the use of metformin in children have been increasing, there is limited information regarding metformin PK in the pediatric population. Thus far, metformin PK information in children and adolescents with type 2 diabetes was summarized in two abstracts,35,36 and also in a small study reporting metformin PK in four non-obese 9-year-old girls with a history of low birth weight.37 Our results showed similar CL/F but different apparent volume of distribution (V/F) values from those of the previous pediatric and adult populations.19,38 The direct comparison of the V/F values between our study and others is not appropriate, however, because we used a model with two absorption sites to describe metformin PK, which could result in V/F values that are different from those obtained from non-compartmental analysis or one-compartment models. In addition, the immediate-release metformin formation that we used was manufactured and packaged in capsules by the NIH Clinical Center Pharmacy Department. Although it had similar dissolution properties compared to the commercial product (data not shown), the bioavailability and PK of the two drug products might differ.
Based upon previous literature findings,39 we added body weight as a covariate into the structural model parameters CL/F and V/F using an allometric approach. Subsequently, using this base structural PK model which includes the body weight covariate, we performed covariate screening and showed that eGFR was a confounding factor for CL/F but SLC22A1 variant genotype was not a significant covariate for metformin PK. These results confirmed renal function as a major factor affecting metformin PK,20, 40–42 since metformin is eliminated primarily via urinary excretion.19,38 However, the lack of SLC22A1 effect is controversial. In a previous study with healthy Caucasians, there was a lack of association between SLC22A1 polymorphisms and metformin CL/F, but metformin CLR rather than CL/F increased significantly with SLC22A1 haplotypes after adjustment for creatinine clearance and age.20 Contrarily, the SLC22A1 reference genotype group had a significantly higher metformin CL/F, but metformin CLR was similar between the two SLC22A1 genotype groups in a study of healthy Caucasians.43 Compared with the healthy volunteer studies,20, 43 the lack of genotype effect in our study may be attributed to the differences in subject populations and the relatively small number of study participants. Because we did not collect urine samples in the current study, the association of SLC22A1 variants with metformin CLR in obese children could not be established.
In our obese subjects, SLC22A1 variant genotype appeared to be associated with trends toward lower reductions in the % total fat, the % trunk fat, and trunk fat mass after six months of metformin therapy. Of note, the % reduction in trunk fat at Month 6 from baseline was small; the median reduction was −2.20 % and −1.20 % for the SCL22A1 genotype reference group and the variant group, respectively. Presumably, the decreased metformin effects were caused by reduced uptake of metformin into the adipose tissue of SCL22A1 variant subjects, leading to attenuated inhibition of intracellular lipid accumulation (via the activation of AMP-activated protein kinase),44 inhibition of adipocyte differentiation,45 and reductions in adipogenic and proinflammatory gene expression.15 Interestingly, SLC22A1 expression correlated significantly with BMI and the percent of fat mass in a previous study,15 suggesting that the genotype effect may be more prominent in obese individuals and may not be observed in healthy subjects. Because the percentage of trunk fat is a good surrogate for visceral adipose tissue, which was associated with obesity-related morbidity and insulin resistance in adults and in children,46 SLC22A1 polymorphisms may have a role in amelioration of obesity-related comorbidities induced by metformin.
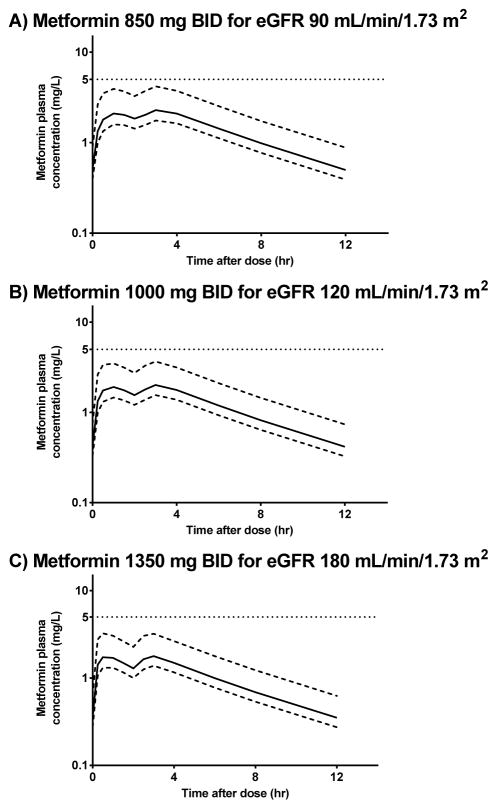
Lactic acidosis is a rare but serious adverse event of metformin that can occur due to metformin accumulation;47 plasma concentrations > 5 mg/L are generally found when metformin is implicated as the cause of lactic acidosis as described by the product labeling.48 Using the results of the final population PK model and a range of body weights generated from a normal distribution of subjects with a mean weight of 78.7 ± 18.2 kg, we performed simulations to determine the maximum metformin doses that would not exceed the maximum plasma concentration of 5 mg/L for individual with varying renal function. The simulation results showed that metformin 850 mg twice a day (bid), 1000 mg bid, and 1350 mg bid can be given to pediatric patients with eGFR of 90, 120, and 180 ml/min/1.73 m2 without exceeding a plasma metformin concentration of 5 mg/L at steady state (Figure 4). These results suggest that the current dosage used in the study (i.e., 1000 mg bid) was appropriate for obese children with normal renal function, and a slightly reduced dosage regimen of 850 bid can be given to pediatric subjects with an eGFR of 90 ml/min/1.73m2. Of note, we performed a simulation with an eGFR value of 180 ml/min/1.73m2 because one of our subjects had such an eGFR value. Because our population PK model was developed using data from obese children without renal dysfunction, dosing in pediatric subjects with renal dysfunction remains to be determined.
Figure 4. Simulations of the metformin plasma concentrations at A) 850 mg bid; B) 1000 mg bid; C) 1350 mg bid for pediatric patients with estimated glomerular filtration rates of 90, 120 and 180 mL/min/1.73 m2, respectively.
The 2.5th (lower dotted line), 50th (solid dotted line) and 97.5th (upper dotted line) percentiles of the simulated concentrations are shown.
There are several limitations in the evaluation of metformin PK and genotype in this study, and our results need to be interpreted with caution. First, the main study was conducted over a period of eight years, and the PK samples were batched analyzed nine years after collecting samples from the first subject. Some of the earliest samples were under storage at −80°C for a long period of time. While the long-term stability of these samples is not known, our concentration data had a range similar to that of the metformin concentrations published in the literature.19 In addition, our sample size was small and multiple comparisons were performed, making the analysis entirely exploratory. The study participants represented only a small subset of children who had age between 7 and 13 years, were obese, and had normal renal function and consisted primarily of Non-Hispanic White and Black subjects. Therefore, our results may not be generalizable to other pediatric subjects of lower ages, of different race/ethnicity, of normal body weight, or who have renal dysfunction. Future studies should be performed to examine metformin PK in these other populations. Finally, the classification of the functional effect of the Trp95Cys, His115Tyr, and Val519Phe using PolyPhen-2 may be misleading. We reassessed the genotype effect with these 3 SNPs classified as non-damaging (i.e., wild type). The analysis results remained similar with lower % trunk fat reduction in the variant group compared to the reference group (p=0.048). Nevertheless, because of our study limitations, it is important to investigate the effects of SLC22A1 genotype on metformin PD in larger samples.
Conclusion
Renal function was a significant covariate of metformin PK in children, a finding that is consistent with data obtained in adults. SLC22A1 polymorphisms were not associated with changes in metformin PK. However, SLC22A1 variants appeared to be associated with lower reductions in % trunk fat after six months of metformin therapy. Future study is warranted to further investigate the role of SLC22A1 polymorphisms in the treatment of obese children with metformin.
Acknowledgments
This work was supported by the Intramural Research Program of National Institutes of Health, Clinical Center Pharmacy Department, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Program in Developmental Endocrinology and Genetics grant 1ZIAHD000641 from NICHD with supplemental funding from the National Institute for Minority Health and Health Disparities (NIMHD), NIH (Dr. Yanovski). There was no commercial sponsorship. JAY is a Commissioned Officer in the U.S. Public Health Service, Department of Health and Human Services. The funding organizations played no role in the design or conduct of the study; the collection, management, analysis, or interpretation of data; or the preparation of the manuscript. The authors thank Dr. Jonathan Krakoff, the participants and their parents for their involvement in this trial.
Footnotes
All authors declared no conflicts of interest.
References
- 1.Lobstein T. Special focus II. The size and risks of the international epidemic of child obesity. Paris: Organization for Economic Cooperation and Development; 2010. [Google Scholar]
- 2.Wang Y, Lobstein T. Worldwide trends in childhood overweight and obesity. Int J Pediatr Obes. 2006;1(1):11–25. doi: 10.1080/17477160600586747. [DOI] [PubMed] [Google Scholar]
- 3.Lobstein T, Baur L, Uauy R. Obesity in children and young people: a crisis in public health. Obes Rev. 2004;5(Suppl 1):4–104. doi: 10.1111/j.1467-789X.2004.00133.x. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan SR, Myers L, Berenson GS. Predictability of childhood adiposity and insulin for developing insulin resistance syndrome (syndrome X) in young adulthood: the Bogalusa Heart Study. Diabetes. 2002;51(1):204–209. doi: 10.2337/diabetes.51.1.204. [DOI] [PubMed] [Google Scholar]
- 5.Sinha R, Fisch G, Teague B, et al. Prevalence of impaired glucose tolerance among children and adolescents with marked obesity. N Engl J Med. 2002;346(11):802–810. doi: 10.1056/NEJMoa012578. [DOI] [PubMed] [Google Scholar]
- 6.Gunnell DJ, Frankel SJ, Nanchahal K, Peters TJ, Davey Smith G. Childhood obesity and adult cardiovascular mortality: a 57-y follow-up study based on the Boyd Orr cohort. Am J Clin Nutr. 1998;67(6):1111–1118. doi: 10.1093/ajcn/67.6.1111. [DOI] [PubMed] [Google Scholar]
- 7.Goodman E, Dolan LM, Morrison JA, Daniels SR. Factor analysis of clustered cardiovascular risks in adolescence: Obesity is the predominant correlate of risk among youth. Circulation. 2005;111(15):1970–1977. doi: 10.1161/01.CIR.0000161957.34198.2B. [DOI] [PubMed] [Google Scholar]
- 8.Freemark M, Bursey D. The effects of metformin on body mass index and glucose tolerance in obese adolescents with fasting hyperinsulinemia and a family history of type 2 diabetes. Pediatrics. 2001;107(4):E55. doi: 10.1542/peds.107.4.e55. [DOI] [PubMed] [Google Scholar]
- 9.Srinivasan S, Ambler GR, Baur LA, et al. Randomized, controlled trial of metformin for obesity and insulin resistance in children and adolescents: improvement in body composition and fasting insulin. J Clin Endocrinol Metab. 2006;91(6):2074–2080. doi: 10.1210/jc.2006-0241. [DOI] [PubMed] [Google Scholar]
- 10.Atabek ME, Pirgon O. Use of metformin in obese adolescents with hyperinsulinemia: A 6-month, randomized, double-blind, placebo-controlled clinical trial. J Pediatr Endocr Met. 2008;21(4):339–348. doi: 10.1515/jpem.2008.21.4.339. [DOI] [PubMed] [Google Scholar]
- 11.Clarson CL, Mahmud FH, Baker JE, et al. Metformin in combination with structured lifestyle intervention improved body mass index in obese adolescents, but did not improve insulin resistance. Endocrine. 2009;36(1):141–146. doi: 10.1007/s12020-009-9196-9. [DOI] [PubMed] [Google Scholar]
- 12.Yanovski JA, Krakoff J, Salaita CG, et al. Effects of metformin on body weight and body composition in obese insulin-resistant children: a randomized clinical trial. Diabetes. 2011;60(2):477–485. doi: 10.2337/db10-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong L, Goswami S, Giacomini KM, Altman RB, Klein TE. Metformin pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet Genomics. 2012;22(11):820–827. doi: 10.1097/FPC.0b013e3283559b22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu Y, Sheardown SA, Brown C, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. The Journal of clinical investigation. 2007;117(5):1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno-Navarrete JM, Ortega FJ, Rodriguez-Hermosa JI, et al. OCT1 Expression in adipocytes could contribute to increased metformin action in obese subjects. Diabetes. 2011;60(1):168–176. doi: 10.2337/db10-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermann LS, Schersten B, Bitzen PO, Kjellstrom T, Lindgarde F, Melander A. Therapeutic comparison of metformin and sulfonylurea, alone and in various combinations. A double-blind controlled study. Diabetes care. 1994;17(10):1100–1109. doi: 10.2337/diacare.17.10.1100. [DOI] [PubMed] [Google Scholar]
- 17.Yin OQ, Tomlinson B, Chow MS. Variability in renal clearance of substrates for renal transporters in chinese subjects. Journal of clinical pharmacology. 2006;46(2):157–163. doi: 10.1177/0091270005283838. [DOI] [PubMed] [Google Scholar]
- 18.Christensen MM, Brasch-Andersen C, Green H, et al. The pharmacogenetics of metformin and its impact on plasma metformin steady-state levels and glycosylated hemoglobin A1c. Pharmacogenet Genomics. 2011;21(12):837–850. doi: 10.1097/FPC.0b013e32834c0010. [DOI] [PubMed] [Google Scholar]
- 19.Graham GG, Punt J, Arora M, et al. Clinical pharmacokinetics of metformin. Clinical pharmacokinetics. 2011;50(2):81–98. doi: 10.2165/11534750-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 20.Tzvetkov MV, Vormfelde SV, Balen D, et al. The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clinical pharmacology and therapeutics. 2009;86(3):299–306. doi: 10.1038/clpt.2009.92. [DOI] [PubMed] [Google Scholar]
- 21.Chae JW, Baek IH, Lee BY, Cho SK, Kwon KI. Population PK/PD analysis of metformin using the signal transduction model. Br J Clin Pharmacol. 2012;74(5):815–823. doi: 10.1111/j.1365-2125.2012.04260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou K, Bellenguez C, Spencer CC, et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nature genetics. 2011;43(2):117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levri KM, Slaymaker E, Last A, et al. Metformin as treatment for overweight and obese adults: A systematic review. Ann Fam Med. 2005;3(5):457–461. doi: 10.1370/afm.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson BJ, Holford NH. Mechanism-based concepts of size and maturity in pharmacokinetics. Annu Rev Pharmacol Toxicol. 2008;48:303–332. doi: 10.1146/annurev.pharmtox.48.113006.094708. [DOI] [PubMed] [Google Scholar]
- 25.Holford N, Heo YA, Anderson B. A pharmacokinetic standard for babies and adults. J Pharm Sci. 2013;102(9):2941–2952. doi: 10.1002/jps.23574. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz GJ, Munoz A, Schneider MF, et al. New equations to estimate GFR in children with CKD. J Am Soc Nephrol. 2009;20(3):629–637. doi: 10.1681/ASN.2008030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang HJ, Song IS, Shin HJ, et al. Identification and functional characterization of genetic variants of human organic cation transporters in a Korean population. Drug metabolism and disposition: the biological fate of chemicals. 2007;35(4):667–675. doi: 10.1124/dmd.106.013581. [DOI] [PubMed] [Google Scholar]
- 28.Zolk O. Disposition of metformin: variability due to polymorphisms of organic cation transporters. Annals of medicine. 2012;44(2):119–129. doi: 10.3109/07853890.2010.549144. [DOI] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirfazaelian A, Mahmoudian M. A simple pharmacokinetics subroutine for modeling double peak phenomenon. Biopharm Drug Dispos. 2006;27(3):119–124. doi: 10.1002/bdd.492. [DOI] [PubMed] [Google Scholar]
- 31.Metsugi Y, Miyaji Y, Ogawara K, Higaki K, Kimura T. Appearance of double peaks in plasma concentration-time profile after oral administration depends on gastric emptying profile and weight function. Pharmaceutical research. 2008;25(4):886–895. doi: 10.1007/s11095-007-9469-z. [DOI] [PubMed] [Google Scholar]
- 32.Singh BN. Effects of food on clinical pharmacokinetics. Clinical pharmacokinetics. 1999;37(3):213–255. doi: 10.2165/00003088-199937030-00003. [DOI] [PubMed] [Google Scholar]
- 33.Stevens JE, Doran S, Russo A, et al. Effects of intravenous fructose on gastric emptying and antropyloroduodenal motility in healthy subjects. American journal of physiology Gastrointestinal and liver physiology. 2009;297(6):G1274–1280. doi: 10.1152/ajpgi.00214.2009. [DOI] [PubMed] [Google Scholar]
- 34.Cheng CL, Yu LX, Lee HL, Yang CY, Lue CS, Chou CH. Biowaiver extension potential to BCS Class III high solubility-low permeability drugs: bridging evidence for metformin immediate-release tablet. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2004;22(4):297–304. doi: 10.1016/j.ejps.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Turner KC, Christensen M, Connor JD, Moore KT, Gao X, Donahue SR. Pharmacokinetics of a glyburide/metformin combination tablet in children and adolescents with type 2 diabetes. Clinical Pharmacology & Therapeutics. 2003;73(2):P66–P66. [Google Scholar]
- 36.Gao X, Christensen M, Burghen GA, et al. Pharmacokinetics of Metformin in Pediatric Type 2 Diabetic and Healthy Adult Subjects. Clinical Pharmacology & Therapeutics. 2003;73(2):P46–P46. [Google Scholar]
- 37.Sanchez-Infantes D, Diaz M, Lopez-Bermejo A, Marcos MV, de Zegher F, Ibanez L. Pharmacokinetics of metformin in girls aged 9 years. Clin Pharmacokinet. 2011;50(11):735–738. doi: 10.2165/11593970-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 38.Scheen AJ. Clinical pharmacokinetics of metformin. Clin Pharmacokinet. 1996;30(5):359–371. doi: 10.2165/00003088-199630050-00003. [DOI] [PubMed] [Google Scholar]
- 39.Bardin C, Nobecourt E, Larger E, Chast F, Treluyer JM, Urien S. Population pharmacokinetics of metformin in obese and non-obese patients with type 2 diabetes mellitus. European journal of clinical pharmacology. 2012;68(6):961–968. doi: 10.1007/s00228-011-1207-0. [DOI] [PubMed] [Google Scholar]
- 40.Goswami S, Yee SW, Stocker S, et al. Genetic variants in transcription factors are associated with the pharmacokinetics and pharmacodynamics of metformin. Clin Pharmacol Ther. 2014;96(3):370–379. doi: 10.1038/clpt.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duong JK, Kumar SS, Kirkpatrick CM, et al. Population pharmacokinetics of metformin in healthy subjects and patients with type 2 diabetes mellitus: simulation of doses according to renal function. Clin Pharmacokinet. 2013;52(5):373–384. doi: 10.1007/s40262-013-0046-9. [DOI] [PubMed] [Google Scholar]
- 42.Hong Y, Rohatagi S, Habtemariam B, Walker JR, Schwartz SL, Mager DE. Population exposure-response modeling of metformin in patients with type 2 diabetes mellitus. J Clin Pharmacol. 2008;48(6):696–707. doi: 10.1177/0091270008316884. [DOI] [PubMed] [Google Scholar]
- 43.Shu Y, Brown C, Castro RA, et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clinical pharmacology and therapeutics. 2008;83(2):273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexandre KB, Smit AM, Gray IP, Crowther NJ. Metformin inhibits intracellular lipid accumulation in the murine pre-adipocyte cell line, 3T3-L1. Diabetes, obesity & metabolism. 2008;10(8):688–690. doi: 10.1111/j.1463-1326.2008.00890.x. [DOI] [PubMed] [Google Scholar]
- 45.Gao Y, Xue J, Li X, Jia Y, Hu J. Metformin regulates osteoblast and adipocyte differentiation of rat mesenchymal stem cells. J Pharm Pharmacol. 2008;60(12):1695–1700. doi: 10.1211/jpp.60/12.0017. [DOI] [PubMed] [Google Scholar]
- 46.Savgan-Gurol E, Bredella M, Russell M, Mendes N, Klibanski A, Misra M. Waist to hip ratio and trunk to extremity fat (DXA) are better surrogates for IMCL and for visceral fat respectively than for subcutaneous fat in adolescent girls. Nutrition & metabolism. 2010;7:86. doi: 10.1186/1743-7075-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lalau JD, Race JM. Lactic acidosis in metformin-treated patients. Prognostic value of arterial lactate levels and plasma metformin concentrations. Drug Saf. 1999;20(4):377–384. doi: 10.2165/00002018-199920040-00006. [DOI] [PubMed] [Google Scholar]
- 48.Clapp JF, 3rd, Kiess W. Cord blood leptin reflects fetal fat mass. J Soc Gynecol Investig. 1998;5(6):300–303. [PubMed] [Google Scholar]