Abstract
Ethylene plays important roles in plant growth, development, and stress responses. Two ethylene receptors, ETR1 from Arabidopsis and NTHK1 from tobacco (Nicotiana tabacum), have been found to have His kinase (HK) activity and Ser/Thr kinase activity, respectively, although both show similarity to bacterial two-component HK. Here, we report the characterization of another ethylene receptor homolog gene, NTHK2, from tobacco. This gene also encodes a HK-like protein and is induced by dehydration and CaCl2 but not significantly affected by NaCl and abscisic acid treatments. The biochemical properties of the yeast (Schizosaccharomyces pombe)-expressed NTHK2 domains were further characterized. We found that NTHK2 possessed Ser/Thr kinase activity in the presence of Mn2+ and had HK activity in the presence of Ca2+. Several lines of evidence supported this conclusion, including hydrolytic stability, phosphoamino acid analysis, mutation, deletion, and substrate analysis. These properties have implications in elucidation of the complexity of the ethylene signal transduction pathway and understanding of ethylene functions in plants.
Ethylene plays important roles in multiple aspects of plant growth, development, and stress responses (Abeles et al., 1992). In the presence of ethylene, plant seedlings exhibit triple responses in the dark, including inhibited root and hypocotyl elongation, radial expansion of the hypocotyl, and exaggerated growth of the apical hook. Taking advantage of the triple responses, a number of Arabidopsis mutants were genetically screened, and a series of components have been identified to be involved in the ethylene signal transduction pathway (Bleecker and Kende, 2000; Schaller and Kieber, 2002; Wang et al., 2002; Guo and Ecker, 2004). These components include ethylene receptors (Chang et al., 1993; Hua et al., 1995, 1998; Sakai et al., 1998), the Raf-like Ser/Thr kinase (STK) CTR1 (Kieber et al., 1993), the membrane protein EIN2 (Alonso et al., 1999), and transcription factors EIN3 and ERF family (Chao et al., 1997; Solano et al., 1998). Additional components in the ethylene-response pathway have been found (Alonso et al., 2003), and an alternative pathway has also been suggested (Moshkov et al., 2003).
In Arabidopsis, five ethylene receptor genes, namely ETR1, ERS1, ETR2, EIN4, and ERS2, have been isolated, and all of the encoded proteins show similarity to bacterial two-component His kinases (HK; Chang et al., 1993; Hua et al., 1995, 1998; Sakai et al., 1998). The two-component signal transduction systems allow bacteria to adapt to changing environmental conditions. Based on the amino acid sequence similarity and structural features, the five Arabidopsis ethylene receptors can be classified into two subfamilies (Bleecker and Kende, 2000; Schaller and Kieber, 2002; Wang et al., 2002). Subfamily I contains ETR1 and ERS1, both of which have three transmembrane domains. Subfamily II contains ETR2, EIN4, and ERS2, and all the three proteins have a putative signal peptide in addition to the three conserved transmembrane domains. Moreover, ETR1, ETR2, and EIN4 have the putative HK domain and the receiver domain, whereas ERS1 and ERS2 only have the HK domain and lack the receiver domain. Compared with the subfamily I members, subfamily II members have a more diverged kinase domain. Among the five ethylene receptors, the subfamily I member ETR1 has been extensively studied. It has been demonstrated that ETR1 has ethylene-binding activity (Schaller and Bleecker, 1995) and a copper cofactor is required for the high-affinity binding (Rodriguez et al., 1999). The ETR1 protein can form a disulfide-linked dimer (Schaller et al., 1995). A HK activity has been detected in this protein (Gamble et al., 1998), and the ETR1 protein is reported to localize to the endoplasmic reticulum (Chen et al., 2002). The ETR1 has been shown to physically interact with CTR1, a component downstream of the ethylene receptors (Clark et al., 1998). The STK activity and the biochemical properties have been investigated for CTR1 (Huang et al., 2003). As to other receptors, ERS1 has been tested for its ethylene-binding activity (Hall et al., 2000). The biochemical properties of the Arabidopsis subfamily II members remain unclear.
Genes encoding homologous ethylene receptors have been isolated from dicots such as tomato (Lycopersicon esculentum) and tobacco (Nicotiana tabacum; Zhang et al., 2001a, 2001b; Klee, 2002, 2004) and monocots such as rice (Oryza sativa; Cao et al., 2003). In tomato, six ethylene receptor genes have been found, and the expressions of these genes in fruit ripening and pathogen response have been well studied (Zhou et al., 1996; Tieman and Klee, 1999; Tieman et al., 2000; Ciardi et al., 2001). In tobacco, four ethylene receptor genes, namely NtETR1, NtERS1, NTHK1, and NTHK2, have been identified (Knoester et al., 1997; Zhang et al., 1999, 2001a, 2001b; Terajima et al., 2001). NTHK1 represents a member of subfamily II. Its transcripts are abundant in flower organs and accumulate in response to wounding, salt, and drought stresses. Upon wounding, the NTHK1 mRNA is first expressed in the palisade cells of a leaf and then spreads to the sponge cells (Zhang et al., 2001a). The protein distribution in tobacco tissues or organs is substantially consistent with its mRNA expression patterns (Xie et al., 2002). Biochemical studies demonstrate that NTHK1 possesses STK activity but no HK activity and is localized to the plasma membrane (Xie et al., 2003).
NTHK2 represents another subfamily II member of ethylene receptors. Its gene expression can be induced by wounding and polyethylene glycol treatment (Zhang et al., 2001b). However, NTHK2 is still partial in sequence, and the biochemical properties of the protein remain unclear. Here, we report the isolation of the full-length sequence of the NTHK2 gene. Different domains or mutant forms of the NTHK2 protein were expressed in yeast (Schizosaccharomyces pombe) and characterized. We found that the NTHK2 possessed dual kinase activities. In the presence of Mn2+, it had STK activity, whereas in the presence of Ca2+, it had HK activity. These properties have significance in elucidation of the complexity of the ethylene signal transduction pathway and understanding of ethylene functions in plants.
RESULTS
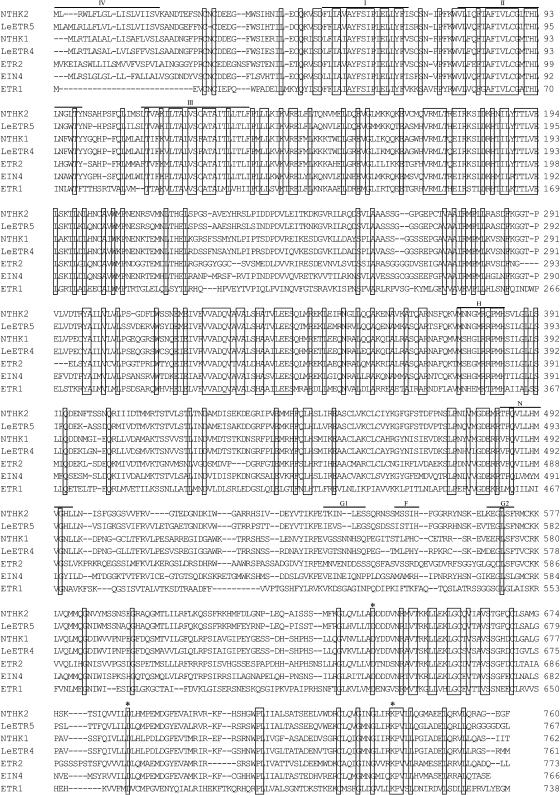
Sequence Analysis of the NTHK2 Gene
By using the 5′-RACE method, the full-length sequence of the NTHK2 was obtained based on the previously isolated 2,038-bp NTHK2 fragment (Zhang et al., 2001b). The full-length NTHK2 gene contained 3,216 bp, including a 5′-untranslated region (UTR) of 509 bp and a 3′-UTR of 427 bp, and encoded a protein of 760 amino acids, with a molecular mass of 85.2 kD and a pI of 7.2. The deduced amino acid sequence of NTHK2 was compared with other known ethylene receptors or homologs (Fig. 1). It is shown that NTHK2 had 78.3% identity with LeETR5 from tomato (Tieman and Klee, 1999). This high identity was in contrast with the relatively low identities obtained when NTHK2 was compared with NTHK1 from tobacco (Zhang et al., 2001a), LeETR4 from tomato (Tieman and Klee, 1999), and EIN4, ETR2, and ETR1 from Arabidopsis (Chang et al., 1993; Hua et al., 1998; Sakai et al., 1998). These identities were 54.3%, 53.8%, 56.3%, 46.4%, and 32.8%, respectively. The domains in NTHK2 were predicted using the SMART program (Schultz et al., 1998) and by comparison with ETR1 (Fig. 1). In the N-terminal end, four hydrophobic regions were identified. Regions I, II, and III may represent transmembrane domains, whereas region IV may be a signal peptide. After the hydrophobic regions, a GAF domain (amino acids 183–341), a putative HK domain (amino acids 367–608), and a receiver domain (amino acids 634–753) were found. These domains resembled the bacterial two-component HK proteins. In the HK domains, motifs corresponding to the Arabidopsis ETR1 conserved boxes H, N, G1, F, and G2 were also recognized. However, except boxes H and N, all other boxes were diverged from those of ETR1. In the H box of NTHK2, the residue corresponding to the ETR1 autophosphorylation site H353 was replaced by an Asn (N377). A potential phosphate receiver site (D686) and two other conserved residues (D640, K738) were also identified in the putative receiver domain (Fig. 1).
Figure 1.
Alignment of the NTHK2 amino acid sequence with other ethylene receptor proteins. LeETR4 and LeETR5 are from tomato (Tieman and Klee, 1999). NTHK1 is from tobacco (Zhang et al., 2001a). ETR1, ETR2, and EIN4 are from Arabidopsis (Chang et al., 1993; Hua et al., 1998; Sakai et al., 1998). Boxes indicate the identity of amino acids. Regions I, II, and III represent the three transmembrane segments. Region IV represents the putative signal peptide. The regions corresponding to the five motifs H, N, G1, F, and G2 from the HK domain of ETR1 were overlined. Asterisks indicate conserved residues in the putative receiver domain. The nucleotide sequence for the NTHK2 gene has been deposited in the GenBank database under GenBank accession number AF203476.
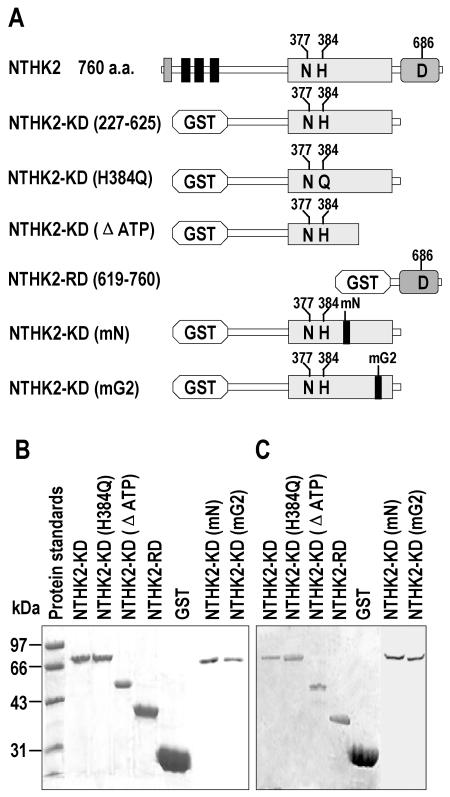
Purification of the NTHK2 Truncated Proteins
NTHK2 had a putative HK domain and a receiver domain. To address the biochemical properties of these domains, six truncated proteins or mutants, including NTHK2-KD (amino acids 227–625), NTHK2-KD (H384Q), NTHK2 (ΔATP; 227–454), NTHK2-RD (619–760), NTHK2-KD (mN), and NTHK2-KD (mG2) were expressed as glutathione S-transferase (GST) fusions in yeast cells (SPQ01; Fig. 2A). NTHK2-KD (227–625) contained most of the GAF domain and the putative HK domain. NTHK2-KD (H384Q) was the same as NTHK2-KD (227–625) except that a mutation of the conserved His (H384) to Gln (Q) was generated. NTHK2-KD (ΔATP) represented a truncated version without the ATP-binding motif, and NTHK2-RD (619–760) represented the NTHK2 receiver domain. For NTHK2-KD (mN), the HMVGHLLN sequence in the N box of NTHK2-KD (227–625) was mutated to QAAAQLLA. For NTHK2-KD (mG2), the EGLS sequence in the G2 box of NTHK2-KD (227–625) was mutated to HAAA. The yeast-expressed proteins were purified by affinity resin and resolved by SDS-PAGE. It can be seen that the fusion proteins exhibited predicted molecular masses of 71, 71, 52, 42, 71, and 71 kD for NTHK2-KD (227–625), NTHK2-KD (H384Q), NTHK2-KD (ΔATP), NTHK2-RD (619–760), NTHK2-KD (mN), and NTHK2-KD (mG2), respectively (Fig. 2B). The GST fusion proteins were further confirmed on western blot using mouse anti-GST monoclonal antibody (Fig. 2C). The GST protein (27 kD) was also expressed and used as a control in later analysis.
Figure 2.
Purification of different domains of NTHK2 from yeast by affinity resin. A, Schematic representation of NTHK2 and various versions expressed in yeast. For NTHK2, the first box indicates the putative signal peptide, the next three boxes the transmembrane segments. The rectangular box represents the putative HK domain. The oval box represents the putative receiver domain. B, SDS-PAGE profiles of purified GST fusion proteins and protein marker. Proteins are stained with Coomassie blue. C, Western-blot analysis of the purified GST fusion proteins using an anti-GST monoclonal antibody.
Kinase Activity of the NTHK2 Kinase Domain
Because both ETR1 from Arabidopsis and NTHK1 from tobacco had kinase activity, we examined if the NTHK2 kinase domain NTHK2-KD (227–625) possessed any kinase activity. The NTHK2-KD protein was incubated with [γ-32P]ATP, and the result was presented in Figure 3. It can be seen that 32P was incorporated into the fusion protein in the presence of Mg2+, Mn2+, and Ca2+. However, the incorporation levels were different. In the presence of Mn2+ and Ca2+, the phosphorylation was stronger, whereas in the presence of Mg2+, the protein was phosphorylated to a lesser degree. The molar percent incorporation was near 4% in the presence of Mn2+ and 3% in the presence of Ca2+. When EDTA was included in the reaction mix, no phosphorylation was detected, indicating the absolute requirement of the divalent cations for the phosphorylation. The GST protein was also incubated with the NTHK2-KD protein in the presence of the three cations. It is shown that the GST protein was not phosphorylated (Fig. 3). In the absence of NTHK2-KD, GST itself cannot be labeled either (data not shown).
Figure 3.
Autophosphorylation of NTHK2-KD. GST fusion protein NTHK2-KD (0.5 μg) was incubated with [γ-32P]ATP in the presence of 5 mm Mg2+, 5 mm Ca2+, 5 mm Mn2+, or 10 mm EDTA. GST (4 μg) was included as a control. The phosphorylated proteins were then separated on SDS-PAGE, transferred onto PVDF membrane, and autoradiographed (top) or Coomassie blue stained (bottom).
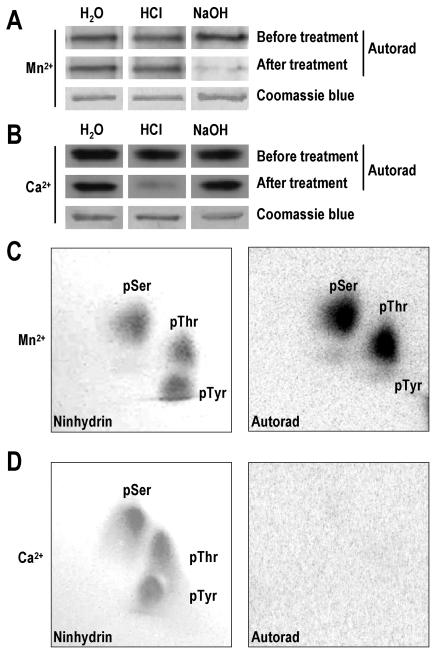
We analyzed what kind of amino acid residues were phosphorylated. The phosphorylated NTHK2-KD proteins were transferred onto polyvinylidene difluoride (PVDF) membranes, and the membranes were treated with HCl or NaOH to examine the stability of the phosphorylated residues. It can be seen that, in the presence of Mn2+, the phosphorylated residues were stable upon acid treatment but sensitive to base treatment (Fig. 4A), indicating that the phosphorylated residues included phospho-Ser, phospho-Thr, or phospho-Tyr but not phospho-His. Similar result has been obtained for the phosphorylated NTHK2-KD in the presence of Mg2+ (data not shown). In the presence of Ca2+, the stability of the phosphorylated residues changed. Under acidic conditions, the phosphorylated residues were unstable, whereas these were stable under basic conditions (Fig. 4B). This result indicated that the phosphorylated residue in the presence of Ca2+ was phospho-His but not other residues. We further performed phosphoamino acid analysis using two-dimensional thin-layer chromatography electrophoresis. The phosphorylated NTHK2-KD was hydrolyzed with concentrated HCl and analyzed together with the phosphoamino acid standards. The result showed that, in the presence of Mn2+, phospho-Ser and phospho-Thr were detected (Fig. 4C), whereas in the presence of Ca2+, no phosphoamino acids were detected, consistent with the assumption that the phospho-His has been hydrolyzed (Fig. 4D). From the analysis above, we propose that NTHK2-KD possesses STK activity in the presence of Mn2+ but has HK activity in the presence of Ca2+.
Figure 4.
Hydrolytic stability and phosphoamino acid analysis of the phosphorylated NTHK2-KD. The NTHK2-KD was autophosphorylated in the presence of Mn2+ (A and C) or Ca2+ (B and D), and then the phosphorylated NTHK2-KD was subjected to treatments with water, HCl, or NaOH (A and B). After autoradiography, the treated protein blots were stained with Coomassie blue. The phosphorylated NTHK2-KD was further subjected to phosphoamino acid analysis (C and D). The positions of the phosphoamino acids were identified by spraying with ninhydrin (left), and the labeled residues were revealed by autoradiography (right).
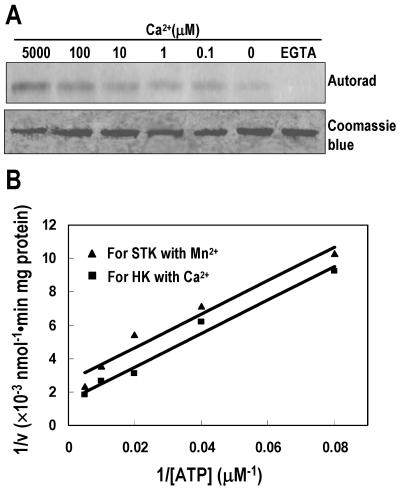
Since the other two ethylene receptors, ETR1 and NTHK1, had no activity in the presence of Ca2+ (Gamble et al., 1998; Xie et al., 2003), we further characterized the HK activity of NTHK2-KD in the presence of various concentrations of Ca2+. When no Ca2+ was added, the NTHK2-KD had some activities, probably due to the trace amount of Ca2+ in the reaction mix. However, when Ca2+-specific chelator EGTA was included, no activity was detected, indicating that this activity of NTHK2-KD is Ca2+ dependent. NTHK2-KD maintained gradually higher activity in the range from 0.1 μm to 5 mm Ca2+ (Fig. 5A).
Figure 5.
Effects of Ca2+ concentration on NTHK2-KD phosphorylation and kinetic analysis. A, The NTHK2-KD was autophosphorylated in the presence of various Ca2+ concentrations or in buffers with 10 mm EGTA. The phosphorylated NTHK2 protein was then resolved on SDS-PAGE, transferred to PVDF membranes, and subjected to autoradiography (top) or Coomassie blue staining (bottom). B, Kinetic analysis of NTHK2-KD phosphorylation. NTHK2-KD protein was incubated with increasing amounts of nonlabeled ATP and [γ-32P]ATP in the presence of Mn2+ or Ca2+. Kinase activities were calculated from the amount of 32P incorporation into the NTHK2-KD. The value of 1/v was plotted versus 1/[ATP] and Km was derived.
To compare the affinity of the NTHK2-KD for ATP in the presence of Mn2+ and Ca2+, the NTHK2-KD was incubated with varying amounts of nonlabeled ATP plus [γ-32P]ATP. Aliquots were removed from each reaction mix at different time points, spotted on filter disc, washed, and then determined for 32P incorporation. The Km was estimated by plotting 1/v versus 1/[ATP] (Fig. 5B). For the STK activity in the presence of Mn2+, the Km for ATP was 38.4 ± 3.5 μm. For the HK activity in the presence of Ca2+, the Km was 70.4 ± 12.1 μm.
Mutation, Deletion, and Substrate Analysis of the NTHK2 Kinase Domain
NTHK2 had HK activity in the presence of Ca2+. However, in the H box of the NTHK2 kinase domain, there was no His at the conserved site for phosphorylation. Instead, another conserved His (H384) was found in the H box, which was seven residues downstream. This His (H384) could potentially function as a phosphorylation site and thus was mutated to a Gln to test this possibility. The result, shown in Figure 6A, indicated that the mutation of His (H384) to Gln did not affect the kinase activity of NTHK2-KD (H384Q), indicating that H384 was not required for the autophosphorylation in the presence of Ca2+. Because no conserved Gly were identified in the G1 box for mutation, we deleted the ATP-binding motif including region of the putative boxes N, G1, F, and G2 to examine its effect on phosphorylation (Fig. 6A). It can be seen that the deletion abolished the autophosphorylation of NTHK2-KD (ΔATP), indicating that the ATP-binding motif was essential for the phosphorylation and the HK activity was likely to be intrinsic to NTHK2-KD.
Figure 6.
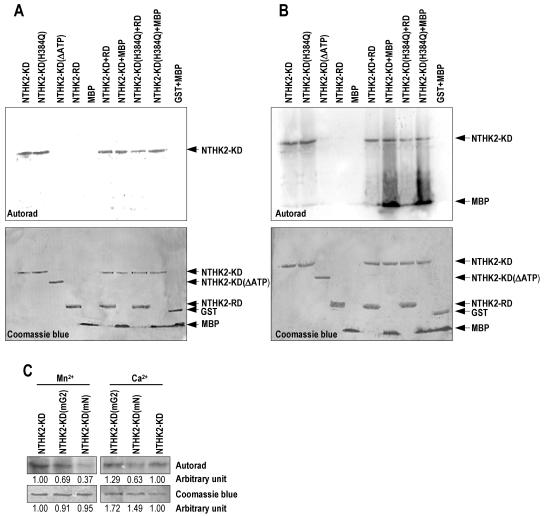
Phosphorylation assay of different versions of NTHK2-KD. The phosphorylation was performed in the presence of Ca2+ (A) or Mn2+ (B). The NTHK2-KD or other versions (1 μg) was incubated under phosphorylating conditions with no substrate or with MBP (2 μg), NTHK2-RD (2 μg), or GST (2 μg). NTHK2-RD, MBP, or GST plus MBP were also incubated in the absence of NTHK2-KD and used as controls. The phosphorylated proteins were resolved on SDS-PAGE gel, transferred to PVDF membranes, and subjected to autoradiography (top) or Coomassie blue staining (bottom). C, Effect of N-box and G2-box mutation on the phosphorylation of NTHK2-KD. NTHK2-KD (mN) with N-box mutation and NTHK2-KD (mG2) with G2-box mutation were assayed for their kinase activities following standard method in the presence of Mn2+ or Ca2+. Quantification of the signal intensity was performed using Alpha Innotech Imaging System (Alpha Innotech, San Leandro, CA).
In bacteria, a phosphorylated HK can transfer its phosphate onto a conserved Asp residue of a response regulator (receiver domain-like; Parkinson and Kofoid, 1992). We further studied if NTHK2-KD can phosphorylate the putative receiver domain and an in vitro substrate myelin basic protein (MBP) when Ca2+ was used as a cofactor. MBP has been used as a substrate to identity new protein kinases (Cicirelli et al., 1988). The results showed that neither NTHK2-KD nor NTHK2-KD (H384Q) could phosphorylate the NTHK2-RD or MBP in the presence of Ca2+ (Fig. 6A). Additionally, the NTHK2-RD, MBP, or GST plus MBP did not exhibit 32P incorporation.
In the presence of Mn2+, NTHK2-KD possessed STK activity. The mutant or the truncated form of the NTHK2-KD was examined for their phosphorylation in the presence of Mn2+ (Fig. 6B). It can be seen that NTHK2-KD (H384Q) still possessed the ability to autophosphorylate, indicating that H384 was not an intermediate phosphorylation site of NTHK2-KD. The ATP-binding motif was still required for the STK activity of NTHK2-KD because deletion of it also destroyed the kinase activity. For the substrate analysis, NTHK2-KD and NTHK2-KD (H384Q) cannot phosphorylate the receiver domain NTHK2-RD. However, both proteins can phosphorylate the in vitro substrate MBP in the presence of Mn2+. This phenomenon was different from that in the case of Ca2+. The NTHK2-RD, MBP, and GST plus MBP did not have the ability to phosphorylate in the presence of Mn2+ (Fig. 6B).
To further investigate the effects of mutation in the ATP-binding motif on the kinase activities, two mutated NTHK2-KD proteins, NTHK2-KD (mN) with six-residue mutation in the N box and NTHK2-KD (mG2) with four-residue mutation in the G2 box, were analyzed for their autophosphorylation ability. The results (Fig. 6C) showed that, in the presence of Mn2+, N-box mutation in NTHK2-KD (mN) reduced autophosphorylation by approximately 61%, whereas G2-box mutation in NTHK2-KD (mG2) reduced autophosphorylation by approximately 24%. In the presence of Ca2+, N-box mutation in NTHK2-KD (mN) reduced autophosphorylation by approximately 58%, whereas G2-box mutation reduced autophosphorylation by approximately 25%. These results indicate that the autophosphorylation of NTHK2-KD is more severely affected by N-box mutation than by G2-box mutation. The facts further support the possibility that the kinase activity is intrinsic to the NTHK-KD.
NTHK2 Gene Expression in Response to Various Treatments
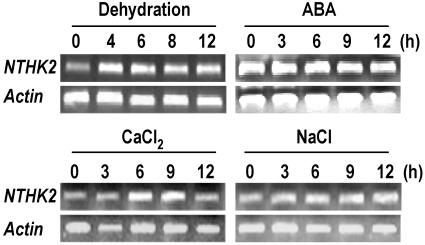
The NTHK2 gene expression was investigated in tobacco seedlings using reverse transcription (RT)-PCR. It can be seen from Figure 7 that the NTHK2 was induced by dehydration and CaCl2 treatments but not significantly affected by abscisic acid (ABA) and NaCl treatments, indicating that NTHK2 may be involved in some specific responses. It should be noted that dehydration-induced NTHK2 expression occurred in roots (Fig. 7) but not in shoots (data not shown), suggesting that the primary site for NTHK2 function during dehydration was in roots.
Figure 7.
RT-PCR analysis of NTHK2 expression upon various treatments. Tobacco seedlings were subjected to dehydration, ABA, CaCl2, and NaCl treatments for the indicated times, and total RNA was used for RT-PCR analysis. For dehydration treatment, only roots were used for analysis. Tobacco actin genes were amplified as a control. The amplified products of NTHK2 and actin are 415 bp and 600 bp, respectively.
DISCUSSION
In bacteria, most two-component proteins have HK activity (Parkinson and Kofoid, 1992). However, a few HK-like proteins from eukaryotes, e.g. phytochromes from higher plants, have diverged and possessed STK activity (Yeh and Lagarias, 1998). Different kinase activities are even found within the plant ethylene receptor family. As a subfamily I member, ETR1 possesses HK activity (Gamble et al., 1998), whereas a subfamily II member, NTHK1, has STK activity (Xie et al., 2003). In this study, we found that NTHK2 had dual kinase activities of STK and HK in the presence of Mn2+ and Ca2+, respectively. Several lines of evidence support this conclusion, including acid/base hydrolysis, phosphoamino acid analysis, mutation, deletion, and substrate analysis. It may be argued that we used relatively low ATP concentration for the phosphorylation assay of NTHK2. However, when ATP concentration was increased in the assay, consistent results were obtained (data not shown).
NTHK2 does not have a His at the conserved phosphorylation site. However, it has a His (H384) at the position seven residues downstream, which is conserved among all the compared proteins. Mutation analysis of the His (H384) to Gln implies that this His is neither an intermediate for Ser/Thr phosphorylation in NTHK2-KD in the presence of Mn2+ nor the phosphorylation site for the HK activity of NTHK2-KD in the presence of Ca2+. Another tobacco ethylene receptor, NTHK1, has the conserved His (H378) at the putative phosphorylation site. However, mutation analysis of H378 to Gln indicates that H378 is not an intermediate for the phosphorylation of NTHK1 either (Xie et al., 2003). On the contrary, Arabidopsis ETR1 needs the His for its HK activity (Gamble et al., 1998). These differences may reflect additional divergence in phosphorylation sites among various ethylene receptors. Although NTHK2 has the ability to autophosphorylate, where the phosphorylated residues are located remains to be identified. Using a NetPhos 2.0 Server program (http://www.cbs.dtu.dk/services/NetPhos/), 17 Ser and 5 Thr in NTHK2-KD are predicted to be the putative phosphorylation sites for the STK activity (data not shown). Mutagenesis of these sites would reveal the important residues for phosphorylation. For the HK activity of NTHK2-KD in the presence of Ca2+, eight more His residues are recognized besides the conserved H384. These residues could potentially serve as the phosphorylation sites for HK activity. Nonconserved His phosphorylation (H48) has been observed in CheA, a two-component protein sensor for chemotaxis in bacteria (Hess et al., 1988). Alternatively, it may still be possible that the alkali-stable phosphorylation in NTHK2 was due to the phosphorylation of an amino acid besides His.
Through deletion analysis of the ATP-binding motif in NTHK2-KD, we found that the kinase activity in NTHK2-KD in the presence of Mn2+ or Ca2+ was completely abolished, probably indicating that the activity is intrinsic to NTHK2-KD. Similar results have been obtained for NTHK1 (Xie et al., 2003). The intrinsic activity may be further supported by the fact that multisite mutation in the N box of NTHK2-KD significantly reduced the kinase activity. However, G2-box mutation led to only a small reduction in kinase activity. The differential influence of the N-box and G2-box mutations on the NTHK2-KD kinase activity may reflect the different roles of the two boxes in forming the ATP-binding pocket. The N box is situated in the pocket wall, whereas the G2 box is part of a large disordered loop analogous to the ATP lid that extends away from the body of the domain (Robinson et al., 2000). We cannot completely abolish the kinase activity by N-box or G2-box mutations, probably indicating that the two mutations only affect the ATP-binding efficiency. Alternatively, the mutations may not significantly change the topological structures of the protein. This fact may also imply that none of the N box or G2 box is absolutely required for the phosphorylation, consistent with the observation that several of the bacterial HKs lack one of these motifs. Examples of missing motifs are N box in FrzE, G1 box in AgrORF2, and G2 box in NarQ and NarX (Parkinson and Kofoid, 1992). A dimeric HPt protein, Spo0B, also contains auxiliary domains similar in topology to the HK ATP-binding domain but lacks the conserved nucleotide-binding motifs (Robinson et al., 2000). Due to the fact that the ethylene receptor ETR1 can bind the kinase CTR1 (Clark et al., 1998), possibility may still exist that NTHK2-KD could bind a contaminating kinase when purified from the yeast cells, although such a case is less likely. Further inhibitor studies may improve our understanding of the kinase activities.
NTHK2 cannot phosphorylate its receiver domain in the in vitro study. The same results have been found for NTHK1 and ETR1 (Gamble et al., 1998; Xie et al., 2003). These studies indicate that the receiver domain may not function in accepting a phosphate as the bacterial response regulators do but rather play roles in regulation or interaction (Bleecker and Kende, 2000).
Most bacterial HKs need divalent cation Mg2+ or Mn2+ for their kinase activity (Hess et al., 1991). However, some HKs, e.g. EnvZ, also possess activity in the presence of Ca2+ (Park et al., 1998), and others may need different divalent cations for their kinase activity (Koo and Stephens, 2003). A bacterial HK PhoQ can even sense Mg2+ and Ca2+ (Regelmann et al., 2002). In plants, the HK-like ethylene receptor ETR1 depends on Mn2+ for its HK activity. It shows limited activity in the presence of Mg2+ and no activity in the presence of Ca2+ (Gamble et al., 1998). Tobacco ethylene receptor NTHK1 requires Mn2+ for its STK activity and does not require Mg2+ or Ca2+ (Xie et al., 2003). In this study, the STK activity of NTHK2 was favored by Mn2+, whereas its HK activity was dependent on Ca2+. It was probably due to the subtle change in protein structure or conformation in the presence of various divalent cations that made NTHK2 exhibit different kinase activities. In plant cells, NTHK2 may show different kinase activities in different microcompartments or microenvironments, such as plasma membrane or vesicles during secretion, where different divalent cations such as Mn2+ or Ca2+ may be transiently dominant. Ca2+ has been found to be involved in many aspects of plant growth, development, and stress responses (Xiong et al., 2002). It is also required for ethylene-dependent responses (Raz and Fluhr, 1992). The NTHK2 gene expression is also induced by CaCl2 treatment. Therefore, the possibility exists that NTHK2 is implicated in Ca2+-regulated processes. This property makes NTHK2 different from another tobacco subfamily II ethylene receptor, NTHK1, and the Arabidopsis subfamily I member ETR1.
Although NTHK2 has dual kinase activities in vitro, whether it has any in vivo kinase activity remains to be elucidated. Recently, it has been reported that the HK activity of ETR1 may not be required for signal transduction as revealed by introducing a kinase-inactivated ETR1 gene into the double mutant ers1-2;etr1-6 or ers1-2;etr1-7 (Wang et al., 2003). Further mutational analysis of the putative phosphorylation sites in NTHK2 and transformation research should reveal the possibility of the necessity for the kinase activity in plants.
The NTHK2 gene has been transformed into Arabidopsis, and the transgenic plants exhibited ethylene less-sensitive phenotypes (Z.G. Zhang, J.S. Zhang, and S.Y. Chen, unpublished data). This observation was consistent with the function of ethylene receptors (Tieman et al., 2000; Klee, 2002; Xie et al., 2002), i.e. ethylene receptors negatively regulate the ethylene responses (Hua and Meyerowitz, 1998). Further research should reveal more detail of the NTHK2 function in plant growth, development, and stress responses.
MATERIALS AND METHODS
Plant Growth and Treatments
One-month-old tobacco (Nicotiana tabacum var. Xanthi) seedlings were grown on wet cheesecloth at 27°C and used in the following treatments. For ABA, NaCl, and CaCl2 treatments, the seedlings were transferred to solutions containing 100 μm ABA, 100 mm NaCl, or 80 mm CaCl2, respectively, and maintained for various times. For dehydration treatment, the plants were carefully pulled out, transferred onto filter paper, and allowed to dry for the indicated times. Samples from all the treatments above were harvested and stored at −70°C for RNA isolation. Total RNA isolation was performed as described previously (Zhang et al., 2001a).
5′-RACE
Based on the partial sequence of NTHK2 (Zhang et al., 2001b), we designed gene-specific primers GSP1 (5′-CAGCATTCTCCTTAGCCTGCTGCAGC-3′) and GSP2 (5′-CCTCCACTGCTTGCAGCTGCAAGAACTG-3′). The 5′-RACE was conducted using SMART RACE cDNA Amplification kit (CLONTECH, Palo Alto, CA). The first-strand cDNAs were synthesized according to the instructions and used as templates for PCR amplification. The PCR products were analyzed on a 1.2% agarose/ethidium bromide gel and the purified DNA fragments were directly sequenced. The full-length NTHK2 gene was then amplified, cloned into T-easy vector (Promega, Madison, WI), and sequenced.
RT-PCR Analysis of NTHK2
The first-strand cDNA was synthesized with 5 μg of total RNA in 20 μL of reaction volume using a cDNA synthesis kit according to the manufacturer's instruction (Promega). One microliter of the cDNA mix was used as template in a 20-μL PCR reaction volume. After 2 min denaturation at 94°C, the amplification was performed at 94°C for 15 s, 56°C for 30 s, and 72°C for 1 min for 28 cycles, with an extension of 10 min at 72°C. The sense and antisense primers used for analysis of NTHK2 expression were 5′-TTGCCAAGTAATTGCTGTG-3′ and 5′-TCCTTGGTTAGAATTCCTG-3′, respectively, and the amplified fragment was 415 bp. The antisense primer was derived from the 3′-UTR of the NTHK2 gene. A 600-bp fragment of tobacco actin gene was also amplified as a control with the sense primer 5′-GATATGGAGAAG/AATC/ATGGCATCAT/CAC-3′ and the antisense primer 5′-GTTTCA/GTGAATT/ACCT/AGCT-3′. The primers were designed according to the consensus sequences of the tobacco actin genes as described (Dhondt et al., 2000).
Recombinant Expression of Truncated NTHK2 Proteins
For expression of different truncated versions of NTHK2 as GST fusions in yeast (Schizosaccharomyces pombe), the pESP-2 vector (Stratagene, La Jolla, CA) was used. DNA fragments corresponding to the putative kinase domain (NTHK2-KD, amino acids 227–625), the kinase domain without the ATP-binding motif [NTHK2-KD (ΔATP), amino acids 227 to 454], and the putative receiver domain NTHK2-RD (amino acids 619–760) were amplified from the original NTHK2 plasmid. For NTHK2-KD, the sense primer is 5′-CCCGGATCCGCTGTAGAATACCATCGTTC-3′ and the antisense primer is 5′-GCTGCTAGCTGCTTGCTCCAAAGGATTTC-3′. For NTHK2-KD (ΔATP), the sense primer is the same as that for NTHK2-KD, and the antisense primer is 5′-CGGGCTAGCAAGACAAGAAGCCTCTCTG-3′. The primers for NTHK2-RD are 5′-CGTGGATCCGGAAATCCTTTGGAGCAAG-3′ and 5′-CGGGCTAGCTCAAAAGCCTTCACCAGCC-3′. A mutant version [NTHK2-KD (H384Q)] of the kinase domain, which has a Gln (CAG) at position 384 instead of the original His (CAC), was generated by using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The mutation was confirmed by sequencing. Two multisite mutations in N box and G2 box were also made by PCR using the original NTHK2 plasmid as template with primers 5′-GCGCAACTATTAGCTATCAGCTTTGGAAGC-3′ and 5′-CGCCGCTTGAAGTAACACCTGAAAC-3′ for the N-box mutation in NTHK2-KD (mN), and 5′-GGCTTTCCGCATGTGCAAAAAGCT-3′ and 5′-GCGGCATGCTTTAACTCTTTGCTGTTAT-3′ for the G2-box mutation in NTHK2-KD (mG2). The underlined sequences represent the mutated sequences. For NTHK2-KD (mN), the amino acid sequence HMVGHLLN (encoded by CATATGGTGGGGCATCTATTAAAT) was mutated to GAAAGLLA (encoded by CAAGCGGCGGCGCAACTATTAGCT). For NTHK2-KD (mG2), the amino acid sequence EGLS (encoded by GAGGGCTTGAGT) was mutated to HAAA (encoded by CATGCCGCGGCT). The PCR products containing the linear mutated plasmid were phosphorylated and self-ligated, and the circular mutated plasmids were transformed into Escherichia coli and confirmed by sequencing. The two mutated plasmids harboring the N-box and G2-box mutations, respectively, were further amplified using primers for NTHK2-KD DNA fragment. Finally, the PCR products for NTHK2-KD, NTHK2-KD (H384Q), NTHK2-KD (ΔATP), NTHK2-RD, NTHK2-KD (mN), and NTHK2-KD (mG2) were digested with BamHI and NheI, cloned into the yeast expression vector pESP-2, and confirmed by sequencing. The recombinant plasmids were transformed into S. pombe SP-Q01 yeast strain (leu1-32h-; Stratagene), and the positive colonies were identified by their ability to grow on Edinburgh minimal medium supplemented with thiamine. The fusion protein was induced by growing the cells in Edinburgh minimal medium without thiamine and purified by Glutathione Sepharose 4B (Amersham, Buckinghamshire, UK) as described (Xie et al., 2003). The expressed GST fusion proteins were examined by western blotting using a mouse anti-GST monoclonal antibody (Amersham). The GST protein itself was also expressed and used as a control.
In Vitro Phosphorylation Assay
The GST fusion proteins NTHK2-KD, NTHK2-KD (H384Q), NTHK2-KD (ΔATP), NTHK2-KD (mN), and NTHK2-KD (mG2) were assayed for their kinase activity. Phosphorylation was carried out in a 25 μL of assay buffer [50 mm Tris-HCl, pH 7.6/50 mm KCl/2 mm dithiothreitol/10% (v/v) glycerol] containing 0.5 μg to 1 μg of GST fusion proteins in the presence of 5 mm MgCl2, 5 mm MnCl2, or 5 mm CaCl2. The phosphorylation was initiated by adding 12.5 μCi of [γ-32P]ATP (30 Ci/mmol), incubated at 22°C for 45 min, and terminated by the addition of EDTA to a final concentration of 10 mm. The phosphorylated proteins were subjected to 10% SDS-PAGE and transferred onto PVDF membranes (Amersham). The incorporated phosphate was visualized by autoradiography. Coomassie blue staining of the same membrane was also performed to verify the protein loading. The stability of the incorporated phosphate was determined by treating the membranes with water, 1 m HCl, or 3 m NaOH for 2 h at room temperature. The treated membranes were then subjected to autoradiography. The fusion protein NTHK2-KD was also tested for its autophosphorylation under various concentrations of Ca2+ or in the presence of 10 mm EGTA. For kinetic assays, reactions were performed with varying amount of nonlabeled ATP plus [γ-32P]ATP at concentrations of 12.5, 25, 50, 100, and 200 μm. At different time points, aliquots were removed. For the STK activity in the presence of Mn2+, the reactions were stopped by 10 mm EDTA. The reaction mix was then spotted onto Whatman GF/C glass fiber filter disc (Clifton, NJ) and plunged into ice-cold 10% (w/v) trichloroacetic acid containing 1% (w/v) sodium pyrophosphate for 30 min (Yeh and Lagarias, 1998). The disc was washed three times, each for 30 min, using the same solution. The discs were rinsed in ethanol, dried, and subjected to 32P determination on a Beckman LS Analyzer (Beckman Instruments, Fullerton, CA). For the HK activity in the presence of Ca2+, the reactions were stopped by 0.5 n NaOH. After incubating at 60°C for 30 min, the reaction mix was spotted onto the Nytran paper disc, which had been immersed in 1 mm ATP (pH 8.5) overnight and dried (Huang et al., 1991). The disc was washed in 1 mm ATP, pH 8.5, four times, each for 30 min. After drying, the disc was subjected to 32P determination. The rate (v) of protein phosphorylation was calculated for each ATP concentration and 1/v was plotted versus 1/[ATP]. Km was derived from the linear regression analysis of the plots. Two independent experiments were performed and one set was presented. The mean value of the Km was calculated. For all the other phosphorylation assays, the experiments were repeated at least twice with different biological samples, and the results were consistent.
Phosphoamino Acid Analysis
The phosphorylated NTHK2-KD was transferred onto PVDF membrane after SDS-PAGE and autoradiographed. The region of the membrane corresponding to the labeled NTHK2-KD was excised and hydrolyzed in 5.7 n HCl at 110°C for 1 h. The supernatant was lyophilized and then dissolved in 5 to 10 μL of distilled water. The samples together with the phosphoamino acid standards (Sigma, St. Louis) were spotted onto 0.1-mm cellulose TLC plates (Merck, Rahway, NJ) and subjected to two-dimensional thin-layer chromatography electrophoresis. The positions of the three phosphoamino acid standards were visualized by spraying the plates with 0.25% ninhydrin in acetone followed by incubation in oven at 65°C until the purple spots of the standards occurred. The plate was then autoradiographed to identify the labeled phosphoamino acids.
Substrate Phosphorylation
For substrate phosphorylation, NTHK2-KD or NTHK2-KD (H384Q) (1 μg) was incubated under phosphorylating conditions with no substrate or with MBP (2 μg), NTHK2-RD (2 μg), or GST (2 μg). GST, MBP, or their combination were also incubated under phosphorylating conditions without NTHK2-KD and used as controls. The phosphorylated proteins were resolved on SDS-PAGE, transferred to PVDF membranes, and subjected to autoradiography or Coomassie blue staining.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AF203476.
Acknowledgments
We thank Dr. Harry Klee (University of Florida, Gainesville) for critical reading of the manuscript.
This work was supported by the National Basic Research Project (grant no. G19990117003), the National High-Tech program of China (grant nos. 2001AA222131, 2002AA2Z1001, and 2001AA212121), the National Transgenic Research Project (grant no. JY03A–10−02), and the National Natural Science Foundation of China (grant no. 30370130).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.034686.
References
- Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in Plant Biology, Ed 2. Academic Press, San Diego
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100: 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Cao WH, Dong Y, Zhang JS, Chen SY (2003) Characterization of an ethylene receptor homolog gene from rice. Sci China 46: 370–378 [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866 [DOI] [PubMed] [Google Scholar]
- Ciardi JA, Tieman DM, Jones JB, Klee HJ (2001) Reduced expression of the tomato ethylene receptor gene LeETR4 enhances the hypersensitive response to Xanthomonas campestris pv. Vesicatoria. Mol Plant Microbe Interact 14: 487–495 [DOI] [PubMed] [Google Scholar]
- Cicirelli MF, Pelech SL, Krebs EG (1988) Activation of multiple protein kinase during the burst in protein phosphorylation that precedes the first meiotic cell division in Xenopus oocytes. J Biol Chem 263: 2009–2019 [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt S, Geoffroy P, Stelmach BA, Legrand M, Heitz T (2000) Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J 23: 431–440 [DOI] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE (1998) Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA 95: 7825–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Hall AE, Findell J, Schaller GE, Sisler EC, Bleecker AB (2000) Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol 123: 1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JF, Bourret RB, Simon MI (1988) Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature 336: 139–143 [DOI] [PubMed] [Google Scholar]
- Hess JF, Bourret RB, Simon MI (1991) Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in Escherichia coli. Methods Enzymol 200: 188–204 [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM (1995) Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269: 1712–1714 [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene-receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wei Y, Kim Y, Osterberg L, Matthews H (1991) Purification of a protein histidine kinase from the yeast Saccharomyces cerevisiae. J Biol Chem 266: 9023–9031 [PubMed] [Google Scholar]
- Huang YF, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldman KA, Ecker J (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Klee HJ (2002) Control of ethylene-mediated processes in tomato at the level of receptors. J Exp Bot 53: 2057–2063 [DOI] [PubMed] [Google Scholar]
- Klee HJ (2004) Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiol 135: 660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoester M, Hennig J, van Loon LC, Bol JF, Linthorst JM (1997) Isolation and characterization of a tobacco cDNA encoding an ETR1 homolog (accession, AF022727) (PGR 97-188). Plant Physiol 115: 1731 [Google Scholar]
- Koo IC, Stephens RS (2003) A developmentally-regulated two-component signal transduction system in Chlamydia. J Biol Chem 278: 17314–17319 [DOI] [PubMed] [Google Scholar]
- Moshkov IE, Mur LAJ, Novikova GV, Smith AR, Hall MA (2003) Ethylene regulates monomeric GTP-binding protein gene expression and activity in Arabidopsis. Plant Physiol 131: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Saha SK, Inouye M (1998) Two-domain reconstitution of a functional protein histidine kinase. Proc Natl Acad Sci USA 95: 6728–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC (1992) Communication modules in bacterial signaling proteins. Annu Rev Genet 26: 71–112 [DOI] [PubMed] [Google Scholar]
- Raz V, Fluhr R (1992) Calcium requirement for ethylene-dependent response. Plant Cell 4: 1123–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regelmann AG, Lesley JA, Mott C, Stokes L, Waldburger CD (2002) Mutational analysis of the Escherichia coli PhoQ sensor kinase: differences with the Salmonella enterica Serovar Typhimurium PhoQ protein and in the mechanism of Mg2+ and Ca2+ sensing. J Bacteriol 184: 5468–5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson VL, Buckler DR, Stock AM (2000) A tale of two components: a novel kinase and a regulatory switch. Nat Struct Biol 7: 626–633 [DOI] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB (1999) A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283: 996–998 [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Kieber JJ (2002) Ethylene. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book, American Society of Plant Biologists, Rockville, MD, doi 10.1199/tab.0071, http://www.aspb.org/publication/arabidopsis/
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB (1995) The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem 270: 12526–12530 [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terajima Y, Nukui H, Kobayashi A, Fujimoto S, Hase S, Yochioka T, Hashiba T, Saton S (2001) Molecular cloning and characterization of a cDNA for a novel ethylene receptor, NT-ERS1, of tobacco (Nicotiana tabacum L.). Plant Cell Physiol 42: 308–313 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Klee HJ (1999) Differential expression of two novel members of the tomato ethylene receptor family. Plant Physiol 120: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ (2000) The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc Natl Acad Sci USA 97: 5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14 (Suppl): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hall AE, O'Malley R, Bleecker A (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang JS, Zhou HL, Li J, Zhang ZG, Wang DW, Chen SY (2003) Serine/threonine kinase activity in the putative histidine kinase-like ethylene receptor NTHK1 from tobacco. Plant J 33: 385–393 [DOI] [PubMed] [Google Scholar]
- Xie C, Zhang ZG, Zhang JS, He XJ, Cao WH, He SJ, Chen SY (2002) Spatial expression and characterization of a putative ethylene receptor protein NTHK1 in tobacco. Plant Cell Physiol 43: 810–815 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought and salt stress. Plant Cell 14 (Suppl): S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC (1998) Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA 95: 13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JS, Xie C, Liu F, Liu FH, Chen SY (1999) A novel tobacco gene coding for a product similar to bacterial two-component regulators. Chin Sci Bull 44: 1025–1029 [Google Scholar]
- Zhang JS, Xie C, Shen YG, Chen SY (2001. a) A two-component gene (NTHK1) encoding a putative ethylene-receptor homolog is both developmentally and stress regulated in tobacco. Theor Appl Genet 102: 815–824 [Google Scholar]
- Zhang JS, Xie C, Du BX, Wu XL, Chen SY (2001. b) Tobacco two-component gene NTHK2. Chin Sci Bull 46: 574–577 [Google Scholar]
- Zhou D, Kalaitzis P, Mattoo AK, Tucker ML (1996) The mRNA for an ETR1 homologue in tomato is constitutively expressed in vegetative and reproductive tissues. Plant Mol Biol 30: 1331–1338 [DOI] [PubMed] [Google Scholar]