Abstract
Incorporation of a reporter peptide in solutions submitted to fast photochemical oxidation of proteins (FPOP) allows for the correction of adventitious scavengers and enables the normalization and comparison of time-dependent results. Reporters will also be useful in differential experiments to control for the inclusion of a radical-reactive species. This incorporation provides a simple and quick check of radical dosage and allows comparison of FPOP results from day-to-day and lab-to-lab. Use of a reporter peptide in the FPOP workflow requires no additional measurements or spectrometers while building a more quantitative FPOP platform. It requires only measurement of the extent of reporter-peptide modification in a LC/MS/MS run, which is performed by using either data-dependent scanning or an inclusion list.
Keywords: FPOP, Reporter, Adventitious radical scavenger, SOD1, mass spectrometry
Graphical Abstract

Introduction
Interest in elucidating protein higher order structures and the conformational changes connecting them continues to grow for both understanding fundamental biophysics and the development of therapeutic proteins. Protein conformational changes can be induced, for example, by posttranslational modifications (PTMs) [1], mutagenesis [2], oligomerization [3], and binding to metal ions, drugs, or to other proteins [4]. Multiple biophysical approaches, including circular dichroism (CD), fluorescence spectroscopy, and isothermal titration calorimetry, can report that a conformational change has occurred [5], but these approaches afford limited structural resolution. X-ray crystallography and NMR can provide site-specific information, but the turnaround is slow and the required sample quantity is large. The mass spectrometry (MS)-based method of hydroxyl-radical footprinting, established by Chance and co-workers [6] and elaborated as fast photochemical oxidation of proteins (FPOP) by Hambly and Gross [7] does offer fast turnaround, high sensitivity, albeit at medium structural resolution.
FPOP utilizes a pulsed laser to photolyze hydrogen peroxide to generate two hydroxy radicals (•OH) that rapidly modify protein side chains in a flow system. The laser provides a spatially small spot but high light flux, maximizing the exposure of small plugs of protein solution to radicals and ensuring that most of the protein in the flow is irradiated only once [8]. FPOP is easily implemented, requiring only a laser, optics, syringe pump, and flow system [9, 10]. By comparing the extent of modification for two samples (e.g., wild-type vs. mutant or apo vs. holo), conformational differences can be established at the peptide and even amino-acid levels [11, 12].
There is always a need to normalize the yields of oxidative modification. For example, the presence of adventitious •OH scavengers (e.g., DTT, ATP, TFE, lipids, DMSO) in a protein solution may significantly reduce the FPOP-induced modification extent. If the scavenger amounts differ between a sample and control, the outcomes may be difficult to compare. In antigen-epitope mapping where one compares modification extent of an antigen with and without an antibody, one can use a non-interacting (“dummy”) antibody to compensate for the many reactions of •OH with the antibody, but reliable comparisons are needed to compensate for changes in the test vs. control solutions or when a non-interacting protein is not available.
Here we report that the use of a reporter peptide to resolve discrepancies in •OH dosage and “normalize” the results. We used leu-enkephalin as the reporter, where the fraction modified is a “ruler” of radical dosage to the protein samples. This enables an important extension of FPOP, namely the acquisition of FPOP time-dependent data acquired by varying the scavenger concentration to modify the primary •OH lifetime. The extent of modification of the sample vs. that of the reporter provides time-dependent data and helps resolve conformational differences from readouts of peptide/residue responses at each time point.
A kinetics-type approach should provide assurance that the modification reactions are occurring normally and also add statistical weight to the data and confidence to comparisons of higher order protein structure, affording an output that is similar to that of HDX. Another approach that accommodates adventitious scavengers is by Xie and Sharp [13], who proposed an adenine-based dosimeter to monitor and correct the radical dose from sample-to-sample in FPOP experiments.
The reporter-peptide approach should be useful for protein therapeutics where higher order structure must be verified [14]. The ALS-linked dimeric protein Cu,Zn superoxide dismutase 1 (SOD1) is used here as test case for developing a robust method for comparing conformational changes between WT and mutant.
Experimental Section
All experiments were performed using recombinant SOD1 protein expressed and purified from BL21-Gold(DE3) PLysS E. coli cells (Stratagene, Inc., Cedar Creek, TX) as previously described [15]. The free cysteines in WT SOD1 were replaced with the C6A/C111S mutations to avoid inter-molecular disulfide-scrambling; the alanine to valine mutation (A4V), the most common ALS variant in North America, was introduced into this pseudoWT background [16]. All materials were obtained from Sigma Aldrich (St. Louis, MO)— see SI.
For footprinting, protein samples were diluted to 40 μM in PBS buffer containing histidine as scavenger and leu-enkephalin. FPOP was performed as described previously [7] (a detailed experimental approach is given in SI. Two control experiments were performed: (1) with all reagents but without laser irradiation and (2) with the protein in PBS buffer without laser irradiation. Modifications were determined at the protein level by using a MaXis 4G (Bruker, Bremen Germany), and at the peptide/residue levels after trypsin digestion and LC/MS/MS analysis with the UltiMate 3000 Nano LC system (Thermo Fisher Scientific, Waltham, MA) and a Thermo Q Exactive Plus Hybrid Quadruple-Orbitrap Mass spectrometer (Thermo Fisher Scientific, Waltham, MA) operated in data-dependent mode (see SI).
For data analysis, the *.mgf files, converted from the *.raw MS files using MassMatrix Mass Spec Data File Converter, were searched using MASCOT (Matrix Science, London, UK) against a custom-built database containing the sequence of WT and A4V SOD1. All known hydroxyl radical side-chain reaction products [17] with a carbamidomethyl modification (57.0214 Da) to Cys-containing peptides were added as variable modifications. Calculation of the extent of modification was performed as previously reported [10] (see SI).
Results and Discussion
FPOP labels rapidly owing to the high reactivity of •OH [7, 8]. Because the rate constants for amino acids reacting with •OH are known [17], the lifetime of •OH is adjustable by either scavenging with different amino acids or with a single amino acid at different concentrations (see Figure S1 for an illustrative numerical simulation of •OH lifetime as a function of time). The FPOP reaction time was varied by using histidine as radical scavenger for the OH primary radical at 0, 0.2, 2.0, 40 mM. We chose histidine because it is a simple amino acid, bio-compatible, soluble, and reactive with radicals. To normalize the results, we introduced leu-enkephalin as a reporter peptide prior to irradiation.
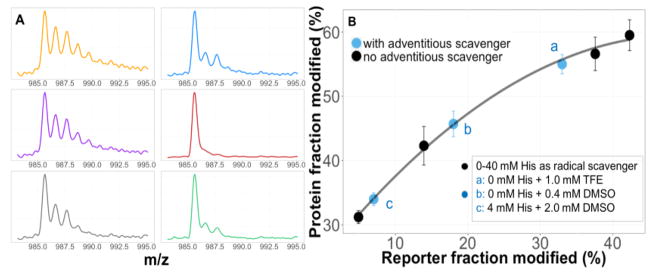
Following FPOP, we analyzed the undigested protein by LC/MS, integrating TI signals over an elution time of 0.5 min. The unmodified protein is the most abundant followed by a singly oxidized species (+15.9949 Da), a doubly oxidized species (+31.9898 Da), etc. (Figure 1A) according to the fraction modified [18]. The kinetics of oxidative modification at the protein level are relative to the oxidative modification of reporter leu-enkephalin (M + 15.9949), determined from the extracted ion chromatograms (EICs) of both unmodified and modified leu-enkephalin (mass tolerance of 15 ppm; see Figure S2). As the •OH lifetime becomes longer, the extent of modification of both the reporter leu-enkephalin and the SOD1 protein increase. Plotting the protein-fraction modified versus the reporter-fraction modified generates a curve whose x-axis reflects the varying •OH lifetime and y-axis tracks the protein response as labeling time changes (Figure 1B).
Figure 1.
(A) Mass spectra from intact measurements of FPOP-labeled WT SOD1 (+16 charge) obtained by adjusting [His]. (B) Intact protein fraction modified vs. reporter fraction modified under various scavenging conditions. The resulting curve is independent of the presence of FPOP adventitious scavengers, it demonstrates that incorporation of reporter peptide could normalize (or correct for) the FPOP outcome, making possible unbiased comparison between one state of protein and another (e.g., apo and holo).
Correction for adventitious scavengers
Protein samples in FPOP are studied in buffer solutions with near-physiological pH, permitting FPOP to be used under reasonably biologically relevant conditions. The lifetime of •OH is tunable and should predominantly be controlled by radical scavenger concentrations [19]. Sometimes, one is unaware of the presence of adventitious •OH scavengers (e.g., DTT, ATP, TFE, lipids, DMSO) that unpredictably scavenge the radical dosage and bias comparisons of the modification extents of the two protein states. Incorporating the reporter peptide into FPOP workflow allows for correction. Although FPOP experiments in the presence of adventitious radical scavengers would show reduced protein modification, the corresponding decrease of reporter-fraction modified maintains the protein-reporter outcome on the curve, correcting for differential scavenging (Figure 1B).
FPOP Kinetics at the peptide/residue levels
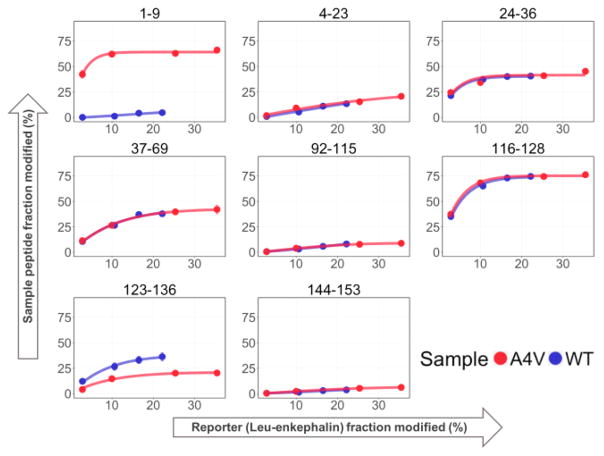
To demonstrate time-dependent measurements, we plotted the fraction of leu-enkephalin modified at each histidine concentration vs. the corresponding fraction modified for each peptide of the protein (here SOD1) on the y-axis. The reporter-fraction modified reports •OH lifetime (i.e., longer times give more modification), and the plot shows the time-dependent SOD1 modification, affording a similar output to that of HDX kinetics data for peptides. The response of each peptide of SOD1 as a function of reporter-fraction modified allow us to distinguish those reactive or solvent-accessible regions from the more protected regions (Figure 2). For example, peptides 24–36, 37–69, and 116–128 exhibit greater fractions modified for both WT and A4V compared to most other peptides, indicating that those regions are more solvent-exposed as expected for these loop regions of SOD1. The curves for peptide regions of WT and A4V SOD1 should overlap if the solvent accessibilities of these peptides are the same for WT and mutant. The nonoverlapping curves for peptides 1–9 and 123–136 (Figure 2) indicate a conformational change in these regions upon mutation, likely due to destabilization of the monomer-dimer equilibrium for the A4V variant [16].
Figure 2.
Response curves of each peptide from WT and A4V SOD1 in an FPOP kinetics experiment performed by adjusting different histidine concentrations. The fraction modified for peptides from WT and A4V SOD1 were normalized by the reporter fraction modified under each scavenging condition.
Although a time dependence is achieved by this simple approach, the increasing yield for the reporter does not have a simple relationship to time (the relationship between yield and time is derived in SI). Nevertheless, the approach is simple and effective for identifying protein regions that change upon perturbation.
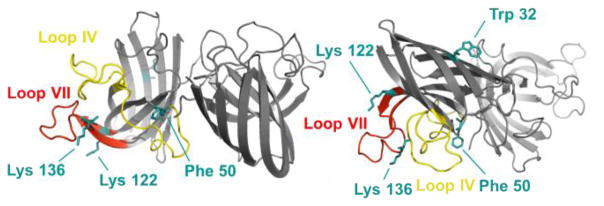
Residue-level analysis can also be normalized by using the reporter peptide. We identified and quantified with high confidence eleven residues that undergo oxidative modification (see Figure S3 and accompanying discussion). Referring to the X-ray crystallography structure (1N18), we see that most peptides/residues that exhibit higher modification extents (F50 in peptide 37–69, K122 in peptide 116–128 and K136 in peptide 123–136) are located on loop IV (Zn-binding loop—residue 49–83) and loop VII (electrostatic loop—residues 121–142) (see Figure 3) [20]. A detailed interpretation of the results will be part of a more extensive biochemical study to be published in the future.
Figure 3.
Crystal structure of thermostable pseudo WT SOD1, containing the C6A/C111S mutations. Loop IV, i.e., the Zinc binding loop (residue 49–83, yellow) and loop VII, i.e., the electrostatic loop (residue 121–142, red) are shown. These two loops exhibited a higher extent of modification by FPOP. Residues that are significantly modified by FROP are labeled (cyan).
In conclusion, an advantage of this simple approach is it allows quick location of conformational changes when comparing two samples, especially when adventitious •OH scavengers are present. Although we used leu-enkephalin, many peptides are candidate reporters. Ideally, they should be (1) mildly reactive towards •OH, so as not to affect the protein radical dosage while remaining sensitive to the •OH lifetime, (2) minimally structured so that its fraction modified won’t be confounded by structural changes; (3) soluble but not too hydrophilic to avoid eluting with the solvent front; (4) non-interacting with the proteins under study, and (5) readily available. A short peptide with one FPOP-reactive residue may be preferred to simplify the quantitation of reporter modifications and the normalization of the extent of modification.
The reporter-peptide approach is the first step to realize our vision to obtain quantitative measures of residue and peptide reactivity, analogous to “protection factors” in HDX. This will require calibrating a reporter peptide’s rate constant against a standard reaction. Chance has identified a similar goal for synchrotron footprinting [21]. This would facilitate cross-laboratory comparisons of FPOP results and enable its use to determine coarse-grained HOS of proteins.
Supplementary Material
Acknowledgments
This research was supported by National Institute of General Medical Sciences (8P41GM103422 and 1S10D0162985-SIG) of the National Institutes of Health and by Genentech to M.L.G. and R01-GM54836 to C.R.M.
References
- 1.Karve TM, Cheema AK. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. Journal of amino acids. 2011;2011:207691. doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furukawa Y, O’Halloran TV. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. The Journal of biological chemistry. 2005;280:17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 3.Kayatekin C, Zitzewitz JA, Matthews CR. Disulfide-reduced ALS variants of Cu, Zn superoxide dismutase exhibit increased populations of unfolded species. Journal of molecular biology. 2010;398:320–331. doi: 10.1016/j.jmb.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 5.Lundblad RL. Chapman and Hall/CRC; Boca Raton, FL: 2009. [Google Scholar]
- 6.Maleknia SD, Brenowitz M, Chance MR. Millisecond radiolytic modification of peptides by synchrotron X-rays identified by mass spectrometry. Analytical chemistry. 1999;71:3965–3973. doi: 10.1021/ac990500e. [DOI] [PubMed] [Google Scholar]
- 7.Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Gau BC, Sharp JS, Rempel DL, Gross ML. Fast photochemical oxidation of protein footprints faster than protein unfolding. Analytical chemistry. 2009;81:6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Gau BC, Jones LM, Vidavsky I, Gross ML. Fast photochemical oxidation of proteins for comparing structures of protein-ligand complexes: the calmodulin-peptide model system. Analytical chemistry. 2011;83:311–318. doi: 10.1021/ac102426d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones LM, JBS, JAC, Gross ML. Fast photochemical oxidation of proteins for epitope mapping. Analytical chemistry. 2011;83:7657–7661. doi: 10.1021/ac2007366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gau B, Garai K, Frieden C, Gross ML. Mass spectrometry-based protein footprinting characterizes the structures of oligomeric apolipoprotein E2, E3, and E4. Biochemistry. 2011;50:8117–8126. doi: 10.1021/bi200911c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Rempel DL, Gau BC, Gross ML. Fast photochemical oxidation of proteins and mass spectrometry follow submillisecond protein folding at the amino-acid level. Journal of the American Chemical Society. 2012;134:18724–18731. doi: 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie B, Sharp JS. Hydroxyl Radical Dosimetry for High Flux Hydroxyl Radical Protein Footprinting Applications Using a Simple Optical Detection Method. Analytical chemistry. 2015;87:10719–10723. doi: 10.1021/acs.analchem.5b02865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan Y, Chen G, Wei H, Huang RY, Mo J, Rempel DL, Tymiak AA, Gross ML. Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. Journal of the American Society for Mass Spectrometry. 2014;25:2084–2092. doi: 10.1007/s13361-014-0993-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svensson AK, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. Mapping the folding free energy surface for metal-free human Cu,Zn superoxide dismutase. Journal of molecular biology. 2006;364:1084–1102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Svensson AK, Bilsel O, Kayatekin C, Adefusika JA, Zitzewitz JA, Matthews CR. Metal-free ALS variants of dimeric human Cu,Zn-superoxide dismutase have enhanced populations of monomeric species. PLoS One. 2010;5:e10064. doi: 10.1371/journal.pone.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chemical reviews. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Rempel DL, Zhang H, Gross ML. An improved fast photochemical oxidation of proteins (FPOP) platform for protein therapeutics. Journal of the American Society for Mass Spectrometry. 2015;26:526–529. doi: 10.1007/s13361-014-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu B, Zhang H, Giblin D, Rempel DL, Gross ML. Dosimetry determines the initial OH radical concentration in fast photochemical oxidation of proteins (FPOP) Journal of the American Society for Mass Spectrometry. 2015;26:843–846. doi: 10.1007/s13361-015-1087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das A, Plotkin SS. SOD1 exhibits allosteric frustration to facilitate metal binding affinity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3871–3876. doi: 10.1073/pnas.1216597110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, Ravikumar KM, Chance MR, Yang S. Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: A protection factor analysis. Biophysical Journal. 2015;108:107–115. doi: 10.1016/j.bpj.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.