Abstract
Non-alcoholic fatty liver disease (NAFLD) is an epidemic metabolic condition driven by an underlying lipid homeostasis disorder. The lipid droplet (LD), the main organelle involved in neutral lipid storage and hydrolysis, is a potential target for NAFLD therapeutic treatment. In this review, we summarize recent progress elucidating the connections between LD-associated proteins and NAFLD found by genome-wide association studies (GWAS), genomic and proteomic studies. Finally, we discuss a possible mechanism by which the protein 17β-hydroxysteroid dehydrogenase 13 (17β-HSD13) may promote the development of NAFLD.
Keywords: non-alcoholic fatty liver disease, lipid droplets, genome-wide association study, proteomics, PNPLA3, 17β-HSD13
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is characterized by a pathological accumulation of triacylglycerol (TAG) in hepatocytes (i.e. hepatic steatosis) without excessive alcohol consumption (Cohen et al., 2011; Hardy et al., 2016). Although the prevalence of NAFLD varies among studies due to the different sensitivities of the instruments used and the ethnic makeup of the populations studied, there is no doubt that the incidence of NAFLD is increasing to epidemic proportions, especially in developing countries. As the incidence of viral infections such as Hepatitis B virus (HBV) and Hepatitis C virus (HCV) is decreasing, NAFLD is becoming the most prevalent liver disease (Wang et al., 2014; Younossi et al., 2015; Rinella and Charlton, 2016). NAFLD encompasses a spectrum of diseases ranging from simple hepatic steatosis, to non-alcoholic steatohepatitis (NASH), to cirrhosis. Severe forms of NAFLD increase the risk of other liver diseases and a portion of patients will ultimately develop hepatocellular carcinoma (HCC) (Cohen et al., 2011; Michelotti et al., 2013). Even though only a small fraction of HCC patients will progress to late stage disease, the increasing prevalence of NAFLD and its sequelae commands attention. In addition to diseases of the liver, NAFLD increases mortality through cardiovascular disease, placing a great burden on public health care systems (Targher et al., 2010; Anstee et al., 2013; Gaggini et al., 2013).
NAFLD is regarded as a manifestation of metabolic syndrome which is influenced by multiple factors (Cohen et al., 2011; Hardy et al., 2016). Genetics play a vital role in the development and progression of the disease, a conclusion which is supported by family cohort studies and ethnic based variations in its incidence and presentation (Anstee and Day, 2013). Increased food intake and a sedentary life style strongly contribute to the establishment of the disease (Anstee et al., 2013). Other metabolic conditions like obesity, insulin resistance, hypertension, and dyslipidemia are also risk factors associated with NAFLD (Smith and Adams, 2011). There are no medications specifically intended to treat NAFLD, but drugs for related conditions including insulin sensitizing agents, antioxidants and lipid-lowering agents are often used in its treatment (Musso et al., 2012; Dowman et al., 2011). It is still unknown why a fraction of NAFLD patients develop NASH or cirrhosis. Therefore, it is vital to uncover the pathogenic mechanisms driving the development of this disease and its sequelae.
Hydrophobic neutral lipids (primarily TAG and cholesteryl ester (CE) in eukaryotic organisms) are mainly stored in an organelle termed the lipid droplet (LD) (Murphy, 2001). The LD consists of a neutral lipid core covered by a monolayer of phospholipids and associated proteins (Tauchi-Sato et al., 2002). Rather than inert energy reservoirs, LDs are actively associated with other organelles and play vital roles in lipid metabolism, membrane trafficking and signal transduction (Martin and Parton, 2006; Zehmer et al., 2009). Dysfunction in this organelle can result in disorders of lipid metabolism, which makes the LD a promising target for research into the pathogenic mechanisms of NAFLD (Greenberg et al., 2011; Krahmer and Farese, 2013; Carr and Ahima, 2016).
The protein complement on LDs which drive the organelle’s functions also likely underlie the metabolic pathology that leads to NAFLD. The Perilipin family (Plin1–5) are the major LD proteins in mammalian cells (Kimmel et al., 2010). Plin1 is hardly detected in normal liver LDs, but its expression is prominently up-regulated in fatty liver LDs (Straub et al., 2008; Fujii et al., 2009). Similarly, Plin2 expression is increased in NAFLD, both in human and rodents, and is associated with oxidative damage (Fujii et al., 2009). Knocking out Plin5 in mice results in reduced hepatic lipid content due to elevated lipolysis and fatty acid oxidation, but also induces lipotoxic injury (Wang et al., 2015). The cell death-inducing DFFA-like effector (CIDE) proteins are located on LDs and the endoplasmic reticulum (ER) and are also involved in fatty liver progression. Cidea and Fsp27 (Cidec) mRNA levels are significantly increased in fatty liver dystrophic mice (Hall et al., 2010). The CIDEB protein is predominately expressed in the liver. Cideb-null mice are resistant to diet-induced obesity and show decreased lipogenesis and increased fatty acid oxidation (Li et al., 2007).
Lipolysis-related proteins, like CGI-58, might also be involved in the pathogenesis of fatty liver. The liver specific ablation of CGI-58 causes NASH and fibrosis (Guo et al., 2013), which is consistent with the clinical presentation of the CGI-58 mutation induced Chanarin-Dorfman syndrome (Srinivasan et al., 2004; Ronchetti et al., 2008). Hypoxia-inducible gene 2 (Hig2), which is primarily localized on LDs, impairs TAG hydrolysis in liver, thereby promoting hepatic lipid accumulation (DiStefano et al., 2015). The loss of LDs and retinoid content in hepatic stellate cell (HSC) is a hallmark of NASH. Blockage of lipolysis by lipases like ATGL in HSC may prevent or alleviate the development of fibrosis (Blaner et al., 2009).
In this review, we focus on LD proteins that may be involved in the pathogenesis of NAFLD as determined by Genome-wide Association Studies (GWAS), genomic and proteomic research. As the mechanisms are still unknown, we also propose a hypothesis based on recent studies.
GWAS reveal pathogenic proteins on lipid droplets like PNPLA3
With the completion of the Human Genome Project, scientists focused on variable regions in the genome, especially single nucleotide polymorphisms (SNPs) associated with diseases. Under the HapMap project, haplotype maps were established using tag SNPs. Disease associations with novel genes can be established by GWAS using a non-candidate driven method (Hardy and Singleton, 2009; Manolio, 2010). GWAS is especially useful in parsing complicated traits not caused by a single genetic mutation but by complex contributions from environmental factors and polygenic components, which describes NAFLD (Anstee and Day, 2013). GWAS studies focused on NAFLD were summarized by Anstee and Day (2013), Sookoian and Pirola (2015). The SNPs involved in NAFLD include PNPLA3 (rs738409), GCKR (rs780094), NCAN (rs2228603), LYPLAL1 (rs12137855), PPP1R3B (rs4240624), CPN1-ERLIN1-CHUK gene cluster, as well as others. These gene polymorphisms may influence NAFLD via hepatic carbohydrate and lipid metabolism, protein modification or signaling pathways (Anstee and Day, 2013; Sookoian and Pirola, 2015).
Patatin-like phospholipid domain containing protein 3 (PNPLA3) is a LD-associated protein that is also distributed on other membranes (He et al., 2010). A GWAS performed as part of the Dallas Heart Study was the first to find an association between PNPLA3 and NAFLD (Romeo et al., 2008). PNPLA3 has the strongest linkage disequilibrium of all hits and has consistently been identified in multiple GWAS using different diagnostic criteria (Anstee and Day, 2013). Moreover, it has been demonstrated by numerous independent candidate-driven and histology-based studies that the I148M polymorphism is associated with and increased risk of NASH, fibrosis, and NAFLD-related HCC (Kotronen et al., 2009; Sookoian et al., 2009; Rotman et al., 2010; Dongiovanni et al., 2013). PNPLA3-I148M may promote liver injury as detected by elevated serum ALT levels (Kollerits et al., 2010). PNPLA3-I148M is proportionally overrepresented in the Hispanic population (0.49 in Hispanics, 0.23 in European-Americans and 0.17 in African-Americans), which may partially explain the higher susceptibility Hispanic people to NAFLD (Romeo et al., 2008).
The PNPLA3 gene is located on chromosome 22 and encodes a 53-kDa protein with 481 amino acids. PNPLA3 in humans is predominantly expressed in the liver, with substantially lower expression level in skin and adipose tissue (Huang et al., 2010). PNPLA3 belongs to the patatin-like phospholipid domain containing (PNPLA) protein family. It is most closely related to PNPLA2, which is also known as adipose triglyceride lipase (ATGL), the major cellular TAG lipase. PNPLA3 has TAG hydrolase activity which is markedly reduced in the I148M variant. Structural analysis of PNPLA3 has demonstrated that the mutation does not affect the catalytic center but rather the groove of the substrate binding domain. Therefore the I148M substitution possibly blocks access of substrate to the catalytic site (Huang et al., 2011).
Much work has been conducted on the physiological and pathological functions of PNPLA3 and the I148M variant. However, the mechanism underlying its association with disease remains enigmatic. PNPLA3 expression is very low in a fasted state, and is strongly induced with feeding. The expression of PNPLA3 is up-regulated by insulin through LXR/RXR signaling to SREBP-1c. This synthesis of fatty acids, which is stimulated by SREBP-1c, increases the half-life of PNPLA3 (Huang et al., 2010). These results indicate that PNPLA3 is regulated by both lipid and carbohydrate metabolism in response to the nutrition environment (Huang et al., 2010). However, the disease associated genetic variants of PNPLA3 are not linked with risk factors such as insulin sensitivity or body mass index (BMI) and do not affect related metabolic syndromes such as dyslipidemia or type 2 diabetes (Romeo et al., 2008; Speliotes et al., 2010). Besides TAG lipase activity, PNPLA3 is also reported as possessing lysophosphatic acid acyltransferase (LPAAT) activity in vitro and retinyl-palmitate lipase activity in human HSCs (Kumari et al., 2012; Pirazzi et al., 2014). Since the development of NASH is accompanied by a loss of retinoid content in HSCs, PNPLA3 may play a role in the hepatic inflammatory process.
Although TAG hydrolysis activity of PNPLA3-I148M is decreased, the association between the genetic variant and NAFLD seems not to be due to the loss of function. The Pnpla3 deletion in mice does not affect liver TAG content even under high-sucrose or high-fat diet conditions (Chen et al., 2010; Basantani et al., 2011) and human PNPLA3 overexpression in mice does not decrease liver steatosis (Li et al., 2012). On the contrary, overexpression of PNPLA3-I148M in mice leads to liver steatosis with elevated lipogenesis and impaired TAG hydrolysis (Li et al., 2012). TAG content and LD size are increased in high-sucrose diet fed Pnpla3-I148M knock-in mice without a significant change of lipogenic genes. However, CGI-58, which facilitates ATGL in hydrolyzing TAG, is increased dramatically on LDs (Smagris et al., 2015). This result suggests that PNPLA3 may alter lipolysis, not by the hydrolysis activity itself, but by inhibition of other lipases. Moreover, PNPLA3-I148M affects VLDL secretion in rat hepatoma McA-RH7777 cells, possibly due to a decreased ability to mobilize TAG in LDs (Pirazzi et al., 2012). PNPLA3-I148M may also enhance TAG synthesis by elevating LPAAT activity (Kumari et al., 2012). A disadvantage of the mouse model in the study of the I148M variant function is the different tissue distribution, compared with humans. PNPLA3 is primarily expressed in liver in humans, while it is mainly expressed in adipose tissue in the mouse (Huang et al., 2010; Hoekstra et al., 2010). Further studies will be required to determine how PNPLA3 is associated with NASH and fibrosis.
There are 21 other SNPs in PNPLA3 identified in the Dallas study which are potentially linked to NAFLD. Among them is PNPLA3-S47A, in which the catalytic serine is substituted by alanine, which results in decreased lipolytic activity and increased hepatic lipid content (He et al., 2010; Huang et al., 2011; Smagris et al., 2015). Variant S543I is more common in African-Americans than in European-Americans or Hispanics and is associated with decreased lipid content in the liver (Romeo et al., 2008). The 434K allele attenuates PNPLA3 protein expression by decreasing PNPLA3 mRNA levels and also attenuates the association of I148M with liver damage (Donati et al., 2016).
Proteomic studies on isolated lipid droplets reveal 17β-HSD13 is involved in THE pathogenesis of NAFLD
Metabolic syndromes like NAFLD are affected not only by genomic alterations but also by complicated factors like environment. As a result, genomic analysis alone is insufficient as it is removed from the actual expression of proteins (Gregorich and Ge, 2014). Chronic diseases always manifest as continuous processes with evolving stages. There are often biomarkers which shift with the development of the disease and identification of diagnostic biomarkers is especially important for detecting early stage disease. Proteomics can be used to identify alterations in protein expression and post translational modifications characteristic of diseases during different stages of their development at the level of tissue, cell, organelle or other subcellular structures. In addition, proteomic analyses of disease states can also identify novel drug targets for drug development (Hanash, 2003). Proteomics can also help reveal protein complexes and signaling networks (Gregorich and Ge, 2014). Tremendous improvements in the sensitivity and throughput of modern mass spectrum technologies have led to a dramatic expansion in protein research in the past decade. Many studies of NAFLD have been conducted at various disease stages, revealing 34 candidate biomarkers in proteomic studies of liver and serum (Lim et al., 2014; Ladaru et al., 2016).
Dozens of proteomic analyses have been performed on isolated LDs in many types of cells and tissues from various organisms (Yang et al., 2012), including liver tissue and hepatocytes from humans and rodents (Yang et al., 2012; Crunk et al., 2013; Su et al., 2014; Khan et al., 2015). Structural proteins, like those of the perilipin family, are prominent in most LD proteomes. Other LD proteins can be categorized into groups including proteins of lipid synthesis and hydrolysis, membrane trafficking, and cell signaling. The presence of these functional classes of proteins on LDs speaks to their central role in lipid metabolism. Apolipoproteins are present on liver LDs suggesting an association between LDs and lipid secretion. Moreover, large numbers of mitochondrial and ER proteins are also found in the liver LD proteome. This suggests the existence of close physical and functional interactions among these organelles, likely involving fatty acid oxidation and steroid metabolism (Crunk et al., 2013; Su et al., 2014; Khan et al., 2015).
Comparative LD proteomic studies reveal that 54 proteins are up-regulated and 35 proteins down-regulated in human livers with simple hepatic steatosis. The up-regulated proteins are involved in the metabolism of retinol, linoleic acid, xenobiotics and drugs (Su et al., 2014). The liver LD proteome of mice with diet-induced hepatic steatosis has also quantitatively analyzed using iTRAQ. As with human liver, the up-regulated proteins included those involved in fatty acid catabolism and xenobiotic metabolism, and also included some ribosomal and ER proteins. Among the down-regulated proteins were the liver X receptor, retinoid X receptor, and proteins involved in glucose metabolism. (Khan et al., 2015). These results provide new insights into the metabolic pathways of hepatic steatosis and may point to possible drug targets for the treatment of NAFLD.
Of the proteins of potential clinical interest identified in proteomic studies, 17β-hydroxysteroid dehydrogenase 13 (17β-HSD13) is possibly the most important. A comparative, quantitative proteomic study in humans detected a dramatic elevation of 17β-HSD13 in patients with NAFLD, compared with healthy controls (Su et al., 2014). Another independent study confirmed this result and also found a slight upregulation of 17β-HSD13 in patients with NASH without fatty liver (Kampf et al., 2014). In a study of fasted and refed mice, 17β-HSD13 was increased markedly on hepatic LDs of mice in the high-fat diet group compared with the low fat group (Crunk et al., 2013).
In parallel, a GWAS study revealed that an intergenic SNP (rs6834314) near HSD17B13 (encoding 17β-HSD13) and MAPK10 (encoding mitogen-activated protein kinase 10, MAPK10) is strongly associated with the concentration of plasma alanine transaminase (ALT) (P = 3.1 × 10−9), the main marker of hepatocellular injury (possibly representing fatty liver disease) (Chambers et al., 2011). Overexpression of 17β-HSD13 in a mouse hepatocyte cell line induced liver steatosis and lipid accumulation. It also lead to increased expression of proteins involved in lipid synthesis such as mature SREBP-1 and FAS, suggesting that 17β-HSD13 is implicated in NAFLD development by promoting lipogenesis (Su et al., 2014). 17β-HSD13 is expressed primarily in the liver, with far less found in the gastrointestinal tract, muscle, spleen and uterus, making 17β-HSD13 an excellent potential therapeutic target for treating fatty liver disease (Horiguchi et al., 2008).
17β-HSD13 was first cloned from a human liver cDNA library in 2007 and was named short-chain dehydrogenase/reductase 9 (SCDR9) (Liu et al., 2007). Now the protein has been grouped with the 17β-hydroxysteroid dehydrogenase (17β-HSD) family, which plays a key role in the final step of estrogen and androgen steroid metabolism. As with the other members of 17β-HSD family (except 17β-HSD5), 17β-HSD13 contains a NAD(P)+/NAD(P)H binding domain (TGxxxGxG) and an enzymatic activation site (YxxxK) at the N-terminus. The 17β-HSDs are regarded as potential therapeutic targets for diseases such as breast cancer, endometriosis, osteoporosis, prostate cancer and even Alzheimer’s disease. All 17β-HSDs can modify the keto and hydroxy groups of steroids at the position C17 in vitro. However, the real physiological function of 17β-HSD13 in vivo is still unknown (Moeller and Adamski, 2006; Marchais-Oberwinkler et al., 2011).
17β-HSD13 is located at the same locus with 17β-HSD11 on chromosome 4q22. Moreover, 17β-HSD13 shares a high sequence similarity (65% identity and 78% similarity) with 17β-HSD11 (Liu et al., 2007). The 1–28 amino acid region of the N-terminus of 17β-HSD11 is necessary and sufficient to target the protein to ER and LD, while similar targeting activity is located in the first 35 amino acids of the N-terminus of 17β-HSD13 (Horiguchi et al., 2008). These results indicate that these two proteins may share features of their function and regulation.
However, the mRNA expression of 17β-HSD11 is strongly induced in liver and intestine by PPARα agonist Wy14643, while 17β-HSD13 is not induced. On the contrary, the 17β-HSD13 expression level is significantly elevated in PPARα knockout mice, which suggests that PPARα may suppress the expression of 17β-HSD13 (Horiguchi et al., 2008). 17β-HSD13 has two putative C/EBP binding sites while 17β-HSD11 has four. Expression of both proteins was induced by overexpression of C/EBPβ in HepG2 cells while only 17β-HSD11 expression was induced by overexpression of C/EBPα (Rotinen et al., 2010). These results suggest 17β-HSD13 and 17β-HSD11 may play different roles under physiological and pathological conditions in liver.
A role for 17β-HSD13 on lipid droplets in the pathogensis of NAFLD?
Sex hormones play important roles in maintaining energy homeostasis and imbalances in their levels can lead to metabolic syndromes like type 2 diabetes (Varlamov et al., 2014). Epidemiological studies have revealed a higher prevalence of NAFLD in men than in women. However, the incidence of NAFLD increases rapidly in postmenopausal women, ultimately erasing the gender difference. This implicates estrogen in regulating hepatic lipid metabolism (Hashimoto and Tokushige, 2011; Brady, 2015; Shen and Shi, 2015). The metabolic protective effect of estrogen in preventing liver steatosis has also been detected in animal models. Female mice treated with ovariectomy (OVX), and a resulting depletion of the majority of circulating estrogens, have increased fat accumulation in the liver than the pair-fed sham operation mice (Shen and Shi, 2015).
Tamoxifen is an estrogen inhibitor that is widely used in the treatment of hormone-sensitive breast cancer. Approximately 43% of the patients treated with this drug develop hepatic steatosis, likely due, primarily, to increased de novo fatty acid synthesis in the liver (Cole et al., 2010). All subtypes of estrogen receptors are expressed in hepatocytes and estrogen receptor-α predominantates (Shen and Shi, 2015). Treatment with the estrogen receptor-α agonist propyl pyrazole triol (PPT) decreases fat content in the liver. Estrogen receptor-α knockout mice display liver steatosis with lipid biosynthesis genes, including SREBP-1c, up-regulated and the lipid transport genes downregulated (Shen and Shi, 2015; Barros and Gustafsson, 2011).
Unlike what has been seen with estrogens, studies examining androgen signaling have found complex effects with inconsistent results and significant variation between genders (Ma et al., 2014). An inverse relationship between circulating testosterone levels and hepatic steatosis has been found in men (Volzke et al., 2010; Kim et al., 2012). However, other studies found that anabolic-androgenic steroids induce hepatotoxicity and result in NAFLD (Schwingel et al., 2011; Awai et al., 2014). These conflicting results may arise from the steroid dose (ranging from physiological to supra-physiological levels) or the type or ratio of androgens studied.
A low plasma level of dehydroepiandrosterone sulfate (DHEA-S), a kind of proandrogen, is associated with advanced NAFLD (Charlton et al., 2008) Dehydroepiandrosterone (DHEA) supplementation of a high-fat plus fructose diet reduced the induction of SREBP-1c and insulin resistance in mice, and protected against steatosis (Aragno et al., 2009). In females, an abnormally high level of androgens is associated with polycystic ovary syndrome (PCOS) with a high risk of NAFLD (Brzozowska et al., 2009; Kelley et al., 2014). Male, but not female, androgen receptor (AR) knockout mice fed a high-fat diet develop hepatic steatosis and insulin resistance with an increased expression of lipogenic genes and a decreased expression of fatty acid oxidation genes (Lin et al., 2008). These results indicate that androgen deficiency is associated with hepatic lipid deposition and the AR may have greater impact on lipid homeostasis in males than in females.
A significant fraction of estrogens and androgens are synthesized locally inside of target peripheral tissue cells, a process described as intracrine regulation. Almost all sex steroids in postmenopausal women and half of androgens in adult men are synthesized in peripheral intracrine tissues (Labrie et al., 2000). Tumor cells adapt the intracrine system to produce high level of estrogens or androgens to stimulate cell proliferation in sex steroid sensitive diseases like breast and prostate cancer (McNamara and Sasano, 2015; Mostaghel, 2013).
Circulating DHEA and DHEA-S originating from the adrenal gland are the major substrates for intracrine sex hormone synthesis and the conversion involves enzymes belonging to 3β-HSD, 17β-HSD, 5α-reductase and aromatase families (Labrie, 2015). Members of the 17β-HSD family are the key enzymes required for synthesizing all active estrogens and androgens. The 17β-HSD proteins are distributed broadly in most tissues where they play a vital role in intracrine sex steroid homeostais (Luu-The and Labrie, 2010). Hepatic progenitor cells in humans and hepatocytes in rats have the ability to activate TGF-β signaling by intracrine signaling, which supports that liver is an intracrine organ (Gressner et al., 2008; Ding et al., 2013). Since more than half of all known types of 17β-HSDs have been found in the liver (Moeller and Adamski, 2009), it is reasonable to speculate that the liver can also generate intracrine sex steroid signaling.
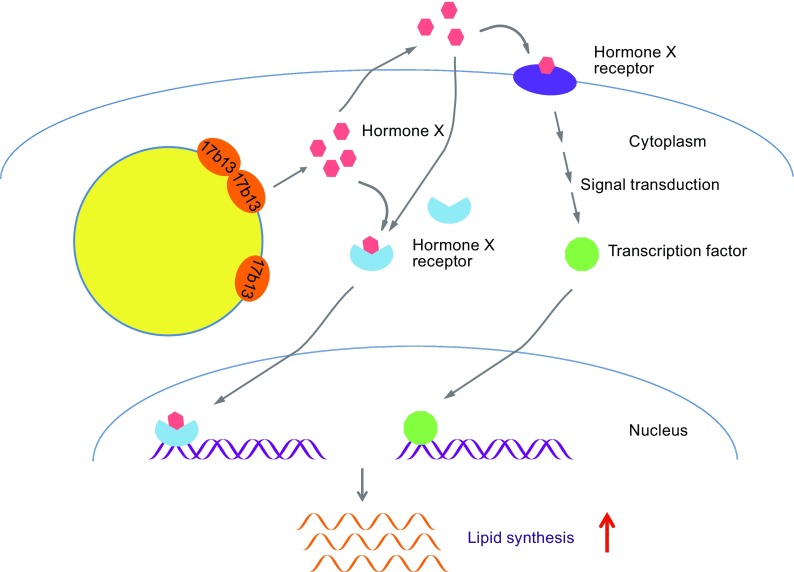
We propose a hypothesis to explain how increased 17β-HSD13 promotes lipid synthesis and expands LDs in hepatocytes (Fig. 1). Excessive expression of 17β-HSD13 proteins results in the production of increased sex steroid hormone intermediates (or possible other, unknown hormones). The locally produced hormones drive intracrine signaling and may also be secreted to the extracellular space signaling to local tissues in a paracrine fashion. This enhanced signaling drives the expression of lipid synthetic genes, driving fat accumulation in LDs.
Figure 1.
Hypothetical mechanism of 17β-HSD13-mediated lipid accumulations. Increased 17β-HSD13 on LDs produces hormone X that binds to a cytosolic receptor by intracrine regulation. The hormone X may also be secreted to extracellular space and bind to a membrane-bound receptor or crosses the plasma membrane and binds to a cytosolic receptor by autocrine or paracrine regulation. The cytosolic ligand-bound receptors thus enter the nucleus and bind to regulate elements. The activated membrane-bound receptors may transduce signals to promote transcription factors enter nucleus. Hence, the expression of genes related to lipid synthesis are eventually up-regulated leading to lipid accumulation and LD expansion in hepatic cells
The mechanisms involved may be more complicated. 17β-HSD enzymes interconvert estrogens in both directions. Therefore, 17β-HSD13 might convert E2 to E1, thereby reducing the E2 concentration in the cellular microenvironment, resulting in a decrease in metabolic protection afforded by E2 and activating lipid anabolic genes. Besides steroid metabolism, some subtypes of 17β-HSDs also play roles in fatty acid elongation and β-oxidation, synthesis of prostaglandin and retinol metabolism. These other activities of 17β-HSD13 may influence fatty liver development via distinct metabolic pathways. In addition, 17β-HSD13 is also present as a major protein on hepatic LDs. Thus it may also act as a structural protein, protecting lipids from hydrolysis by preventing lipases from gaining access to the interior lipids. The mechanisms regulating 17β-HSD13 and its involvement in the progression of NAFLD still need to be elucidated.
CONCLUSION
The LD is the central organelle regulating lipid homeostasis and disorders of LD-associated proteins induce metabolic diseases like NAFLD. GWAS studies reveal that SNPs linked with PNPLA3 are strongly associated with NAFLD, especially the PNPLA3-I148M variant. PNPLA3-I148M seems to be a loss-of-function mutation, but its pathogenic effect appears to be mediated through a gain in function resulting in a suppression of lipase activity. The 17β-HSD13 enzyme was identified by a comparative proteomic study of patients with normal and simple hepatic steatosis liver. Increased 17β-HSD13 on LDs greatly elevates lipid content in both hepatocytes and mouse liver. Since 17β-HSD family proteins catalyze the key step in sex hormone synthesis, we speculate that 17β-HSD13 may promote lipid synthesis by producing hormones which regulate lipogenic gene expression. However, the detailed underlying mechanisms by which LDs and LD-associated proteins induce NAFLD remain to be elucidated.
ACKNOWLEDGEMENTS
The authors thank Dr. John Zehmer for his critical reading and useful suggestions. This work was supported by the National Natural Science Foundation of China (Grant No. 31100854 and U1402225), the Ministry of Science and Technology of China (Grant No. 2016YFA0500100), the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (CIT&TCD201504086).
ABBREVIATIONS
17β-HSD13, 17β-hydroxysteroid dehydrogenase 13; AR, androgen receptor; BMI, body mass index; CE, cholesteryl ester; DHEA, Dehydroepiandrosterone; GWAS, genome-wide association studies; HBV, Hepatitis B virus; HCC, hepatocellular carcinoma; HCV, Hepatitis C virus; LD, lipid droplet; LPAAT, lysophosphatic acid acyltransferase; NAFLD, non-alcoholic fatty liver disease; NASH. non-alcoholic steatohepatitis; OVX, ovariectomy; PCOS, polycystic ovary syndrome; PNPLA, Patatin-like phospholipid domain containing; PPT, propyl pyrazole triol; SCDR9, short-chain dehydrogenase/reductase 9; SNP, single nucleotide polymorphism; TAG, triacylglycerol.
COMPLIANCE WITH ETHICS GUIDELINES
Xuelin Zhang, Yang Wang and Pingsheng Liu declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
Contributor Information
Xuelin Zhang, Email: zhangxuelin@cupes.edu.cn.
Pingsheng Liu, Email: pliu@ibp.ac.cn.
REFERENCES
- Anstee QM, Day CP. The genetics of NAFLD. Nat Rev Gastroenterol Hepatol. 2013;10:645–655. doi: 10.1038/nrgastro.2013.182. [DOI] [PubMed] [Google Scholar]
- Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330–344. doi: 10.1038/nrgastro.2013.41. [DOI] [PubMed] [Google Scholar]
- Aragno M, Tomasinelli CE, Vercellinatto I, Catalano MG, Collino M, Fantozzi R, Danni O, Boccuzzi G. SREBP-1c in nonalcoholic fatty liver disease induced by Western-type high-fat diet plus fructose in rats. Free Radic Biol Med. 2009;47:1067–1074. doi: 10.1016/j.freeradbiomed.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Awai HI, Yu EL, Ellis LS, Schwimmer JB. Liver toxicity of anabolic androgenic steroid use in an adolescent with nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr. 2014;59:e32–e33. doi: 10.1097/MPG.0b013e3182952e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros RP, Gustafsson JA. Estrogen receptors and the metabolic network. Cell Metab. 2011;14:289–299. doi: 10.1016/j.cmet.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Basantani MK, Sitnick MT, Cai L, Brenner DS, Gardner NP, Li JZ, Schoiswohl G, Yang K, Kumari M, Gross RW, Zechner R, Kershaw EE. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52:318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaner WS, O’Byrne SM, Wongsiriroj N, Kluwe J, D’Ambrosio DM, Jiang H, Schwabe RF, Hillman EM, Piantedosi R, Libien J. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim Biophys Acta. 2009;1791:467–473. doi: 10.1016/j.bbalip.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CW. Liver disease in menopause. World J Gastroenterol. 2015;21:7613–7620. doi: 10.3748/wjg.v21.i25.7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowska MM, Ostapowicz G, Weltman MD. An association between non-alcoholic fatty liver disease and polycystic ovarian syndrome. J Gastroenterol Hepatol. 2009;24:243–247. doi: 10.1111/j.1440-1746.2008.05740.x. [DOI] [PubMed] [Google Scholar]
- Carr RM, Ahima RS. Pathophysiology of lipid droplet proteins in liver diseases. Exp Cell Res. 2016;340:187–192. doi: 10.1016/j.yexcr.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, Coin LJ, Deng G, Gieger C, Heard-Costa NL, Hottenga JJ, Kuhnel B, Kumar V, Lagou V, Liang L, Luan J, Vidal PM, Mateo Leach I, O’Reilly PF, Peden JF, Rahmioglu N, Soininen P, Speliotes EK, Yuan X, Thorleifsson G, Alizadeh BZ, Atwood LD, Borecki IB, Brown MJ, Charoen P, Cucca F, Das D, de Geus EJ, Dixon AL, Doring A, Ehret G, Eyjolfsson GI, Farrall M, Forouhi NG, Friedrich N, Goessling W, Gudbjartsson DF, Harris TB, Hartikainen AL, Heath S, Hirschfield GM, Hofman A, Homuth G, Hypponen E, Janssen HL, Johnson T, Kangas AJ, Kema IP, Kuhn JP, Lai S, Lathrop M, Lerch MM, Li Y, Liang TJ, Lin JP, Loos RJ, Martin NG, Moffatt MF, Montgomery GW, Munroe PB, Musunuru K, Nakamura Y, O’Donnell CJ, Olafsson I, Penninx BW, Pouta A, Prins BP, Prokopenko I, Puls R, Ruokonen A, Savolainen MJ, Schlessinger D, Schouten JN, Seedorf U, Sen-Chowdhry S, Siminovitch KA, Smit JH, Spector TD, Tan W, Teslovich TM, Tukiainen T, Uitterlinden AG, Van der Klauw MM, Vasan RS, Wallace C, Wallaschofski H, Wichmann HE, Willemsen G, Wurtz P, Xu C, Yerges-Armstrong LM, Alcohol Genome-wide Association, C. Diabetes Genetics, R. Meta-analyses, S. Genetic Investigation of Anthropometric Traits, C. Global Lipids Genetics, C. Genetics of Liver Disease, C. International Consortium for Blood, P. Meta-analyses of, G. Insulin-Related Traits, C. Abecasis GR, Ahmadi KR, Boomsma DI, Caulfield M, Cookson WO, van Duijn CM, Froguel P, Matsuda K, McCarthy MI, Meisinger C, Mooser V, Pietilainen KH, Schumann G, Snieder H, Sternberg MJ, Stolk RP, Thomas HC, Thorsteinsdottir U, Uda M, Waeber G, Wareham NJ, Waterworth DM, Watkins H, Whitfield JB, Witteman JC, Wolffenbuttel BH, Fox CS, Ala-Korpela M, Stefansson K, Vollenweider P, Volzke H, Schadt EE, Scott J, Jarvelin MR, Elliott P, Kooner JS. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton M, Angulo P, Chalasani N, Merriman R, Viker K, Charatcharoenwitthaya P, Sanderson S, Gawrieh S, Krishnan A, Lindor K. Low circulating levels of dehydroepiandrosterone in histologically advanced nonalcoholic fatty liver disease. Hepatology. 2008;47:484–492. doi: 10.1002/hep.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Chang B, Li L, Chan L. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology. 2010;52:1134–1142. doi: 10.1002/hep.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole LK, Jacobs RL, Vance DE. Tamoxifen induces triacylglycerol accumulation in the mouse liver by activation of fatty acid synthesis. Hepatology. 2010;52:1258–1265. doi: 10.1002/hep.23813. [DOI] [PubMed] [Google Scholar]
- Crunk AE, Monks J, Murakami A, Jackman M, Maclean PS, Ladinsky M, Bales ES, Cain S, Orlicky DJ, McManaman JL. Dynamic regulation of hepatic lipid droplet properties by diet. PLoS One. 2013;8:e67631. doi: 10.1371/journal.pone.0067631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZY, Jin GN, Liang HF, Wang W, Chen WX, Datta PK, Zhang MZ, Zhang B, Chen XP. Transforming growth factor beta induces expression of connective tissue growth factor in hepatic progenitor cells through Smad independent signaling. Cell Signal. 2013;25:1981–1992. doi: 10.1016/j.cellsig.2013.05.027. [DOI] [PubMed] [Google Scholar]
- DiStefano MT, Danai LV, Roth Flach RJ, Chawla A, Pedersen DJ, Guilherme A, Czech MP. The lipid droplet protein hypoxia-inducible gene 2 promotes hepatic triglyceride deposition by inhibiting lipolysis. J Biol Chem. 2015;290:15175–15184. doi: 10.1074/jbc.M115.650184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati B, Motta BM, Pingitore P, Meroni M, Pietrelli A, Alisi A, Petta S, Xing C, Dongiovanni P, Del Menico B, Rametta R, Mancina RM, Badiali S, Fracanzani AL, Craxi A, Fargion S, Nobili V, Romeo S, Valenti L. The rs2294918 E434K variant modulates patatin-like phospholipase domain-containing 3 expression and liver damage. Hepatology. 2016;63:787–798. doi: 10.1002/hep.28370. [DOI] [PubMed] [Google Scholar]
- Dongiovanni P, Donati B, Fares R, Lombardi R, Mancina RM, Romeo S, Valenti L. PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol. 2013;19:6969–6978. doi: 10.3748/wjg.v19.i41.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowman JK, Armstrong MJ, Tomlinson JW, Newsome PN. Current therapeutic strategies in non-alcoholic fatty liver disease. Diabetes Obes Metab. 2011;13:692–702. doi: 10.1111/j.1463-1326.2011.01403.x. [DOI] [PubMed] [Google Scholar]
- Fujii H, Ikura Y, Arimoto J, Sugioka K, Iezzoni JC, Park SH, Naruko T, Itabe H, Kawada N, Caldwell SH, Ueda M. Expression of perilipin and adipophilin in nonalcoholic fatty liver disease; relevance to oxidative injury and hepatocyte ballooning. J Atheroscler Thromb. 2009;16:893–901. doi: 10.5551/jat.2055. [DOI] [PubMed] [Google Scholar]
- Gaggini M, Morelli M, Buzzigoli E, DeFronzo RA, Bugianesi E, Gastaldelli A. Non-alcoholic fatty liver disease (NAFLD) and its connection with insulin resistance, dyslipidemia, atherosclerosis and coronary heart disease. Nutrients. 2013;5:1544–1560. doi: 10.3390/nu5051544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorich ZR, Ge Y. Top-down proteomics in health and disease: challenges and opportunities. Proteomics. 2014;14:1195–1210. doi: 10.1002/pmic.201300432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressner OA, Lahme B, Siluschek M, Rehbein K, Herrmann J, Weiskirchen R, Gressner AM. Activation of TGF-beta within cultured hepatocytes and in liver injury leads to intracrine signaling with expression of connective tissue growth factor. J Cell Mol Med. 2008;12:2717–2730. doi: 10.1111/j.1582-4934.2008.00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Ma Y, Kadegowda AK, Betters JL, Xie P, Liu G, Liu X, Miao H, Ou J, Su X, Zheng Z, Xue B, Shi H, Yu L. Deficiency of liver comparative gene identification-58 causes steatohepatitis and fibrosis in mice. J Lipid Res. 2013;54:2109–2120. doi: 10.1194/jlr.M035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AM, Brunt EM, Chen Z, Viswakarma N, Reddy JK, Wolins NE, Finck BN. Dynamic and differential regulation of proteins that coat lipid droplets in fatty liver dystrophic mice. J Lipid Res. 2010;51:554–563. doi: 10.1194/jlr.M000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy T, Oakley F, Anstee QM, Day CP. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annu Rev Pathol. 2016;11:451–496. doi: 10.1146/annurev-pathol-012615-044224. [DOI] [PubMed] [Google Scholar]
- Hashimoto E, Tokushige K. Prevalence, gender, ethnic variations, and prognosis of NASH. J Gastroenterol. 2011;46(Suppl 1):63–69. doi: 10.1007/s00535-010-0311-8. [DOI] [PubMed] [Google Scholar]
- He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, Cohen JC, Hobbs HH. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra M, Li Z, Kruijt JK, Van Eck M, Van Berkel TJ, Kuiper J. The expression level of non-alcoholic fatty liver disease-related gene PNPLA3 in hepatocytes is highly influenced by hepatic lipid status. J Hepatol. 2010;52:244–251. doi: 10.1016/j.jhep.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Horiguchi Y, Araki M, Motojima K. 17beta-Hydroxysteroid dehydrogenase type 13 is a liver-specific lipid droplet-associated protein. Biochem Biophys Res Commun. 2008;370:235–238. doi: 10.1016/j.bbrc.2008.03.063. [DOI] [PubMed] [Google Scholar]
- Huang Y, He S, Li JZ, Seo YK, Osborne TF, Cohen JC, Hobbs HH. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci USA. 2010;107:7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286:37085–37093. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf C, Mardinoglu A, Fagerberg L, Hallstrom BM, Edlund K, Lundberg E, Ponten F, Nielsen J, Uhlen M. The human liver-specific proteome defined by transcriptomics and antibody-based profiling. FASEB J. 2014;28:2901–2914. doi: 10.1096/fj.14-250555. [DOI] [PubMed] [Google Scholar]
- Kelley CE, Brown AJ, Diehl AM, Setji TL. Review of nonalcoholic fatty liver disease in women with polycystic ovary syndrome. World J Gastroenterol. 2014;20:14172–14184. doi: 10.3748/wjg.v20.i39.14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA, Wollaston-Hayden EE, Markowski TW, Higgins L, Mashek DG. Quantitative analysis of the murine lipid droplet-associated proteome during diet-induced hepatic steatosis. J Lipid Res. 2015;56:2260–2272. doi: 10.1194/jlr.M056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kwon H, Park JH, Cho B, Kim D, Oh SW, Lee CM, Choi HC. A low level of serum total testosterone is independently associated with nonalcoholic fatty liver disease. BMC Gastroenterol. 2012;12:69. doi: 10.1186/1471-230X-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51:468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollerits B, Coassin S, Kiechl S, Hunt SC, Paulweber B, Willeit J, Brandstatter A, Lamina C, Adams TD, Kronenberg F. A common variant in the adiponutrin gene influences liver enzyme values. J Med Genet. 2010;47:116–119. doi: 10.1136/jmg.2009.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotronen A, Johansson LE, Johansson LM, Roos C, Westerbacka J, Hamsten A, Bergholm R, Arkkila P, Arola J, Kiviluoto T, Fisher RM, Ehrenborg E, Orho-Melander M, Ridderstrale M, Groop L, Yki-Jarvinen H. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52:1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- Krahmer N, Farese RV., Jr Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:905–915. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M, Schoiswohl G, Chitraju C, Paar M, Cornaciu I, Rangrez AY, Wongsiriroj N, Nagy HM, Ivanova PT, Scott SA, Knittelfelder O, Rechberger GN, Birner-Gruenberger R, Eder S, Brown HA, Haemmerle G, Oberer M, Lass A, Kershaw EE, Zimmermann R, Zechner R. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab. 2012;15:691–702. doi: 10.1016/j.cmet.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie F. All sex steroids are made intracellularly in peripheral tissues by the mechanisms of intracrinology after menopause. J Steroid Biochem Mol Biol. 2015;145:133–138. doi: 10.1016/j.jsbmb.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Labrie F, Luu-The V, Lin SX, Simard J, Labrie C, El-Alfy M, Pelletier G, Belanger A. Intracrinology: role of the family of 17 beta-hydroxysteroid dehydrogenases in human physiology and disease. J Mol Endocrinol. 2000;25:1–16. doi: 10.1677/jme.0.0250001. [DOI] [PubMed] [Google Scholar]
- Ladaru A, Balanescu P, Stan M, Codreanu I, Anca IA. Candidate proteomic biomarkers for non-alcoholic fatty liver disease (steatosis and non-alcoholic steatohepatitis) discovered with mass-spectrometry: a systematic review. Biomarkers. 2016;21:102–114. doi: 10.3109/1354750X.2015.1118542. [DOI] [PubMed] [Google Scholar]
- Li JZ, Ye J, Xue B, Qi J, Zhang J, Zhou Z, Li Q, Wen Z, Li P. Cideb regulates diet-induced obesity, liver steatosis, and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes. 2007;56:2523–2532. doi: 10.2337/db07-0040. [DOI] [PubMed] [Google Scholar]
- Li JZ, Huang Y, Karaman R, Ivanova PT, Brown HA, Roddy T, Castro-Perez J, Cohen JC, Hobbs HH. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest. 2012;122:4130–4144. doi: 10.1172/JCI65179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JW, Dillon J, Miller M. Proteomic and genomic studies of non-alcoholic fatty liver disease–clues in the pathogenesis. World J Gastroenterol. 2014;20:8325–8340. doi: 10.3748/wjg.v20.i26.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HY, Yu IC, Wang RS, Chen YT, Liu NC, Altuwaijri S, Hsu CL, Ma WL, Jokinen J, Sparks JD, Yeh S, Chang C. Increased hepatic steatosis and insulin resistance in mice lacking hepatic androgen receptor. Hepatology. 2008;47:1924–1935. doi: 10.1002/hep.22252. [DOI] [PubMed] [Google Scholar]
- Liu S, Huang C, Li D, Ren W, Zhang H, Qi M, Li X, Yu L. Molecular cloning and expression analysis of a new gene for short-chain dehydrogenase/reductase 9. Acta Biochim Pol. 2007;54:213–218. [PubMed] [Google Scholar]
- Luu-The V, Labrie F. The intracrine sex steroid biosynthesis pathways. Prog Brain Res. 2010;181:177–192. doi: 10.1016/S0079-6123(08)81010-2. [DOI] [PubMed] [Google Scholar]
- Ma WL, Lai HC, Yeh S, Cai X, Chang C. Androgen receptor roles in hepatocellular carcinoma, fatty liver, cirrhosis and hepatitis. Endocr Relat Cancer. 2014;21:R165–R182. doi: 10.1530/ERC-13-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- Marchais-Oberwinkler S, Henn C, Moller G, Klein T, Negri M, Oster A, Spadaro A, Werth R, Wetzel M, Xu K, Frotscher M, Hartmann RW, Adamski J. 17beta-Hydroxysteroid dehydrogenases (17beta-HSDs) as therapeutic targets: protein structures, functions, and recent progress in inhibitor development. J Steroid Biochem Mol Biol. 2011;125:66–82. doi: 10.1016/j.jsbmb.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- McNamara KM, Sasano H. The intracrinology of breast cancer. J Steroid Biochem Mol Biol. 2015;145:172–178. doi: 10.1016/j.jsbmb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10:656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- Moeller G, Adamski J. Multifunctionality of human 17beta-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2006;248:47–55. doi: 10.1016/j.mce.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Moeller G, Adamski J. Integrated view on 17beta-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2009;301:7–19. doi: 10.1016/j.mce.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Mostaghel EA. Steroid hormone synthetic pathways in prostate cancer. Transl Androl Urol. 2013;2:212–227. doi: 10.3978/j.issn.2223-4683.2013.09.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/S0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- Musso G, Cassader M, Rosina F, Gambino R. Impact of current treatments on liver disease, glucose metabolism and cardiovascular risk in non-alcoholic fatty liver disease (NAFLD): a systematic review and meta-analysis of randomised trials. Diabetologia. 2012;55:885–904. doi: 10.1007/s00125-011-2446-4. [DOI] [PubMed] [Google Scholar]
- Pirazzi C, Adiels M, Burza MA, Mancina RM, Levin M, Stahlman M, Taskinen MR, Orho-Melander M, Perman J, Pujia A, Andersson L, Maglio C, Montalcini T, Wiklund O, Boren J, Romeo S. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol. 2012;57:1276–1282. doi: 10.1016/j.jhep.2012.07.030. [DOI] [PubMed] [Google Scholar]
- Pirazzi C, Valenti L, Motta BM, Pingitore P, Hedfalk K, Mancina RM, Burza MA, Indiveri C, Ferro Y, Montalcini T, Maglio C, Dongiovanni P, Fargion S, Rametta R, Pujia A, Andersson L, Ghosal S, Levin M, Wiklund O, Iacovino M, Boren J, Romeo S. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet. 2014;23:4077–4085. doi: 10.1093/hmg/ddu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinella M, Charlton M. The globalization of non-alcoholic fatty liver disease -prevalence and impact on world health. Hepatology. 2016;64(1):19–22. doi: 10.1002/hep.28524. [DOI] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchetti A, Prati D, Pezzotta MG, Tavian D, Colombo R, Callea F, Colli A. Severe steatohepatitis in a patient with a rare neutral lipid storage disorder due to ABHD5 mutation. J Hepatol. 2008;49:474–477. doi: 10.1016/j.jhep.2008.05.027. [DOI] [PubMed] [Google Scholar]
- Rotinen M, Villar J, Celay J, Encio I. Type 10 17beta-hydroxysteroid dehydrogenase expression is regulated by C/EBPbeta in HepG2 cells. J Steroid Biochem Mol Biol. 2010;122:164–171. doi: 10.1016/j.jsbmb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ, Nash CRN. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwingel PA, Zoppi CC, Cotrim HP. Increased liver steatosis in anabolic-androgenic steroid users: more evidence towards toxicant-associated fatty liver disease development. Liver Int. 2011;31:1240–1241. doi: 10.1111/j.1478-3231.2011.02552.x. [DOI] [PubMed] [Google Scholar]
- Shen M, Shi H. Sex Hormones and Their Receptors Regulate Liver Energy Homeostasis. Int J Endocrinol. 2015;2015:294278. doi: 10.1155/2015/294278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagris E, BasuRay S, Li J, Huang Y, Lai KM, Gromada J, Cohen JC, Hobbs HH. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61:108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BW, Adams LA. Non-alcoholic fatty liver disease. Crit Rev Clin Lab Sci. 2011;48:97–113. doi: 10.3109/10408363.2011.596521. [DOI] [PubMed] [Google Scholar]
- Sookoian S, Pirola CJ. Liver enzymes, metabolomics and genome-wide association studies: from systems biology to the personalized medicine. World J Gastroenterol. 2015;21:711–725. doi: 10.3748/wjg.v21.i3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sookoian S, Castano GO, Burgueno AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Butler JL, Palmer CD, Voight BF, Consortium G, Consortium MI, Nash CNR, Hirschhorn JN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Hadzic N, Fischer J, Knisely AS. Steatohepatitis and unsuspected micronodular cirrhosis in Dorfman-Chanarin syndrome with documented ABHD5 mutation. J Pediatr. 2004;144:662–665. doi: 10.1016/j.jpeds.2004.01.036. [DOI] [PubMed] [Google Scholar]
- Straub BK, Stoeffel P, Heid H, Zimbelmann R, Schirmacher P. Differential pattern of lipid droplet-associated proteins and de novo perilipin expression in hepatocyte steatogenesis. Hepatology. 2008;47:1936–1946. doi: 10.1002/hep.22268. [DOI] [PubMed] [Google Scholar]
- Su W, Wang Y, Jia X, Wu W, Li L, Tian X, Li S, Wang C, Xu H, Cao J, Han Q, Xu S, Chen Y, Zhong Y, Zhang X, Liu P, Gustafsson JA, Guan Y. Comparative proteomic study reveals 17beta-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA. 2014;111:11437–11442. doi: 10.1073/pnas.1410741111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targher G, Day CP, Bonora E. Risk of cardiovascular disease in patients with nonalcoholic fatty liver disease. N Engl J Med. 2010;363:1341–1350. doi: 10.1056/NEJMra0912063. [DOI] [PubMed] [Google Scholar]
- Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- Varlamov O, Bethea CL, Roberts CT., Jr Sex-specific differences in lipid and glucose metabolism. Front Endocrinol (Lausanne) 2014;5:241. doi: 10.3389/fendo.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volzke H, Aumann N, Krebs A, Nauck M, Steveling A, Lerch MM, Rosskopf D, Wallaschofski H. Hepatic steatosis is associated with low serum testosterone and high serum DHEAS levels in men. Int J Androl. 2010;33:45–53. doi: 10.1111/j.1365-2605.2009.00953.x. [DOI] [PubMed] [Google Scholar]
- Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099–2108. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhao Y, Gao X, Li L, Yuan Y, Liu F, Zhang L, Wu J, Hu P, Zhang X, Gu Y, Xu Y, Wang Z, Li Z, Zhang H, Ye J. Perilipin 5 improves hepatic lipotoxicity by inhibiting lipolysis. Hepatology. 2015;61:870–882. doi: 10.1002/hep.27409. [DOI] [PubMed] [Google Scholar]
- Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, Ma Y, Liu P. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res. 2012;53:1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of non-alcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence and outcomes. Hepatology. 2015;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]