Despite the importance of cell walls to the biology of plants, not much is known about the biosynthesis and function of their major macromolecular components. From the known complexity of the cell wall structure, we can predict that its synthesis requires hundreds of enzymes, but biochemical approaches have been unsuccessful thus far in characterizing more than a few of them (Henrissat et al., 2001; Reiter, 2002; Vincken et al., 2003). Likewise, many proteins involved in cell wall reorganization, degradation, and the signaling pathways that control cell wall metabolism remain functionally uncharacterized. Comparative molecular genetic studies have not been useful because the walls of other organism groups, such as bacteria and yeast, are fundamentally different in composition, structure, and function from those of plants. In addition, the wide evolutionary distances between the kingdoms create serious limitations for sequence similarity-based approaches. Recent advances in genomics make it possible to identify quickly large numbers of genes as being putatively involved in particular plant cell processes. These new resources provide unexplored opportunities for integrative systems biology studies. In addition, the availability of plant genome sequences and large expressed sequence tag (EST) sets from cell wall model species, like cotton (Gossypium sp.) and poplar (Populus sp.), are excellent tools for comparative studies. However, with the new resources for identifying candidate genes encoding biosynthetic enzymes and regulatory proteins comes the challenge of extracting the critical information from complex data sets to guide the functional analysis of these genes and the proteins they encode. Many Web services are available for individual cell wall-related protein families, primarily from Arabidopsis. Yet, efforts to consolidate the knowledge concerning the different enzyme and structural protein families for a wide spectrum of plant and non-plant species in one interface are missing. Our objective is to fill this gap by creating and maintaining Cell Wall Navigator (CWN; http://bioinfo.ucr.edu/projects/Cellwall/index.pl), a Web-based database that integrates cell wall-related protein families and allows easy comparison among sequences derived from fully sequenced plant genomes plus the known protein sequences from other species. Databases with global family cluster information for all known proteins are available (Krause et al., 2002), but their interfaces typically lack the flexibility required for process-oriented databases (Tchieu et al., 2003), which are more suited for organizing detailed functional and annotation information. The unique features of the CWN database are (1) its adaptable design for organizing complex protein families across many organisms to cover the complete space of known sequences, (2) its flexible architecture for rapid integration of new families, (3) its automated update and analysis pipeline to maintain current information, and (4) its numerous visualization and interactive mining tools.
INDIVIDUAL FAMILIES AND THEIR EXPECTED FUNCTION
The primary cell wall of higher plants is a complex structure that includes cellulose, the major polysaccharide forming the backbone of plant cell walls, embedded in a matrix composed of hemicellulosic polysaccharides, pectic polysaccharides, and several types of glycoproteins. Because walls determine the size and shape of plant cells, they need to be both rigid and flexible. In young cells that are capable of growth, the wall needs to be adjustable to allow rapid expansion, sometimes resulting in more than a 100-fold increase in surface area while still maintaining cell integrity. Later in development, the wall is modified in specific ways, including selected degradation of certain components, leading to the specific walls present in the many different cell types characteristic of a higher plant. Adding to the complexity of wall metabolism is the fact that not all components are made in the same cellular compartments. Cellulose is synthesized at the plasma membrane and deposited directly into the wall, whereas glycoproteins, cell wall metabolic enzymes, pectic polysaccharides, and hemicelluloses are synthesized in the Golgi and delivered to the wall for subsequent assembly. The precise regulation of all of these biosynthesis, assembly, deposition, and reorganization events requires precise hormonal, temporal, and environmental control for proper completion of plant development plans.
Because our knowledge of cell wall metabolism is still incomplete, it is currently not possible to create a complete list of enzymes and structural proteins that are required during normal plant development. For Arabidopsis and rice (Oryza sativa) alone, we can estimate that roughly 100 protein families with more than a thousand members are involved in cell wall-related processes (Henrissat et al., 2001). Among those that are currently known or predicted are the sugar nucleotide synthesis and interconversion enzymes, the glycan synthases and glycosyltransferases for polysaccharide biosynthesis and glycoprotein modification, as well as enzymes for wall assembly, reorganization, and selective degradation (Table I). Examples of later types of enzymes are the methyl esterases that remove methyl ester groups from pectin causing a change in their physical properties.
Table I.
Important protein families involved in primary wall metabolism
| CWN Abr | CAZy | ||||
|---|---|---|---|---|---|
| 1 | Monosaccharide activation and interconversion | ||||
| 1.1 | Sugar 1-kinases (Ara1) | S1K | |||
| 1.2 | Nucleotide-sugar pyrophosphorylases | NSPP | |||
| 1.2.1 | GDP-Man pyrophosphorylase (CYT1) | GMP | |||
| 1.2.2 | UDP-Glc pyrophosphorylase | UGP | |||
| 1.3 | Nucleotide-sugar interconversion: NAD-dependent epimerase/dehydratase superfamily | ||||
| 1.3.1 | UDP-d-Glc 4-epimerases (UGE1-5) | UGE | |||
| 1.3.2 | UDP-d-GalUA 4-epimerases (GAE1-6) | GAE | |||
| 1.3.3 | UDP-l-rhamnose synthases (RHM1-3, UER1) | RHM | |||
| 1.3.4 | UDP-d-apiose/Xyl synthases (AXS1-2, AUD1-3, SUD1-3) | UXS | |||
| 1.3.5 | UDP-d-Xyl 4-epimerase (UXE1-4; MUR4) | UXE | |||
| 1.3.6 | GDP-d-Man 3,5-epimerase | GME | |||
| 1.3.7 | GDP-4-keto-6-deoxy-d-Man-3,5-epimerase-4-reductase (GER1-2) | GER | |||
| 1.3.8 | GDP-d-Man-4,6-dehydratase (GMD1-2; MUR1) | GMD | |||
| 1.4 | Nucleotide-sugar interconversion: nucleotide sugar dehydrogenase superfamily | ||||
| 1.4.1 | UDP-d-Glc dehydrogenases (UGD1-4) | UGD | |||
| 2 | Polysaccharide synthesis | ||||
| 2.1 | Cellulose and galactomannan | ||||
| 2.1.1 | Cellulose synthases (CESAx) | CES | GT2 | ||
| 2.1.2 | Cellulose and β-mannan synthase-like (CSLx) | CSL | GT2 | ||
| 2.2 | Hemicellulose | ||||
| 2.2.1 | Reversibly glycosylated polypeptides | RGP | |||
| 2.2.2 | Xyloglucan galactosyltransferases (MUR3) | XGT | GT47 | ||
| 2.2.3 | Xyloglucan fucosyltransferase (MUR2) | XFT | GT37 | ||
| 2.2.4 | Xyloglucan xylosyltransferases and galactomannan gal-transferases | XXT | GT34 | ||
| 2.2.5 | Glucuronoarabinoxylan* | GAX | |||
| 2.3 | Pectin (selection) | ||||
| 2.3.1 | Homogalacturonan (HG, involvement of GT8) | ||||
| 2.3.2 | Xylogalacturonan (XGA)* | ||||
| 2.3.3 | Rhamnogalacturonan I (RG-I, involvement of RHM)* | ||||
| 2.3.4 | Rhamnogalacturonan II (RG-II, involvement of XGT; GUT1) | ||||
| 2.3.5 | Arabinogalactan I (AG-I)* | ||||
| 2.3.6 | Arabinogalactan II (AG-II)* | ||||
| 2.3.7 | Arabinan (A)* | ||||
| 2.4 | Callose | ||||
| 2.4.1 | Glucan synthase-like | GSL | GT48 | ||
| 2.5 | Other glycosyl transferases | ||||
| 2.5.1 | Glycosyltransferases (Qua1; associated with HG biosynthesis) | GT8 | GT8 | ||
| 3 | Reassembly and Degradation | ||||
| 3.1 | Cell expansion | ||||
| 3.1.1 | Expansins (EXP10) | EXP | |||
| 3.1.2 | Yieldins | GH18 | GH18 | ||
| 3.2 | Hemicellulose reassembly | ||||
| 3.2.1 | Xyloglucan endotransglucosylases/hydrolases | XTH | GH16 | ||
| 3.3 | Glycoside hydrolases | ||||
| 3.3.1 | β-Galactosidase | BGAL | GH35 | ||
| 3.3.2 | Endo-1,4-β-glucanase | GH9 | GH9 | ||
| 3.3.3 | Glucan 1,3-β-glucosidase | GH17 | GH17 | ||
| 3.3.4 | Glycoside hydrolase: polyGalAase, Rha-GalAase | GH28 | GH28 | ||
| 3.3.5 | Xylan degradation | GH10 | GH10, 43 & 51 | ||
| 3.4 | Lyases | ||||
| 3.4.1 | Pectate and pectin lyases | PL1 | PL1 | ||
| 3.4.2 | Rhamnogalacturonan I lyases | PL4 | PL4 | ||
| 3.5 | Esterases | ||||
| 3.5.1 | Pectin methyl esterases | PME | CE8 | ||
| 3.5.2 | Pectin acetylesterases | PAE | CE13 | ||
| 3.5.3 | Feruloyl esterases | FE | CE1 | ||
| 4 | Structural proteins | ||||
| 4.1 | Hyp-rich glycoproteins or extensins | HRGP | |||
| 4.2 | Leu-rich repeat extensins | LRX | |||
| 4.3 | Pro-rich proteins | PRP | |||
| 4.4 | Gly-rich proteins | GRP | |||
| 4.5 | Arabinogalactan proteins | AGP | |||
| 5 | Glycoprotein glycosyltransferases | ||||
| 5.1 | Glycoprotein fucosyltransferases | GFT | GT10 | ||
| 5.2 | Glycosyltransferases | GT31a/b | GT31 | ||
The protein classes in this table are primarily organized by sequence similarity. The superimposed product classifications are not intended to be complete. *, Several listed polysaccharide classes are still lacking sequence information associated with the biosynthesis of their backbone structure. Additionally, the boundaries of sequence- and product-based categorization are often not in alignment. For instance, the β-mannan synthase is functionally involved in hemicellulose synthesis, and by sequence similarity it is a member of the CSL superfamily. CWN Abr, Family abbreviations used in CWN. The corresponding family identifiers of the CAZy database are listed in the last column: GH, glycoside hydrolase; GT, glycosyltransferase; PL, polysaccharide lyase; CE, carbohydrate esterase.
GENERAL FUNCTIONALITY OF CWN
CWN is an integrated database and mining tool for protein families that are involved in plant cell wall biogenesis. Its interface provides comprehensive query and visualization functions for mining sequence features, exploring evolutionary relationships, and retrieving biological information within and across families. It is also an annotation forum that allows registered users to share their expert knowledge about cell wall-associated genes with the community by uploading annotations and comments to the database. Most of the tools in CWN are based on modular open-source code that is shared with the public via the BioPerl project (Stajich et al., 2002). This open modular architecture maximizes the versatility and portability of the computational backend of this resource.
INCLUDED FAMILIES, ORGANISM SPECTRUM, AND ANALYSIS PIPELINE
At this time, CWN contains more than 30 gene families with more than 5,000 members coding for enzymes and structural proteins known to be involved in sugar substrate generation and primary cell wall metabolism. Upon request from researchers, we will include additional protein families involved in these or related processes, such as secondary wall biosynthesis, wall-associated defense, and regulatory mechanisms. The sequences in CWN are exclusively clustered by sequence similarity and the families themselves are currently organized by classes of polysaccharides, structural proteins, assembly/disassembly stages, and Carbohydrate-Active enZYmes (CAZy) families (Table I). The categorization by polysaccharide class is often in disagreement with the similarity-based clustering. This is largely due to the fact that sequences from one family can be involved in the formation of different classes of polysaccharides. An additional factor for this misalignment is the tendency of many glycosyltransferase and glycoside hydrolase families to exhibit variable substrate specificity while maintaining conserved stereochemistry within one family (Henrissat et al., 2001). A more enzyme specificity-oriented classification approach, similar to the one used by the CAZy database, resolves this conflict to some extent, but it lacks information about the broader biological processes, which we intend to maintain in CWN. In case of polyfunctional families, the similarity approach can occasionally incorporate members that are involved in other processes than cell wall metabolism.
To provide a comprehensive and organism-unbiased overview of protein families, CWN incorporates sequences from three different resources: (1) the genome sequences from Arabidopsis and O. sativa spp. japonica provided by The Institute for Genomic Research (TIGR); (2) the UniProt database (Apweiler et al., 2004); and (3) the EST database from the National Center for Biotechnology Information (NCBI; EST_Others). The inclusion of completed plant genomes offers a broad baseline for comparative gene family studies while allowing maximum integration with the rich genomics and bioinformatics resources available for these organisms. Additional plant genomes will be included in the future when they become available in annotated format (e.g. Chlamydomonas reihnardtii, Populus). The incorporated UniProt database currently provides access to more than 1.2 million highly annotated, low-redundant protein sequences from a broad spectrum of organisms. Sequence duplications between the UniProt and plant genome databases are prevented by removing the Arabidopsis and rice entries from the UniProt database prior to sequence searching. This combinatorial database selection allows comprehensive coverage of the known sequence space while maintaining the completeness of entirely sequenced plant genomes.
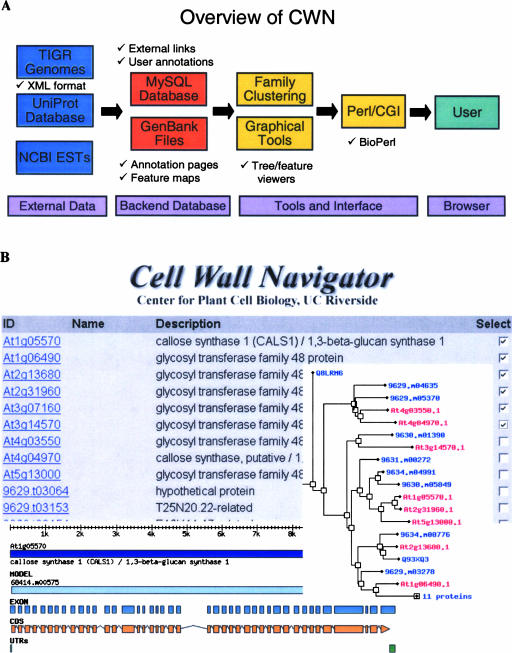
To provide up-to-date sequence and annotation information, an automated analysis pipeline of CWN reformats new releases of TIGR's plant genome annotations and the UniProt database into GenBank format, performs additional protein domain/motif predictions, and uploads the information into a relational MySQL database (Fig. 1A). An iterative search and alignment model building pipeline executes sensitive BLAST (Altschul et al., 1990) and HMM-based (http://hmmer.wustl.edu/) searches to populate the protein families with closely and remotely related members from the genome and UniProt databases. After this family population process is saturated with all detectable members, the final multiple alignments are generated with ClustalW (Thompson et al., 1994) and hmmalign (http://hmmer.wustl.edu/), quality filtered with Perl scripts, and used to calculate similarity trees with the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html). A more detailed description of the different search and analysis steps is available online (http://bioinfo.ucr.edu/projects/Cellwall/Documents/README.html). In addition, detailed and standardized annotation pages for each gene/protein are available through the Web interface (Fig. 1). The use of this automated pipeline combined with a coherent data management strategy makes it very easy to incorporate new protein members and families into the database. Users can simply send one or several protein accession numbers to the curator for populating a new family in the database with all related sequences and their annotations.
Figure 1.
Outline of data flow (A) and graphical interfaces (B) in CWN.
EST data are incorporated into CWN by performing BLAST searches of all family members, retrieved from the above databases, against the latest EST_Others set from NCBI in quarterly intervals. The obtained search results constitute by far the largest data domain in CWN due to the fast-growing number of entries and the extensive sequence redundancy in unclustered EST data. In spite of their complexity, organism-unspecific EST searches can provide very useful information for collecting evidence of the expression of genes and for guiding the discovery of novel gene functions by comparative studies between organisms with distinctive cell wall compositions. Due to their fragmentary nature and low sequence quality, the EST data in CWN are excluded from certain analysis tools that depend on complete, high-quality sequence information, as for instance multiple alignments and similarity trees (see below). As an alternative, the obtained EST results can be queried, filtered, and sorted via the Web interface by many criteria or viewed in graphical mode. In addition, the number of EST hits within each family is recorded on a statistics page along with the number of proteins included in each family.
QUERY, VISUALIZATION, AND MINING TOOLS IN CWN
The Web interface of CWN provides numerous query and visualization tools for exploring sequence families (Fig. 1B). It allows users to find sequences and other information in the database by providing accession numbers, key words, and user annotations or by performing sequence searches using the BLAST algorithm. All sequences, alignments, and trees can be downloaded in different formats as single or batch units. Alternatively, entire data sets are downloadable in a single step via an ftp page. Gene and protein domain structures can be displayed graphically for individual entries or for a group of entries to allow comparative studies between family members. Up-to-date structure images are provided by dynamically creating them upon client request from the latest annotation data using the biographics module from the BioPerl project (Stajich et al., 2002).
For effectively mining complex similarity trees with hundreds of members, we have designed an interactive tree viewer that allows users to browse through tree structures by collapsing or expanding tree branches, highlighting selected members in color, and opening their annotation pages via encoded hyperlinks in the tree images. Additionally, the trees can be imported into local viewing applications (e.g. TreeView). The underlying tree files are calculated with the PHYLIP package using a robust distance-based neighbor-joining approach for tree construction and the midpoint method for defining root positions. The corresponding multiple alignments can be displayed in form of graphical overview plots, as HTML-encoded text with colorful residue shading, or in plain text format for import into local alignment editors.
The extensive annotation pages in CWN provide detailed information about each family member and allow external databases to link to them or their associated family resources by using a uniform URL structure. The pages include interactive viewing options for sequence features, annotation data, and many links to external resources for accessing additional information about gene function, mutants, and expression profiles from different profiling techniques. In detail, a clickable gene structure viewer highlights exon, intron, and UTR elements in the corresponding sequences by clicking on the graphical features. A similar tool can visualize domains and motifs in the provided protein sequences. The current set of external links guides to the following resources: (1) the sequence annotation pages from TIGR, The Arabidopsis Information Resource (TAIR), the Munich Information Center for Protein Sequences (MIPS), and UniProt; (2) the functional gene categorizations from the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al., 2004), AraCyc (Mueller et al., 2003), and the Gene Ontology (GO) consortium (Ashburner et al., 2000); (3) the CAZy site (Coutinho and Henrissat, 1999); (4) the membrane domain predictions from the Aramemnon database (Schwacke et al., 2003); (5) the insertional knockout collections from the Salk Institute Genomic Analysis Laboratory (SIGnAL; Alonso et al., 2003) and the Genome Analysis of the Plant Biological System–Knockout of Arabidopsis thaliana (GABI-Kat) project (Rosso et al., 2003); and (6) the genome-wide expression data from the Arabidopsis Functional Genomics Consortium (AFGC) cDNA microarray project (Finkelstein et al., 2002), the Nottingham Arabidopsis Stock Centre (NASC) Affymetrix Chip Facility (http://ssbdjc2.nottingham.ac.uk/), and the Delaware Biotechnology Institute Massively Parallel Signature Sequencing (DBI MPSS) database (Meyers et al., 2004). Since many of these external resources exist only for Arabidopsis, it is not possible to provide all of these links for the other organisms. Additional links are available to various cell wall-related Web sites and an internally maintained collection of cell wall-specific literature. Beyond information retrieval, the CWN interface allows registered users to upload important information from their research about sequences, mutants, phenotypes, antibodies, protein functions, and other valuable information. The uploaded data will be reviewed frequently by the database curator. Authors can edit or remove their provided information at any time.
FUTURE PERSPECTIVE OF CWN
The protein family database CWN is unique by integrating a wide range of cell wall-related families from an unrestricted number of organisms in a single interface that contains numerous interactive visualization functions. Its rich family annotations are a valuable tool to aid future functional predictions and characterizations of individual members. For instance, diverged catalytic consensus sequences are often a strong indicator for a distinct function within families of glycosyltransferase (e.g. substrate specificity). We will further maintain and improve this resource by including additional cell wall families and adding new features to enhance its functionality for the community. Links to Web sites about mutants, antibodies, and publications will be extended and new technology resources incorporated. We will also continue to work on data interoperability and sharing of data with the CAZy, TAIR, and other databases.
This work was supported by the National Science Foundation (plant genome grant no. DBI–0211797).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32 (Database issue): D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Henrissat B (1999) Carbohydrate-Active enZYmes. AFMB-CNRS. http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html (June, 2004)
- Finkelstein D, Ewing R, Gollub J, Sterky F, Cherry JM, Somerville S (2002) Microarray data quality analysis: lessons from the AFGC project. Arabidopsis Functional Genomics Consortium. Plant Mol Biol 48: 119–131 [DOI] [PubMed] [Google Scholar]
- Henrissat B, Coutinho PM, Davies GJ (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47: 55–72 [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32 (Database issue): D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause A, Haas SA, Coward E, Vingron M (2002) SYSTERS, GeneNest, SpliceNest: exploring sequence space from genome to protein. Nucleic Acids Res 30: 299–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD (2004) Arabidopsis MPSS. An online resource for quantitative expression analysis. Plant Physiol 135: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD (2002) Biosynthesis and properties of the plant cell wall. Curr Opin Plant Biol 5: 536–542 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff GE, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12: 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu JH, Fana F, Fink JL, Harper J, Nair TM, Niedner RH, Smith DW, Steube K, Tam TM, Veretnik S, (2003) The PlantsP and PlantsT Functional Genomics Databases. Nucleic Acids Res 31: 342–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken JP, Schols HA, Oomen RJ, Mccann MC, Ulvskov P, Voragen AG, Visser RG (2003) If homogalacturonan were a side chain of rhamnogalacturonan I. Implications for cell wall architecture. Plant Physiol 132: 1781–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]