Abstract
Growing evidence suggests that the bacteria present in our gut may play a role in mediating the effect of genetics and lifestyle on obesity and metabolic diseases. Most of the current literature on gut bacteria consists of cross‐sectional and correlative studies, rendering it difficult to make any causal inferences as to the influence of gut bacteria on obesity and related metabolic disorders. Interventions with germ‐free animals, treatment with antibiotic agents, and microbial transfer experiments have provided some evidence that disturbances in gut bacteria may causally contribute to obesity‐related insulin resistance and adipose tissue inflammation. Several potential mediators have been hypothesized to link the activity and composition of gut bacteria to insulin resistance and adipose tissue function, including lipopolysaccharide, angiopoietin‐like protein 4, bile acids and short‐chain fatty acids. In this review we critically evaluate the current evidence related to the direct role of gut bacteria in obesity‐related metabolic perturbations, with a focus on insulin resistance and adipose tissue inflammation. It is concluded that the knowledge base in support of a role for the gut microbiota in metabolic regulation and in particular insulin resistance and adipose tissue inflammation needs to be strengthened.
Keywords: adipose tissue inflammation, angiopoietin‐like protein 4, bile acids, gut bacteria, insulin resistance, lipopolysaccharide, short‐chain fatty acids
Abbreviations
- ANGPTL4
angiopoietin‐like protein 4
- FXR
farnesoid X receptor
- GLP‐1
glucagon‐like peptide‐1
- GPCR
G protein‐coupled receptor
- LPS
lipopolysaccharide
- PYY
peptide YY
- SCFA
short‐chain fatty acid
- TGR5
G protein‐coupled bile acid receptor 1
Introduction
Obesity is associated with a variety of metabolic complications including dyslipidaemia and insulin resistance. Evidence is accumulating that the development of insulin resistance and other complications of obesity may be driven by a heightened inflammatory state of the adipose tissue. Elevated adipose tissue inflammation is characterized by the influx of various immune cells, including macrophages, and the upregulation of numerous inflammatory cytokines (Boutens & Stienstra, 2016).
Obesity and related metabolic diseases are believed to be the result of an interaction between genetics and lifestyle factors, such as diet and physical activity. In the last decade, there has been growing realization that part of the effect of genetics and lifestyle on obesity and metabolic diseases may be mediated by the microorganisms residing in our gastrointestinal tract, which are referred to as the gut microbiota (Janssen & Kersten, 2015; Wu et al. 2015). Indeed, changes in gut microbiota composition have been observed in people with obesity (Ley et al. 2006; Schwiertz et al. 2010), and obesity‐related diseases such as type 2 diabetes (Qin et al. 2012), non‐alcoholic fatty liver disease (Mouzaki et al. 2013; Jiang et al. 2015) and cardiovascular disease (Karlsson et al. 2012).
The gut microbiota predominantly consists of bacteria but also includes viruses, fungi, protozoa, and archaea. The composition of the gut microbiota varies greatly between individuals, reflecting the impact of the host genome and environmental factors, such as lifestyle, hygiene, the use of antibiotics and especially diet. In addition, the intestinal microbiota composition may be affected by specific disease states (Benson et al. 2010; Sommer & Bäckhed, 2013). Conversely, the intestinal microbiota may impact the host and contribute to certain diseases (Rabot et al. 2010; Ridaura et al. 2013; Gregory et al. 2015; Schaubeck et al. 2016). Most of the contemporary literature on the relation between the composition of the gut microbiota and obesity and related parameters consists of cross‐sectional and correlative studies. Because of the complicated interaction between the host, environmental factors and the gut microbiota, no causal inferences can be drawn from these studies about the influence of gut bacteria on obesity and obesity‐related disturbances, which represents a major limitation. In this review we discuss the current evidence related to the direct role of gut bacteria in obesity‐related metabolic perturbations, with a focus on insulin resistance and adipose tissue inflammation. In the first part, we describe the results of various interventions that addressed the causal role of gut bacteria in metabolic diseases. In the second part, we describe mechanistic studies on potential mediators that may link changes in gut bacteria to obesity‐related insulin resistance and adipose tissue inflammation.
Possible interventions to study the role of gut bacteria in metabolic diseases
To investigate whether the gut bacteria contribute to insulin resistance and adipose tissue inflammation, interventions with germ‐free animals, treatment with antibiotic agents, and bacterial transfer experiments are conducted. Germ‐free animals are maintained free from any microorganisms throughout life and are therefore useful to elucidate the role of the gut microbiota in metabolic disorders. Compared to conventionalized mice, germ‐free mice fed a high‐fat diet were found to have an improved glucose tolerance, improved insulin sensitivity, and reduced adipose tissue inflammation (Bäckhed et al. 2004; Rabot et al. 2010; Caesar et al. 2012). However, it is unclear whether the elevated insulin resistance and adipose tissue inflammation in conventionalized mice as compared to germ‐free mice are directly mediated by the gut microbiota, or indirectly via a higher body weight gain. To circumvent the effects of body weight and adiposity on adipose tissue inflammation, Caesar et al. (2015) compared germ‐free and body weight‐matched conventionally raised mice. Interestingly, adipose tissue inflammation was improved in germ‐free mice as compared with conventionally raised mice. While this study thus suggests that the gut microbiota promote adipose tissue inflammation, it is important to realize that interventions with germ‐free mice have certain limitations. Indeed, germ‐free mice have large defects in the development of the immune system and antibody production, show morphological defects in the intestine, and may suffer from a vitamin deficiency, which may significantly affect the experimental outcome (Smith et al. 2007; Yi & Li, 2012).
To overcome these limitations, administration of antibiotics and microbial transfer are a popular alternative to modulate gut bacterial composition. Antibiotics suppress the gut bacteria (Cani et al. 2008; Carvalho et al. 2012). Similar to the observations in germ‐free mice, treatment with antibiotics improved the glucose tolerance and reduced the infiltration of macrophages in adipose tissue in mice (Cani et al. 2008; Membrez et al. 2008; Carvalho et al. 2012). In contrast, in humans, administration of a cocktail of antibiotics for 4 days had no effect on glucose tolerance (Mikkelsen et al. 2015), while treatment with vancomycin for 1 week decreased the peripheral insulin sensitivity (Vrieze et al. 2014). One disadvantage of antibiotics is that they do not suppress all gut bacteria, which might result in the overgrowth of non‐targeted bacteria (Walsh et al. 2014; Morgun et al. 2015), or outgrowth of intestinal fungi (Mulligan et al. 1982; Dollive et al. 2013). Furthermore, it is important to note that antibiotics possess direct anti‐inflammatory properties and may cause mitochondrial dysfunction, independent of their bactericidal or bacteriostatic effects (Tauber & Nau, 2008; Steel et al. 2012; Wang et al. 2015).
Microbial transfer can be achieved by oral gavage or intrarectal administration of bacteria, or by co‐housing animals. Transfer of microbiota from obesity‐prone – but not from obesity‐resistant – mice to germ‐free mice increased weight gain, increased homeostasis model assessment of insulin resistance (HOMA‐IR), and stimulated the infiltration of macrophages in adipose tissue (Duca et al. 2014). Another study showed that mice receiving microbiota from obese mice at weaning had an improved glucose tolerance as compared with mice receiving microbiota from lean mice, but not when the microbial transfer was performed at 8 weeks of age (Ellekilde et al. 2014). The role of the gut microbiota in human obesity and insulin resistance has been investigated by transplanting faecal microbiota from female adult twins discordant in obesity to germ‐free mice. While an increase in obesity and adiposity was observed in mice receiving the gut microbiota from the obese twin as compared with mice receiving the gut microbiota from the lean twin, only a trend towards impaired glucose tolerance (P = 0.06) was observed (Ridaura et al. 2013). Interestingly, the transplantation of gut microbiota from lean individuals to patients with metabolic syndrome improved insulin sensitivity (Vrieze et al. 2012). Of importance, the effectiveness of bacterial transfer depends on whether bacteria are able to colonize in the recipient's intestinal microbial niche.
Microbial transfer can also be achieved by co‐housing, which is exclusively applied in animals. Importantly, cohousing of mice with different genotypes will equalize intestinal bacterial composition in the co‐housed animals. Therefore, only if the gut microbiota have profound effects on the phenotype might differences between the two co‐housed groups yield significant results (Laukens et al. 2016).
The interventions mentioned above are commonly used to investigate the role of the gut bacteria in metabolic health. However, these interventions cannot discriminate between pathogenic and beneficial bacteria. Bacteria also confer health benefits related to, for example, vitamin synthesis, tissue homeostasis and immune function (Smith et al. 2007; Sommer & Bäckhed, 2013). As a result, the loss of beneficial bacteria can have detrimental effects on the host. A more targeted approach to modulate the intestinal bacteria, for example by administrating specific bacterial species, is expected to give more specific insight into which and how bacteria can affect the health of the host. For instance, administration of Escherichia coli to germ‐free mice has been shown to impair glucose tolerance (Caesar et al. 2012), whereas administration of Akkermansia muciniphila to conventional mice was shown to improve glucose tolerance (Everard et al. 2013). To successfully administer bacterial species, bacteria need to remain viable during storage, survive the passage through the gastrointestinal tract, and be able to colonize the intestine. Although the number of bacteria that can be cultured has gone up significantly, still many gastrointestinal bacteria cannot be cultured (Li et al. 2014; Rajilić‐Stojanović & de Vos, 2014). Hence, due to this limitation, the more targeted approach is only applicable to a selection of bacterial strains.
Overall, current evidence lends some credence to the notion that changes in the gut bacterial composition contribute to insulin resistance and adipose tissue inflammation. However, additional human and animal studies are needed to bolster the causal link between the gut bacteria and obesity‐related metabolic parameters.
Potential mediators
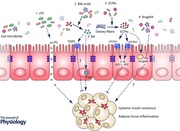
Several potential mediators have been hypothesized to link the activity and composition of the gut microbiota to insulin resistance and adipose tissue function, including lipopolysaccharide (LPS), angiopoietin‐like protein 4 (Angptl4), bile acids and short‐chain fatty acids (SCFAs).
Lipopolysaccharides
LPS or so‐called endotoxin is a major component of the gram‐negative bacterial cell wall. The first indications that LPS may be involved in obesity and related metabolic disorders were reported by Cani et al. (2007). Continuous subcutaneous infusion of LPS for 4 weeks induced weight gain, insulin resistance, and adipose tissue inflammation to a similar extent as high‐fat feeding. Importantly, high‐fat feeding was found to elevate plasma LPS levels, which was associated with an increased gut permeability and a decreased expression of the tight junction proteins zonula occludens‐1 and occludin. Treatment of mice fed a high‐fat diet with antibiotics did not increase the gut permeability and plasma LPS levels, suggesting a role for the gut bacteria in this process. In addition to reduced LPS levels, antibiotic‐treated mice also displayed less macrophage infiltration in adipose tissue and improved glucose and insulin tolerance (Cani et al. 2008). That the suppression of the gut bacteria improves insulin resistance and adipose tissue inflammation concurrent with a reduction in plasma LPS levels has also been found in other studies that used mice fed a high‐fat diet (Membrez et al. 2008; Carvalho et al. 2012), ob/ob mice (Cani et al. 2008), or germ‐free mice (Caesar et al. 2012). Interestingly, supplementing A. muciniphila during high‐fat feeding reduced fat mass gain and improved glucose tolerance in association with reduced serum LPS levels. These findings were not observed with heat‐killed A. muciniphila, suggesting that live A. muciniphila has a profound role in gut barrier function (Everard et al. 2013).
It is important to note that not all mouse studies have provided supportive evidence for a role of LPS in insulin resistance (Lichtenstein et al. 2010; Caesar et al., 2012, 2015; Laugerette et al. 2012). Whereas administration of the gram‐negative bacterium E. coli W3110 to germ‐free mice increased plasma LPS, administration of the isogenic mutant MLK1067 did not. However, both strains increased adipose tissue weight and impaired glucose tolerance, suggesting that gut‐derived LPS is not essential for adiposity and glucose and insulin tolerance in mice (Caesar et al. 2012). In another study by the same authors, obesity, adipose tissue inflammation and insulin resistance in mice fed a lard‐based high‐fat diet were not accompanied by a significant elevation in plasma LPS levels as compared with mice fed a fish oil‐based high‐fat diet (Caesar et al. 2015). However, lard‐feeding significantly increased toll‐like receptor 4 activation (Caesar et al. 2015), which might reflect an increased sensitivity of the mice to LPS, a notion that was also raised in another study (Huang et al. 2007).
Whereas the original publication by Cani et al. (2008) in mice reported a stimulatory effect of LPS on insulin resistance, subsequent studies in rats have failed to find an effect of LPS administration on insulin resistance (Liu et al. 2010; Dudele et al. 2015). In humans, LPS injection in healthy subjects was shown to induce insulin resistance and adipose tissue inflammation (Dandona et al. 2010; Mehta et al. 2010). To investigate whether the changes in gut bacterial composition associated with overfeeding and obesity can elevate plasma LPS levels in humans, healthy humans were placed on high‐fat/hypercaloric diets in two independent studies. Laugerette et al. (2014) reported that although overfeeding for 8 weeks caused marked weight gain, it did not influence the fasting plasma LPS levels. By contrast, Pendyala et al. (2012) observed that 4 weeks of Western diet raised the plasma LPS levels. Interestingly, the increased LPS levels were not accompanied by higher serum pro‐inflammatory cytokine levels.
Taken together, the current literature on LPS as a potential causal link between disturbances in the gut microbiota and obesity‐related insulin resistance and adipose tissue inflammation is inconsistent. One possible explanation for the inconsistent results might be the difficulty of accurately measuring LPS in blood. Most studies have used the FDA‐approved Limulus amoebocyte lysate (LAL) assay, which measures LPS activity via an enzymatic reaction. One drawback of the LAL assay is that several factors in plasma – such as bile salts and lipoproteins – can interact and thereby inactivate LPS, rendering LPS undetectable. In addition, anticoagulants, including EDTA and heparin, might interfere with the LAL assay (Hurley, 1995; Boutagy et al. 2016). Another drawback is the risk of contaminations, which may give rise to false‐positive results.
Angiopoietin‐like protein 4
Angiopoietin‐like protein 4 (ANGPTL4) is a ubiquitously expressed glycoprotein that plays an important role in lipid metabolism by inhibiting the activity of the enzyme lipoprotein lipase (Dijk & Kersten, 2014). Lipoprotein lipase catalyses the hydrolysis of circulating triglycerides along the capillary lumen of muscle and fat tissue. Bäckhed et al. (2004, 2007) first identified ANGPTL4 as a potential link between the gut microbiota and fat storage. Whereas germ‐free mice were protected against diet‐induced obesity, germ‐free mice lacking the Angptl4 gene were not. Conventionalization of germ‐free mice resulted in the suppression of Angptl4 expression in the intestines but not in adipose tissue. Consequently, it was suggested that downregulation of intestinal Angptl4 expression by gut microbiota may promote adipose tissue lipoprotein lipase activity and thereby peripheral fat storage (Bäckhed et al. 2004, 2007; El Aidy et al. 2013). In contrast, although Fleissner et al. similarly observed increased intestinal Angptl4 expression in germ‐free mice, conventional mice were leaner than their germ‐free counterparts (Fleissner et al. 2010).
Currently, it is still unclear to what extent ANGPTL4 produced in the intestine has an endocrine function and is able to lower LPL activity in adipose tissue. Indeed, there is growing evidence suggesting that ANGPTL4 acts more locally instead of systemically (Dijk & Kersten, 2014). In the intestine, ANGPTL4 may primarily target pancreatic lipase and thereby reduce fat absorption (Mattijssen et al. 2014). Consequently, the inhibition of intestinal Angptl4 upon conventionalization may promotes fat storage via elevated pancreatic lipase activity. In terms of glucose metabolism, while some studies have found an effect of ANGPTL4 overexpression on glucose metabolism (Xu et al. 2005; Lichtenstein et al. 2007), it remains to be determined whether ANGPTL4 plays an important regulatory role in insulin sensitivity and glucose homeostasis.
In apparent contrast to the finding of reduced intestinal Angptl4 expression upon conventionalization, specific bacterial species and short‐chain fatty acids potently induce ANGPTL4 in colonic cell lines (Are et al. 2008; Aronsson et al. 2010; Alex et al. 2013; Korecka et al. 2013). To further investigate the role of intestinal ANGPTL4 as a potential link between the gut microbiota and metabolic health, future studies using intestinal‐specific Angptl4 knockout mice should be performed.
Bile acids
Alterations in the gut bacterial composition have profound consequences for bile acid metabolism. For instance, it is known that the conversion of primary bile acids to secondary bile acids is carried out by the gut bacteria. Besides having an important role in the emulsification and absorption of dietary lipids, bile acids also serve as important signalling molecules that act through the farnesoid X receptor (FXR) and G protein‐coupled bile acid receptor 1 (TGR5). By activating FXR and TGR5, bile acids can influence a variety of biological processes including bile acid metabolism, intestinal hormone secretion, inflammation, and lipid, glucose and energy metabolism. Accordingly, disturbances in the gut bacteria are suggested to affect metabolic parameters and pathways via changes in bile acid metabolism (Fiorucci & Distrutti, 2015).
Experiments in germ‐free and antibiotic‐treated mice (Swann et al. 2011; Sayin et al. 2013), as well as in humans treated with antibiotics (Vrieze et al. 2014), have indicated that the gut bacteria play an important role in the conversion of primary into secondary bile acids. In addition, germ‐free and antibiotic‐treated mice have an increased bile acid pool, increased biliary bile acid secretion and intestinal reabsorption, and decreased faecal bile acid excretion, indicating that the gut microbiota have a major impact on bile acid homeostasis (Sayin et al. 2013; Out et al. 2015). In turn, bile acids may impact the gut bacteria via their bactericidal properties, illustrating the complex relationship between the gut bacteria and bile acids (Mikkelsen et al. 2016).
The nuclear bile acid receptor FXR is expressed at high levels in the liver and small intestine, which are both tissues characterized by high concentrations of bile acids. Intestinal FXR signalling has been shown to protect against the development of obesity and to improve insulin resistance (Li et al. 2013). The impact of the microbiota on FXR signalling has been studied using germ‐free and conventionally raised wild‐type and FXR knockout mice. While the effect of the gut microbiota on insulin tolerance was found to be dependent on FXR, the influence of the gut microbiota on glucose tolerance was not (Parséus et al. 2016). Adipose tissue inflammation was significantly increased upon conventionalization in wild‐type mice, but not in FXR‐deficient mice, suggesting that the microbiota promote adipose tissue inflammation in an FXR‐dependent manner (Parséus et al. 2016). Expression of FXR is relatively low in adipose tissue suggesting that effects of bile acids on adipose tissue inflammation are likely to be mediated via intestinal FXR.
The membrane bile acid receptor TGR5 is also mainly expressed in the intestine and has been implicated in glucose metabolism. Specifically, activation of TGR5 by bile acids was shown to improve insulin sensitivity and glucose tolerance via enhanced glucagon‐like peptide‐1 (GLP‐1) secretion, which was blunted in mice lacking TGR5 (Harach et al. 2012; Potthoff et al. 2013).
Different bile acids are known to have a different potency towards FXR and TGR5 (de Aguiar Vallim et al. 2013). Accordingly, it is difficult to predict how changes in the gut bacterial composition and hence faecal bile acid composition affect bile acid signalling and subsequently impact metabolic processes. Moreover, the bile acid composition is substantially different between mice and humans, including differences in the production of the various primary bile acids and their conjugation with glycine in humans versus taurine in mice (Chiang, 2013). For this reason, the results obtained in studies on mice cannot easily be extrapolated to humans.
Short‐chain fatty acids
SCFAs are the main intestinal bacterial fermentation end products of indigestible dietary components, such as dietary fibres. It has been hypothesized that part of the beneficial health effects of dietary fibres is mediated by SCFAs (Jakobsdottir et al. 2013; den Besten et al. 2014; Chassaing et al. 2015). In line with this notion, supplementation of SCFAs to a high‐fat diet protected against diet‐induced obesity and improved insulin sensitivity (Gao et al. 2009; Lin et al. 2012; den Besten et al. 2015a). Although these data support a direct impact of SCFA on metabolic health, absorption of orally ingested SCFAs takes place already in the small intestine and not in the caecum or colon. Different hormones with various physiological functions are produced along the gastrointestinal tract (Murphy & Bloom, 2006) and as a result, SCFAs absorbed in the small intestine may give rise to different metabolic effects as compared to SCFA produced by bacterial fermentation and absorbed in the large intestine (den Besten et al. 2015b).
The effects of SCFAs on glucose homeostasis are thought to be mediated via the secretion of GLP‐1 and peptide YY (PYY) from enteroendocrine L‐cells by activation of the G protein‐coupled receptors (GPCR) 41 and 43. In vitro primary colonic cells lacking either GPCR41 or ‐43 were shown to secrete less GLP‐1 and PYY after SCFA stimulation (Tolhurst et al. 2012; Psichas et al. 2015). Targeted delivery of the SCFA propionate to the colon increased GLP‐1 and PYY in the portal vein in wild‐type mice but not in mice lacking GPCR43 (Psichas et al. 2015). Additionally, mice lacking GPCR41 and ‐43 had impaired glucose tolerance (Tolhurst et al. 2012; Kimura et al. 2013). In humans, rectal propionate infusions have been shown to increase serum glucose levels, consistent with the hypothesis of propionate being a substrate for gluconeogenesis (Wolever et al. 1991). By contrast, while targeted delivery of propionate via an inulin‐propionate ester effectively increased propionate in the colon and increased the postprandial plasma PYY and GLP‐1 concentrations, no acute effects on plasma glucose levels were found (Chambers et al. 2015). Interestingly, inulin‐propionate supplementation for 24 weeks prevented a deterioration in the glycaemic response in overweight adults (Chambers et al. 2015). However, it is unclear whether these effects are directly mediated by propionate impacting gut hormones, or indirectly via the reduction in body weight gain and adiposity in the inulin‐propionate group as compared to the control group.
The direct effects of SCFAs on adipose tissue inflammation have been investigated in only a few studies. Since the effects of SCFAs on metabolic health frequently involve changes in obesity development (Canfora et al. 2015), which can be predicted to lead to changes in adipose tissue inflammation, the direct effects of SCFAs on adipose tissue inflammation are difficult to assess in vivo.
In vitro, SCFAs have been shown to exert anti‐inflammatory effects by affecting cytokine production and chemotaxis (Cox, 2009; Maa et al. 2010; Liu et al. 2012). The effect of SCFAs on inflammation seems to be dependent on the concentration and the type of SCFA as well as on the type of immune cell (Vinolo et al. 2009; Bailón et al. 2010; Meijer et al. 2010). For example, Al‐Lahham et al. (2012) showed that propionate reduced several inflammatory cytokines and chemokines in human omental adipose tissue explants, which could only be achieved using supraphysiological concentrations (3 mmol l−1).
Taken together, there is some evidence that SCFAs affect insulin resistance and adipose tissue inflammation, but further research is necessary to better clarify the impact of intestinally derived SCFAs on insulin resistance and adipose tissue inflammation in vivo.
Conclusion
In conclusion, several potential mediators, including LPS, ANGPTL4, bile acids and SCFAs, have been proposed to link disturbances in the gut bacteria to obesity‐related insulin resistance and adipose tissue inflammation. Currently, the literature on LPS as a potential causal link between the gut microbiota and obesity‐related insulin resistance and adipose tissue inflammation is inconsistent, which may be related to the difficulty of accurately measuring LPS in blood. Bile acids are also suggested as possible mediators, but evidence from in vivo studies is limited. Since the gut bacteria impact bile acid composition and vice versa, it is difficult to investigate the role of bile acids as causal mediators of the gut bacteria. ANGPTL4 has been proposed as a link between the gut microbiota and obesity, but whether regulation of (intestinal) Angptl4 by the gut microbiota also affects insulin resistance and adipose tissue inflammation is completely unknown. In contrast to LPS, ANGPTL4 and bile acids, SCFAs have been suggested to improve glucose tolerance and reduce adipose tissue inflammation, although further research is necessary to better clarify the impact of intestinally derived SCFAs on insulin resistance and adipose tissue inflammation in vivo.
It should be noted that the observed effects of the gut microbiota and its potential mediators on insulin resistance and adipose inflammation are often confounded by changes in obesity development, which automatically impact insulin resistance and adipose tissue inflammation. Therefore, studies should be designed to better tease out the direct effect of the gut microbiota on insulin resistance and adipose tissue inflammation independent of obesity.
Studies on potential mediators linking disturbances in gut bacteria to insulin resistance and adipose tissue inflammation are mostly performed in mice. Although murine models provide a powerful tool to study host‐gut microbe interactions, they do not always very well recapitulate the human situation, partly because the composition of the gut bacteria is quite dissimilar between humans and mice (Ley et al. 2005). Pre‐clinical studies exploring the role of gut bacteria in obesity and metabolic regulation should therefore ideally be undertaken in a variety of mouse models on different diets. Moreover, as the gut bacterial composition can vary substantially in the same mouse strain depending on the animal facility and background diet, it is worthwhile to try to repeat existing studies in different animal facilities and using different diets. Finally, a more targeted approach involving modification of only one bacterial strain may provide more useful insight into the role of the gut bacteria in obesity and metabolic regulation, as compared to interventions in which nearly the entire bacterial population is modulated.
Additional information
Competing interests
The authors report no conflict of interest.
Author contributions
All authors have approved the final version of the manuscript and agree to be accountable for all aspects of the work. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This work was supported by The Netherlands Heart Foundation IN‐CONTROL grant (CVON2012‐03).
Biographies
Aafke Janssen is a PhD student in the laboratory of Sander Kersten. She received her MSc degree in Biomedical Sciences from Utrecht University in 2013. Her PhD research aims to explore the potential impact of the gut microbiota on liver function and non‐alcoholic fatty liver disease.

Sander Kersten is a Professor of Molecular Nutrition and Chair of Nutrition, Metabolism and Genomics at Wageningen University in the Netherlands. He received his PhD from Cornell University, USA, and performed his postdoctoral research at the University of Lausanne, Switzerland. The work in his laboratory is aimed at gaining more insight into the molecular mechanisms of action of dietary nutrients and their impact on human health and metabolism. His main focus is on lipids and the key organs relevant for metabolism: the intestine, liver, adipose tissue and muscle, as well as the interplay between these organs and other important systems such as the microbiome.
This is an Editor's Choice article from the 15 January 2017 issue.
References
- Alex S, Lange K, Amolo T, Grinstead JS, Haakonsson AK, Szalowska E, Koppen A, Mudde K, Haenen D, Al‐Lahham S, Roelofsen H, Houtman R, van der Burg B, Mandrup S, Bonvin AMJJ, Kalkhoven E, Müller M, Hooiveld GJ & Kersten S (2013). Short‐chain fatty acids stimulate angiopoietin‐like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator‐activated receptor γ. Mol Cell Biol 33, 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Lahham S, Roelofsen H, Rezaee F, Weening D, Hoek A, Vonk R & Venema K (2012). Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur J Clin Invest 42, 357–364. [DOI] [PubMed] [Google Scholar]
- Are A, Aronsson L, Wang S, Greicius G, Lee YK, Gustafsson J‐A, Pettersson S & Arulampalam V (2008). Enterococcus faecalis from newborn babies regulate endogenous PPARgamma activity and IL‐10 levels in colonic epithelial cells. Proc Natl Acad Sci USA 105, 1943–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson L, Huang Y, Parini P, Korach‐André M, Håkansson J, Gustafsson JÅ, Pettersson S, Arulampalam V & Rafter J (2010). Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin‐like 4 protein (ANGPTL4). PLoS One 5, e13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF & Gordon JI (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101, 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Manchester JK, Semenkovich CF & Gordon JI (2007). Mechanisms underlying the resistance to diet‐induced obesity in germ‐free mice. Proc Natl Acad Sci USA 104, 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailón E, Cueto‐Sola M, Utrilla P, Rodríguez‐Cabezas ME, Garrido‐Mesa N, Zarzuelo A, Xaus J, Gálvez J & Comalada M (2010). Butyrate in vitro immune‐modulatory effects might be mediated through a proliferation‐related induction of apoptosis. Immunobiology 215, 863–873. [DOI] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA & Pomp D (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107, 18933–18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutagy NE, McMillan RP, Frisard MI & Hulver MW (2016). Metabolic endotoxemia with obesity: Is it real and is it relevant? Biochimie 124, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutens L & Stienstra R (2016). Adipose tissue macrophages: going off track during obesity. Diabetologia 59, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R, Reigstad CS, Bäckhed HK, Reinhardt C, Ketonen M, Lundén GÖ, Cani PD & Bäckhed F (2012). Gut‐derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut 61, 1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R, Tremaroli V, Kovatcheva‐Datchary P, Cani PD & Bäckhed F (2015). Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab 22, 658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfora EE, Jocken JW & Blaak EE (2015). Short‐chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 11, 577–591. [DOI] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Sulpice T, Chamontin B, Gibson GR, Casteilla L, Delzenne NM & Alessi MC (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. [DOI] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Neyrinck AM & Delzenne NM (2008). Changes in gut microbiota control metabolic endotoxemia‐induced inflammation in high‐fat diet‐induced obesity and diabetes in mice. Diabetes 57, 1470–1481. [DOI] [PubMed] [Google Scholar]
- Carvalho BM, Guadagnini D, Tsukumo DML, Schenka AA, Latuf‐Filho P, Vassallo J, Dias JC, Kubota LT, Carvalheira JBC & Saad MJA (2012). Modulation of gut microbiota by antibiotics improves insulin signalling in high‐fat fed mice. Diabetologia 55, 2823–2834. [DOI] [PubMed] [Google Scholar]
- Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac‐Varghese SE, MacDougall K, Preston T, Tedford C, Finlayson GS, Blundell JE, Bell JD, Thomas EL, Mt‐Isa S, Ashby D, Gibson GR, Kolida S, Dhillo WS, Bloom SR, Morley W, Clegg S & Frost G (2015). Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64, 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B, Miles‐Brown J, Pellizzon M, Ulman E, Ricci M, Zhang L, Patterson AD, Vijay‐Kumar M & Gewirtz AT (2015). Lack of soluble fiber drives diet‐induced adiposity in mice. Am J Physiol Gastrointest Liver Physiol 309, G528–G541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JYL (2013). Bile acid metabolism and signaling. Compr Physiol 3, 1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MA (2009). Short‐chain fatty acids act as antiinflammatory mediators by regulating prostaglandin E2 and cytokines. World J Gastroenterol 15, 5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandona P, Ghanim H, Bandyopadhyay A, Korzeniewski K, Lia CL, Dhindsa S & Chaudhuri A (2010). Insulin suppresses endotoxin‐induced oxidative, nitrosative, and inflammatory stress in humans. Diabetes Care 33, 2416–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aguiar Vallim TQ, Tarling EJ & Edwards PA (2013). Pleiotropic roles of bile acids in metabolism. Cell Metab 17, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, Oosterveer MH, Jonker JW, Groen AK, Reijngoud D‐J & Bakker BM (2015a). Short‐chain fatty acids protect against high‐fat diet‐induced obesity via a PPARγ‐dependent switch from lipogenesis to fat oxidation. Diabetes 64, 2398–2408. [DOI] [PubMed] [Google Scholar]
- den Besten G, Gerding A, van Dijk TH, Ciapaite J, Bleeker A, van Eunen K, Havinga R, Groen AK, Reijngoud D‐J & Bakker BM (2015b). Protection against the metabolic syndrome by guar gum‐derived short‐chain fatty acids depends on peroxisome proliferator‐activated receptor γ and glucagon‐like peptide‐1. PLoS One 10, e0136364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G, Havinga R, Bleeker A, Rao S, Gerding A, van Eunen K, Groen AK, Reijngoud D‐J & Bakker BM (2014). The short‐chain fatty acid uptake fluxes by mice on a guar gum supplemented diet associate with amelioration of major biomarkers of the metabolic syndrome. PLoS One 9, e107392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk W & Kersten S (2014). Regulation of lipoprotein lipase by Angptl4. Trends Endocrinol Metab 25, 146–155. [DOI] [PubMed] [Google Scholar]
- Dollive S, Chen Y‐Y, Grunberg S, Bittinger K, Hoffmann C, Vandivier L, Cuff C, Lewis JD, Wu GD & Bushman FD (2013). Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One 8, e71806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca FA, Sakar Y, Lepage P, Devime F, Langelier B, Doré J & Covasa M (2014). Replication of obesity and associated signaling pathways through transfer of microbiota from obese‐prone rats. Diabetes 63, 1624–1636. [DOI] [PubMed] [Google Scholar]
- Dudele A, Fischer CW, Elfving B, Wegener G, Wang T & Lund S (2015). Chronic exposure to low doses of lipopolysaccharide and high‐fat feeding increases body mass without affecting glucose tolerance in female rats. Physiol Rep 3, e12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Aidy S, Merrifield CA, Derrien M, van Baarlen P, Hooiveld G, Levenez F, Doré J, Dekker J, Holmes E, Claus SP, Reijngoud DJ, Kleerebezem M, Dore J, Dekker J, Holmes E, Claus SP, Reijngoud DJ & Kleerebezem M (2013). The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut 62, 1306–1314. [DOI] [PubMed] [Google Scholar]
- Ellekilde M, Selfjord E, Larsen CS, Jakesevic M, Rune I, Tranberg B, Vogensen FK, Nielsen DS, Bahl MI, Licht TR, Hansen AK & Hansen CHF (2014). Transfer of gut microbiota from lean and obese mice to antibiotic‐treated mice. Sci Rep 4, 5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM & Cani PD (2013). Cross‐talk between Akkermansia muciniphila and intestinal epithelium controls diet‐induced obesity. Proc Natl Acad Sci USA 110, 9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S & Distrutti E (2015). Bile acid‐activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med 21, 702–714. [DOI] [PubMed] [Google Scholar]
- Fleissner CK, Huebel N, Abd El‐Bary MM, Loh G, Klaus S & Blaut M (2010). Absence of intestinal microbiota does not protect mice from diet‐induced obesity. Br J Nutr 104, 919–929. [DOI] [PubMed] [Google Scholar]
- Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT & Ye J (2009). Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58, 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, Didonato JA, Lusis AJ, Hazen SL, Gregory JC, Buffa JA, Org E, Wang Z, Levison BS & Zhu W (2015). Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem 290, 5647–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harach T, Pols TWH, Nomura M, Maida A, Watanabe M, Auwerx J & Schoonjans K (2012). TGR5 potentiates GLP‐1 secretion in response to anionic exchange resins. Sci Rep 2, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Liu T, Rose JL, Stevens RL & Hoyt DG (2007). Sensitivity of mice to lipopolysaccharide is increased by a high saturated fat and cholesterol diet. J Inflamm (Lond) 4, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JC (1995). Endotoxemia: Methods of detection and clinical correlates. Clin Microbiol Rev 8, 268–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsdottir G, Xu J, Molin G, Ahrné S & Nyman M (2013). High‐fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fibre counteracts these effects. PLoS One 8, e80476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen AWF & Kersten S (2015). The role of the gut microbiota in metabolic health. FASEB J 29, 3111–3123. [DOI] [PubMed] [Google Scholar]
- Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, Hu Y, Li J & Liu Y (2015). Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non‐alcoholic fatty liver disease. Sci Rep 5, 8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F & Nielsen J (2012). Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 3, 1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, Terasawa K, Kashihara D, Hirano K, Tani T, Takahashi T, Miyauchi S, Shioi G, Inoue H & Tsujimoto G (2013). The gut microbiota suppresses insulin‐mediated fat accumulation via the short‐chain fatty acid receptor GPR43. Nat Commun 4, 1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korecka A, de Wouters T, Cultrone A, Lapaque N, Pettersson S, Doré J, Blottière HM & Arulampalam V (2013). ANGPTL4 expression induced by butyrate and rosiglitazone in human intestinal epithelial cells utilizes independent pathways. Am J Physiol Gastrointest Liver Physiol 304, G1025–G1037. [DOI] [PubMed] [Google Scholar]
- Laugerette F, Alligier M, Bastard JP, Drai J, Chanséaume E, Lambert‐Porcheron S, Laville M, Morio B, Vidal H & Michalski MC (2014). Overfeeding increases postprandial endotoxemia in men: Inflammatory outcome may depend on LPS transporters LBP and sCD14. Mol Nutr Food Res 58, 1513–1518. [DOI] [PubMed] [Google Scholar]
- Laugerette F, Furet J‐P, Debard C, Daira P, Loizon E, Geloen A, Soulage CO, Simonet C, Lefils‐Lacourtablaise J, Bernoud‐Hubac N, Bodennec J, Peretti N, Vidal H & Michalski M‐C (2012). Oil composition of high‐fat diet affects metabolic inflammation differently in connection with endotoxin receptors in mice. Am J Physiol Endocrinol Metab 302, E374–E386. [DOI] [PubMed] [Google Scholar]
- Laukens D, Brinkman BM, Raes J, De Vos M & Vandenabeele P (2016). Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol Rev 40, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD & Gordon JI (2005). Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102, 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein SGJ (2006). Human gut microbes associated with obesity. Nature 444, 1022–1023. [DOI] [PubMed] [Google Scholar]
- Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD & Gonzalez FJ (2013). Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun 4, 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Mendis N, Trigui H, Oliver JD & Faucher SP (2014). The importance of the viable but non‐culturable state in human bacterial pathogens. Front Microbiol 5, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein L, Berbée JFP, van Dijk SJ, van Dijk KW, Bensadoun A, Kema IP, Voshol PJ, Müller M, Rensen PCN & Kersten S (2007). Angptl4 upregulates cholesterol synthesis in liver via inhibition of LPL‐ and HL‐dependent hepatic cholesterol uptake. Arterioscler Thromb Vasc Biol 27, 2420–2427. [DOI] [PubMed] [Google Scholar]
- Lichtenstein L, Mattijssen F, de Wit NJ, Georgiadi A, Hooiveld GJ, van der Meer R, He Y, Qi L, Köster A, Tamsma JT, Tan NS, Müller M & Kersten S (2010). Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metab 12, 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HV, Frassetto A, Kowalik EJ, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G & Marsh DJ (2012). Butyrate and propionate protect against diet‐induced obesity and regulate gut hormones via free fatty acid receptor 3‐independent mechanisms. PLoS One 7, e35240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Kao C, Chung C & Hsieh P (2010). Chronic hepatic inflammation induced by mild portal endotoxemia is not associated with systemic insulin resistance in rats. J Med Sci 30, 15–20. [Google Scholar]
- Liu T, Li J, Liu Y, Xiao N, Suo H, Xie K, Yang C & Wu C (2012). Short‐chain fatty acids suppress lipopolysaccharide‐induced production of nitric oxide and proinflammatory cytokines through inhibition of NF‐κB pathway in RAW264.7 cells. Inflammation 35, 1676–1684. [DOI] [PubMed] [Google Scholar]
- Maa MC, Chang MY, Hsieh MY, Chen YJ, Yang CJ, Chen ZC, Li YK, Yen CK, Wu RR & Leu TH (2010). Butyrate reduced lipopolysaccharide‐mediated macrophage migration by suppression of Src enhancement and focal adhesion kinase activity. J Nutr Biochem 21, 1186–1192. [DOI] [PubMed] [Google Scholar]
- Mattijssen F, Alex S, Swarts HJ, Groen AK, van Schothorst EM & Kersten S (2014). Angptl4 serves as an endogenous inhibitor of intestinal lipid digestion. Mol Metab 3, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta NN, McGillicuddy FC, Anderson PD, Hinkle CC, Shah R, Pruscino L, Tabita‐Martinez J, Sellers KF, Rickels MR & Reilly MP (2010). Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes 59, 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer K, de Vos P & Priebe MG (2010). Butyrate and other short‐chain fatty acids as modulators of immunity: what relevance for health? Curr Opin Clin Nutr Metab Care 13, 715–721. [DOI] [PubMed] [Google Scholar]
- Membrez M, Blancher F, Jaquet M, Bibiloni R, Cani PD, Burcelin RG, Corthesy I, Macé K & Chou CJ (2008). Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J 22, 2416–2426. [DOI] [PubMed] [Google Scholar]
- Mikkelsen KH, Allin KH & Knop FK (2016). Effect of antibiotics on gut microbiota, glucose metabolism and bodyweight regulation – a review of the literature. Diabetes Obes Metab 18, 444–453. [DOI] [PubMed] [Google Scholar]
- Mikkelsen KH, Frost M, Bahl MI, Licht TR, Jensen US, Rosenberg J, Pedersen O, Hansen T, Rehfeld JF, Holst JJ, Vilsbøll T & Knop FK (2015). Effect of antibiotics on gut microbiota, gut hormones and glucose metabolism. PLoS One 10, e0142352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgun A, Dzutsev A, Dong X, Greer RL, Sexton DJ, Ravel J, Schuster M, Hsiao W, Matzinger P & Shulzhenko N (2015). Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut 64, 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID & Allard JP (2013). Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58, 120–127. [DOI] [PubMed] [Google Scholar]
- Mulligan ME, Citron DM, McNamara BT & Finegold SM (1982). Impact of cefoperazone therapy on fecal flora. Antimicrob Agents Chemother 22, 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KG & Bloom SR (2006). Gut hormones and the regulation of energy homeostasis. Nature 444, 854–859. [DOI] [PubMed] [Google Scholar]
- Out C, Patankar JV, Doktorova M, Boesjes M, Bos T, de Boer S, Havinga R, Wolters H, Boverhof R, van Dijk TH, Smoczek A, Bleich A, Sachdev V, Kratky D, Kuipers F, Verkade HJ & Groen AK (2015). Gut microbiota inhibit Asbt‐dependent intestinal bile acid reabsorption via Gata4. J Hepatol 63, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parséus A, Sommer N, Sommer F, Caesar R, Molinaro A, Ståhlman M, Greiner TU, Perkins R & Bäckhed F (2016). Microbiota‐induced obesity requires farnesoid X receptor. Gut (in press; doi: 10.1136/gutjnl-2015-310283). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendyala S, Walker JM & Holt PR (2012). A high‐fat diet is associated with endotoxemia that originates from the gut. Gastroenterology 142, 1100–1101.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Potts A, He T, Duarte JAG, Taussig R, Mangelsdorf DJ, Kliewer SA & Burgess SC (2013). Colesevelam suppresses hepatic glycogenolysis by TGR5‐mediated induction of GLP‐1 action in DIO mice. Am J Physiol Gastrointest Liver Physiol 304, G371–G380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psichas A, Sleeth ML, Murphy KG, Brooks L, Bewick GA, Hanyaloglu AC, Ghatei MA, Bloom SR & Frost G (2015). The short chain fatty acid propionate stimulates GLP‐1 and PYY secretion via free fatty acid receptor 2 in rodents. Int J Obes (Lond) 39, 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K & Wang J (2012). A metagenome‐wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. [DOI] [PubMed] [Google Scholar]
- Rabot S, Membrez M, Bruneau A, Gérard P, Harach T, Moser M, Raymond F, Mansourian R & Chou CJ (2010). Germ‐free C57BL/6J mice are resistant to high‐fat‐diet‐induced insulin resistance and have altered cholesterol metabolism. FASEB J 24, 4948–4959. [DOI] [PubMed] [Google Scholar]
- Rajilić‐Stojanović M & de Vos WM (2014). The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38, 996–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC & Gordon JI (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H‐U, Bamberg K, Angelin B, Hyötyläinen T, Orešič M & Bäckhed F (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro‐beta‐muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17, 225–235. [DOI] [PubMed] [Google Scholar]
- Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, Basic M, Dupont A, Hornef M, Von Bergen M, Bleich A & Haller D (2016). Dysbiotic gut microbiota causes transmissible Crohn's disease‐like ileitis independent of failure in antimicrobial defence. Gut 65, 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C & Hardt PD (2010). Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18, 190–195. [DOI] [PubMed] [Google Scholar]
- Smith K, McCoy KD & Macpherson AJ (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 19, 59–69. [DOI] [PubMed] [Google Scholar]
- Sommer F & Bäckhed F (2013). The gut microbiota–masters of host development and physiology. Nat Rev Microbiol 11, 227–238. [DOI] [PubMed] [Google Scholar]
- Steel HC, Theron AJ, Cockeran R, Anderson R & Feldman C (2012). Pathogen‐ and host‐directed anti‐inflammatory activities of macrolide antibiotics. Mediators Inflamm 2012, 584262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK & Holmes E (2011). Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA 108 Suppl, 4523–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber SC & Nau R (2008). Immunomodulatory properties of antibiotics. Curr Mol Pharmacol 1, 68–79. [PubMed] [Google Scholar]
- Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F & Gribble FM (2012). Short‐chain fatty acids stimulate glucagon‐like peptide‐1 secretion via the G‐protein‐coupled receptor FFAR2. Diabetes 61, 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinolo MAR, Rodrigues HG, Hatanaka E, Hebeda CB, Farsky SHP & Curi R (2009). Short‐chain fatty acids stimulate the migration of neutrophils to inflammatory sites. Clin Sci (Lond) 117, 331–338. [DOI] [PubMed] [Google Scholar]
- Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga‐Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB & Nieuwdorp M (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916. [DOI] [PubMed] [Google Scholar]
- Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, van Nood E, Holleman F, Knaapen M, Romijn JA, Soeters MR, Blaak EE, Dallinga‐Thie GM, Reijnders D, Ackermans MT, Serlie MJ, Knop FK, Holst JJ, van der Ley C, Kema IP, Zoetendal EG, de Vos WM, Hoekstra JB, Stroes ES, Groen AK & Nieuwdorp M (2014). Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol 60, 824–831. [DOI] [PubMed] [Google Scholar]
- Walsh CJ, Guinane CM, O'Toole PW & Cotter PD (2014). Beneficial modulation of the gut microbiota. FEBS Lett 588, 4120–4130. [DOI] [PubMed] [Google Scholar]
- Wang X, Ryu D, Houtkooper RH & Auwerx J (2015). Antibiotic use and abuse: A threat to mitochondria and chloroplasts with impact on research, health, and environment. Bioessays 37, 1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolever T, Spadafora P & Eshuis H (1991). Interaction between colonic and propionate in humans. Am J Clin Nutr 53, 681–687. [DOI] [PubMed] [Google Scholar]
- Wu H, Tremaroli V & Bäckhed F (2015). Linking microbiota to human diseases: a systems biology perspective. Trends Endocrinol Metab 26, 758–770. [DOI] [PubMed] [Google Scholar]
- Xu A, Lam MC, Chan KW, Wang Y, Zhang J, Hoo RLC, Xu JY, Chen B, Chow W, Tso AWK & Lam KSL (2005). Angiopoietin‐like protein 4 decreases blood glucose and improves glucose tolerance but induces hyperlipidemia and hepatic steatosis in mice. Proc Natl Acad Sci USA 102, 6086–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P & Li L (2012). The germfree murine animal: An important animal model for research on the relationship between gut microbiota and the host. Vet Microbiol 157, 1–7. [DOI] [PubMed] [Google Scholar]


