Abstract
Amino acids and amino acid analogs have been used in numerous genetic screens to isolate mutants deficient in amino acid biosynthetic pathways or in the regulation of amino acid metabolism. Several of these mutants exhibit relaxed feedback control of branched amino acid biosynthetic pathways and are thus resistant to accumulation of pathway end products. For example, feedback-regulated enzymes of the shikimate pathway are anthranilate synthase on the branch leading to Trp and chorismate mutase on the branch leading to Phe and Tyr. A feedback-insensitive mutant of anthranilate synthase α, trp5-1, is resistant to toxic Trp analogs. Mutants resistant to Phe have not previously been reported, and this article describes the isolation of the recessive Arabidopsis Phe insensitive growth mutant pig1-1 by a forward genetic screen. pig1-1 was not only tolerant to Phe, Tyr, and Trp, but also to other, not biosynthetically related amino acids. Amino acid contents in pig1-1 were significantly elevated with respect to wild-type controls but, in contrast to the wild type, dramatically decreased when plants were supplemented with 2 mm Phe. Protein contents were similar in the mutant and the wild type at all tested conditions. Phe catabolism was similar to the wild type in pig1-1 roots but was significantly increased in pig1-1 shoots. Phenylalanine uptake into the root, its root-to-shoot translocation, and Phe and phenylpropanoid contents were unaltered in pig1-1, indicating that pig1-1 is not affected in amino acid translocation or the shikimate pathway. Instead, the response of pig1-1 toward amino acid feeding indicates that amino acid metabolism is generally deregulated in pig1-1.
End product inhibition is frequently observed at branching points of branched amino acid biosynthetic pathways to balance the flux between the different amino acid end products in one pathway. External supply of feedback-competent amino acids to the plant growth medium disturbs amino acid homeostasis by inhibiting feedback-regulated enzymes and thus causes shortages of other amino acids resulting from these biosynthetic pathways. Naturally, such induced amino acid shortages are toxic to the plant.
These toxic effects of externally applied amino acids have been employed to isolate feedback-insensitive Asp kinase mutants (Rognes et al., 1983; Arruda et al., 1984; Miao et al., 1988; Heremans and Jacobs, 1995, 1997) and acetohydroxy acid synthase mutants (Hervieu and Vaucheret, 1996). Alternatively, herbicidal substrate analogs have been used to screen for mutants in amino acid biosynthetic pathways (Last et al., 1991; Mourad and King, 1995; Li and Last, 1996; Radwanski et al., 1996).
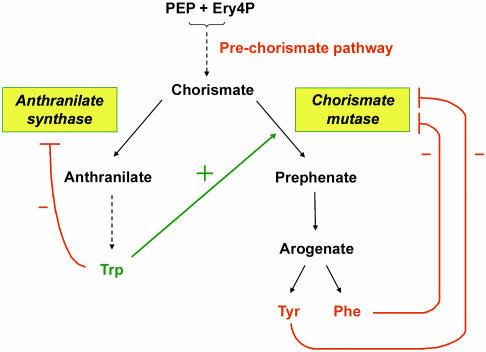
Feedback regulation in the plant shikimate pathway occurs at the branching point chorismate (Schmid and Amrhein, 1995), where the chorismate mutase and anthranilate synthase (AS) branches compete for the common substrate chorismate (Fig. 1). Chorismate mutase is the target for negative feedback control by Phe and Tyr, while AS is inhibited by Trp (Schmid and Amrhein, 1995). Arabidopsis mutants with a relaxed feedback control of Trp were isolated in the 1990s by either screening for accumulation of the intermediate anthranilate or for resistance to toxic Trp analogs such as 6-methyltryptophan (Kreps et al., 1996; Li and Last, 1996; Bender and Fink, 1998). It was demonstrated that mutations in the AS α-subunit gene (Kreps et al., 1996; Li and Last, 1996) and in a transcriptional regulator of AS (Bender and Fink, 1998) convey Trp insensitivity. The trp5-1 mutant, which is defective in AS α, contains 3-fold more soluble Trp than the wild type (Li and Last, 1996).
Figure 1.
Allosteric feedback regulation of chorismate utilization in plants. Chorismate mutase constitutes the committed and regulated step toward Phe and Tyr, and AS catalyses the committed step toward Trp. Phe and Tyr feedback inhibit chorismate mutase, which is activated by Trp. In turn, AS is feedback inhibited by Trp.
Elevated contents of particular free amino acids could also be attained by introducing feedback-insensitive isoforms into plants. For example, overexpressing feedback-insensitive orthologs of AS, Asp kinase, acetohydroxy acid synthase, or dihydrodipicolinate synthase from plants or bacteria (Falco et al., 1995; Cho et al., 2000; Galili et al., 2000; Lee and Duggleby, 2002; Zhu and Galili, 2003) have resulted in substantially increased free Trp, Thr, Val, and Lys contents, respectively.
Here, we describe the isolation of an Arabidopsis Phe insensitive growth (pig) mutant, which is able to germinate in the presence of 10 mm Phe, while 6 mm Phe completely inhibits the germination of wild-type Arabidopsis. When grown on Murashige and Skoog (MS) agar plates, free amino acids in pig1-1 are elevated by 1.4- to 2.3-fold compared to the wild type. However, when grown on media supplemented with 2 mm Phe, free amino acid contents, except Phe and Tyr, are up to 4-fold lower in pig1-1 than in the wild type. In addition to this concomitant decrease of all free amino acids upon external Phe supply, pig1-1 displays an increased tolerance to a range of externally applied, feedback-competent amino acids. This indicates that a regulatory mechanism of amino acid metabolism, which may resemble or correspond to the general control in yeast (Saccharomyces cerevisiae), is deregulated in pig1-1. To the best of our knowledge, pig1-1 is the first Arabidopsis mutant that is characterized by constitutively increased foliar free amino acid contents.
RESULTS
Isolation of Phe-Insensitive Arabidopsis Mutants
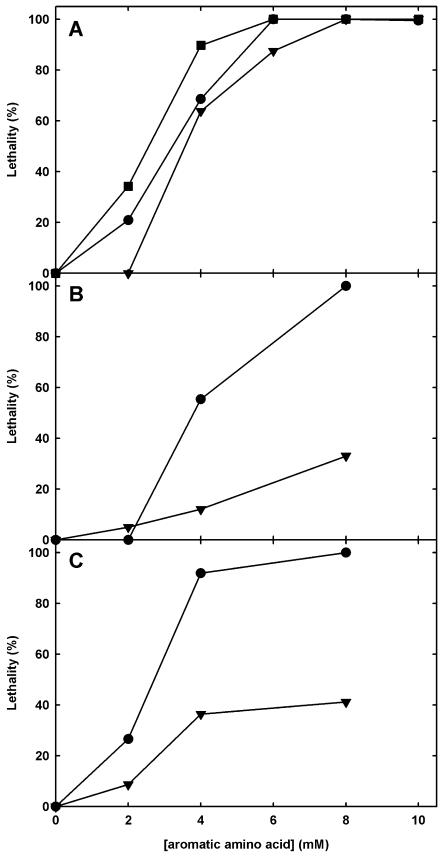
To establish a robust screen for Phe-resistant Arabidopsis mutants, we assessed the impact of aromatic amino acids on seed germination in dose-response curves for the wild-type accessions Col-0 (data not shown) and Ws-2 (Fig. 2A). Aromatic amino acid concentrations higher than 6 mm completely inhibited germination of the Arabidopsis wild types (for Ws-2, see Fig. 2A). Seedlings were not viable on soil if they had been preincubated on aromatic amino acids for more than 14 d before they were transplanted to soil (data not shown). When intermediate concentrations of 2 to 4 mm aromatic amino acid were applied, seedling lethality was most pronounced in media containing Trp but less severe in media containing Tyr (Fig. 2), irrespective of the accession tested. The calculated I50 values for the Ws-2 accession were 2.3 mm Trp, 3.7 mm Tyr, and 4.0 mm Phe. Remarkably, the lethality of all three amino acids to Arabidopsis seedlings decreased with increasing Suc supplements, indicating a cross-talk between carbon availability and aromatic amino acid toxicity.
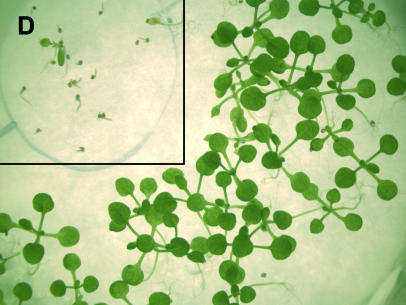
Figure 2.
Toxicity of aromatic acids (•, Phe; ▾, Tyr; ▪, Trp) to wild-type Arabidopsis (cv Ws-2) in percent (A), dose-response curves for wild type (•) and pig1-1 (▾) after 14 d on different Tyr (B) and Trp (C) concentrations, and comparison of pig1-1 and Ws-2 after 14 d of growth on half-strength MS, 10 mm Phe, and 0.5% Suc (D). Per individual treatment, 100 to 200 seeds of the wild-type Ws-2 and pig1-1 were germinated on half-strength MS, 0.5% Suc supplemented with each of the given amino acid concentrations, and the survival rate was scored after 14 d of culture at 50 μmol m−2 s−1. A toxic effect was judged to be present when the plantlets died before the emergence of primary leaves or failed to germinate. The data were corrected for the germination rates on half-strength MS plates. Because of limiting Tyr solubility, the data point for 8 mm Tyr has been extrapolated according to the dose-response curves and the calculated I50 (see “Results”) for Phe and Trp. Note that the toxicity of 10 mm Phe to pig1-1 was 18.8% ± 3.2% (data not shown).
We employed half-strength MS plates supplemented with 10 mm Phe and 0.5% Suc to screen an M2 population of 200,000 ethyl methanesulfonate-treated Arabidopsis (cv Ws-2) individuals as described in more detail in “Materials and Methods.” In this screening, eight mutant lines were isolated. Hence, the pig mutation appears at a frequency of 1/25,000. Among the isolated mutants, the growth of pig1-1 was least affected on medium containing 10 mm Phe; therefore, pig1-1 was selected for further analysis. The Phe-insensitive phenotype of pig1-1 was confirmed in the M3 and M4 generations before pig1-1 was backcrossed three times to the wild type to clean the genetic background from second site mutations. Backcrosses of pig1-1 pollen to wild-type stigmata demonstrated that the pig1-1 phenotype is caused by a single nuclear recessive mutation because of two observations: (1) the F1 progeny of backcrosses to the wild type was uniformly Phe intolerant when grown on MS-Phe, and (2) the ratio of Phe-susceptible to Phe-tolerant F2 individuals of these backcrosses was 204 to 55, which best fits a 3:1 segregation according to χ2 tests.
pig1-1 Confers Tolerance to High External Aromatic Amino Acid Concentrations
While 10 mm Phe is lethal to both tested wild-type accessions, pig1-1 seedlings displayed a germination rate of 81.2% ± 3.2% on 10 mm Phe containing MS. In addition, pig1-1 also tolerated elevated Tyr and Trp concentrations (Fig. 2). Eight millimolar of either Tyr or Trp were 100% lethal to the wild type, whereas 66% of the pig1-1 seedlings survived on 8 mm Tyr and 59% survived on 8 mm Trp. The values for 8 mm Tyr are extrapolated because Tyr solubility in the medium was limited to slightly less than 8 mm.
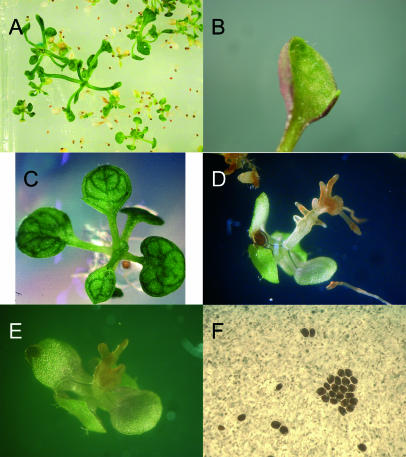
While the dose-response curves of Phe, Tyr, and Trp for wild-type seeds did not indicate pronounced differences between the treatments (Fig. 2), specific effects for individual aromatic amino acids could be observed upon feeding intermediate concentrations (4 mm) to wild-type (cv Ws-2) seedlings (Fig. 3). To demonstrate that the degree of aromatic amino acid toxicity depended on carbon supply, Figure 3 depicts growth effects observed on half-strength MS, 4 mm Phe, and 2% Suc, i.e. milder selection conditions than in the mutant screen where 0.5% Suc and 10 mm Phe were employed. Interestingly, feeding of 4 mm Phe or Tyr to the wild type induced a reticulate leaf phenotype with darker paraveinal tissue and light green to chlorotic intercostal regions (Fig. 3, A and C). A similar phenotype has been reported for the cue1 mutant that is deficient in a plastidic phosphoenolpyruvate/phosphate translocator PPT (Li et al., 1995; Streatfield et al., 1999) and that displays a disturbed secondary metabolism (Streatfield et al., 1999; Voll et al., 2003). Supplying Phe induced anthocyanin accumulation (Fig. 3B), while Trp feeding resulted in extremely dwarfish rosette growth and almost completely abrogated root growth, utterly altering root morphology (Fig. 3D). Feeding a cocktail of all three aromatic amino acids (4 mm each) phenocopied Trp feeding (Fig. 3E), whereas a 10 mm cocktail was toxic to seeds (Fig. 3F). However, these pronounced and specific effects of externally applied aromatic amino acids on wild-type plants were never observed in the pig1-1 mutant, even at aromatic amino acid concentrations up to 10 mm. Feeding 4 mm aromatic amino acids to pig1-1 provoked some minor effects in root growth (data not shown): Positive gravitropism was abrogated in pig1-1 roots; they grew stunted and developed only a few root hairs, which were confined to small regions on the stunted primary root or the hypocotyls.
Figure 3.
Growth habit of 21-d-old Ws-2 plants on half-strength MS, 2% Suc and sublethal aromatic amino acid concentrations. A and B, 4 mm Phe; C, Tyr; D, Trp; E, 4 mm of each aromatic amino acid. F depicts the complete growth inhibition of Ws-2 on half-strength MS, 0.5% Suc and a cocktail of 10 mm of each aromatic amino acid.
pig1-1 Does Not Share Phenotypic Features of trp Mutants
The pathway for aromatic amino acid biosynthesis is bifurcated with one branch leading to Trp and the other one leading to Phe and Tyr. The loci encoding enzymes of the biosynthetic branch toward Trp, TRP1 to TRP5, are covered with mutants (for summary, see Li and Last, 1996) that are characterized by Trp auxotrophy (trp2-1 and trp3-1, trp4-1/trp1-100 double mutants), resistance to Trp analogs (trp1-100 and trp5-1), and Trp accumulation (for trp5-1).
Because Phe and Trp are biosynthetically related, it was investigated whether pig1-1 shared one of the already described trp phenotypes. pig1-1 and the trp mutants were subjected to the screening conditions that had been previously employed to isolate the individual trp mutants (Table I). Alternatively, pig1-1 was grown alongside the Trp auxotrophic mutants trp2-1 and trp3-1 on Trp-deplete medium (Table I). pig1-1 did not share any of the examined traits with the individual trp mutants, and, vice versa, the trp mutants trp1-100, trp2-1, trp3-1, and trp5-1 did not display tolerance to 10 mm Phe. In summary, this suggests that the mutation in pig1-1 differs from the mutations in trp1-100, trp2-1, trp3-1, and trp5-1.
Table I.
Compilation of conducted tests to check whether the pig1-1 phenocopies one of the trp mutants
| trp Allele | Affected Gene | Reference | Identified By |
|---|---|---|---|
| trp1-100 | Phosphoribosyl anthranilate transferase (PAT) | Rose et al. (1992), Niyogi et al. (1993) | UV-A fluorescence on 50 μm Trp/Phe/Tyr/PABAa |
| trp2-1 | Trp synthase β | Last et al. (1991) | Resistance to 50 μm 5-Methyl-Trpa, 50 Trp auxotrophy (±50 μm Trp)a |
| trp3-1 | Trp synthase α | Radwanski et al. (1996) | Resistance to 50 μm 5-methyl-Trpa, Trp auxotrophy (±50 μm Trp)a |
| trp5-1 | AS α | Li and Last (1996) | Resistance to 50 μm 5-methyl-Trpa |
The indicated substances were supplemented to half-strength MS, 2% Suc plates, and growth or UV fluorescence of the plantlets was scored after 21 d. Nonsupplemented plates served as controls.
pig1-1 Displays a Deregulated Amino Acid Biosynthesis
Because of its Phe-insensitive growth phenotype, examining amino acid contents in pig1-1 was of paramount interest. Therefore, the free amino acid contents of wild type and pig mutants grown on MS agar plates with and without 2 mm Phe were determined (Table II). An external Phe concentration of 2 mm was chosen to elucidate the physiological effects of Phe supply, as 2 mm Phe did not cause severe growth retardation in the wild type (Fig. 2). It should be noted that the free amino acid pools of plants grown on MS closely resembled that of plants cultivated on soil at standard growth conditions (at a photon flux density [PFD] of 100 μmol m−2 s−1, 12/12-h light/dark cycle, 21°/18°C; data not shown for simplicity).
Table II.
Free amino acid contents in pig1-1 and wild-type (Ws-2) plantlets grown on nonsupplemented MS and MS + 2 mm Phe agar plates
| Amino Acid | Amino Acid Ratio on MS
|
Amino Acid Ratio on 2 mm Phe
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| MS
|
MS + 2 mm Phe
|
MS + 2 mm Phe
|
Fold Increase in pig1-1+
|
Fold Decrease in pig1-1+
|
Repression Ratio*
|
||||
| Ws-2 | pig1-1 | Ws-2 | pig1-1 | Ws-2 | pig1-1 | pig1-1/Ws-2 | Ws-2/pig1-1 | pig1-1/Ws-2 | |
| nmol g fw−1 | mol % | ||||||||
| Glu | 250 ± 11 | 403 ± 26a | 580 ± 35 | 336 ± 25a | 2.6 ± 0.1 | 3.7 ± 0.5a | 1.6 | 1.7 | 2.8 |
| Gln | 1,207 ± 61 | 2,650 ± 345a | 9,372 ± 933 | 3,251 ± 458a | 38.6 ± 1.3 | 34.8 ± 0.7a | 2.2 | 2.9 | 6.3 |
| Asp | 146 ± 9 | 223 ± 11a | 296 ± 8 | 208 ± 10a | 1.4 ± 0.1 | 2.3 ± 0.3a | 1.5 | 1.4 | 2.2 |
| Asn | 624 ± 65 | 1,132 ± 164a | 3,427 ± 353 | 1,379 ± 157a | 15.3 ± 0.3 | 14.9 ± 0.4 | 1.8 | 2.5 | 4.5 |
| Ser | 457 ± 13 | 665 ± 33a | 785 ± 35 | 684 ± 19a | 3.6 ± 0.3 | 7.6 ± 0.9a | 1.5 | 1.1 | 1.7 |
| Gly | 430 ± 27 | 756 ± 80a | 717 ± 85 | 459 ± 9a | 3.2 ± 0.2 | 6.5 ± 0.5a | 1.8 | 1.6 | 2.7 |
| Ala | 147 ± 4 | 203 ± 16a | 452 ± 37 | 177 ± 3a | 2.0 ± 0.1 | 2.3 ± 0.1a | 1.4 | 2.6 | 3.5 |
| Cys | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| His | 16 ± 1 | 23 ± 3a | 147 ± 9 | 53 ± 9a | 0.7 ± 0.03 | 0.6 ± 0.03a | 1.5 | 2.8 | 4.0 |
| Arg | 777 ± 49 | 635 ± 18a | 760 ± 56 | 719 ± 37a | 3.4 ± 0.2 | 7.9 ± 0.5a | 0.8 | 1.1 | 0.9 |
| Pro | 162 ± 13 | 375 ± 48a | 445 ± 51 | 307 ± 3a | 1.8 ± 0.2 | 3.9 ± 0.2a | 2.3 | 1.4 | 3.4 |
| Lys | 24 ± 2 | 14.6 ± 0.6a | 38 ± 1 | 29 ± 2a | 0.2 ± 0.02 | 0.3 ± 0.02a | 0.6 | 1.3 | 0.8 |
| Met | 4.4 ± 0.3 | 7.8 ± 0.7a | 31 ± 2 | 8.1 ± 0.6a | 0.1 ± 0.0 | 0.1 ± 0.01a | 1.8 | 3.9 | 6.9 |
| Thr | 307 ± 33 | 377 ± 38 | 984 ± 37 | 624 ± 60a | 4.5 ± 0.2 | 6.8 ± 0.3a | 1.2 | 1.6 | 1.9 |
| Val | 29 ± 1 | 35 ± 3a | 122 ± 9 | 42 ± 4a | 0.6 ± 0.03 | 0.5 ± 0.02a | 1.2 | 2.9 | 3.5 |
| Ile | 8.1 ± 0.7 | 7.0 ± 0.4 | 20 ± 1 | 14 ± 1a | 0.1 ± 0.01 | 0.2 ± 0.01a | 0.9 | 1.4 | 1.2 |
| Leu | 35 ± 6 | 35 ± 3 | 124 ± 10 | 85 ± 9a | 0.6 ± 0.1 | 0.8 ± 0.06a | 1.0 | 1.5 | 1.4 |
| Phe | 13.6 ± 0.7 | 12.2 ± 0.8 | 4,654 ± 373 | 400 ± 98a | 21.0 ± 1.0 | 6.7 ± 1.3a | 0.9 | 11.6 | 10.5 |
| Tyr | 7.6 ± 0.6 | 4.8 ± 0.6a | 91 ± 5 | 17 ± 3a | 0.4 ± 0.02 | 0.2 ± 0.01a | 0.6 | 5.3 | 3.3 |
| Trp | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Total | 4,559 ± 229 | 8,356 ± 1,134a | 22,328 ± 2,009 | 7,819 ± 344a | 100 | 100 | 1.8 | 2.9 | 5.2 |
Ws-2 and pig1-1 seeds had been germinated and grown at a PFD of 150 μmol m−2 s−1 on half-strength MS, 0.5% Suc with or without 2 mm Phe prior to harvesting whole plantlets after 14 d. From left to right: The four columns on the left represent absolute free amino acid contents in nmol g fw−1, while the next two columns give the relative amino acid abundance for Phe-replete grown plantlets in mol %. Based on the absolute values shown, ratios between free amino acid contents in pig1-1 and the wild type were calculated (right three columns, ratios are either given as pig1-1 to Ws-2 or Ws-2 to pig1-1 to indicate an increase or decrease, respectively). n.d., Not determined. +, In comparison to the wild-type Ws-2. *, The repression ratio (rr) expresses how much more the content of any given amino acid (aa) is diminished in pig1-1 compared to the wild type upon feeding 2 mm Phe. It was calculated as rr = ([aa]pig on soil/[aa]Ws on soil) × ([aa]Ws on Phe/[aa]pig on Phe). The data represent the mean values ± sd of five independent replicates (n). The data were tested for significant differences between the wild type and the mutants using the Welsh method.
Significant differences with P < 0.01 are indicated.
When grown on MS, the total amino acid content in pig1-1 was 1.8-fold increased as compared to the Ws-2 controls (Table II). Major amino acids were elevated between 1.4- and 2.3-fold in pig1-1, whereas some minor amino acids, including Phe, were essentially unaltered in comparison to the wild type (Table II).
By contrast, free amino acid contents except for Phe and Tyr decreased up to 4-fold in pig1-1 but not in the wild type, when plants were grown on 2 mm Phe (Table II). In pig1-1, Phe and Tyr decreased 5- and 11-fold under these conditions in comparison to the wild type, respectively. Taken together, Phe, Gln, and Asn accounted for more than 85% of the total amino acid discrepancy between pig1-1 and the wild type. However, the protein content in pig1-1 (5.0 ± 0.4 mg protein g fw−1) was similar to wild type (5.1 ± 0.4 mg protein g fw−1), indicating a specific effect on the free amino acid pool.
Compared to plantlets grown on MS, the usually low-abundant amino acid Phe accumulated 340-fold on 2 mm Phe in the wild type but only 30-fold in pig1-1 (Table II). This suggests that Phe uptake and/or catabolism could be altered in pig1-1. Nevertheless, both the mutant and the wild type showed a 25% increase of soluble protein when Phe was provided (from approximately 3.8 mg protein g fw−1 on MS to 5.0 mg protein g fw−1 on 2 mm Phe), indicating that organic nitrogen in form of Phe was allocated into protein in both lines.
Expressing the data for 2 mm Phe as molar percentages to correct for the large differences in total amino acid contents between pig1-1 and the wild type evinced that the relative compositions of the amino acid pools in pig1-1 and the control were similar, except for a massive decrease of Phe in pig1-1 (Table II). Nevertheless, the total free amino acid pool in pig1-1 apparently decreased relative to the control on 2 mm Phe (see above). To express the different effects of Phe feeding on pig1-1 and the wild type in numbers, a repression ratio was calculated that is indicative of the relative decrease of a particular amino acid content in pig1-1 in comparison to the wild type upon Phe supply (Table II). Phe displayed the strongest repression ratio, 10.5, on 2 mm Phe (Table II). Gln and Asn displayed repression ratios of 6.3 and 4.5, respectively, which is close to the average repression ratio (5.2) for all amino acids (Table II). Most of the other amino acids were only up to 4-fold deregulated in pig1-1, underlining the fact that changes in Phe, Gln, and Asn account for the largest portion of the decrease of the total amino acid pool in pig1-1.
Importantly, the Glu-to-Gln and Asp-to-Asn ratios remained constant in pig1-1 in all experimental conditions, while these ratios exhibited a considerable variation in the wild type (Table III). In consequence, a nitrogen overflow with a more than 5-fold increase in total amino acid content, as observed in the wild type, is prevented in pig1-1 (Table II). In milder conditions, i.e. MS or MS + 2 mm Phe at low light (LL; 50 μmol m−2 s−1), pig1-1 exhibited a 3- to 4-fold lower Glu-to-Gln ratio, whereas the Pro-to-Glu and the Asp-to-Asn ratios were similar to the wild type (Table III). This suggests a specific imbalance between Glu and Gln and a nitrogen surplus in pig1-1 at LL. In these conditions, the Ser-to-Gly ratio was decreased by 30% to 40% in pig1-1 (Table III), indicating a lower flux through the photorespiratory C2 cycle in LL, especially as Ser and Gly contents were substantially diminished compared to pig1-1 or wild type at all other conditions (data not shown).
Table III.
Ratios of selected amino acids in Ws-2 and pig1-1 after 14 d of growth on agar plates
| MS
|
MS + 2 mm Phe
|
MS + 2 mm Phe (LL)
|
||||
|---|---|---|---|---|---|---|
| Ws-2 | pig1-1 | Ws-2 | pig1-1 | Ws-2 | pig1-1 | |
| Glu/Gln | 0.21 ± 0.02 | 0.14 ± 0.02a | 0.07 ± 0.0 | 0.11 ± 0.02a | 0.43 ± 0.07 | 0.1 ± 0.01a |
| Pro/Glu | 0.65 ± 0.06 | 0.94 ± 0.14a | 0.68 ± 0.08 | 1.09 ± 0.15a | 0.56 ± 0.08 | 0.56 ± 0.07 |
| Asp/Asn | 0.25 ± 0.04 | 0.19 ± 0.04 | 0.09 ± 0.01 | 0.16 ± 0.02a | 0.29 ± 0.03 | 0.19 ± 0.02a |
| Ser/Gly | 1.08 ±.0.07 | 0.9 ± 0.05a | 1.13 ± 0.1 | 1.21 ± 0.21 | 1.72 ± 0.04 | 0.98 ± 0.01a |
The depicted ratios for MS and MS + 2 mm Phe were calculated from the same data set as the values given in Table II. LL replicates were grown at a PFD of 50 μmol m−2 s−1 instead of 150 μmol m−2 s−1. The data represent the mean values ± sd of four to five independent replicates (n). The data were tested for significant differences between the wild type and the mutants using the Welsh method.
Significant differences with P < 0.01 are indicated.
The effects of 2 mm Phe feeding at a PFD of 50 μmol m−2 s−1 were similar to the results at a PFD of 150 μmol m−2 s−1 (data not shown). However, three interesting aspects were exacerbated by low incident light:
(1) The decrease of amino acid contents was more pronounced when pig1-1 was grown on 2 mm Phe at LL (data not shown) compared to elevated light conditions (Table II). Compared to the wild type, the total free amino acid pool was decreased 4-fold and some individual free amino acid pools declined up to 9-fold at LL in pig1-1, while these changes were less pronounced (3-fold and 4-fold, respectively) at higher PFD. Consequently, the calculated repression ratios ranged between 5 and 10 for the majority of the amino acids at LL (data not shown), while most of the amino acids displayed a repression ratio from 1 to 5 at elevated light (Table II). A higher repression ratio indicates a stronger deregulation of a particular amino acid in pig1-1. Thus, the amino acid deregulation in pig1-1 is dampened with increasing PFD.
(2) The wild type did not exhibit a substantial increase of Gln and Asn on 2 mm Phe at LL (data not shown), while Gln and Asn accounted for more than 75% of the amino acid surplus in the wild type in comparison to the mutant at elevated PFD (Table II). In addition, Gln contents remained comparatively constant in LL-grown pig1-1 with respect to the wild type (data not shown), while Gln accounted for more than 50% of the total amino acid decrease observed in pig1-1 at elevated PFD (Table II).
(3) Consequently, the decrease of Phe, Asn, and Gln in pig1-1 relative to the wild type at LL (data not shown) was less pronounced than at a higher PFD (Table II).
pig1-1 Displays Increased Tolerance to a Broad Range of Externally Supplied Amino Acids
As outlined above, pig1-1 was not only resistant to high concentrations of all three aromatic amino acids but also displayed a general deregulation of amino acid homeostasis. This raised the question as to whether pig1-1 would also display increased tolerance to nonaromatic amino acids. We therefore tested the effects of all proteinogenic amino acids on germination and growth of pig1-1 and the wild type.
The dose-response curves of individual nonaromatic amino acids (data not shown) resembled those of aromatic amino acids (Fig. 2). Pronounced and characteristic effects were usually observed at amino acid concentrations of 10 mm, hence all further experiments were carried out at this concentration (Table IV).
Table IV.
Leaf rosette fresh weight of pig1-1 and wild type on 10 mm nonaromatic amino acids after 14 d of growth, given in percent of nonsupplemented control plates (half-strength MS, 0.5% Suc)
| In % of MS Controls
|
||
|---|---|---|
| Ws-2 | pig1-1 | |
| Glu | 107 ± 5 | 106 ± 6 |
| Gln | 127 ± 14 | 132 ± 10 |
| Asp | 0 ± 0 | 0 ± 0 |
| Asn | 95 ± 5 | 122 ± 8a |
| Ser | 8 ± 4 | 43 ± 6a |
| Gly | 66 ± 11 | 173 ± 5a |
| Ala | 131 ± 12 | 116 ± 5 |
| Cys | 13 ± 3 | 42 ± 5a |
| His | 28 ± 3 | 56 ± 5a |
| Arg | 5 ± 1 | 5 ± 3 |
| Pro | 87 ± 2 | 79 ± 4 |
| Lys | 6 ± 3 | 5 ± 5 |
| Met | 12 ± 4 | 36 ± 2a |
| Thr | 19 ± 5 | 87 ± 10a |
| Val | 27 ± 7 | 49 ± 8a |
| Ile | 40 ± 2 | 64 ± 2a |
| Leu | 22 ± 2 | 69 ± 7a |
The presented data are taken from one representative experiment out of two to three independent experiments. In every replicate experiment; the four to six largest individuals from a population of 60 plantlets were scored and averaged. The data were tested for significant differences between the wild type and the mutants using the Welsh method.
Significant differences with P < 0.01 are indicated.
Similar to aromatic amino acids, 10 mm Asp, Lys, and Arg were toxic to the wild type (Table IV). Although pig1-1 is insensitive toward aromatic amino acids, it exhibited a similar susceptibility to Asp, Lys, and Arg as the wild type (Table IV). At concentrations up to 10 mm, all other amino acids did not inhibit germination but either positively or negatively affected the fresh weight production of seedlings. The major amino acids Glu, Gln, and Ala enhanced the fresh weight production of both the wild type and pig1-1 to a similar extent, whereas Asn or Gly were beneficial only to the mutant (Table IV). Asn had no effect on the wild type, and Gly significantly reduced its fresh weight production by 40%. Similar to the wild type, the fresh weight production of pig1-1 was impeded by most minor amino acids (Table IV) but significantly less so than the wild type (for example, see the effects of Thr, Ser, and His; Table IV). In summary, pig1-1 grew equally well or, in most cases, better than the wild type on all tested amino acids.
Phe Uptake and Root-to-Shoot Transport Are Not Affected in pig1-1
In this work, we have demonstrated that external supply of 10 mm Phe caused a substantial concomitant decrease of all free amino acid contents in pig1-1 (Table II). An impaired amino acid uptake or a compromised long-distance amino acid transport in pig1-1 could explain this observation. Therefore, it was tested whether uptake or long-distance transport of Phe are impaired in pig1-1. To this end, [U-14C]-labeled Phe was fed to roots of 14-d-old intact pig1-1 and wild-type plantlets, and the incorporation of 14C into the root and shoot was determined. The experiment was conducted in the light to stimulate the transpiration stream.
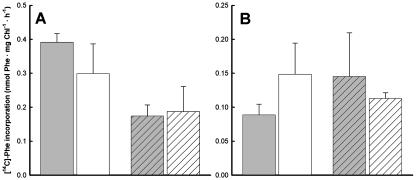
Phe accumulation in roots (Fig. 4A) and shoots (Fig. 4B) were comparable in pig1-1 and the wild type, irrespective of whether the plantlets had been adapted to Phe on Phe-replete medium (Fig. 4, solid bars) or not (Fig. 4, hatched bars), indicating that the uptake of Phe into pig1-1 roots, rather by active uptake than by diffusion, and the transport of Phe with the transpiration stream in pig1-1 were not altered. It should be noted that Phe uptake by pig1-1 and wild-type roots was stimulated when the plantlets had been grown on 1 mm Phe before the experiment (Fig. 4A), while the root-to-shoot transport was not subject to adaptation (Fig. 4B). Based on these results, we conclude that the uptake and long-distance transport of Phe in pig1-1 is unaltered with respect to the wild type.
Figure 4.
In vivo incorporation rates of [14C]Phe into roots (A) and rosettes (B) of intact wild-type (gray bars) and pig1-1 (white bars) plantlets. Roots of 2-week-old plantlets were incubated in half-strength MS, 0.5% Suc, 1 mm Phe (specific activity 23.1 MBq mmol−1) at a PFD of 60 μmol m−2 s−1 for 3 and 6 h. For the plotted data, the calculated incorporation rates after 3 and 6 h have been averaged and are given ±sd. The plantlets had been germinated and grown for 14 d on half-strength MS, 0.5% Suc plates with (hatched bars) or without (solid bars) 1 mm unlabeled Phe prior to the experiment. The radiolabel detected in the rosette corresponds to the amount of Phe transported from the root with the xylem stream. No significant differences between pig1-1 and the wild type were detected with a Welsh test. Three replicates consisting of two plantlets each were conducted for each time point. For simplicity reasons, the data for roots are also normalized to the chlorophyll content of the corresponding rosette sample, by which the qualitative result was retained in comparison to normalization to fresh weight (data not shown).
Phe Catabolism Is Increased in pig1-1 Shoots
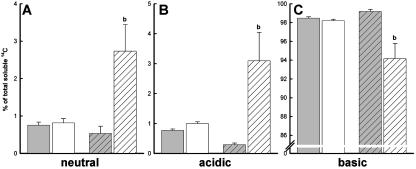
After ruling out that Phe allocation in the cormus was altered in pig1-1, it was examined if an increased Phe catabolism could account for the tolerance of pig1-1 toward externally applied amino acids. To this end, ethanol extracts of [14C]Phe-fed wild-type and mutant plants were separated into soluble and insoluble fractions, and the soluble fraction was subsequently further separated into neutral (e.g. sugars; Fig. 5A), acidic (e.g. phosphorylated intermediates, organic acids; Fig. 5B), and basic (e.g. amino acids; Fig. 5C) fractions by ion-exchange chromatography.
Figure 5.
Partitioning of 14C into neutral (A), acidic (B), and basic (C) compounds in Arabidopsis roots (solid bars) and shoots (hatched bars) in the wild-type Ws-2 (gray bars) and pig1-1 (white bars) after a 6-h pulse with [14C]Phe. These data were obtained by separating ethanol extracts resulting from the uptake and transport assay (Fig. 5) by ion-exchange chromatography. For simplicity, only experimental data for plantlets grown on half-strength MS, 0.5% Suc plates with 1 mm Phe are shown. The incorporation into insoluble fractions was negligible. Each bar represents the mean values ± sd of three independent replicates. Significant differences (P < 0.02) are indicated above the error bar (b).
The incorporation of label from [14C]Phe into the ethanol insoluble fraction was negligible (data not shown), indicating that carbon derived from externally applied Phe was not allocated to structural carbohydrates, lignin, or starch. In roots of both mutant and wild type, a predominant portion of the 14C label (>98%) was retained in the cationic fraction, and the allocation of label to the acidic and neutral fractions was low and not significantly different between mutant and wild type. However, a significantly (P < 0.02) larger portion of the label was allocated to the acidic and neutral fractions in pig1-1 shoots but not in the wild type (Fig. 5, A and B). Accordingly, a smaller portion of the label was retained in the basic fraction of pig1-1 shoots (Fig. 5C). The 10-fold increase of 14C found in the neutral and acidic fractions of pig1-1 shoots indicates a 10-fold higher catabolism of the Phe carbon backbone in pig1-1 shoots in comparison to the wild type.
Phenylpropanoid Production Is Not Enhanced in pig1-1
As reasoned initially, tolerance to high external Phe concentrations may involve a Phe-feedback insensitive shikimate pathway. As it has been observed previously that the flux through Phe ammonia lyase (PAL), the committed step of phenylpropanoid biosynthesis, can be restricted by Phe availability (Da Cunha, 1987), a mutant that tolerates high Phe levels may consequently have higher symplastic Phe contents and concomitantly display an elevated phenylpropanoid production.
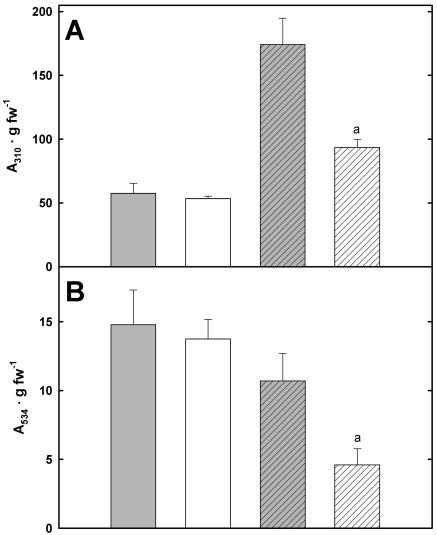
To test this hypothesis, the flavonoid content was measured in crude extracts of plantlets grown on 2 mm Phe and in standard growth conditions on soil. Although the contents of UV-absorbing phenylpropanoids were increased 3-fold in the wild type when Phe was supplied, this increase was less pronounced (1.75-fold) in pig1-1 (Fig. 6A). Furthermore, phenylpropanoid contents were similar in wild type and pig1-1 at standard growth conditions (Fig. 6).
Figure 6.
Contents of soluble phenylpropanoids in crude extracts of wild type (gray bars) and pig1-1 (white bars) grown on soil (solid bars) or on 2 mm Phe (hatched bars) normalized on fresh weight. A, Absorbance at λ = 310 nm, representing UV-absorbing compounds such as flavonols and hydroxycinnamic acid derivatives. B, Absorbance at λ = 534 nm, representing the anthocyanin content. The values represent the means of three independent samples ± sd. Significant differences between the wild type and pig1-1 (P < 0.01) are indicated above the error bars (a) and were calculated according to the Welsh method. Soil, Leaves were sampled from plants that had initially been grown for 18 d on MS plates and then were further cultivated on soil for 7 d prior to harvest. 2 mm Phe, Whole plantlets were harvested after 18 d of growth on 2 mm Phe, half-strength MS, 0.5% Suc (see “Materials and Methods”).
Surprisingly, anthocyanin contents were generally higher in soil-grown plants as when 2 mm Phe was supplied. In addition, pig1-1 mutants grown on 2 mm Phe displayed the lowest anthocyanin contents (Fig. 6B). These results will be discussed in more detail; however, it seems clear that phenylpropanoid metabolism is not stimulated in pig1-1.
DISCUSSION
The Shikimate Pathway and Phenylpropanoid Metabolism Are Not Positively Affected in pig1-1
In this article, we have described the Arabidopsis pig1-1 mutant that is insensitive to high external Phe concentrations. The aromatic amino acid Phe serves as building block for secondary metabolites of the phenylpropanoid class and Phe supply to the committed step of phenylpropanoid biosynthesis (PAL) can be rate limiting (Da Cunha, 1987). Thus, increasing the intracellular Phe contents could potentially promote the production of phenylpropanoids.
Although pig1-1 exhibits tolerance to high external Phe, Phe did not accumulate in pig1-1 when grown on MS (Table II) or soil (data not shown). Accordingly, phenylpropanoid contents were similar in pig1-1 and the wild type on soil (Fig. 6). However, the contents of UV-absorbing phenylpropanoids were increased 3-fold in the wild type on 2 mm Phe as compared to standard growth conditions (Fig. 6), indicating that the provided Phe can partially be funneled into phenylpropanoids as observed previously (Da Cunha, 1987). Interestingly, this stimulation of phenylpropanoid production by external Phe was significantly less pronounced in pig1-1 (Fig. 6). Likely, the lower contents of UV-absorbing phenylpropanoids in pig1-1 are caused by a lower rate of phenylpropanoid synthesis from internal Phe, indicating that Phe availability for PAL is possibly decreased in pig1-1 when Phe is supplied. This could be due to (1) an elevated Phe turnover, (2) an increased Phe allocation into the vacuole, or (3) by a reduced Phe uptake in pig1-1. This latter point will be further discussed below.
Many approaches to enhance phenylpropanoid production in plants have demonstrated that anthocyanins could be regarded as an overfall destination for excess carbon directed into phenylpropanoid metabolism in different angiosperm species as tomato (Lycopersicon esculentum), maize (Zea mays), and Arabidopsis (Borevitz et al., 2000; Bruce et al., 2000; Bovy et al., 2002; Mathews et al., 2003; Yu et al., 2003). Coherently, feeding intermediate levels of Phe also resulted in a visible accumulation of anthocyanins in the wild type (Fig. 3B) but not in pig1-1. The anthocyanin accumulation in the wild type may however be judged as a stress response toward the high Phe concentration in the medium. This stress response is absent in pig1-1 because pig1-1 obviously does not suffer from stress when Phe is supplied. In support of this interpretation, transitory starch accumulated in wild type but not in pig1-1 on 2 mm Phe (data not shown). A concomitant increase of anthocyanin and starch levels has previously been described as a stress syndrome in Arabidopsis (e.g. Voll et al., 2003). However, the total anthocyanin content in the wild type was lower on Phe-supplemented medium than at standard growth conditions (Fig. 6B), presumably because anthocyanin biosynthesis is developmentally up-regulated in the soil-grown plants.
Which Locus Is Then Likely To Be Affected in pig1-1?
The Shikimate Pathway
As discussed above, the pig1-1 mutation is unlikely to affect a Phe-feedback response in the shikimate pathway because Phe contents are not elevated in pig1-1 at standard growth conditions (Fig. 4). By contrast, feedback-relaxed mutants are known to accumulate the amino acid end product (Heremans and Jacobs, 1995, 1997; Mourad and King, 1995; Li and Last, 1996).
If the chorismate mutase branch was affected in pig1-1, Tyr levels would predominantly be altered together with Phe in the mutant. Indeed, we observed that Phe (11-fold) and Tyr (5-fold) contents declined strongest in pig1-1 on 2 mm Phe (Table II). However, the calculated repression ratio, an indicator for the deregulation of free amino acid abundance, for Tyr was comparable to the majority of the amino acids, while the repression ratio for Phe was much greater (Table II). This indicates the absence of a specific effect of Phe supply on Tyr contents in pig1-1. Phe feeding also had no specific effect on Tyr contents in the wild type. Furthermore, it is unlikely that a locus promoting phenylpropanoid biosynthesis downstream of Phe is affected in pig1-1 because phenylpropanoid contents were not increased in pig1-1 (see above).
Phe Uptake or Long-Distance Transport
As examined in our initial experiments, carbon availability ameliorated growth constraints exerted by exogenous feeding of aromatic amino acids (Fig. 3). Hence, the degree of amino acid toxicity depends on the ratio of provided carbon to provided nitrogen. A metabolization of the supplied amino acids may thus be enhanced when carbon skeletons are abundant, relieving their toxic effects.
We found that total free amino acid contents were 2-fold elevated when Ws-2 was grown on 2 mm Phe at 150 μmol m−2 s−1 as compared to 50 μmol m−2 s−1 (data not shown). It has been observed previously that sugar abundance enhanced amino acid uptake in Scots pine (Pinus sylvestris; Persson and Näsholm, 2003), and the increased amino acid content in Ws-2 at higher PFD could thus be explained by a stimulation of amino acid uptake by photosynthetically produced carbohydrates. However, because Suc is abundant in the growth medium and the overall carbon supply should thus not be a limiting factor at both PFDs, it is more likely that an enhanced provision of Calvin cycle intermediates and reducing equivalents to amino acid biosyntheses could account for the higher amino acid contents at higher PFD. Phe and the nitrogen shuttles Gln and Asn account for 85% of the total amino acid increase in the wild type (Table II), suggesting that the changes in the free amino acid pool size depend on Phe uptake from the medium. This stimulatory effect is less pronounced in pig1-1 (Table II), indicating that the correlation between carbon assimilation and amino acid uptake is uncoupled in the mutant.
It might thus be hypothesized that amino acid transport in pig1-1 is disturbed in such a fashion that the externally supplied amino acids are either not taken up from the medium or not transported throughout the plant body as in the wild type and hence do not exert the same effects on amino acid metabolism in pig1-1. Therefore, we tested as to whether amino acid (i.e. [14C]Phe) uptake by the root or amino acid translocation from roots to the shoot are altered in pig1-1, but both were comparable in pig1-1 and the wild type (Fig. 4). When root extracts were further separated into neutral, acidic, and basic fractions, 98% of the label in both wild-type and pig1-1 roots were retained in the cationic fraction (Fig. 5), indicating that Phe metabolization rates are similar in pig1-1 and wild-type roots. However, Phe metabolization in pig1-1 shoots was elevated 10-fold compared to the wild type (Fig. 5) in plants grown in permissive Phe-replete conditions. As discussed below, we assume that not only Phe catabolism but also free amino acid metabolization in general are increased in pig1-1.
Amino Acid Metabolization
The pig1-1 mutation confers increased tolerance of pig1-1 to many unrelated—but not to all—amino acids (Table IV), although the I50 values for the individual amino acids deviate considerably. Whereas Tyr or Phe concentrations as low as 4 mm are already lethal to 50% of the wild-type individuals (Fig. 2) and 10 mm of aromatic amino acids do not permit survival of wild-type seedlings, between 20% and 40% of wild-type individuals survive and thrive on 10 mm of branched-chain amino acids (Table IV). Thus, the tolerance of the pig1-1 mutant toward externally supplied amino acids extends beyond Phe and aromatic amino acids, indicating the presence of a rather nonspecific tolerance mechanism in pig1-1.
Likewise, the observed effects of Phe feeding on amino acid metabolism in pig1-1 are nonspecific (Table II). Ammonium (data not shown) and free amino acid contents mutually declined in the mutant upon Phe supply (Table II), while soluble protein contents even increased when Phe was abundant in the medium (from approximately 3.8–5.0 mg protein g fw−1). This broad effect argues against a specific elevation of Phe breakdown in pig1-1.
However, we found a 10-fold increased Phe metabolization rate in pig1-1 shoots during a 6-h pulse with [14C]Phe, while the partitioning of 14C label was similar in pig1-1 and wild-type roots (Fig. 5). Physiological counterbalances in response to excess amounts of one particular amino acid are frequently observed in genetic engineering approaches that aim at modifying amino acid breakdown or biosynthesis (e.g. Karchi et al., 1994; Li et al., 2003). For example, an induction of Lys catabolism had been observed when Lys biosynthesis was increased or Lys was externally supplied in tobacco (Nicotiana tabacum) seeds (Karchi et al., 1994). The endogenous induction of Lys breakdown could, however, not completely compensate for the artificially elevated Lys production, eventually leading to increased Lys contents in the transgenic plants (Karchi et al., 1994). In another example, overexpression of a bacterial branched-chain α-keto acid dehydrogenase complex subunit (BCKDC), which is involved in branched-chain amino acid breakdown, even led to a 2- to 3-fold increase of branched-chain amino acid contents in Arabidopsis (Li et al., 2003). Likewise, the response of pig1-1 to external Phe supply seems to be inducible. All free amino acids declined concomitantly upon Phe provision (Table II), while the free amino acid pool in pig1-1 was almost 2-fold larger than in the wild type on MS medium (Table II). As indicated by the stimulated [14C]Phe catabolism rates in pig1-1 shoots (Fig. 5), the inability to repress Phe and amino acid metabolization when they are abundant is the likely reason for the observed decrease in free amino acids in pig1-1 on 2 mm Phe. Although this has not been tested, we hypothesize that external supply of other amino acids would result in a similar reduction of free amino acid levels in pig1-1 as caused by Phe.
In conclusion, the mechanism by which pig1-1 achieves tolerance toward Phe apparently involves a general stimulation of amino acid breakdown, which is presumably more relaxed in pig1-1 at standard growth conditions in comparison to Phe-replete conditions.
Amino Acid Homeostasis
A recent study of wheat (Triticum aestivum), potato (Solanum tuberosum), and barley (Hordeum vulgare) revealed that the abundance of minor amino acids relative to each other remains extremely constant over a range of different growth conditions; their ratio is even constant between a photorespiratory barley mutant and wild-type barley (Noctor et al., 2002). Paralleling the observations by Noctor et al. (2002), the relative abundance of free minor amino acids also remained relatively constant in pig1-1 throughout the tested conditions (Table II). This is illustrated by similar repression ratios for most amino acids (Table II). Nevertheless, the pig1-1 mutation seems to prohibit an accumulation of Phe and of the nitrogen shuttles Gln and Asn on 2 mm Phe (Table II), thereby balancing the total free amino acid pool. On 2 mm Phe, the diminished accumulation of amino acids in pig1-1 is obviously effectuated by derepressed amino acid catabolism in the shoot but not in roots (Fig. 5). Apparently, amino acid catabolism in pig1-1 can better compensate for amino acid uptake at lower PFDs, leading to lower free amino acid pools in LL (see “Results”) than at a PFD of 150 μmol m−2 s−1 (Table II). It was previously found that amino acid catabolism is a key player in the control of free minor amino acids such as Lys (Zhu and Galili, 2003) or Thr (Jander et al., 2004).
A flexible response seems to be affected by the pig1-1 mutation because the total free amino acid pool in pig1-1 is almost 2-fold elevated on MS medium and substantially decreased upon Phe supply when compared to the wild type (Table II). Secondly, this flexible response toward exogenous amino acid supply is nonspecific, as pig1-1 displays a higher tolerance toward a number of exogenously supplied nonbiosynthetically related minor and major amino acids (Table IV). In addition, amino acid catabolism seems to be affected in pig1-1. Hence, we hypothesize that the integration of amino acid catabolism into a superseding regulatory network that governs amino acid homeostasis is deregulated or defective in pig1-1. Such a control mechanism, the general amino acid control, has been reported in Escherichia coli and yeast (Hinnebusch, 1988). In contrast to the observed physiological effects in pig1-1, the general amino acid control mechanism mediates the concomitant up-regulation of amino acid biosynthesis and nucleic acid biosynthesis in response to a limitation of, e.g. one single amino acid in yeast (Hinnebusch, 1988). Evidence for the operation of a general amino acid control also exists in plants (Guyer et al., 1995; Bonner and Jensen, 1997). First, impairing His biosynthesis by an inhibitor leads to a coordinated up-regulation of both the shikimate pathway and purine biosynthesis in Arabidopsis (Guyer et al., 1995). Second, it could be demonstrated that Gln feeding relieves the feedback inhibition caused by supplying branched-chain amino acids to tobacco suspension culture cells (Bonner and Jensen, 1997). In our study, Gln and Ala proved to be beneficiary for both the wild type and the mutant, while Asn and Gly supplements improved only the growth performance of the mutant (Table IV). The observation of additional effects in our experimental system most likely relate to the fact that the response of a multicellular organism is generally more intricate than that of cultivated cells. Furthermore, our results provide further evidence that the balance and homeostasis of free amino acids in plants is achieved by regulating both amino acid turnover and biosynthesis (Karchi et al., 1994; Li et al., 2003; Zhu and Galili, 2003; Jander et al., 2004). We infer that this mechanism constitutes the general amino acid inhibition observed by Bonner and Jensen (1997).
CONCLUSION
The response of free amino acid contents in pig1-1 to Phe feeding exhibits four important features: (1) The Phe-induced decline of free amino acid contents is not specific to aromatic amino acids but extends to all proteinogenic amino acids, (2) it is light-dependent, (3) soluble protein contents remain unaffected, and (4) Phe and likely amino acid catabolism in general are increased in pig1-1 shoots but not in pig1-1 roots. In addition, pig1-1 is resistant to a large number of biosynthetically nonrelated amino acids, and the uptake of Phe by roots and its translocation to the shoot is similar to the wild type. These results suggest that the deregulated amino acid metabolism in pig1-1 might be caused by a defect in a regulatory mechanism that integrates into a network comparable to the general amino acid control in yeast. Testing this hypothesis will require the molecular identification of the defective gene in pig1-1 that is currently in progress.
MATERIALS AND METHODS
Plant Material
A population of 200,000 ethyl methanesulfonate-mutagenized M2 individuals (cv Ws-2) was obtained from Lehle Seeds (Round Rock, TX) and screened for the Phe-tolerant phenotype as described below.
Seed material for the lines trp1-100, trp2-1, trp3-1, and trp5-1 was obtained from Arabidopsis Biological Resource Center stock center (Ohio State University, Columbus, OH).
Screening Procedures
Aliquots of the seed populations were vapor sterilized according to Clough and Bent (1998) and grown on half-strength MS plates, pH 5.7, containing 0.5% (w/v) Suc and 10 mm Phe (MS-Phe) at a PFD of 50 μmol m−2 s−1 in a 12/12-h light/dark cycle. Survivors were transplanted to soil after 2 to 3 weeks of growth on MS-Phe when the first three primary leaves had emerged. The plants were grown to maturity, allowed to self, and the seeds of the M3 generation were harvested. Selection on MS-Phe was repeated with the M3 and M4 generations to eliminate false positives. The M4 progeny were uniformly Phe tolerant.
Selected individuals of the uniform M4 pig mutant populations were backcrossed to the wild type three times prior to further physiological analysis.
Determination of Growth Phenotypes and Cross-Tolerance to Tyr and Trp
To evaluate cross-tolerance of pig mutants to Trp and Tyr, pig1-1 and control seeds were sterilized and dispersed on sterile plates as outlined in “Screening Procedures,” except that 10 mm Phe was substituted by other aromatic amino acids as indicated. Germination rates on the individual plates were scored after 14 d of growth at a PFD of 50 μmol m−2 s−1 in a 12/12-h light/dark cycle. Germination was judged to have failed when growth of the plantlets was aborted after the cotyledons and the radicle had emerged, and the plants subsequently died.
To better visualize growth effects of aromatic amino acid feeding to the wild type, seeds were also germinated and grown on half-strength MS, 2% Suc, and 4 mm of the respective aromatic amino acid.
Growth Inhibition Assays for Nonaromatic Amino Acids
Concentration-dependent growth inhibition effectuated by nonaromatic amino acids was essentially determined as the germination rates on aromatic amino acids described in the section above, except for two differences. First, 2.5 mm MES, pH 5.7, was added to the plates to avoid pH-dependent effects that might be induced by acidic or basic amino acids. Secondly, the fresh weight of the four to six biggest individuals from populations consisting of 60 individuals per replicate was scored after 14 d of growth at the same conditions indicated above. Table IV only displays the results for 10 mm amino acid supplements because this concentration was judged to be the most representative among the tested amino acid concentrations.
Examination of Phenotypic Similarities between pig Mutants and trp Mutants
Surface-sterilized pig1-1, trp1-100, trp2-1, trp3-1, trp5-1, and Ws-2 control seeds (see “Screening Procedures”) were sown on plates of four different compositions as indicated in Table I. After 21 d at a PFD of 50 μmol m−2 s−1 in a 12/12-h light/dark cycle, the individual plant lines were tested for the growth or fluorescence responses indicated in Table I. The observed effects were compared to plants sown on half-strength MS, 2% Suc control plates.
Determination of Free Amino Acid and Protein Contents
Free amino acid and protein contents were assayed in Arabidopsis plantlets grown on sterile agar plates. Vapor-sterilized seeds were germinated on half-strength MS, 0.5% Suc, and 2 mm Phe, and whole plantlets were harvested after 14 d of growth at a PFD of 150 μmol m−2 s−1 in a 12/12-h light/dark cycle.
For the determination of free amino acids, the plant samples were extracted in distilled water as described previously (Voll et al., 2003), and the contents of free amino acids were essentially assayed according to van Wandelen and Cohen (1997).
To assess total soluble proteins, plant samples were extracted in a buffer containing 50 mm HEPES/NaOH, pH 7.5, 5 mm MgCl2, 1 mm EDTA, and 0.5% (v/v) Triton X-100 with a rotating pestle and quantified according to Bradford (1976), applying the modifications of Zor and Selinger (1996).
Determination of Soluble Phenylpropanoids in Crude Extracts
The contents of UV-absorbing compounds and anthocyanins were determined in methanol extracts of plant material grown on 2 mm Phe (see paragraph above) and in leaves of soil-grown plants as described by Voll et al. (2003).
To obtain soil-grown material, seeds of pig1-1 and the Ws-2 wild-type control were germinated on half-strength MS plates, transplanted to soil after 18 d, and fully expanded leaves were harvested 4 h before the end of the light period after 7 d of growth on soil at a PFD of 100 μmol m−2 s−1 in a 12/12-h light/dark cycle.
[14C]Phe Uptake Assays and 14C Partitioning
Arabidopsis seeds were germinated and grown on half-strength MS, 0.5% Suc at a PFD of 50 μmol m−2 s−1 for 14 d, either in the presence or absence of 1 mm Phe. For the uptake experiment, individual wells of a 48-well plate were loaded with 800 μL of liquid medium (half-strength MS, 0.5% Suc, 1 mm [U-14C]Phe; 23.1 MBq mmol−1 specific activity). The roots of two plantlets were submerged in each well with the rosettes attaching the edges of the wells, not shading each other, and the plate was illuminated at room temperature at a PFD of 60 μmol m−2 s−1. Three replicate samples for each pretreatment (+Phe, −Phe) were harvested after 3 and 6 h as follows: The roots were rinsed twice with distilled water to remove adhering label and root and rosettes were then separated by cutting the hypocotyls with a razor blade. The samples were snap-frozen in liquid nitrogen, and the fresh weight was recorded in the frozen state. The samples were extracted by boiling the seedlings in 600 μL of 80% ethanol at 95°C for 45 min. The ethanol extracts were transferred to fresh tubes, and 50-μL aliquots were subjected to (1) liquid scintillation counting and (for rosette samples) to (2) spectrophotometric chlorophyll determination according to Lichtenthaler (1987). The remaining volume was dried in a vacuum concentrator, resuspended in 300 μL of distilled water, and 280 μL were separated into neutral, acidic, and basic fractions by coupled chromatography on Dowex anion (AG 1 × 8) and cation (AG 50W × 8) exchange resins (Bio-Rad, Munich), as described by Quick et al. (1989). The radioactivity in each fraction was quantified by scintillation counting.
Acknowledgments
We thank Momoko Minakawa and Samuel Vandenberg for plant culture work and the Michigan State University Macromolecular Structure Analysis Facility for help with amino acid analysis.
This work was supported by the Deutsche Forschungsgemeinschaft (postdoctoral fellowship to L.M.V.), by the National Science Foundation (REU-supplement MCB–0348074 to A.P.M.W.), and by the Michigan State University Center for Plant Products and Technologies (grant to A.P.M.W.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.047506.
References
- Arruda P, Bright SWJ, Kueh JSH, Lea PJ, Rognes SE (1984) Regulation of aspartate kinase isoenzymes in barley mutants resistant to lysine plus threonine—construction and analysis of combinations of the LT1A, LT1B and LT2 mutant-genes. Plant Physiol 76: 442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J, Fink GR (1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95: 5655–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner CA, Jensen RA (1997) Recognition of specific patterns of amino acid inhibition of growth in higher plants, uncomplicated by glutamine-reversible ‘general amino acid inhibition’. Plant Sci 130: 133–143 [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, de Vos R, Kemper M, Schijlen E, Pertejo MA, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, et al (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B (2000) Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12: 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Brotherton JE, Song HS, Widholm JM (2000) Increasing tryptophan synthesis in a forage legume Astragalus sinicus by expressing the tobacco feedback-insensitive anthranilate synthase (ASA2) gene. Plant Physiol 123: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Da Cunha A (1987) The estimation of L-phenylalanine ammonia-lyase shows phenylpropanoid biosynthesis to be regulated by L-phenylalanine supply and availability. Phytochemistry 26: 2723–2727 [Google Scholar]
- Falco SC, Guida T, Locke M, Mauvais J, Sanders C, Ward RT, Webber P (1995) Transgenic canola and soybean seeds with increased lysine. Biotechnology (N Y) 13: 577–582 [DOI] [PubMed] [Google Scholar]
- Galili G, Guenoune D, Wininger S, Hana B, Schupper A, Ben-Dor B, Kapulnik Y (2000) Enhanced levels of free and protein-bound threonine in transgenic alfalfa (Medicago sativa L.) expressing a bacterial feedback-insensitive aspartate kinase gene. Transgenic Res 9: 137–144 [DOI] [PubMed] [Google Scholar]
- Guyer D, Patton D, Ward E (1995) Evidence for cross-pathway regulation of metabolic gene-expression in plants. Proc Natl Acad Sci USA 92: 4997–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heremans B, Jacobs M (1995) Threonine accumulation in a mutant of Arabidopsis thaliana (L.) Heynh. with an altered aspartate kinase. J Plant Physiol 146: 249–257 [Google Scholar]
- Heremans B, Jacobs M (1997) A mutant of Arabidopsis thaliana (L) Heynh with modified control of aspartate kinase by threonine. Biochem Genet 35: 139–153 [DOI] [PubMed] [Google Scholar]
- Hervieu F, Vaucheret H (1996) A single amino acid change in acetolactate synthase confers resistance to valine in tobacco. Mol Gen Genet 251: 220–224 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (1988) Mechanisms of gene regulation and general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev 52: 248–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu SX, Li LL, Last RL (2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening; evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39: 465–475 [DOI] [PubMed] [Google Scholar]
- Karchi H, Shaul O, Galili G (1994) Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc Natl Acad Sci USA 91: 2577–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Ponappa T, Dong WQ, Town CD (1996) Molecular basis of α-methyltryptophan resistance in amt-1, a mutant of Arabidopsis thaliana with altered tryptophan metabolism. Plant Physiol 110: 1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Bissinger PH, Mahoney DJ, Radwanski ER, Fink GR (1991) Tryptophan mutants in Arabidopsis: the consequences of duplicated tryptophan synthase β genes. Plant Cell 3: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YT, Duggleby RG (2002) Regulatory interactions in Arabidopsis thaliana acetohydroxyacid synthase. FEBS Lett 512: 180–184 [DOI] [PubMed] [Google Scholar]
- Li H, Culligan K, Dixon RA, Chory J (1995) CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Thipyapong P, Breeden DC, Steffens JC (2003) Overexpression of a bacterial branched-chain α-keto acid dehydrogenase complex in Arabidopsis results in accumulation of branched-chain acyl-CoAs and alteration of free amino acid composition in seeds. Plant Sci 165: 1213–1219 [Google Scholar]
- Li LY, Last RL (1996) The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble tryptophan. Plant Physiol 110: 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco DJ, Wagoner W, Lightner J, et al (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15: 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao S, Duncan DR, Widholm J (1988) Selection of regenerable maize callus-cultures resistant to 5-methyl-DL-tryptophan, S-2-aminoethyl-L-cysteine and high levels of L-lysine plus L-threonine. Plant Cell Tiss Org 14: 3–14 [Google Scholar]
- Mourad G, King J (1995) L-O-Methylthreonine-resistant mutant of Arabidopsis defective in isoleucine feedback-regulation. Plant Physiol 107: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Last RL, Fink GR, Keith B (1993) Suppressors of trp1 fluorescence identify a new Arabidopsis gene, TRP4, encoding the anthranilate synthase β-subunit. Plant Cell 5: 1011–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Novitskaya L, Lea PJ, Foyer CH (2002) Co-ordination of leaf minor amino acid contents in crop species: significance and interpretation. J Exp Bot 53: 939–945 [DOI] [PubMed] [Google Scholar]
- Persson J, Näsholm T (2003) Regulation of amino acid uptake by carbon and nitrogen in Pinus sylvestris. Planta 217: 309–315 [DOI] [PubMed] [Google Scholar]
- Quick P, Siegl G, Neuhaus E, Feil R, Stitt M (1989) Short-term water stress leads to a stimulation of sucrose synthesis by activating sucrose-phosphate synthase. Planta 177: 535–546 [DOI] [PubMed] [Google Scholar]
- Radwanski ER, Barczak AJ, Last RL (1996) Characterization of tryptophan synthase α subunit mutants of Arabidopsis thaliana. Mol Gen Genet 253: 353–361 [DOI] [PubMed] [Google Scholar]
- Rognes SE, Bright SWJ, Miflin BJ (1983) Feedback-insensitive aspartate kinase isoenzymes in barley mutants resistant to lysine plus threonine. Planta 157: 32–38 [DOI] [PubMed] [Google Scholar]
- Rose AB, Casselman AL, Last RL (1992) A phosphoribosylanthranilate transferase gene is defective in blue fluorescent Arabidopsis thaliana tryptophan mutants. Plant Physiol 100: 582–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J, Amrhein N (1995) Molecular organization of the shikimate pathway in higher plants. Phytochemistry 4: 737–749 [Google Scholar]
- Streatfield SJ, Weber A, Kinsman EA, Häusler RE, Li J, Post-Beittenmiller D, Kaiser WM, Pyke KA, Flügge U-I, Chory J (1999) The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development and plastid-dependent nuclear gene expression. Plant Cell 11: 1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wandelen C, Cohen SA (1997) Using quarternary high-performance liquid chromatography eluent systems for separating 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate-derivatized amino acid mixtures. J Chromatogr A 763: 11–22 [Google Scholar]
- Voll L, Häusler RE, Hecker R, Weber A, Weissenbock G, Fiene G, Waffenschmidt S, Flügge U-I (2003) The phenotype of the Arabidopsis cue1 mutant is not simply caused by a general restriction of the shikimate pathway. Plant J 36: 301–317 [DOI] [PubMed] [Google Scholar]
- Yu O, Shi J, Hession AO, Maxwell CA, McGonigle B, Odell JT (2003) Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63: 753–763 [DOI] [PubMed] [Google Scholar]
- Zhu X, Galili G (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zor T, Selinger Z (1996) Linearization of the Bradford protein assay increases its sensitivity, theoretical and experimental studies. Anal Biochem 236: 302–308 [DOI] [PubMed] [Google Scholar]