Abstract
The gravitropism defective 2 (grv2) mutants of Arabidopsis show reduced shoot phototropism and gravitropism. Amyloplasts in the shoot endodermal cells of grv2 do not sediment to the same degree as in wild type. The GRV2 gene encodes a 277-kD polypeptide that is 42% similar to the Caenorhabditis elegans RME-8 protein, which is required for endocytosis. We hypothesize that a defect in endocytosis may affect both the initial gravity sensing via amyloplasts sedimentation and the subsequent more general tropic growth response.
Higher plants sense their orientation with respect to the gravity vector through the sedimentation of starch-filled organelles, the amyloplasts (Sack et al., 1986). In stems this occurs in the endodermis (Fukaki et al., 1998), and in roots it occurs in the columella cells of the root cap. Changes in amyloplast sedimentation are thought to trigger a signal transduction cascade that mediates the relocalization of auxin transporters. Altered auxin flow causes a compensatory asymmetric growth response (for review, see Sack, 1991).
Several agravitropic mutants have been identified in which various aspects of this process are altered (Masson et al., 2002). The sgr1/scr (shoot gravitropism/scarecrow) and sgr7/shr (short-root) mutants both lack the endodermal cell layer in shoots. The absence of the tissue layer in which amyloplast sedimentation occurs results in agravitropic responses in both hypocotyls and inflorescence stems (Fukaki et al., 1998). SGR1 /SCR and SGR7/SHR both encode putative transcription factors (Di Laurenzio et al., 1996; Helariutta et al., 2000) that are required for the formation of the endodermal cell layer.
In the sgr2, sgr3, and zig (zigzag)/sgr4 mutants, amyloplasts in the endodermal cells of the shoot do not sediment according to the gravity vector, while amyloplasts in the columella cells of the root sediment normally (Morita et al., 2002; Yano et al., 2003). Strong alleles of sgr2 and zig/sgr4 cause loss of gravitropic responses in both hypocotyls and inflorescence stems but not in roots (Fukaki et al., 1996a; Yamauchi et al., 1997; Kato et al., 2002). Mutations in sgr3 cause defects in the gravitropic response in inflorescence stems but not in hypocotyls or roots (Fukaki et al., 1996a; Yano et al., 2003). SGR2 encodes a putative phospholipase A1, SGR3 encodes the vacuolar t-SNARE AtVAM3, and ZIG/SGR4 encodes the vacuolar v-SNARE AtVTI11. In each case, the effect on gravitropism is thought to be indirect. The primary defect in the mutants is a change in the vacuolar membrane system that may mechanically constrain the movement of amyloplasts (Morita et al., 2002; Kato et al., 2002).
Auxin is transported basipetally, from the shoot tips to the roots, through the polar localization of efflux carriers in the plasma membrane (Galweiler et al., 1998). These carriers are cycled between the plasma membrane and the Golgi (Geldner et al., 2001), thereby allowing for relocalization upon gravity or light stimuli (Friml et al., 2002). Mutants of PIN3, which encodes a putative auxin efflux carrier, have defects in both gravitropic and phototropic responses. In stems, PIN3 is normally localized to the lateral, inner sides of endodermal cells. The localization of PIN3 in endodermal cells upon gravity stimulation has not been determined. However, in columella cells, PIN3 is uniformly distributed, and upon gravity stimulation PIN3 is quickly relocalized via endocytosis followed by exocytosis to the lateral sides of the columella cells closest to the gravity vector (Friml et al., 2002). This relocalization of a putative auxin efflux carrier likely results in a change in the flow of auxin, mediating a change in growth orientation.
We have isolated mutations in a novel gene, termed GRAVITROPISM DEFECTIVE 2 (GRV2), that result in a reduction of gravitropic responses in the hypocotyls and shoots. Based on its sequence similarity to the Caenorhabditis elegans protein RME-8 (Zhang et al., 2001), GRV2 likely functions in endocytosis.
RESULTS
grv2 Mutations Affect Gravitropic Responses of the Shoots But Not the Roots
The grv2 mutants were identified in two independent mutant screens as plants with pronounced shoot agravitropism. In wild-type plants, primary inflorescence shoots grow upright in the opposite direction to the gravity vector. The lateral shoots also grow in a vertical direction (Fig. 1A). By contrast, grv2 primary inflorescence shoots curve slightly, and the lateral shoots grow in a horizontal direction (Fig. 1B). This growth pattern is similar to that of the sgr3, sgr5, and sgr6 mutants (Fukaki et al., 1996a; Yamauchi et al., 1997). The extent of irregular growth of the grv2-2 to grv2-4 alleles (Columbia [Col]) is less than that of grv2-1 (Landsberg erecta [Ler]).
Figure 1.
Growth habit and gravitropic response of Ler wild type and the grv2-1 mutant. A and C, Wild type; B and D, grv2-1 mutant. The pots in C and D were placed in a horizontal orientation for 3 h in darkness before the photograph was taken.
In addition, the grv2 mutant plants are slightly smaller and have reduced vigor compared to wild type. The hypocotyl elongation rates of the mutants were lower than in the corresponding wild-type strains (Table I). The final hypocotyl lengths of the mutants were also reduced relative to the wild types. In the Col alleles, a reduction in cell size accounts for the overall reduction in hypocotyl lengths. By contrast, in the Ler allele the reduced size of the hypocotyl cells was not, by itself, sufficient to account for the reduced hypocotyl length. Thus, in the grv2-1 allele, there is a defect in cell division as well. The Ler allele also has a reduction in fertility due to variable defects in stamen growth and pollen dehiscence. Manual pollination circumvents this defect and restores fertility.
Table I.
Hypocotyl and root growth rates and cell sizes
| Hypocotyl Growth Rate | Hypocotyl Length | Hypocotyl Cell Size | Root Growth Rate | |
|---|---|---|---|---|
| mm/h | mm | mm | mm/h | |
| Ler | 0.28 | 32.43 ± 0.56 (50) | 0.43 ± 0.02 (52) | 0.15 |
| grv2-1 | 0.10 | 13.99 ± 0.86 (23) | 0.36 ± 0.03 (43) | 0.13 |
| Col | 0.29 | 31.71 ± 0.90 (15) | 0.53 ± 0.03 (44) | 0.15 |
| grv2-2 | 0.21 | 25.12 ± 0.88 (39) | 0.31 ± 0.02 (23) | 0.17 |
| grv2-3 | 0.19 | 22.35 ± 0.74 (49) | 0.32 ± 0.02 (39) | 0.19 |
| grv2-4 | 0.19 | 21.85 ± 0.65 (47) | 0.38 ± 0.02 (32) | 0.17 |
Sample sizes are indicated in parentheses for hypocotyl length and cell size. Sample sizes for hypocotyl and root growth rate are: Ler, n = 44–52; grv2-1, n = 22–47; Col, n = 15–55; grv2-2, n = 31–50; grv2-3, n = 27–53; and grv2-4, n = 30–58.
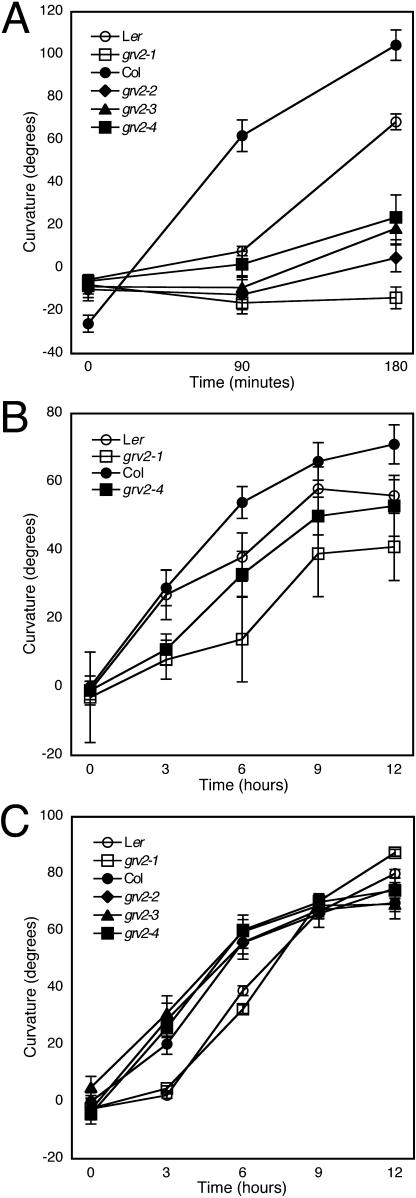
To measure the gravitropic response of grv2 shoots, bolting stems of wild-type and grv2 plants were placed in a horizontal orientation. Both wild type and grv2 initially hung downward toward the gravity vector. The upward curvature of Ler (wild-type) plants reached 70 degrees within 180 min (Figs. 1C and 2A). By contrast, grv2-1 shoots showed little curvature in 180 min and even after 24 h failed to respond (Figs. 1D and 2A). Col (wild-type) shoots showed faster tropic response than Ler shoots, curving 60 degrees within 90 min. grv2-2, -3, and -4 (Col) alleles showed little gravitropic curvatures within 90 min, but they showed weak gravitropic curvature within 180 min (Fig. 2A).
Figure 2.
Measurements of gravitropism in shoots (A; Ler n = 15, grv2-1 n = 12, Col n = 8, grv2-2 n = 14, grv2-3 n = 12, grv2-4 n = 12), hypocotyls (B; Ler, n = 15–37; grv2-1, n = 10–29; Col, n = 19; grv2-4, n = 21), and roots (C; Ler, n = 148; grv2-1, n = 132; Col, n = 19; grv2-2, n = 16; grv2-3, n = 15; grv2-4, n = 21) in grv2 mutants and wild types. Error bars represent the se of the mean.
Since many agravitropic mutations affect gravity responses specifically in some organs but not in others, we also examined the gravitropic responses of grv2 hypocotyls and roots. Hypocotyls of 3-d-old etiolated wild-type seedlings showed negative gravitropism and, when placed horizontally, curved upward. Etiolated hypocotyls from grv2-1 and grv2-4 showed a slight reduction in negative gravitropism (Fig. 2B), whereas grv2-2 and grv2-3 responded to the same degree as wild type (supplemental data, available at www.plantphysiol.org). The reduction in gravitropism in grv2 hypocotyls is sufficiently weak that we are not able to exclude the possibility that it is simply due to a growth defect rather than a gravitropic defect. Wild-type roots showed positive gravitropism, growing downwards toward the gravity vector. All four grv2 alleles also showed normal positive gravitropism (Figs. 2C and 3). Taken together, the gravitropic responses of grv2 mutants, reduced gravitropism of hypocotyls and shoots but normal gravitropism of roots, are similar to the phenotypes observed in sgr1, sgr2, sgr4, and sgr7.
Figure 3.
Gravitropic response of wild type (A) and the grv2-1 mutant (B). Three-day-old seedlings, germinated on vertically oriented agar-solidified medium in darkness, were turned 90 degrees and photographed after 12 h.
Phototropism
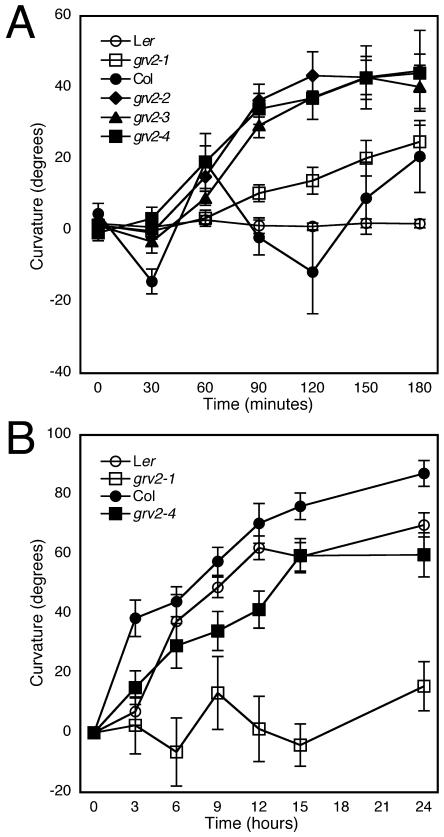
To test if grv2 mutations have an effect on tropic growth in general, we examined the phototropic response of grv2 shoots and hypocotyls. Wild-type Col shoots showed circumnutational motion, i.e. growth with oscillating direction. By contrast, Col alleles of grv2 caused an absence of circumnutation but a strong phototropic response (Fig. 4A). Ler wild-type stems showed no detectable circumnutation or phototropism at the light intensities used here. The Ler allele grv2-1 also did not display detectable circumnutation but did show a slight curvature toward the light. These results indicate that grv2 shoots are not deficient in phototropic responses but, to the contrary, have a tendency to respond more strongly than wild type. This can perhaps be attributed to their dampened gravitropic responses, which might not be as effectively counterbalancing phototropic stimuli as in wild type.
Figure 4.
Measurements of phototropism in shoots (A; Ler, n = 9; grv2-1, n = 10; Col, n = 7; grv2-2, n = 6; grv2-3, n = 6; grv2-4, n = 6) and hypocotyls (B; Ler, n = 27–34; grv2-1, n = 23–34; Col, n = 11–23; grv2-4, n = 24–32) in grv2 mutants and wild types. Error bars represent the se of the mean.
Etiolated wild-type hypocotyls, both from Ler and Col seedlings, curve toward the light when exposed to a lateral light source. By contrast, grv2-1 (Ler background) and grv2-4 (Col background) hypocotyls showed reduced curvature toward the light (Fig. 4B), and grv2-2 and -3 (Col) showed a slightly reduced response (supplemental data). The reduced phototropic response in hypocotyls is in sharp contrast to the enhanced phototropic phenotype in inflorescence stems. It is not clear if this is due to fundamental differences between the phototropic response in the two organs or to differences in how phototropism is assayed in each organ.
Localization of Amyloplasts
Within the organs of the shoot, gravity is sensed in a specialized layer of cells, termed endodermis. Endodermal cells contain amyloplasts, which are denser than the cytoplasm and sediment with gravity (Fig. 5A). The position of the amyloplasts within the cell is thought to provide the directional clue for sensing the gravity vector.
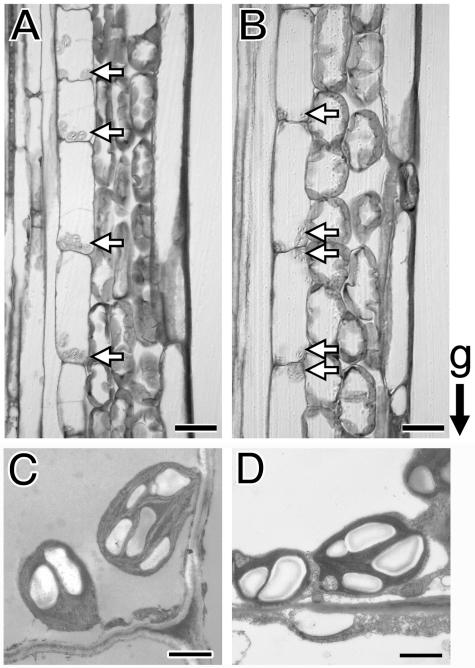
Figure 5.
Microscopic image of thin sections of primary inflorescence shoots (3–4 cm below the apex) from wild type (A and C) and the grv2-1 mutant (B and D). The stems were fixed with the gravity direction maintained. The arrows point to amyloplasts. The bars indicate 20 μm (A and B) and 1 μm (C and D).
To test if the grv2 phenotype could be attributed to a defect in the process of gravity sensing, the position of the amyloplasts in the endodermal cells was determined. Longitudinal segments of inflorescence shoots were fixed, maintaining an upright orientation. In wild type, nearly all amyloplasts (74.6% Ler and 94.1% Col) were localized toward the basal end of endodermal cells (Table II) and accumulated at a distance from the cell wall (Fig. 5, A and C). In the grv2-1 mutant, a significant number of amyloplasts (48.7%) were localized to the top of the endodermal cells and especially accumulated at the corners of the cells appressed to the cell wall (Fig. 5, B and D). Amyloplast sedimentation was also reduced in grv2-2, -3, and -4 but to a lesser degree than grv2-1. Altogether, the localization of amyloplasts in grv2 endodermal cells is abnormal, as it is in the agravitropic mutants sgr2, sgr3, and zig/sgr4 (Kato et al., 2002; Morita et al., 2002; Yano et al., 2003). It seems possible that in all these mutants agravitropism might result from a defect in gravity sensing.
Table II.
The position of amyloplasts in shoot endodermal cells
| Top | Middle | Bottom | Total | |
|---|---|---|---|---|
| % | % | % | ||
| Ler | 5.4 | 20.0 | 74.6 | 130 (25) |
| grv2-1 | 48.7 | 19.9 | 31.4 | 191 (28) |
| Col | 0.9 | 5.0 | 94.1 | 221 (24) |
| grv2-2 | 8.4 | 42.6 | 49.0 | 237 (33) |
| grv2-3 | 22.4 | 31.9 | 45.7 | 232 (33) |
| grv2-4 | 10.4 | 35.5 | 54.0 | 211 (28) |
Amyloplasts were counted in longitudinal sections. Total indicates the total number of the amyloplasts with the number of the examined endodermal cell in parentheses. The amyloplasts that touched the top and bottom sides of the cell were respectively counted as Top and Bottom. All others were counted as Middle.
In the root, gravity is sensed through the sedimentation of amyloplasts in the columella cells. We found no difference between wild type and grv2 in the position of root amyloplasts (data not shown). This result is consistent with the observation that grv2 roots respond to gravity to the same degree as wild type.
Mapping and Cloning of GRV2
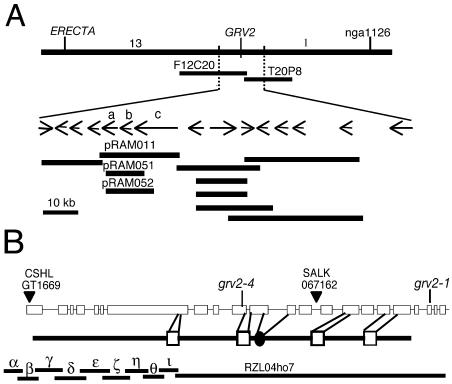
The GRV2 gene was cloned based on its map position. The grv2 mutation is closely linked to erecta, enabling selection of recombination events in the vicinity of grv2 based on the visible phenotype conferred by erecta. Using PCR-based markers, grv2 was mapped to an approximately 70-kb interval in the bacterial artificial chromosome clones F12C20 and T20P8. A contig of binary T-DNA cosmids spanning this region (Fig. 6A) was established and the cosmids transformed into grv2 mutant plants. A single cosmid, pRAM011, containing three genes (At2g26870, At2g26880, and At2g26890) complemented the grv2 phenotype. One of the three genes, a MADS box gene (At2g26880), was completely contained on two other cosmids, pRAM051 and pRAM052, which did not complement grv2. The grv2-1 and grv2-4 alleles of the remaining two genes were sequenced. No mutations were found in At2g26870, a putative phospholipase. However, mutations in At2g26890 were found in both alleles (see below). A 15.3-kb genomic fragment spanning from 2 kb upstream to 1 kb downstream of the predicted At2g26890 coding sequence complemented the grv2 mutant phenotype. We conclude that GRV2 corresponds to At2g26890.
Figure 6.
Structure of the GRV2 locus. A, The region of chromosome II containing the GRV2 gene was spanned by bacterial artificial chromosomes F12C20 and T20P8 (www.arabidopsis.org). A cosmid contig spanning the GRV2 gene is shown as overlapping lines beneath The Arabidopsis Information Resource annotation of the region. The three genes in the complementing cosmid, pRAM011, are At2g26870 (a), At2g26880 (b), and At2g26890 (c). B, The intron-exon structure of the GRV2 gene (At2g26890) is shown as lines and boxes, respectively. The locations of various mutations are indicated by lines and arrowheads. grv2-1 has an 85-residue truncation, grv2-4 has a frame shift resulting in a stop codon in exon 11, and GT1669 and SALK 067162 are T-DNA insertions. A schematic drawing of the GRV2 protein is shown below the transcript schematic. White boxes represent IWN repeat domains, and the black circle represents the J domain. The expressed sequence tag, RZL04ho7, and the reverse transcription-PCR products that were sequenced to determine the intron-exon structure (α, β, γ, δ, ɛ, ξ, η, θ, ι) are shown beneath the protein structure.
To determine the intron/exon structure of GRV2, we analyzed the sequence of a partial cDNA clone (RZL04ho7) and nine reverse transcription-PCR products (Fig. 6B). Our results confirmed the predicted intron/exon structure. GRV2 transcripts are 11.8 kb long with a 7,665-bp coding region predicted to encode a 277-kD protein. The GRV2 gene contains 21 introns (Fig. 6B).
Domain searches revealed that the GRV2 gene product contains a single Dna-J domain (amino acid position 1,500–1,588) and four IWN repeats (Fig. 6B; supplemental material). GRV2 shows 26% identity and 42% similarity over the entire protein with the C. elegans gene product RME-8 (Zhang et al., 2001). The J domain and the IWN repeats in particular are well conserved between the two proteins. CeRME-8 was identified in a screen for mutants with reduced receptor-mediated endocytosis of yolk protein in developing embryos (Zhang et al., 2001). DmRme-8 was identified in a screen for enhancers of shi, an endocytosis mutant (Chang et al., 2004). Putative RME-8 homologs have also been identified in rice (Oryza sativa), rats, and humans (Zhang et al., 2001; Chang et al., 2004), but their functions remain to be determined. BLAST searches of the GRV2 nucleotide sequence, as well as the deduced protein sequence, failed to identify any other Arabidopsis gene with significant sequence similarity to GRV2 over the entire length of the coding region.
The grv2-1 allele, which was induced by ethyl methane sulfonate, has a G-to-A transition resulting in a stop codon 85 amino acids from the native stop codon. In the grv2-4 allele, which was induced by gamma rays, 13 bp in exon 10 (ATGCGAGCAGAAA) are substituted by a stretch of 14 unrelated bp (CTATCACTTTCAAT). The resulting frame shift creates a stop codon in exon 11. Two T-DNA insertions in the coding region of GRV2 were identified in the Cold Spring Harbor Laboratory (Sundaresan et al., 1995) and Salk Institute for Biological Studies collections (Alonso et al., 2003; Fig. 6B). Both insertion alleles show similar phenotypes as the other grv2 alleles (data not shown), suggesting that the mutant phenotype described above is caused by a complete inactivation of GRV2.
Expression of GRV2
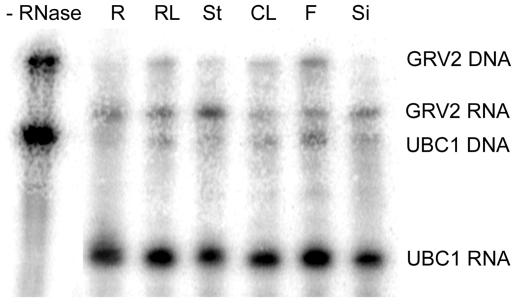
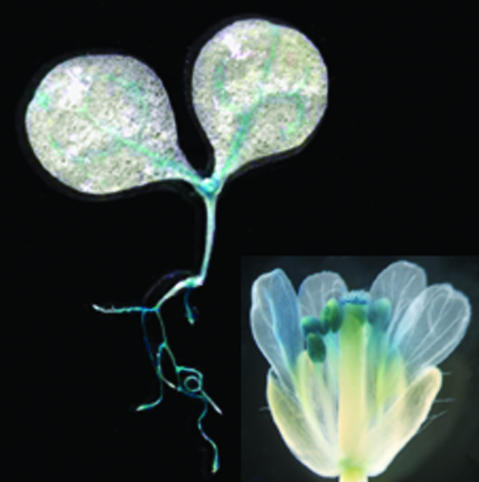
RNAse protection assays indicated that GRV2 is expressed at similar levels in roots, rosette leaves, stems, cauline leaves, flowers, and siliques (Fig. 7). This result is corroborated by publicly available data from whole-genome microarrays (supplemental data). Plants expressing the reporter gene β-glucuronidase (GUS) under the control of the putative GRV2 promoter, a 1.5-kb DNA fragment at the 5′ end of the GRV2 coding region, showed GUS expression in all organs analyzed (roots, hypocotyls, leaf vascular tissue, petals, stigmas, and pollen; Fig. 8).
Figure 7.
RNAse protection assays of GRV2 mRNA from various tissues of wild type. The four fragments correspond to GRV2 DNA (484 bp), GRV2 RNA (387 bp), UBC1 DNA (245 bp), and UBC1 RNA (150 bp) Lanes: R, roots; RL, rosette leaves; St, stems; CL, cauline leaves; F, flowers; Si, siliques.
Figure 8.
Expression pattern of GRV2 promoter GUS fusion in wild-type seedling and flower (inset).
DISCUSSION
Gravitropic responses can be broken down into three consecutive steps: perception of the gravity vector in specialized cells, transduction of the resulting signal to the surrounding tissue, and coordinated asymmetric growth of the organ. Perception of the gravity vector is mediated by the sedimentation of amyloplasts that results in mechanical pressure on cellular components such as the endoplasmic reticulum, cytoskeleton, or internal membranes. This mechanical stimulus is thought to trigger an unknown signal transduction cascade and, ultimately, to regulate auxin flux. Mutants lacking starch have more buoyant plastids with different sedimentation properties and show slower and less pronounced gravitropic responses than wild type (Caspar and Pickard, 1989).
The inflorescence shoots of grv2 mutants exhibit three abnormalities related to tropisms: reduced response to gravity, extended horizontal growth of lateral shoots, and enhanced response to light. A related syndrome of abnormalities was also observed in the sgr mutants (Fukaki et al., 1996b) and is correlated with an abnormal localization of amyloplasts within the cells of the endodermis of sgr2, sgr3, and zig/sgr4 (Morita et al., 2002; Yano et al., 2003). Similarly, the amyloplasts of grv2 endodermis cells are not distributed in the same way as in wild type, indicating that their sedimentation is altered. It seems plausible that the main reason for the reduced gravitropic responses of grv2 shoots is a defect in gravity sensing due to abnormal sedimentation of amyloplasts.
These reduced gravitropic responses might also indirectly cause the enhanced phototropic responses of shoots of grv2 mutants. Gravitropism is thought to be one of the factors responsible for circumnutational movements, the other being most likely an internal oscillator (Hatakeda et al., 2003). In Col wild-type inflorescence shoots exposed to lateral white light, net growth predominantly contributes to the circumnutational movement of the apex, whereas relatively little net growth contributes to tropic responses toward the light. Col mutant alleles of grv2 show no detectable circumnutation, presumably because gravity sensing or gravitropic growth responses are essential for circumnutational movements. Thus, loss of circumnutation in grv2 mutants might make a larger portion of the net growth available for the shoot phototropic response.
Hypocotyls of grv2 mutants also exhibit a slight defect in the gravitropic response, as well as, in contrast to shoots, a reduction in the phototropic response. Given the reduction in the growth rates of grv2 hypocotyls, it is possible that these tropic defects may actually be due to general growth defects rather than specific gravity or light response defects.
Cloning of GRV2 revealed that it is similar to CeRME-8, which encodes a protein required for endocytosis in C. elegans. The first allele of CeRME-8 was isolated in a screen for temperature-sensitive mutations. At the permissive temperature, this allele causes a reduction in receptor-mediated endocytosis of yolk protein into oocytes. At the restrictive temperature, fluid-phase endocytosis from the body cavity, the pseudocoelom, into scavenger cells, coelomocytes, is also impaired. CeRME-8 localizes to the periphery of large vesicles that appear to be endosomes, but its specific biochemical function is not known (Zhang et al., 2001). The Drosophila homolog of RME-8 was identified in a screen for enhancers of shiK39A, a dominant negative mutant of dynamin. DmRme-8 mutants are defective in receptor-mediated endocytosis of BOSS, a membrane ligand, and fluid phase clathrin-dependent uptake of Texas red-conjugated avidin, an endocytic tracer. DmRme-8 localizes to multiple endocytic organelles. The J domain of DmRme-8 interacts with ADP-bound Hsc 70-4 in vitro. This indicates that DmRme-8 functions in endocytosis through an interaction with Hsc 70-4. Hsc70 has several diverse roles in endocytosis, including promoting the release of coat proteins from vesicles, priming clathrin for future rounds of endocytosis, and binding to dynamin in an early step of clathrin-coated vesicle formation. It is not clear which of these Hsc70 functions may require DmRme-8 (Chang et al., 2004).
A complete block in endocytosis in a eukaryotic cell is expected to be lethal because it would prevent recycling of membrane components required for maintaining vesicle-mediated secretory processes. Indeed, the temperature-sensitive rme-8 mutant is lethal at the restrictive temperature indicating that RME-8 is essential in worms. Likewise, all 17 alleles of DmRme-8 are also embryonic lethal (Chang et al., 2004). Based on the type of DNA changes associated with several of the grv2 alleles, we consider it likely that they are null mutations. Thus, it is somewhat surprising that the mutations do not affect viability. The mutants have significantly smaller cells than the wild type, suggesting that some aspect of cell expansion is partially compromised. Because cell wall modifications are required for cell expansion, a defect in plasma membrane recycling would be expected to have a deleterious nonspecific effect on this process.
There are no structurally similar genes to GRV2 in the Arabidopsis genome that could compensate for the loss of GRV2 function. Thus, since it seems unlikely that a plant can survive with a complete loss of endocytosis, grv2 mutations likely could result either in generally inefficient endocytosis and membrane recycling or in the loss of a specific membrane transport pathway. Both scenarios could perceivably have a dramatic impact on the endomembrane system. Preliminary results using the dye FM1-43 as a general marker for endocytosis do not show any dramatic differences between grv2-1 and the wild type (data not shown). The sgr2, sgr3, and zig/sgr4 mutations are thought to primarily affect the tonoplast membrane, causing alterations in vacuole morphology and dynamics (Morita et al., 2002; Yano et al., 2003). Because amyloplasts must somehow sediment around the periphery of the vacuole or through cytoplasmic strands that transverse the vacuole, similar alterations in grv2 could explain the abnormal distribution of amyloplasts in the mutants.
An alternative hypothesis is that a defect in grv2 prevents proper cycling of proteins from the plasma membrane to the internal membrane network. This could, for example, affect the normal turnover of auxin efflux carriers via endocytosis and polarized secretion (Geldner et al., 2001) as well as their relocalization kinetics in a gravitropic response. Another agravitropic mutant, arg1 (altered response to gravity 1), also encodes a protein containing a Dna-J domain (Sedbrook et al., 1999). ARG1 localizes to compartments of the secretory pathway and is thought to regulate the localization or function of membrane proteins involved in the early phases of gravity signal transduction (Boonsirichai et al., 2003).
Based on our current data, it is not possible to distinguish between the two models. It is conceivable that a defect in endocytosis could affect gravitropism in both the sensing step and the asymmetric growth response. This may be the case for zig/sgr4, which has a defect in amyloplasts sedimentation and a 35% decrease in auxin transport in hypocotyls, though it does not dramatically change the localization of PIN1 or PIN3 (Surpin et al., 2003). It is not clear if these two defects both contribute to the reduction in gravitropism or if the amount of auxin that is transported is sufficient for a normal gravitropic response. It will be interesting to see if the cycling of auxin efflux carriers or other membrane proteins is affected in the grv2 mutant.
MATERIALS AND METHODS
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Plant Materials and Growth Conditions
The grv2-1 mutant was isolated from an ethyl methane sulfonate-mutagenized population in the Ler ecotype. The grv2-2 (renamed from sgr8-1), grv2-3 (sgr8-2), and grv2-4 (sgr8-3) alleles were isolated in the Col ecotype. The grv2-2 and grv2-3 alleles were isolated from a fast neutron-mutagenized population, and the grv2-4 allele was isolated from a gamma ray-mutagenized population. Additional T-DNA insertions in GRV2 were obtained from publicly available collections at the Cold Spring Harbor Laboratory (GT1669; Sundaresan et al., 1995) and the Salk Institute (SALK 067162; Alonso et al., 2003). Plants were grown in soil under constant white light 70 to 100 μmol m−2 s−1 at 23°C.
To determine the growth rates, seedlings were grown on the surface of agar medium in vertically oriented petri plates containing Murashige and Skoog medium with 1.5% Suc and 0.8% agar in the dark at 23°C. Hypocotyl lengths were measured every 12 h for 7 d. Cell lengths were measured in the fully elongated hypocotyls on day 7.
Gravitropic Assays
Intact stems of 4-week-old plants, 4 to 8 cm tall, were placed horizontally in the dark at 23°C. In 90-min intervals from 0 to 180 min, the curvature of the stem was measured as the angle between the basal region of a primary inflorescence shoot and the growing tip. To assay the hypocotyl and root gravitropic response, seedlings were grown on Murashige and Skoog medium as described above. Three days after germination, the orientation of the seedlings with respect to gravity was changed to horizontal. The curvature of both hypocotyls and roots were measured at various times thereafter as described previously (Fukaki et al., 1996a).
Phototropic Assays
White light (25–35 μmol m−2 s−1) was applied from the side of the inflorescence shoots in 4-week-old plants (4–8 cm). The degree of curvature of stems was measured every 30 min for 3 h. To assay the hypocotyl phototropic response, seedlings were grown on Murashige and Skoog medium as described above. Three-day-old etiolated seedlings growing vertically were illuminated with horizontal white light 10 to 40 μmol m−2 s−1 for 24 h, and the curvature of the hypocotyls was measured at intervals as described previously (Fukaki et al., 1996a).
Histology
Stem segments (2–3 cm below the apex) cut from primary inflorescence stems were fixed in 4% paraformaldehyde in phosphate buffer, pH 6.8, overnight at 4°C. The growth orientation of the stem was maintained during fixation. Stem segments were dehydrated through an ethanol series and embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) according to the manufacturer's instructions. Sections (4 μm) were stained for 5 min with 0.5% toluidine blue. Amyloplasts were viewed with a compound microscope by 400× magnification and counted at the top and bottom of cells.
Electron Microscopy
Stem segments (2–3 cm below the apex) cut from primary inflorescence stems were fixed in 4% formaldehyde, 2% glutaraldehyde in 0.01 m sodium phosphate buffer, pH 6.8, overnight at 4°C. The growth orientation of the stem was maintained during fixation. Samples were postfixed in 1% osmium tetroxide for 1.5 h, dehydrated in an ethanol series, and embedded in Spurr's resin. Silver-gold sections (60–90 nm) were stained with 2% uranyl acetate (7 min), lead citrate (1 min), and viewed with a JEOL 1230 transmission electron microscope, manufactured in Japan.
Mapping and Cloning
The grv2-1 mutant was crossed to Col wild-type plants to generate an F2 mapping population. Analysis with PCR-based markers indicated that grv2 mapped approximately 2 cM south of erecta. Using the visible phenotype of erecta mutants, recombinants in the grv2-1 erecta interval were selected for fine mapping. Polymorphisms between Col and Ler were identified by sequencing and from the Cereon collection (Jander et al., 2002). New polymorphic markers were deposited in The Arabidopsis Information Resource.
A cosmid library made from the Col ecotype in the binary cosmid vector pBIC20 (Meyer et al., 1994) was screened with probes from the 70-kb mapping interval. Alignment of the four IWN domain repeats was performed with the EMBL CLUSTALW program.
Expression Analysis
Total RNA was isolated from Landsberg wild-type plants using Trizol reagent (Invitrogen, Carlsbad, CA). cDNAs were synthesized from flower RNA using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) and primers specific for GRV2, 5′-CACGGCTGAGGACCTTG-3′ and 5′-CCACAACACAACCTTCG-3′. Nine cDNA fragments were amplified from the primary transcripts by PCR using the following primer pairs: 5′-GGGAGGCCTTAGTAAGAATAAGC-3′ and 5′-GCATTTGCACCGGAGAGACACCG-3′; 5′-CATCGGGACGTGTATGC-3′ and 5′-ATCTGCGGACTCTGCTGGGTTTGTTCTGT-3′; 5′-TCGACATCACCAGGGGAA-3′ and 5′-GTTGAGAGACGAATAG-3′; 5′-CCCTCCACCAAAACCATCAAGCCTACTCA-3′ and 5′-CAGTGATGGATGTCCAGTTC-3′; and 5′-CATCCATTCCTTAGGTGGTTCC-3′ and 5′-CCACACGGAGAGACTACCC-3′. The PCR products, as well as a truncated expressed sequence tag, were sequenced to determine the intron/exon structure of the GRV2 transcripts.
Radioactively labeled GRV2 and UBC1 (Sullivan and Viersta, 1991) antisense probes were transcribed from PCR products. A T7 promoter module (5′-ccaagcttctaatacgactcactatagggag-3′, core promoter underlined) was added to the reverse primers. Primers for assaying GRV mRNA were 5′-cgtgaacaacagaagggttc-3′ and 5′-T7+cttcagccttcacttcgcc-3′. Primers for the control mRNA (At1g14400) were 5′-ctgctatacttacctccatcc-3′ and 5′-T7+agtcagcagtccagcttgc-3′. Ribonuclease protection assays were performed using an RPAII kit (Ambion, Austin, TX) according to the manufacturer's instructions. RNaseA digestion was carried out at room temperature for 20 min.
The sequence of GRV2 is deposited with the EMBL/GenBank data libraries under accession number NM_128246.
Supplementary Material
Acknowledgments
We thank Rob Martienssen, Joe Ecker, and colleagues for providing indexed T-DNA insertion lines, Kazusa DNA Research Institute for providing expressed sequence tag clones, and P. Poindexter for technical assistance. We thank D. Bergmann and W. Briggs for comments on the manuscript.
This work was supported in part by grants from the U.S. Department of Energy (grant no. DOE–FG02–00ER20133), the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant no. CSREES 00–35304–9394), the National Institutes of Health Cellular and Molecular Biology Training Program (grant no. 2–T32–GM007276 to R.A.S.), and the Human Frontiers Science Project organization (grant no. LT–594–96 to W.L.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.050583.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Boonsirichai K, Sedbrook JC, Chen R, Gilroy S, Masson P (2003) ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell 15: 2612–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Pickard BG (1989) Gravitropism in a starchless mutant of Arabidopsis. Planta 177: 185–197 [PubMed] [Google Scholar]
- Chang HC, Hull M, Mellman I (2004) The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J Cell Biol 164: 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Fujisawa H, Tasaka M (1996. a) SGR1, SGR2, and SGR3: novel genetic loci involved in shoot gravitropism in Arabidopsis thaliana. Plant Physiol 110: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Fujisawa H, Tasaka M (1996. b) How do plant shoots bend up? The initial step to elucidate the molecular mechanisms of shoot gravitropism using Arabidopsis thaliana. J Plant Res 109: 129–137 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M (1998) Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J 14: 425–430 [DOI] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof Y, Jürgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Hatakeda Y, Kamada M, Goto N, Fukaki H, Tasaka M, Suga H, Takahashi H (2003) Gravitropic response plays an important role in the nutational movements of the shoots of Pharbitis nil and Arabidopsis thaliana. Physiol Plant 118: 464–473 [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Morita MT, Fukaki H, Yamauchi Y, Uehara M, Niihama M, Tasaka M (2002) SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson PH, Tasaka M, Morita MT, Guan C, Chen R, Boonsirichai K (2002) Arabidopsis thaliana: a model for the study of root and shoot gravitropism. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0043, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2c involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Morita MT, Kato T, Nagafusa K, Saito C, Ueda T, Nakano A, Tasaka M (2002) Involvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell 14: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack FD (1991) Plant gravity sensing. Int Rev Cytol 127: 193–252 [DOI] [PubMed] [Google Scholar]
- Sack FD, Suyemoto MM, Leopold AC (1986) Amyloplast sedimentation and organelle saltation in living corn columella cells. Am J Bot 73: 1692–1698 [PubMed] [Google Scholar]
- Sedbrook JC, Chen R, Masson PH (1999) ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc Natl Acad Sci USA 96: 1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan ML, Viersta RD (1991) Cloning of a 16-kDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J Biol Chem 266: 23878–23885 [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Hong M, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al (2003) The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Fukaki H, Fujisawa H, Tasaka M (1997) Mutations in the SGR4, SGR5, and SGR6 loci of Arabidopsis thaliana alter the shoot gravitropism. Plant Cell Physiol 38: 530–535 [DOI] [PubMed] [Google Scholar]
- Yano D, Sato M, Saito C, Sato MH, Morita MT, Tasaka M (2003) A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc Natl Acad Sci USA 100: 8589–8594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Grant B, Hirsh D (2001) RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol Biol Cell 12: 2011–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.