Abstract
Together with recent advances in the processing and culture of human tissue, bioengineering, xenotransplantation and genome editing, Induced pluripotent stem cells (iPSCs) present a range of new opportunities for the study of human cancer. Here we discuss the main advantages and limitations of iPSC modeling, and how the method intersects with other patient-derived models of cancer, such as organoids, organson-chips and patient-derived xenografts (PDXs). We highlight the opportunities that iPSC models can provide beyond those offered by existing systems and animal models and present current challenges and crucial areas for future improvements toward wider adoption of this technology.
The reprogramming of somatic cells into induced pluripotent stem cells (iPSCs)—cells that can be maintained in a self-sustaining pluripotent state equivalent to that of embryonic stem cells (ESCs)—is a technology that has infiltrated almost all areas of biomedical research1–3. iPSCs are derived from somatic cells through the transient exogenous expression of a set of transcription factors (TFs) and have two unique properties: they can be maintained indefinitely in culture in an undifferentiated pluripotent state, and they can be directed to differentiate into any cell type of the human body. Thus, the derivation of iPSCs from primary human cells offers unprecedented opportunities for creating disease models that capture the primary human cell genome. Although multiple iPSC-based models of monogenic and complex diseases have been created in the past few years4,5, the potential of iPSC modeling in cancer research is just beginning to be explored.
Both basic and translational cancer research rely on model systems to recapitulate the malignant state at the molecular, cellular, tissue, organ and organism level. In recent years, the interest of the scientific community in the development of patient-derived models of cancer has been renewed by increasing concerns regarding the low translation rates of basic research findings and the realization that cancer is a much more complex disease than previously appreciated, along with recent advances expanding the usage of human tissues. Although preclinical cancer research has, in recent years, used primarily immortalized cell lines and genetically engineered mouse models, patient-derived models, including conditional reprogramming (CR)6,7, 3D organotypic cultures (organoids, cell-aggregate cultures, spheres, tissue explants and slices)8–12, patient-derived xenografts (PDXs)13,14 and organs-on-chips15 are increasingly gaining in popularity. In this Perspective, we posit that iPSCs derived from malignant cells can offer yet another tool in the armamentarium of modern cancer research.
iPSCs and cancer modeling
Current iPSC models of cancer and premalignancy
Early studies using transplantation of nuclei from mouse cancer cells showed that cancer genomes can be reprogrammed toward pluripotency16,17. More recently, iPSCs and ‘iPSC-like’ cells have been generated from immortalized human cell lines18–23. Although such studies can address questions pertaining to the reversibility of the cancer phenotype and its epigenetic determinants24, by erasing most of the latter through the reprogramming process, the most exciting application of induced pluripotency is perhaps the reprogramming of primary cells isolated directly from patients.
So far, only a few studies have succeeded at deriving iPSCs from primary malignant or premalignant cells. These are limited to myeloid malignancies, such as myeloproliferative neoplasms (MPNs)—including chronic myeloid leukemia, polycythemia vera and primary myelofibrosis—myelodysplastic syndromes (MDSs) and the MDS–MPN overlap syndrome, juvenile myelomonocytic leukemia25–32. iPSCs from patients with these disorders have shown cellular and molecular phenotypes characteristic of the underlying disorders, such as altered differentiation potential, hematopoietic cell colony formation, cell proliferation and viability, gene expression changes, signaling aberrations and drug sensitivities. Incompletely reprogrammed ‘iPSC-like’ cells—cells that have not attained independence from exogenous expression of reprogramming TFs—have been generated from patients with pancreatic adenocarcinoma33. iPSCs have also been generated from patients with familial cancer predisposition syndromes resulting from germline mutations: Li–Fraumeni syndrome (TP53 mutation)34, Fanconi anemia (FANCA and FANCC mutations)35, familial platelet disorder (FPD) with a predisposition to acute myeloid leukemia (AML) (FPD/AML; RUNX1 mutation)36 and breast cancer predisposition (BRCA1 mutation)37. Li–Fraumeni syndrome iPSCs showed defective osteoblastic differentiation and tumorigenic potential, and they captured gene signatures of primary osteosarcomas, a tumor type that commonly develops in these patients34.
General principles of cancer modeling with iPSCs
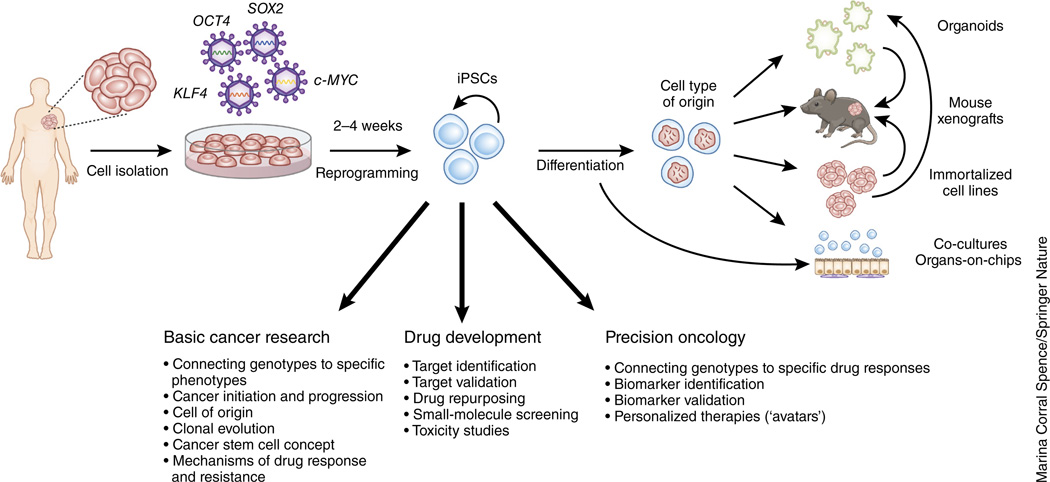
The derivation of iPSCs from cancer cells starts with the isolation and culture of malignant cells from a primary or metastatic tumor specimen obtained surgically, through biopsy or—in the case of hematologic malignancies—from a bone marrow aspirate or a blood sample (Fig. 1). Normal iPSCs that genetically match the malignant iPSCs can be derived from the same cancer patients to provide paired tumor and normal iPSCs that share the same genetic background29,31,33. These can be derived in parallel, through the same reprogramming experiment from normal cells that frequently ‘contaminate’ a tumor specimen, and identified retrospectively through genetic analyses29,31. Alternatively, matched normal iPSCs can be derived in independent reprogramming experiments from normal tissue separately obtained from an area adjacent to the tumor, from a skin biopsy or from the blood (in the case of nonhematologic malignancies)24. For reprogramming, gene transfer of the four TFs OCT4, SOX2, KLF4 and c-MYC, also known as Yamanaka factors, or of alternative factor cocktails is performed using various methods, such as retroviral, lentiviral, episomal or Sendai viruses38. Exogenous expression of these TFs induces massive epigenetic remodeling through processes poorly understood at the molecular level, which leads ultimately to the activation of an endogenous network of pluripotency regulators39. These establish a self-sustaining circuitry of autoregulatory loops that, thereafter, maintain a stable cell state that no longer requires the expression of the exogenous TFs40. TF independence—demonstrated either by the loss of the TF transgenes from the cells (if integration-free methods are used) or by their profound silencing (in the case of integrating vectors)—is thus a defining characteristic of fully reprogrammed, high-quality iPSCs2,40–42. Established iPSC lines can in principle be maintained in culture indefinitely, expanded and cryopreserved without any loss of their genetic, epigenetic or phenotypic properties43–46. Defined media and well-established culture conditions either on feeder cells (most commonly, mitotically inactivated mouse embryonic fibroblasts) or feeder-free exist both for the derivation and the maintenance of human iPSCs47.
Figure 1.
An overview of iPSCs and cancer modeling. Tumor cells are isolated and gene transfer of the four TFs OCT4, SOX2, KLF4 and c-MYC (also known as Yamanaka factors) or of alternative factor cocktails is performed using various methods, such as retroviral, lentiviral, episomal or Sendai viruses. After a period of 2–4 weeks, colonies with the characteristic morphology of pluripotent cells appear and are manually re-plated and expanded to create lines. These are then differentiated into the cell type of origin of the initial tumor. Tumor-derived iPSCs intersect with other model systems of cancer in a variety of ways. iPSC-derived differentiated cells can be used to derive organoids, immortalized cell lines, xenografts, co-cultures and organs-on-chips. Co-cultures and organs-on-chips may also incorporate other cell types that can be derived from iPSCs. ‘Secondary’ iPSC-derived cell lines could theoretically be derived from cancer iPSCs, and in turn, used to generate organoid cultures. Either of these could then be transplanted into xenograft models.
For most experimental applications, the iPSCs need to be differentiated into the cell type that corresponds to the cancer of interest. This is informed by, first, our understanding of the cell of origin of a given tumor, and second, our ability to differentiate the iPSCs into this cell type. Most differentiation protocols harness insights from normal development to recapitulate this process ‘in a dish’ by introducing cocktails of growth factors and/or small molecules at defined times and concentrations. Evaluation of successful derivation of the desired cell type relies on the expression of lineage-specific surface markers or genes or on other tissue-defining properties. Cellular and molecular analyses hinge on comparisons with normal cells, differentiated from normal iPSC lines in the same way as the cancer-derived lines, which, ideally, should be isogenic or nearly isogenic—that is, derived, respectively, through genome editing or from a different cell type of the same individual, so that they share all germline (but not somatic) variants. If none of these controls is available, multiple unrelated normal lines should be used to account for phenotypic differences resulting from genetic background48–51.
Current challenges and limitations of iPSC modeling of human cancers
Technical challenges
A decade after iPSCs were first generated, why don’t we have more iPSC-based cancer models? The two most crucial bottlenecks in the establishment of iPSC cancer models are the efficiency of malignant-cell reprogramming and the ability to differentiate iPSCs into the cell type of interest. A few published studies and anecdotal reports suggest that cancer cells are generally more refractory to reprogramming than normal cells29. However, the success rate of iPSC-line establishment might well be dependent on the cancer type, given that iPSC lines have been derived very successfully from several patients with MPNs25–28,30–32.
Cancer-associated mutations or genetic alterations can have strong—positive or negative—effects on reprogramming efficiency. For example, TP53 inactivation enhances reprogramming, whereas mutations in genes in the Fanconi-anemia pathway have detrimental effects on reprogramming efficiency35,52–56. The possibility that some cancer-related genetic lesions might be incompatible with iPSC generation cannot be excluded, because such lesions may affect pathways that are required for the induction or maintenance of pluripotency. It is also conceivable that the cancer cell type might influence reprogramming efficiency owing to reasons related to the biology of the cell (epigenetic aberrations, impaired DNA damage response, accumulated DNA damage, genetic instability or oncogene-induced senescence) or because of technical impediments related to the inability to dissociate or induce the outgrowth of viable cancer cells from a tumor specimen. Not surprisingly, the first iPSC-based cancer models were derived from hematologic malignancies and premalignancies25–29, in which cancer cells can easily be obtained from blood or bone marrow aspirates as single cells in suspension. Although extended ex vivo growth—required for the derivation of conventional cancer cell lines—is not a prerequisite for reprogramming, some cell division is almost always required57–59. The prediction would thus be that iPSC lines might be difficult or impossible to derive from certain cancers, and the reasons will be difficult to pinpoint. The development of more standardized procedures for the processing of tumor tissues to maximize the recovery of viable cells, as well as optimized culture conditions to propagate them effectively, could make more tumor types and individual samples amenable to reprogramming. Circulating tumor cells (CTCs)—cells shed into the bloodstream by solid tumors—could also provide an alternative, more accessible source of cells for reprogramming, although one that might be skewed toward the most evolved clones, because CTCs often represent the tumor cells that give rise to metastases. New reprogramming methods affording very high efficiency and the use of reprogramming factors alternatively or in addition to the Yamanaka cocktail (for example, LIN28, NANOG, TP53 shRNA), ideally tailored to each cancer’s genetic composition, might help to overcome refractoriness to reprogramming in some cases. Integration-free methods, i.e., methods that do not require the integration of foreign genetic material into the cell’s genome, ensure the derivation of iPSC lines independent of the exogenous factors because these are lost from the cells in the absence of integration, and they eliminate concerns of insertional mutagenesis that might confound cellular phenotypes. However, vectors that integrate into the genome and can subsequently be excised can afford higher efficiency of derivation of cells than nonintegrating vectors, as well as analysis of clonality, given that viral integration sites facilitate the tracking of line provenance from distinct cells of the original tumor29,60.
The second limiting step is the derivation of cells of a defined cell type from iPSCs, which involves their directed differentiation to first specify a germ layer and then a specific lineage, often followed by terminal differentiation. Given that most, if not all, cancers arise from tissue-specific stem and progenitor cells61, iPSC cancer-modeling applications will mostly require the—easier and faster—derivation of these rather than of terminally differentiated functional cell types that are often required in nonmalignant-disease-modeling applications5,29. On the other hand, the evaluation of successful differentiation might be complicated by the fact that cancers often show aberrant expression of lineage markers. Improved differentiation protocols that afford high efficiency, scalability, use fully defined reagents and, ideally, are amenable to automation are still needed.
Because iPSC differentiation protocols mimic developmental processes, a general challenge of iPSC disease modeling is the discrimination of disease phenotypes from phenotypes of earlier developmental stages. This issue could, at least in part, be addressed by the use of conditional alleles that enable the introduction of a specific genetic modification at a later differentiation stage62. Alternatively, careful characterization of the stage of differentiation in which a cellular phenotype first manifests can be pursued by detailed step-wise phenotyping. Another potentially confounding factor is that most differentiation procedures give rise to cells that are developmentally more immature than their naturally existing counterparts, i.e., more similar to embryonic or fetal than adult cell types63–65. Although our ability to drive cell maturation in culture with improved protocols constantly increases, efforts should, in parallel, be made to thoroughly characterize disease-relevant phenotypes in the derivative cell types to determine which are dependent on developmental maturation and which, potentially, are not.
Genetic stability
Early studies raised concerns that iPSCs might be genetically unstable, as compared to other pluripotent or somatic cell types66,67. However, sensitive methods using next-generation sequencing subsequently showed that most copy-number and single-nucleotide variants detected in iPSCs are pre-existent in the starting somatic cells, and that they are merely captured and amplified during iPSC derivation68–70. Most of the remaining mutations were shown to have been acquired during expansion in culture or during differentiation, and at a rate similar to that of normal adult somatic cells that is consistent with estimates of spontaneous mutation acquisition during cell division70–74. It is therefore now clear that reprogramming per se is not mutagenic, and that iPSCs are not inherently genetically unstable44,75. However, these studies examined normal iPSCs and cannot exclude genomic instability as a feature of iPSCs from cancers. Even iPSCs from genetically stable cancers, as with all cultured cells, can acquire spontaneous genetic abnormalities that can be subjected to selection, if they confer a growth advantage, or cause genetic drifts if they are neutral75. The latter can often be induced inadvertently by manual manipulations that introduce clonal bottlenecks and are common practice in human pluripotent stem cell culture, such as single-cell subcloning or even routine manual passaging. On the other hand, because iPSCs can easily be grown as single cells and expanded, frequent monitoring using standard karyotyping and/or more sensitive molecular techniques, and in the case of a genetic drift, retrieval of the original genome, is feasible by single-cell subcloning of earlier passage cryopreserved cells.
Epigenetic instability, incomplete reprogramming and line-to-line variation
The reprogramming of the cancer genome to pluripotency induces dramatic global changes to the epigenome and transcriptome of the somatic cell39,40. Occasionally, incomplete or aberrant reprogramming can produce cells with altered epigenomes and functional properties76,77. Recent reports suggest that the reprogramming of established cancer cell lines or primary cells might often give rise to partially reprogrammed ‘iPSC-like’ cells, characterized by continuous dependency on exogenous TF expression20–23. In one study, by reprogramming epithelial cells from primary pancreatic ductal adenocarcinoma, the authors were able to derive only ‘iPSC-like’ cell lines with features of pluripotency, such as the ability for tri-lineage differentiation in vitro and in vivo, but the cells remained dependent on TF expression33. Because partially reprogrammed cells exhibit variable functional and epigenetic properties, they should be clearly identified as such, and results obtained by using them should be interpreted with caution. Fully reprogrammed, high-quality iPSCs should be selected after stringent characterization according to well-established criteria of pluripotency and the demonstration of factor independence46. Bona fide iPSCs, by these criteria, have thus far been derived only from hematologic malignancies25,26,28,29,31,32. Incomplete reprogramming can also be observed at early passages of iPSC derivation and manifest as transcriptional, epigenetic and chromatin-conformation ‘memory’ of the donor cell type and altered differentiation propensity45,78–80. Importantly, several studies have now shown that iPSCs do not exhibit greater phenotypic or transcriptional line-to-line variation than that of human embryonic stem cell (ESC) lines, and are thus not inherently epigenetically unstable43,81,82. In fact, accumulating evidence indicates that the major determinant of the transcriptional, epigenetic and functional (differentiation propensity) properties of iPSCs is their genetic makeup, validating them as models for genotype-to-phenotype cancer genetics studies29,48–51,83. By contrast, their value as models of cancer epigenetics is less certain because epigenetic aberrations of the starting cancer cell are likely to have been reset or erased during reprogramming39. However, epigenetic alterations that are genetically determined (for example, owing to mutation of an epigenetic regulator) might persist or become re-established upon differentiation. Studies characterizing the epigenetic landscape of iPSC-derived cells, as compared to that of the starting primary tumor cells, might help to understand the limitations of these models and to interrogate the interrelationship between genetic and epigenetic lesions in specific cancers.
Applications of iPSCs in basic and translational cancer research
Basic cancer research
The characteristics that give iPSCs a unique status as cancer models are intimately linked to their properties as pluripotent cells. First, their unlimited self-renewal offers the opportunity to capture the entire intact genome of a single cancer cell and amplify it into an unlimited number of copies. Unlimited expansion can, at least theoretically, be achieved without phenotypic drifts (often observed in organoids or PDXs), because the pluripotent state is uniform, stable and self-sustaining in vitro40,46. Second, their broad developmental potential offers the opportunity to examine the effects of a specific cancer genotype or a specific driver mutation, a mutation thought to drive oncogenesis, in different cell types and developmental stages, providing a unique system through which to address questions such as whether a specific tissue-restricted epigenetic environment is required for an oncogene to exert its tumorigenic potential, and whether common or distinct pathways operate downstream of a common oncogene in different tissues. It can also empower studies on the cell of origin of cancers, i.e., the normal cell type in which the transformation process was initiated by the acquisition of the first cancer-promoting mutation61, and aid the identification of commonalities among different types of cancer driven by a common oncogene in the search for broad therapeutic targets.
iPSCs are particularly well suited to in-depth mechanistic studies because they provide a scalable source of homogeneous cellular material that allows for well-controlled experiments (Fig. 2). Because they enable the study of phenotypes at the molecular and cellular level—phenotypes that are more proximal to disease mechanisms than phenotypes at the level of the organ or whole organism—they offer a reductionist approach for investigations into the underlying mechanisms of tumorigenesis, drug response and drug resistance (Fig. 2a). The effects of specific genetic lesions on the cellular and molecular phenotype and the cooperation between co-occurring mutations can be studied in depth using genome-wide molecular analyses, because large cell numbers can be obtained relatively easily. For the same reason, genetic and chemical screens and other high-throughput analyses are possible.
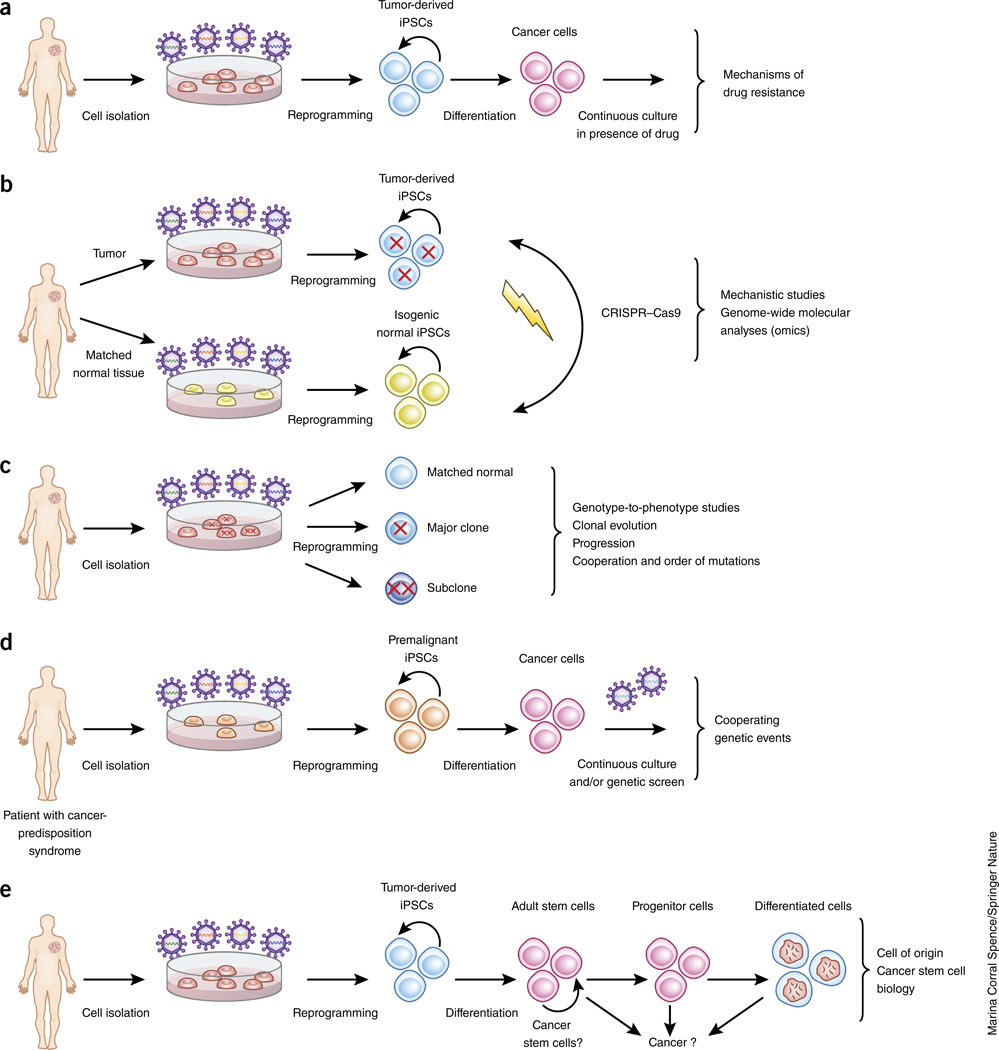
Figure 2.
Potential uses of iPSCs in basic cancer research. (a) Cancer-derived iPSCs treated with a cancer drug over prolonged periods of time can be used to study resistance mechanisms. (b) Cancer-derived iPSCs and isogenic normal iPSCs, derived either from matched normal tissue of the same patient or through genome-editing alteration of the cancer genes, can be used for mechanistic studies and genome-wide molecular analyses. (c) iPSCs capturing distinct clones (major and minor), representing different stages of cancer, of a single tumor can be used to model cancer progression and to study the principles underlying cooperation among coexisting genetic lesions. (d) Premalignant iPSCs can be used to interrogate recurrent genetic events required for progression. (e) iPSCs differentiated into adult (or tissue-specific) stem and progenitor cells can be used to interrogate the cell of origin of a given cancer (from adult stem, progenitor or more differentiated cells). Tumor-derived iPSCs can also aid investigations of the cancer stem cell concept (positing that at least some cancers are organized hierarchically and sustained by a subpopulation of self-renewing cells that exist at a distinct epigenetic state within a genetically homogeneous cancer cell population).
Owing to the ease of single-cell subcloning, iPSCs are highly amenable to the introduction of precise genetic modifications by the CRISPR–Cas9 system or other genome-editing tools84. Specific gene mutations, individually or in combinations, can thus be engineered into normal iPSCs or, conversely, corrected in cancer iPSCs, to provide complementary isogenic systems (Fig. 2b). The two most common types of mutation that can be engineered using CRISPR–Cas9 are gene disruption through the introduction of frame-shifting indels, mediated by nonhomologous end-joining (NHEJ) in a gene-coding region, and point mutations through homology-directed repair (HDR), mediated through the co-delivery of a donor DNA template85,86. The former can be used to mono-allelically or bi-allelically inactivate haplo-insufficient or classical tumor-suppressor genes, respectively, and the latter to model hotspot point mutations in cancer-promoting genes. The CRISPR–Cas9 system thus enables modeling of the two main classes of cancer mutations in their natural genomic context. The CRISPR–Cas9 system can also be used to introduce large-scale genetic lesions often found in cancers, including chromosomal deletions, inversions and translocations29,87. Finally, conditional or inducible epigenetic editing using CRISPR–Cas9 can also provide a valuable tool with which to study the effects of specific gene-expression alterations at defined stages of cell differentiation88.
Modeling cancer progression
Because reprogramming is a clonal process, iPSCs that capture distinct clones at different stages of cancer evolution, including dominant and minor clones, theoretically, can be generated by reprogramming a tumor specimen, and can be retrospectively identified on the basis of the distribution—clonal (present in most cells) or subclonal (present only in a fraction of the cells)—of their mutations in the original tumor29 (Fig. 2c). Another approach to modeling multistage cancer might involve the prospective modeling of the acquisition of co-operating events in iPSCs harboring premalignant or early-stage genetic lesions (Fig. 2d). An ‘iPSC-like’ line from a human pancreatic ductal adenocarcinoma generated precancerous lesions that resembled pancreatic intraepithelial neoplasia in mice and progressed to invasive cancer33. This suggests that, at least in some cases, reprogramming might yield cells that ‘revert’ to a less aggressive cancer stage, presumably by altering the epigenetic state of the original cancer cells or selecting for cancer cells with a less-aggressive phenotype. More cancer-reprogramming studies are needed to clarify whether this is a common phenomenon and whether it reflects the dependence of particular cancers on epigenetic aberrations. Finally, by capturing early cancer stages, iPSC models offer a new platform through which to revisit and explore basic concepts and fundamental principles in cancer biology, such as cells of origin of cancers and cancer stem cells61,89 (Fig. 2e).
Complex cancer genetics
Because iPSCs capture a cancer-cell genome, regardless of the extent to which we understand its drivers, they can be used to study cancers in which the genetic drivers are incompletely understood. Similarly, iPSC technology enables the capture and study of genomes harboring disease predisposition variants identified through linkage or association studies and, as such, enables their functional characterization90. iPSCs also provide a faster means of modeling complex genotypes (for example, chromosomal rearrangements, aneuploidy or cooperating gene mutations) that cannot easily be engineered in model organisms. Kotini et al.29 used iPSC technology to capture the genomes of two patients with MDS harboring large deletions of chromosome 7q, which are found frequently in this cancer and span large chromosomal fragments that contain hundreds of genes, which are, in turn, divided among different chromosomes in the mouse. Through functional studies and a genetic rescue screen, the authors were thus able to functionally map a critical deleted chromosomal region and pinpoint specific genes with a role in del(7q)-MDS. Finally, increasingly larger collections of iPSC lines from different genetic backgrounds with linked single-nucleotide polymorphism (SNP) or whole-genome sequencing data are becoming available, and these can enable studies on the influence of genetic variation in the tumorigenicity of a given oncogene and the interplay between genetic background and environmental factors in the malignant process.
Drug development
iPSC cancer modeling could also empower more translational-research areas, such as the identification and validation of therapeutic targets, preclinical efficacy and safety studies and compound screening for drug discovery and drug repurposing91–95. iPSC-derived cell types can provide a more relevant platform for high-throughput drug screening than can immortalized cell lines, fibroblasts or lymphoblastoid cells. A particularly attractive use of iPSC technology in drug development is toxicology testing, given that it is possible to generate a variety of healthy cells from normal (matched isogenic or nonisogenic) iPSCs, including hepatocytes, cardiomyocytes and hematopoietic cells, as well as organoids and organs-onchips derived from them, composed of tissues such as kidney, lung, intestine, liver or cholangiocytes96–105. Cardiomyocytes derived from iPSCs from patients with breast cancer were shown recently to model at the cellular level the doxorubicin-induced cardiotoxicity manifested in the patients106. Such models of specific drug-induced toxicities can aid the understanding of the genetic basis and molecular mechanisms of these side effects. iPSC modeling also presents an opportunity for phenotype-based drug discovery91,107. Phenotype-based drug testing and drug screening can be particularly attractive in cancers with no previously defined targets, and it is based on the identification of cellular phenotypes or other functional assays—for example, proliferation, apoptosis, activation of a particular signaling pathway or metabolic changes—that correlate with patient responses and can be used as surrogate assays predictive of a therapeutic response108. For example, a hyperexcitability phenotype in neurons derived from iPSCs from patients with bipolar disorder was found to be responsive to lithium treatment and to correlate with patient responses109.
Precision oncology
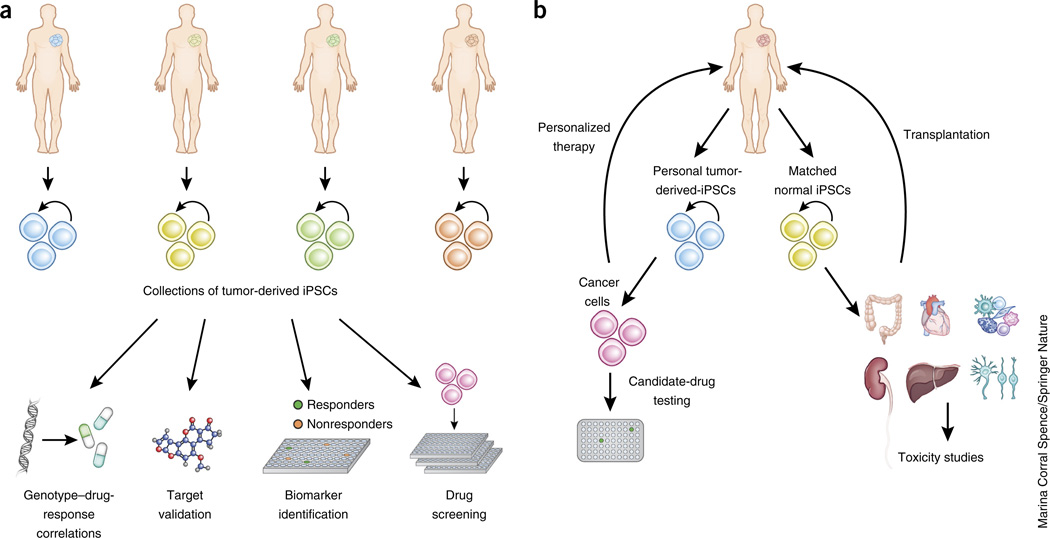
iPSCs can be used to draw associations between genotype and drug responses and to identify biomarkers to inform patient selection for enrollment in clinical trials109–111. For example, the pharmacological reversal, using a selective sodium-channel blocker, of a hyperexcitability phenotype in sensory neurons derived from iPSCs from patients with the pain disorder inherited erythromelalgia was found to correlate with patient responses and specific patient mutations111. For such applications to materialize, large and diverse collections of cancer-derived iPSCs, capturing a range of cancer types and genotypes, will need to be assembled. This will require the formation of collaborative initiatives to support their derivation and characterization, as well as repositories for banking, documentation, authentication and distribution. Guidelines and good practices to safeguard against line misidentification and cross-contamination will be essential. Regulatory issues will need to be addressed to enable the sharing of materials while protecting donors’ privacy, given that genetic information will be obtained from the lines. Finally, personal iPSCs, as companion diagnostics or as avatars to guide treatment tailored to individual patients, can also be envisioned112 (Fig. 3). A big challenge in these applications will be to align the timescales of model establishment to those of clinical decision-making. The establishment of iPSCs could be advantageous in this respect, because the time from harvesting the tissue to deriving the relevant cell type is, on average, 2–4 months, as compared to 2–6 months for CR and organoids and longer than 6 months for PDX models. Finally, with advances in regenerative-medicine applications, tissue and organ regeneration using normal autologous or histocompatible iPSCs might be possible for patients with cancer who are in need of tissue-replacement therapies owing to surgical resection or infiltration by malignant cells.
Figure 3.
Applications of iPSCs in translational cancer research. (a) Large collections of iPSCs capturing cancer diversity can be used for drug discovery and precision oncology. (b) Applications in personalized cancer treatment toxicology studies and regenerative medicine.
iPSCs and other patient-derived cancer models
iPSCs versus immortalized cell lines
Conventional cell lines represent fully transformed and mostly aggressive cancer cells from advanced-stage cancers, derived either spontaneously through selection for rapid growth in culture or through exogenous expression of viral or mammalian oncogenes113. By contrast, the immortalization of iPSCs is not conferred by transformation but instead by the induction of pluripotency. Although the pluripotent state shares some cellular properties and molecular pathways with cellular transformation, pluripotency and transformation are two clearly distinct processes46. This makes the derivation of iPSCs from cells that are not fully transformed—and hence, from which conventional cell lines cannot be derived—feasible, thus enabling the capture of premalignant cells, cells in early stages of the malignant process and cells with cancer predisposition mutations that have not yet initiated the process of transformation (Table 1). iPSCs are independent of exogenous genes, whereas most cancer cell lines rely on continuous expression of strong exogenous oncogenes, which might alter their cellular behavior and gene expression in nonphysiological ways2,41,114,115. Reprogramming to pluripotency has been proven to be a universal property shared by all cells irrespective of cell type, developmental stage, species or gene expression116. Thus, although this remains to be shown, iPSCs can potentially be derived from a wide range of genotypes and cell types, including those for which very few or no cancer cell lines exist (for example, prostate cancer, pancreatic cancer, glioblastomas and cancers with isocitrate dehydrogenase (IPH)-1 or IPH2 mutations). Finally, it is conceivable, although currently hypothetical, that immortal cell lines, similarly to conventional cell lines, could be derived from cells differentiated from iPSCs. Such ‘secondary’ cell lines could be propagated in nonpluripotent conditions and would therefore be more accessible to investigators without stem cell culture expertise. They could be especially useful in cancers from which direct derivation of cell lines from the original primary tumor is not technically feasible.
Table 1.
Comparison of 2D iPSC modeling to other patient-derived models of cancer
| Immortalized cell lines | Conditional reprogramming | 3D organotypic cultures | PDX | Tumor-derived iPSCs | |
|---|---|---|---|---|---|
| Rate of initial establishment | Moderate | Moderate (only epithelial cancers) | Moderate | ~2/3 of samples | Low/moderate |
| Derivation time | 2–4 months | 2–6 months | 2–6 months | >6 months | 2–4 months* |
| Ease of maintenance/required expertise | High/low | Moderate/high | Low/high | Moderate/high | Low/high |
| Expansion/scalability | Very high | Unknown | High | High | Very high |
| Cost | Low | Moderate | High | High | High |
| Amenability to high throughput | High | Unknown | Moderate | Moderate | High |
| Modeling early-stage cancer and premalignancy |
No | Unknown | Yes | No | Yes |
| Amenability | High | Unknown | High | Low | High |
| Autologous | No | Unknown | Yes | No | Yes |
| Isogenic | Yes | Unknown | Yes | No | Yes |
| Genetic | Yes | Unknown but likely | Unknown but likely | Yes | Yes |
| Microenvironmental | No | No | Yes | Xenogeneic | Possible (co-culture) |
Asterisk (*) indicates that it includes differentiation to the target cell type.
iPSCs versus conditional reprogramming
A method of establishing 2D cultures of patient-derived tumor cells in the presence of Rho kinase (Rock) inhibitor and feeder cells, empirically developed recently by Schlegel and colleagues and referred to as CR, gives rise to cultures of expandable cells that are thought to acquire some features of adult stem cells6,7,117. CR is technically more simple than reprogramming to pluripotency, but is confined to epithelial tumors and gives rise to polyclonal populations. The cell state that CR induces and the properties of the cells that are produced using this method are not well defined. Several important aspects of this model, such as the ability for cell expansion, the degree of genetic and phenotypic drift over passage and its permissiveness to clonal growth and genetic manipulation, have not yet been explored. In contrast to CR, reprogramming to pluripotency can be—and typically is—performed in conditions preserving clonality (whereby individual iPSC lines are derived from a single starting cell) and thus gives rise to genetically homogeneous lines.
Intersection with organoids, organs-on-chips and PDX technologies
iPSCs can also offer a window to higher-order modeling when combined with 3D organoid or organ-like culture or xenotransplantation. Simple, heterotypic 2D or 3D culture systems that incorporate niche elements of the tumor microenvironment can also be developed through co-culture with primary autologous, allogeneic or iPSC-derived stromal cells. Organoid cultures consist of 3D structures that recapitulate aspects of the architecture of the tissue from which they were derived, and they can be initiated using differentiated cells, tissue-specific (or adult) stem and progenitor cells or human pluripotent stem cells (hPSCs, including ESCs and iPSCs)118–120. Organoids derived from primary resected tumors or biopsies have been recently reported121–125. iPSCs can be differentiated in embryoid body culture, and subsequently, embedded in extracellular matrix (ECM) to initiate organoid cultures with media appropriate for the respective tissue9. Organoids have been derived from normal iPSCs and offer advantages over organoids derived from tissue-specific stem cells because the latter typically contain only the epithelial component of the tissue, whereas the former can contain multiple tissue cell types, including stroma and endothelium126,127.
Organs-on-chips are microfluidic devices composed of cell culture chambers and channels that allow for continuous perfusion. Organs-onchips have been made using primary cells or transformed cell lines and, very recently, human ESCs and iPSCs128. The transplantation of iPSC-derived cells into mice can further expand their experimental capabilities in an in vivo setting33. Such secondary PDX models, derived through an intermediate iPSC stage, could be useful in the approximately one-third of patients with cancer from which PDXs cannot be derived directly from primary cells. An intermediate iPSC stage would also enable the genetic manipulation of the cells before transplantation and the establishment of xenografts to, for example, express luciferase or a fluorescent protein to facilitate tracking or to introduce specific genetic modifications to study their effects on tumor growth in vivo.
Concluding remarks
A new era in preclinical cancer research is emerging, in which human-based models are taking center stage and patient-derived cells are increasingly being used as primary discovery platforms. In this modern era of basic cancer research and precision oncology, iPSCs derived from patients with cancer can substantially expand the experimental repertoire applicable to human cells in ways that were hitherto restricted to model organisms. We envision that models for at least some cancers can be developed using iPSC technologies, and that these will occupy a unique place in this new era, bridging primary cells with immortalized cell lines by combining the physiological relevance of the former with the amenability to experimentation of the latter. Interdisciplinary collaborations between stem cell researchers, cancer researchers, physicians, translational scientists, bioengineers and drug developers will be paramount to harness the full potential of iPSCs as a new tool in this modern era of cancer research.
Acknowledgments
E.P.P. is supported by US National Institutes of Health (NIH) grants R00 DK087923 and R01 HL121570; by the Lawrence Ellison Foundation; by the Damon Runyon Cancer Research Foundation; by the Edward Evans Foundation; and by the Taub Foundation for MDS research.
Footnotes
COMPETING FINANCIAL INTERESTS
The author declares no competing financial interests.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Papapetrou EP. Induced pluripotent stem cells, past and future. Science. 2016;353:991–992. doi: 10.1126/science.aai7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onder TT, Daley GQ. New lessons learned from disease modeling with induced pluripotent stem cells. Curr. Opin. Genet. Dev. 2012;22:500–508. doi: 10.1016/j.gde.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeltner N, Studer L. Pluripotent stem cell-based disease modeling: current hurdles and future promise. Curr. Opin. Cell Biol. 2015;37:102–110. doi: 10.1016/j.ceb.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crystal AS, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 10.Ridky TW, Chow JM, Wong DJ, Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat. Med. 2010;16:1450–1455. doi: 10.1038/nm.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaira V, et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl. Acad. Sci. USA. 2010;107:8352–8356. doi: 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majumder B, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun. 2015;6:6169. doi: 10.1038/ncomms7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gould SE, Junttila MR, de Sauvage FJ. Translational value of mouse models in oncology drug development. Nat. Med. 2015;21:431–439. doi: 10.1038/nm.3853. [DOI] [PubMed] [Google Scholar]
- 14.Day CP, Merlino G, Van Dyke T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell. 2015;163:39–53. doi: 10.1016/j.cell.2015.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell. 2016;164:1105–1109. doi: 10.1016/j.cell.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 16.Hochedlinger K, et al. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18:1875–1885. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blelloch RH, et al. Nuclear cloning of embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA. 2004;101:13985–13990. doi: 10.1073/pnas.0405015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carette JE, et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 2009;122:3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi N, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. USA. 2010;107:40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Cruz FD, Terry M, Remotti F, Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013;32:2249–2260. doi: 10.1038/onc.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stricker SH, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corominas-Faja B, et al. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell Cycle. 2013;12:3109–3124. doi: 10.4161/cc.26173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Zaret KS. Reprogramming of human cancer cells to pluripotency for models of cancer progression. EMBO J. 2015;34:739–747. doi: 10.15252/embj.201490736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu K, et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandre-Babbe S, et al. Patient-derived induced pluripotent stem cells recapitulate hematopoietic abnormalities of juvenile myelomonocytic leukemia. Blood. 2013;121:4925–4929. doi: 10.1182/blood-2013-01-478412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumano K, et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–6242. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- 28.Hosoi M, et al. Generation of induced pluripotent stem cells derived from primary and secondary myelofibrosis patient samples. Exp. Hematol. 2014;42:816–825. doi: 10.1016/j.exphem.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Kotini AG, et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat. Biotechnol. 2015;33:646–655. doi: 10.1038/nbt.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Z, et al. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Z, et al. Differential sensitivity to JAK inhibitory drugs by isogenic human erythroblasts and hematopoietic progenitors generated from patient-specific induced pluripotent stem cells. Stem Cells. 2014;32:269–278. doi: 10.1002/stem.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulero-Navarro S, et al. Myeloid dysregulation in a human induced pluripotent stem cell model of PTPN11-associated juvenile myelomonocytic leukemia. Cell Rep. 2015;13:504–515. doi: 10.1016/j.celrep.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3:2088–2099. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee DF, et al. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161:240–254. doi: 10.1016/j.cell.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller LU, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antony-Debré I, et al. Level of RUNX1 activity is critical for leukemic predisposition but not for thrombocytopenia. Blood. 2015;125:930–940. doi: 10.1182/blood-2014-06-585513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soyombo AA, et al. Analysis of induced pluripotent stem cells from a BRCA1 mutant family. Stem Cell Reports. 2013;1:336–349. doi: 10.1016/j.stemcr.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 39.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papapetrou EP, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 43.Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7:263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tapia N, Schöler HR. Molecular obstacles to clinical translation of iPSCs. Cell Stem Cell. 2016;19:298–309. doi: 10.1016/j.stem.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 45.Krijger PH, et al. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell. 2016;18:597–610. doi: 10.1016/j.stem.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Los Angeles A, et al. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- 47.Chen KG, Mallon BS, McKay RD, Robey PG. Human pluripotent stem cell culture: considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14:13–26. doi: 10.1016/j.stem.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouhani F, et al. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kyttälä A, et al. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Reports. 2016;6:200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Féraud O, et al. Donor dependent variations in hematopoietic differentiation among embryonic and induced pluripotent stem cell lines. PLoS One. 2016;11:e0149291. doi: 10.1371/journal.pone.0149291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burrows CK, et al. Genetic variation, not cell type of origin, underlies the majority of identifiable regulatory differences in iPSCs. PLoS Genet. 2016;12:e1005793. doi: 10.1371/journal.pgen.1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marión RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raya A, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruiz S, et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo S, et al. Nonstochastic reprogramming from a privileged somatic cell state. Cell. 2014;156:649–662. doi: 10.1016/j.cell.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papapetrou EP, Sadelain M. Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat. Protoc. 2011;6:1251–1273. doi: 10.1038/nprot.2011.374. [DOI] [PubMed] [Google Scholar]
- 61.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 62.Chen Y, et al. Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hrvatin S, et al. Differentiated human stem cells resemble fetal, not adult, β cells. Proc. Natl. Acad. Sci. USA. 2014;111:3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van den Berg CW, et al. Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development. 2015;142:3231–3238. doi: 10.1242/dev.123810. [DOI] [PubMed] [Google Scholar]
- 65.Rowe RG, Mandelbaum J, Zon LI, Daley GQ. Engineering hematopoietic stem cells: lessons from development. Cell Stem Cell. 2016;18:707–720. doi: 10.1016/j.stem.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hussein SM, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 67.Ji J, et al. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- 68.Young MA, et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abyzov A, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng L, et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mayshar Y, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 72.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varela C, et al. Recurrent genomic instability of chromosome 1q in neural derivatives of human embryonic stem cells. J. Clin. Invest. 2012;122:569–574. doi: 10.1172/JCI46268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson SE, Loring JF. Genomic instability in pluripotent stem cells: implications for clinical applications. J. Biol. Chem. 2014;289:4578–4584. doi: 10.1074/jbc.R113.516419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Polo JM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Polo JM, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghosh Z, et al. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bock C, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guenther MG, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi J, et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sterneckert JL, Reinhardt P, Schöler HR. Investigating human disease using stem cell models. Nat. Rev. Genet. 2014;15:625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 86.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mandegar MA, et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell. 2016;18:541–553. doi: 10.1016/j.stem.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 90.Soldner F, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Engle SJ, Puppala D. Integrating human pluripotent stem cells into drug development. Cell Stem Cell. 2013;12:669–677. doi: 10.1016/j.stem.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 92.Engle SJ, Vincent F. Small molecule screening in human induced pluripotent stem cell-derived terminal cell types. J. Biol. Chem. 2014;289:4562–4570. doi: 10.1074/jbc.R113.529156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee G, et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat. Biotechnol. 2012;30:1244–1248. doi: 10.1038/nbt.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wainger BJ, et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Naryshkin NA, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 96.Ogawa M, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat. Biotechnol. 2015;33:853–861. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 97.Sampaziotis F, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 99.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 100.Morizane R, et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 2015;33:1193–1200. doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 102.McCracken KW, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Watson CL, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dye BR, et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015;4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schwartz MP, et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl. Acad. Sci. USA. 2015;112:12516–12521. doi: 10.1073/pnas.1516645112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burridge PW, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eggert US. The why and how of phenotypic small-molecule screens. Nat. Chem. Biol. 2013;9:206–209. doi: 10.1038/nchembio.1206. [DOI] [PubMed] [Google Scholar]
- 108.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer. 2015;15:747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mertens J, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garbes L, et al. VPA response in SMA is suppressed by the fatty acid translocase CD36. Hum. Mol. Genet. 2013;22:398–407. doi: 10.1093/hmg/dds437. [DOI] [PubMed] [Google Scholar]
- 111.Cao L, et al. Pharmacological reversal of a pain phenotype in iPSC-derived sensory neurons and patients with inherited erythromelalgia. Sci. Transl. Med. 2016;8:335ra56. doi: 10.1126/scitranslmed.aad7653. [DOI] [PubMed] [Google Scholar]
- 112.Terrenoire C, et al. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J. Gen. Physiol. 2013;141:61–72. doi: 10.1085/jgp.201210899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maqsood MI, Matin MM, Bahrami AR, Ghasroldasht MM. Immortality of cell lines: challenges and advantages of establishment. Cell Biol. Int. 2013;37:1038–1045. doi: 10.1002/cbin.10137. [DOI] [PubMed] [Google Scholar]
- 114.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Henderson E, Miller G, Robinson J, Heston L. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology. 1977;76:152–163. doi: 10.1016/0042-6822(77)90292-6. [DOI] [PubMed] [Google Scholar]
- 116.Maherali N, Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 117.Suprynowicz FA, et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA. 2012;109:20035–20040. doi: 10.1073/pnas.1213241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 119.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 120.Wu J, Izpisua Belmonte JC. Stem cells: a renaissance in human biology research. Cell. 2016;165:1572–1585. doi: 10.1016/j.cell.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 121.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang L, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 124.Gao D, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Passier R, Orlova V, Mummery C. Complex tissue and disease modeling using hiPSCs. Cell Stem Cell. 2016;18:309–321. doi: 10.1016/j.stem.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 127.Guye P, et al. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Giobbe GG, et al. Functional differentiation of human pluripotent stem cells on a chip. Nat. Methods. 2015;12:637–640. doi: 10.1038/nmeth.3411. [DOI] [PubMed] [Google Scholar]