Abstract
Natural products have traditionally served as a dominant source of therapeutic agents. They are produced by dedicated biosynthetic gene clusters that assemble complex, bioactive molecules from simple precursors. Recent genome sequencing efforts coupled with advances in bioinformatics indicate that the majority of biosynthetic gene clusters are not expressed under normal laboratory conditions. Termed ‘silent’ or ‘cryptic’, these gene clusters represent a treasure trove for discovery of novel small molecules, their regulatory circuits and their biosynthetic pathways. In this review, we assess the capacity of exogenous small molecules in activating silent secondary metabolite gene clusters. Several approaches that have been developed are presented, including coculture techniques, ribosome engineering, chromatin remodeling and high-throughput elicitor screens. The rationale, applications and mechanisms attendant to each are discussed. Some general conclusions can be drawn from our analysis: exogenous small molecules comprise a productive avenue for the discovery of cryptic metabolites. Specifically, growth-inhibitory molecules, in some cases clinically used antibiotics, serve as effective inducers of silent biosynthetic gene clusters, suggesting that old antibiotics may be used to find new ones. The involvement of natural antibiotics in modulating secondary metabolism at subinhibitory concentrations suggests that they represent part of the microbial vocabulary through which inter- and intraspecies interactions are mediated.
Keywords: secondary metabolites, antibiotics, interspecies interaction, ribosome engineering, chromatin remodeling, HiTES
Microbial secondary metabolites represent a significant source of potential drug leads; however, the majority of the corresponding biosynthetic genes are not expressed under normal laboratory conditions. In this review, we assess the capacity of exogenous small molecules, especially antibiotics, to activate these silent gene clusters.
Graphical Abstract Figure.
Microbial secondary metabolites represent a significant source of potential drug leads; however, the majority of the corresponding biosynthetic genes are not expressed under normal laboratory conditions. In this review, we assess the capacity of exogenous small molecules, especially antibiotics, to activate these silent gene clusters.
INTRODUCTION
Natural products have had a profound impact on the discovery of new therapeutic agents. Newman and Cragg have highlighted their critical roles in modern medicine, and in a series of reports, compiled comprehensive data sets for all major groups of drugs (Newman and Cragg 2009, 2016; Cragg and Newman 2013). Accordingly, in the period between 1981 and the end of 2014, 60% of all small-molecule drugs approved were derived from natural products. These molecules have been especially effective as antibiotics and anticancer agents. Of the 175 small molecule anticancer agents approved since the 1940s, 49% are either natural products or directly derived thereof. A staggering 73% of small molecule antibiotics are natural products or their derivatives. Soil-dwelling actinomycetes have been major contributors in this regard as over 50% of clinical antibiotics are produced by this group of prolific bacteria (Berdy 2005).
Most antibiotic scaffolds in use today were discovered between 1940 and the early 1960s, a period now referred to as the golden age of antibiotics discovery. This productive era was followed by a ∼40 year innovation gap, during which no new antibiotic scaffolds were introduced (Walsh and Clatworthy, Pierson and Hung 2007; Fischbach and Walsh 2009). The consequences of this gap have been dire, especially considering the emergence of multidrug-resistant bacterial pathogens such as methicillin-resistant Staphylococcus aureus, extensively drug-resistant Mycobacterium tuberculosis or pan-drug-resistant Pseudomonas aeruginosa, to name a few. Resistance has been detected to every antibiotic that is currently on the market, and this observation coupled with our inability to identify new effective antibiotics has catapulted infectious diseases to one of the main health challenges of this century. The Center for Disease Control and Prevention recently reported that annually over 2 million Americans are infected by drug-resistant pathogens; over 23 000 of these are fatal. If new methods for discovery of bioactive molecules are not developed, infectious disease may again become the leading cause of death worldwide.
Traditional methods, now often referred to as ‘grind and find’, were for decades an excellent source of diverse, potent and inspirational new molecules (Miller and Clardy 2009). However, their application often leads to labor-intensive dereplication of previously known compounds. Due to diminishing returns and the relatively limited profit margin of antibiotics, major pharmaceutical companies have for the most part abandoned antibiotics discovery pipelines (Projan 2003). This decision might have been premature, not only because restocking our antibiotic supplies is a public health priority, but also because modern advances in genome sequencing technologies and the development of bioinformatics have reinvigorated antibiotic discovery. The abundance of bacterial genome sequences now available enables genome-mining approaches for finding new bioactive small molecules, while simultaneously facilitating cheminformatic dereplication methods (Gaudêncio and Pereira 2015; Mohamed, Nguyen and Mamitsuka 2016).
Microbial natural products are generated by dedicated biosynthetic gene clusters. The corresponding biosynthetic enzymes assemble, in a stepwise fashion, architecturally complex secondary metabolites from simple building blocks. One of the main insights from the multitude of microbial genome sequences has been that most secondary metabolite biosynthetic gene clusters are inactive during normal laboratory fermentation (Bentley et al. 2002; Ikeda et al. 2003; Oliynyk et al. 2007; Nett, Ikeda and Moore 2009). Saccharopolyspora erythraea, the industrial producer of erythromycin, provides an illustrative case. While its genome revealed 27 biosynthetic clusters, decades of research have only uncovered products for five of these. The remaining majority, termed ‘silent’ or ‘cryptic’ gene clusters, represent a treasure trove for the discovery of novel small molecules, their biosynthetic pathways and the regulatory circuits underlying their expression. These data lead to two profound conclusions: (1) our current arsenal of naturally derived drugs has been acquired from a small fraction of constitutively expressed biosynthetic gene clusters and (2) methods that access the silent majority would have a deep impact on drug discovery and increase our collection of bioactive molecules.
The research community has recognized that silent gene clusters represent a large reservoir of therapeutic molecules, and several methods have been developed for activating them (Chiang et al. 2011; Rutledge and Challis 2015). These include expression of the entire gene cluster in a suitable heterologous host, chromosomal insertion of constitutively active promoters, overexpression of pathway-specific regulators and the OSMAC (One Strain Many Compounds) approach (Bode et al. 2002). Herein, we consider the use of exogenous small molecules in modulating the expression of silent gene clusters. We present the major approaches that have been developed—including coculture techniques, ribosome engineering, chromatin remodeling and high-throughput elicitor screens—and discuss each in terms of rationale, applications and mechanism. We exclude quorum-sensing-regulated pathways from our analysis, as the autoinducers are known and largely produced by the host, and consequently the corresponding gene clusters are not silent. We also exclude OSMAC from our discussion. While successful, it is in most cases unclear what media component(s) induce expression of silent gene clusters. Our analysis underlines the power of exogenous small molecules in stimulating secondary metabolism and highlights the function of elicitors, especially antibiotics, in mediating microbial interactions and inducing the production of cryptic secondary metabolites.
COCULTURE
Motivation
Traditional bacterial natural products discovery often involves growth of a selected strain in a nutrient-rich monoculture. Though effective, this culturing method is in stark contrast to the complex, nutrient-limited environment in which bacteria naturally grow and evolve. One gram of soil, for example, contains 10 000 different species of bacteria (Curtis, Sloan and Scannell 2002). As such, inter- and intraspecies interactions are prevalent in the wild, and it is perhaps unsurprising that growth of a bacterium in coculture with a potential competitor would alter or enhance the secondary metabolic output. It would stand to reason then that systematic coculture screens could lead to new, perhaps cryptic, bioactive molecules that cannot be accessed otherwise. Early support for this idea came from Peipp and Sokolova (Iakovleva and Sokolova 1978; Sonnenbichler, Dietrich and Peipp 1994). Peipp, for example, found that constitutive, albeit low-level, production of a toxin by the fungus Heterobasidion annosum stimulated the biosynthesis of several toxins by a second fungus, Gloeophyllum abietinum, in liquid coculture. These overproduced toxins included oosponol (∼92-fold) and oospoglycol (∼14-fold) (Sonnenbichler, Dietrich and Peipp 1994). While numerous applications have followed, this method largely remains an underexplored strategy for the discovery of new and cryptic small molecules (Kolter and van Wezel 2016).
Applications
Relevant applications of coculture may be traced back to an entirely different area: foods and beverages such as vinegar, beer, chocolate and cheese have long been produced in cocultures, which take advantage of interactions between multiple microorganisms. The first example of coculture in natural products discovery, though also ‘unintended’, is that of penicillin, produced by Penicillium notanum on a petri dish containing Staphylococcus aureus. The observation of a halo of staphylococcal growth inhibition by Alexander Fleming and further developments by Walter Florey and Ernst Chain revolutionized the treatment of bacterial infections. However, neither of these examples represents a methodical microbial coculture experiment. Testing of bacterial and fungal extracts against other bacteria continued for the following decades, but systematic coculture studies only became popular toward the end of the century (Iakovleva and Sokolova 1978; Sonnenbichler, Dietrich and Peipp 1994; Mearns-Spragg et al. 1998; Ueda et al. 2000). Broadly speaking, induction of silent gene clusters as a function of coculture can result from one of three interaction modes: (1) provision of nutrients, (2) competition via production of an antibiotic or a signaling molecule and (3) cell–cell contact. Examples in each of these categories will be discussed in turn.
Application: provision of nutrients
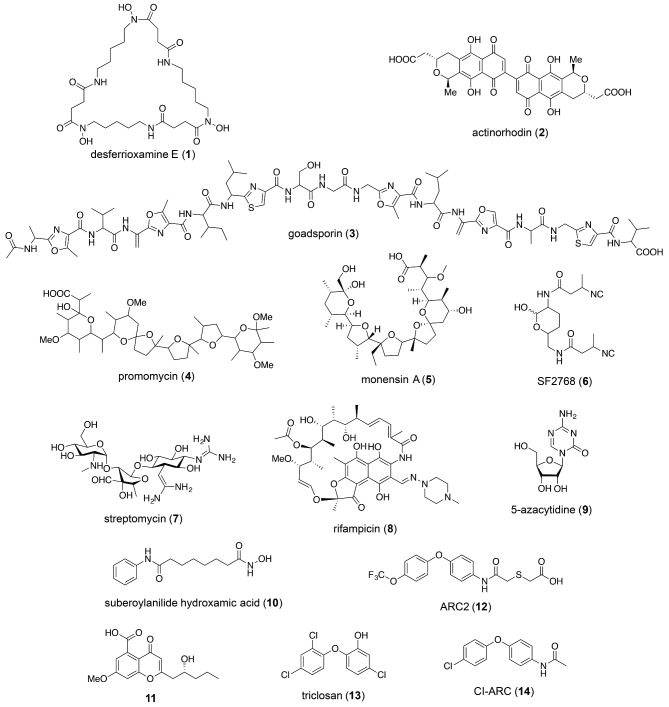
Many of the earliest coculture experiments monitored readily apparent phenotypic changes, such as pigment production, sporulation or antibiotic synthesis. In numerous cases, diffusible small molecules from one strain elicit production of secondary metabolites from a second strain. These diffusible molecules act as either nutrients, signals or antibiotics. Examples of the first category were presented by Ueda and coworkers in a broad binary screen with 76 strains of Streptomyces. In one set of experiments, they found that 34% of the Streptomycetes tested were active in inducing antibiotic production and/or sporulation in neighboring Streptomycetes (Ueda et al. 2000). These results indicated that Streptomyces spp. harbored cryptic antibiotics well before the genome sequence of the first actinomycete was reported. Additional studies with one of the interacting pairs identified the siderophore desferrioxamine E (Fig. 1, 1) as the stimulatory molecule (Yamanaka et al. 2005). Other siderophores did not display this phenotype. These results indicated that iron, delivered by specific siderophores, plays an important role in Streptomycete development. Consistent with this model, siderophores that monopolize iron rather than provide it to the neighboring strain retard developmental pathways, as shown in the case of Amycolatopsis sp. AA4 and Streptomyces coelicolor (Seyedsayamdost et al. 2011b; Traxler et al. 2012).
Figure 1.
Selected structures, including desferrioxamine E (1), actinorhodin (2), goadsporin (3), promomycin (4), monensin A (5), SF2768 (6), streptomycin (7), rifampicin (8), 5-AC (9), SAHA (10), bortezomib-induced metabolite (11), ARC2 (12), triclosan (13) and CI-ARC (14).
An early intrakingdom case in this category involved a binary fungal interaction with immediate industrial relevance: coculture of Rhodotorula glutinis and Debaryomyces castellii resulted in increased carotenoid pigment production that was not seen in either monoculture (Buzzini 2001). The authors suspected that D. castellii hydrolyzes the oligosaccharides in the medium to mono- and disaccharides, which R. glutinis can use more effectively in carotenoid synthesis.
Application: competition and signaling
While nutrient provision, in the form of iron or sugar, affords one means of interaction, competition and antibiotic production itself can underlie other bacterial or fungal interactions. Many examples fall into this category. An early bacterial–bacterial example was the induction of the blue S. lividans pigment, actinorhodin (Fig. 1, 2) by another Streptomyces strain, found by screening supernatants from 405 strains of actinomycetes (Onaka et al. 2001). The signaling compound was isolated and identified as a thiazole/oxazole-containing linear oligopeptide, which the authors named goadsporin (Fig. 1, 3). Goadsporin proved to be a more general inducer of Streptomycete secondary metabolism: it was tested against 42 randomly selected strains and elicited pigment production in 20 strains and sporulation in 32 strains. Interestingly, goadsporin was found to be a potent Streptomyces-specific antibiotic: it did not show bioactivity against selected proteobacteria, firmicutes or fungi, but exhibited minimal inhibitory concentrations of 0.2, 3.2 and 6.4 μg mL−1 against S. scabies, S. coelicolor and S. lividans, respectively. At subinhibitory concentrations, goadsporin stimulated sporulation and antibiotic production, while at high concentrations, it served as a potent toxin. This phenomenon of low-dose stimulation and high-dose toxicity by the same molecule is referred to as hormesis, and the following sections will show that it is a prevalent interaction paradigm between microbes and molecules.
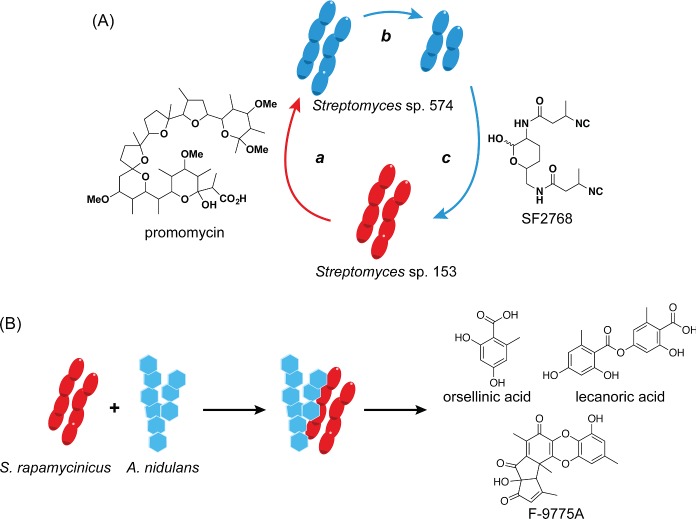
A case similar to goadsporin was provided by promomycin (Fig. 1, 4). It was discovered as an elicitor in an interaction where one Streptomyces strain promoted production of an otherwise cryptic antibiotic in another (Amano et al. 2010). The eliciting molecule, promomycin, was shown to be a polyether antibiotic, structurally similar to lonomycin. Like goadsporin, promomycin displayed hormetic properties. At low concentrations, it served a stimulatory role for antibiotic production, while at high concentrations, it displayed antibacterial properties (Fig. 2A). The similarity to polyether antibiotics led the authors to use monensin A (Fig. 1, 5) as an elicitor, which eventually resulted in the identification of the antibiotic elicited from the second strain (Amano et al. 2011). Although the level of induction appeared weak, the authors elucidated its structure as the diisonitrile-bearing antibiotic, SF2768 (Fig. 1, 6).
Figure 2.
Stimulation of secondary metabolism by coculture techniques. (A) Promomycin, a polyether antibiotic, is released by one Streptomyces strain (a), which causes growth inhibition (b) and production of SF2768, a diisonitrile antibiotic (c), from a second Streptomycete. (B) A physical interaction between S. rapamycinicus and A. nidulans induces the fungus to produce orsellinic acid, lecanoric acid and F-9775A.
Of note in the category of toxin-inducing-toxin cocultures are also results by Shin et al. (1998), which showed that a fungal Monascus species, grown with either Saccharomyces cerevisiae or Aspergillus oryzae, exhibited ∼11-fold enhanced pigment production and morphological differentiation. Remarkably, the inducer provided by Sa. cerevisiae was shown to be a chitinase enzyme. Among a number of degradases tested, the Sa. cerevisiae chitinase was shown to be the most effective at hydrolyzing Monascus cell walls. The authors suggested that partial hydrolysis of Monascus cell walls served as a signal for stimulation of pigment production, which they proposed was a form of chemical defense.
A similar example involving a fungal–bacterial interaction was reported by Patterson and Bolis. They showed that fungal cell wall homogenates from P. notatum elicited a 2.5-fold upregulation of tolytoxin by the cyanobacterium Scytonema ocellatum (Patterson and Bolis 1997). Further, they identified the inducer as variable oligomers of N-acetylglucosamine, that is, chitin polymers of differing lengths. Interestingly, the inducer again displayed hormetic properties: high concentrations of the homogenate and of chitin oligomers were toxic, while low concentrations led to tolytoxin overproduction. The authors concluded that tolytoxin is an inducible chemical defense agent, known as a phytoalexin. Much like Streptomycetes, this cyanobacterium also responds to N-acetylglucosamine by turning on toxin production pathways (Rigali et al. 2008). Similar interactions have been shown in marine bacterial–bacterial interactions as well as algal–bacterial interactions (Long and Azam 2001; Seyedsayamdost et al. 2011a).
Examples where the elicitors are not growth inhibitory have also been reported. Akin to the studies of Patterson and Bolis above, stimulation of Streptomyces sp. US80 with heat-killed fungi led to a modest overproduction of antifungal metabolites, including the potent toxin irumamycin (Fourati-Ben Fguira et al. 2008). In this case, no growth inhibition of the bacterial strain was reported. Further, Proksch and colleagues have reported a 1.5- to 78-fold overproduction of bioactive metabolites from the ascomycete Fusarium tricinctum when cocultured with Bacillus subtilis (Ola et al. 2013). These studies led to identification of four cryptic metabolites, three of which are known to be produced by the fungus. Interestingly, these results depended on the duration of pre-incubation of B. subtilis on agar media before introduction of the fungus. Best results were obtained with a 6-day pre-incubation of B. subtilis, perhaps suggesting a growth phase-dependent production of a signal by the bacterium.
As was alluded to above, a study in the bacterial–bacterial interactions category showed that exposing unidentified, surface-associated marine bacteria to St. aureus, Pseudomonas aeruginosa or Escherichia coli, resulted in increased antibiotic synthesis by the marine bacteria. This study, much like those by Ueda, reported the production of cryptic antibiotics from marine bacteria and indicated that the observed metabolic changes depended on an unidentified signal (Mearns-Spragg et al. 1998). In a similar approach, Slattery, Rajbhandari and Wesson (2001) examined the effect of 53 diverse bacterial species in coculture with the marine istamycin producer, Streptomyces tenjimariensis. They reported that 23% of interactions tested led to a ∼2-fold overproduction of istamycin. Their results were interpreted as an inducible defense mechanism in the marine microbe elicited by Gram-positive and Gram-negative bacteria. Despite the low level of overproduction, these studies were fully in line with an altered metabolism in response to coculture.
Application: cell–cell contact
The last category in the mode of coculture interactions mentioned above is physical cell–cell contact. Perhaps the first example in this category was demonstrated by Clardy and colleagues in the induction of a cryptic metabolite through coculture of a marine fungus with an unidentified Gram-negative marine bacterium (Cueto et al. 2001). These studies led to isolation and structural elucidation of pestalone, a benzophenone antibiotic, and demonstrated that it was not detectable in discrete fungal and bacterial controls. Pestalone displayed potent antibacterial activities with MICs of 37 and 78 ng mL−1 against MRSA and VRE, respectively. A similar study from the Fenical lab showed that Libertella, a marine fungus, could be induced by a marine proteobacterium to produce the otherwise cryptic libertellenones (Oh et al. 2005). This new series of pimarane diterpenoids demonstrated moderate to potent cytotoxicity against a cancer cell line. In both of these studies, attempts to replicate the interaction by substituting dead bacterial cells, cell-free supernatant or ethyl acetate extracts failed, consistent with a requirement for contact in the induction of pestalone and libertellenone biosyntheses.
A more recent study on a bacterial–fungal interaction utilized a clever secondary metabolite gene microarray of A. nidulans in coculture with 58 soil-dwelling actinomycetes to assess activation of silent fungal biosynthetic gene clusters. The studies revealed that coculture with S. rapamycinicus induced production of the cryptic metabolite orsellinic acid and its derivatives (Schroeckh et al. 2009). Additional experiments addressing the mode of induction showed that orsellinic acid was not produced when the two species were separated by a dialysis tube or even when the bacteria were grown with a knockout strain of A. nidulans, and the resulting supernatant applied to the wt fungus. These experiments indicated that a diffusible signal was not involved. When the authors switched tactics and examined scanning electron micrographs of coculture biomass, they saw that the bacteria nested inside of the fungus, anchoring themselves to the fungal hyphae. Thus, in these cases, the physical interaction between the microorganisms was responsible for their altered metabolic output (Fig. 2B). For many more examples of mixed fermentation, we refer the reader to two comprehensive reviews by Pettit and Proksch (Pettit 2009; Marmann et al. 2014).
Mechanism
Mechanistic considerations for the various vignettes presented above will be discussed in regard to the mode of interaction, though it should be mentioned at the outset that relatively few cocultures have been studied in enough detail to implicate a specific regulatory induction pathway. The stimulatory role in antibiotic production and sporulation ascribed to desferrioxamine E (Fig. 1, 1) can be rationalized in light of the crucial requirement of iron in Streptomycete development. Indeed, studies by Traxler et al. (2012) have demonstrated that expression of genes involved in the developmental program of S. coelicolor are strongly altered in response to changes in iron availability: abundance of iron leads to accelerated developmental pathways, while iron sequestration or limitation slows them down. Complete removal of iron causes growth retardation and failure to produce aerial hyphae. As desferrioxamine is one of the most common siderophores among terrestrial actinomycetes, the results by Ueda and colleagues indicated that provision of this siderophore by one strain supplemented the amount of iron available to the receiving strain in the binary interaction, thus promoting development including sporulation and secondary metabolite production (Yamanaka et al. 2005). This conclusion is consistent with the arrested development that occurs when S. coelicolor is provided with a rare siderophore that it cannot utilize.
Many cocultures involve a growth-inhibitory molecule produced by one microbe, which stimulates secondary metabolite production, often antibiotics, from the neighboring microbe. These cases quite explicitly demonstrate the hormetic properties of antibiotics. At a subinhibitory concentration, the antibiotic acts as an elicitor or inducer of silent biosynthetic gene clusters resulting in production of cryptic metabolites. At higher concentrations, that same antibiotic has toxic effects on the receiving strain. These experiments seem to indicate that in some interactions, bacteria respond to exogenous antibiotics with antibiotics. Undoubtedly, in some cases, where the elicitor was not an antibiotic, nutrient limitation may be the trigger for induced secondary metabolism. Both conditions, diminished growth from exposure to antibiotics or from nutrient limitation, elicit the same response of sporulation and secondary metabolite biosynthesis.
In the case of promomycin, which is presumably produced constitutively, the mode of induction was examined further. A key observation was provided by studies of Suh and colleagues, which showed a strong inverse relationship between secondary metabolite production and intracellular ATP levels in Streptomyces spp. (Meng et al. 2011). We posit that because promomycin and monensin are ionophores, which abolish the proton motive force that is utilized to synthesize ATP, they elicit secondary metabolite production by diminishing levels of intracellular ATP. Further studies will certainly test this hypothesis and enhance our understanding of regulatory pathways that activate secondary metabolite biosynthesis.
A number of fungal–bacterial interactions have been found to be mediated not by diffusible small molecules but rather by physical cell-to-cell contact. Brakhage and colleagues showed that interactions of A. nidulans and S. rapamycinicus fell into this category, and scanning electron micrographs beautifully visualized the process. (Schroeckh et al. 2009). Strikingly, when this mode of induction was examined in more detail, it was revealed that the bacteria were causing alterations to fungal chromatin by histone acetylation via the Saga/Ada complex, which contains the histone acetyl transferase GcnE (Nützmann et al. 2011). This provided the first example of Saga/Ada-mediated histone acetylation triggered by a bacterial interaction. As will be discussed more fully in the discussion on chromatin remodeling, fungal secondary metabolism can be highly sensitive to histone modifications.
RIBOSOME ENGINEERING
Motivation
Ribosome engineering approaches the problem of silent gene clusters from a unique angle: How can the producers themselves be improved? While numerous methods for strain improvement exist, they are typically hampered by high costs and/or laborious procedures (Santos and Stephanopoulos 2008). In contrast, ribosome engineering has emerged as a simple and rapid secondary metabolism-focused alternative. This technique is predicated on the use of certain antibiotics to produce mutations in component(s) of the ribosome or RNA polymerase (RNAP) that result in increased production of secondary metabolites. The inventors of the method were inspired by the observation that a mutated, streptomycin-resistant strain of Streptomyces lividans produced the blue pigment actinorhodin (Act, Fig. 1, 2), whereas the parent strain, from which the mutant was derived, did not (Shima et al. 1996). They mapped the mutation to a ribosomal protein and showed that only the mutant ribosome led to activation of the silent Act gene cluster (act), thus demonstrating translational control in Act biosynthesis. The method has been extended to other antibiotics and many successful applications have followed (Ochi 2007; Ochi and Hosaka 2013).
Applications
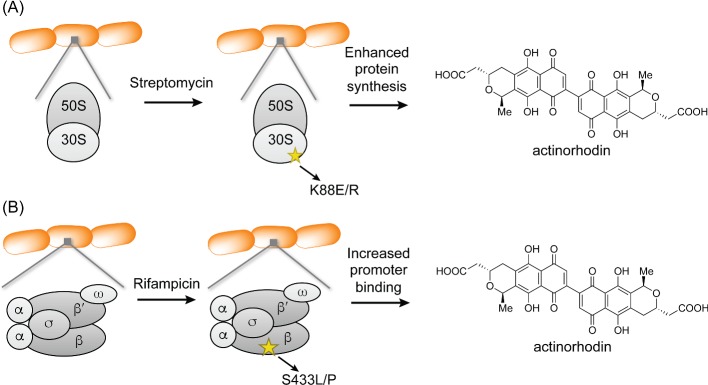
Ochi and Hosaka (2013) have recently provided a comprehensive summary of the applications of ribosome engineering. Here we focus on the original studies that provided the impetus for the field as well as a molecular rationale for ribosome engineering. As alluded to above, wt S. lividans TK21 normally does not produce Act, even though it contains a complete act biosynthetic gene cluster (Shima et al. 1996). The original study by Ochi and colleagues investigated how the streptomycin-resistant S. lividans strain TK24 produced abundant quantities of Act (Fig. 3A). Resistance to streptomycin (Fig. 1, 7) frequently results from point mutations in rpsL, which encodes the S12 ribosomal protein. When rpsL from strains TK21 and TK24 was sequenced, Shima et al. found a K88E mutation in TK24. Selection of additional streptomycin mutants from TK21 showed that Act production often accompanied resistance. Convincingly, introduction of the K88E-S12 into the parent strain gave the same result, indicating that the K88E mutation was linked with the ability to induce the act gene cluster. These results established that activation of act in S. lividans can occur at the translational level and is dependent on a mutated or ‘engineered’ ribosome.
Figure 3.
Stimulation of secondary metabolism by ribosome engineering. (A) Streptomycin treatment produces a resistant strain with a mutation in the S12 protein, a component of the 30S subunit of the ribosome. The mutated ribosome endows the streptomycin-resistant strain with enhanced protein synthesis, resulting in increased actinorhodin production. (B) Rifampicin treatment produces a resistant strain with a mutation in the β-subunit of RNAP. The mutated protein exhibits enhanced promoter binding, resulting in increased actinorhodin production.
The correlation between streptomycin resistance, which arises through a specific mutation in S12, and the induction of a biosynthetic gene cluster has been extended to additional Streptomycetes, other bacterial genera and even fungi. Studies have shown that selection of streptomycin resistance in S. coelicolor, S. antibioticus, S. chattanoogensis and S. lavendulae led to a 5- to 6-fold overproduction of Act, actinomycin, fredericamycin and formycin, respectively (Hosoya et al. 1998). Notably, the mutants resulted not in global overproduction of secondary metabolites but rather in pathway-specific stimulatory effects (see below). Further studies have helped extend this method to Bacillus subtilis, B. cereus and Pseudomonas pyrrocinia, where selection of streptomycin-resistant mutants (str) resulted in a 5- to 10-fold overproduction of a peptidic antibiotic, FR900493, and pyrrolnitrin, respectively.
Ochi and coworkers have expanded the streptomycin-induced ribosome remodeling paradigm to other antibiotics. Among these, experiments with rifampicin have been the most successful (Hu, Zhang and Ochi 2002; Lai et al. 2002; Xu et al. 2002). Rifampicin (Fig. 1, 8) is an inhibitor of RNAP, and accordingly rifampicin resistance is typically conferred by a mutation in the gene encoding its β-subunit, rpoB (Fig. 3B). When Hu et al. selected for rifampicin-resistant mutants (rif) in S. lividans, they observed that the mutant phenotype occurred concomitant with overproduction of Act (up to 10-fold), undecylprodigiosin (Red, up to 5-fold) and calcium-dependent antibiotic (CDA, fold-change not reported), three secondary metabolites that are minimally produced in the wt strain. Again, through mutant mapping and gene replacement studies, they showed that two classes of mutants could arise from rifampicin treatment: (1) those that provided resistance to rifampicin but did not enhance secondary metabolite production, and (2) those that provided resistance and upregulation of Act and Red. Interestingly, increased secondary metabolite production in the mutants was dependent on the location and nature of the amino acid substitution.
Perhaps the best application of ribosome engineering has been in the discovery of new cryptic metabolites (Hosaka et al. 2009). A screen of 1068 soil actinomycete isolates showed that 43% of non-antibiotic producing Streptomycetes and 6% of non-antibiotic-producing non-Streptomycetes acquired the ability to synthesize antibacterial compounds after a selection step that generated spontaneous rif or str mutants. Investigation of one of these isolates, which was determined to be closest to S. mauvecolor by 16S rRNA sequencing, led to the identification of a new cryptic antibiotic piperidamycin. Piperidamycin showed potent growth-inhibitory activity against Gram-positive bacteria, e.g. Staphylococcus aureus, S. pyogenes and Enterococcus faecalis, with MICs of 0.8–1.6 μg mL−1. Thus, ribosome engineering can be used to improve production strains and induce silent biosynthetic gene clusters.
Mechanism
The onset of secondary metabolite production in Streptomycetes typically occurs at the end of exponential growth phase and the start of stationary phase, a period sometimes referred to as idiophase. A key regulatory system in this growth period is the stringent response, which is initiated by nutrient limitation (Starosta et al. 2014; Gaca, Colomer-Winter and Lemos 2015). When uncharged tRNAs, which accumulate under conditions of amino acid scarcity, bind to the A-site of the ribosome, RelA uses ATP and GTP to synthesize the hyperphosphorylated nucleotide ppGpp (guanosine-5′-diphosphate-3′-diphosphate). This so-called alarmone acts directly on RNAP, thus changing its transcriptional activity. A structure of the RNAP-ppGpp complex has been solved and a model for how ppGpp exerts its effect has been proposed at the molecular level (Artsimovitch et al. 2004). At the cellular level, production of ppGpp is associated with upregulation of stress response genes, such as those involved in amino acid uptake and biosynthesis as well as the induction of some secondary metabolites.
Upon observing the enhanced production of certain secondary metabolites using the str and rif phenotypes, Ochi and colleagues suspected that ppGpp was involved. However, numerous lines of evidence showed that the stringent response did not play a role (Ochi et al. 2004; Ochi 2007). Most importantly, direct measurement of the levels of ppGpp showed that they did not change significantly in the wt and mutant strains. In fact, the str strain showed slightly reduced levels of ppGpp. Further, relA rif and relA str double mutants, which cannot generate ppGpp, also exhibited stimulated production of secondary metabolites much like the single str and rif mutants. Thus, the str and rif mutants can circumvent ppGpp and stimulate production of certain secondary metabolites; that is, a ‘stringent phenotype’ can be induced in the absence of ppGpp.
The authors have proposed that the rifampicin-resistant rpoB mutant behaves like a ‘stringent’ RNAP, even in the absence of the alarmone ppGpp (Lai et al. 2002). Consistent with this idea, the rif mutants display a lower rate of RNA synthesis, a stringent phenotype, even in nutritionally rich medium. Further, the ppGpp-binding domain on the β-subunit of RNAP is close to the site of point mutations that render the rif phenotype. These mutations are only several angstroms removed from the active site, suggesting that they could have a major impact on the activity of RNAP. In the model proposed, the mutated RNAP provides rifampicin resistance and behaves like a stringent RNAP. It demonstrates an enhanced affinity for promoters, increasing expression of actII-ORF4, which encodes an act-pathway-specific positive transcriptional regulator, whose expression levels are directly correlated with Act production (Ochi and Hosaka 2013).
In the case of str mutants, ppGpp was also shown not to be required (Hosoya et al. 1998). By comparing a number of properties of wt and streptomycin-resistant ribosomes, the authors proposed that the mutant ribosome was structurally more stable under conditions of stress, such as amino acid starvation, and that it exhibited higher levels of protein synthesis in the stationary phase (Hosaka, Xu and Ochi 2006; Hosaka et al. 2009; Ochi and Hosaka 2013). In the case of Act biosynthesis, this increased protein synthesis activity in the stationary phase resulted in stimulated production of the positive transcriptional regulator actII-ORF4, as determined by western blot analysis, which led to augmented levels of the small molecule product.
Additional antibiotics have been used successfully for augmented secondary metabolite production by Ochi and coworkers. These include erythromycin and gentamicin, both of which target the ribosome (Chai et al. 2012; Imai et al. 2012). While further studies with these antibiotics are necessary, initial mechanistic experiments indicate that their mode of induction is different from those of streptomycin and rifampicin. It seems likely that antibiotics with modes of action alternative to streptomycin and rifampicin will engage in new modes of induction of secondary metabolism. The ribosome, consisting of three rRNA molecules and over 50 proteins, provides an abundance of molecular targets that can be inhibited by specific antibiotics (Davies, Spiegelman and Yim 2006). The mechanistic details of how inhibition or mutation is linked to upregulated secondary metabolite production with other antibiotics will be of great interest.
A final interesting observation made by Ochi and coworkers was the induction of secondary metabolism in the presence of exogenous antibiotics but without development of a resistant phenotype. This was the case with tetracycline and lincomycin (Shima et al. 1996; Imai et al. 2015). It will be interesting to see what the mechanism of induced secondary metabolite production is in these non-resistant strains, a case that is akin to examples of goadsporin or promomycin in coculture experiments.
CHROMATIN REMODELING
Motivation
Chromatin remodeling is one of the newest techniques for induction of secondary metabolism. Its roots lie in a study from Keller and colleagues. While investigating the regulatory pathway of sterigmatocystin, a toxin produced by Aspergillus nidulans, Keller identified several mutant strains that showed suppressed production of the toxin (Butchko, Adams and Keller 1999). One of the mutations was in LaeA, a methyltransferase, implicating it as a regulator of sterigmatocystin production. In addition, LaeA had pleiotropic effects on secondary metabolism: its deletion blocked expression of several biosynthetic gene clusters, while its overexpression triggered penicillin and lovastatin production (Bok and Keller 2004). LaeA has homology to histone methyltransferases, which led Keller to propose a chromatin-based model of regulation. Indeed, deletion of hdaA, an Aspergillus histone deacetylase (HDAC), also led to increased production of two telomere-proximal secondary metabolite gene clusters. In contrast, transcription of a telomere-distal cluster was unchanged (Shwab et al. 2007). These studies clearly implicated a chromatin modification-based regulatory system in Aspergillus (Palmer and Keller 2010). Building on these results, Keller and Cichewicz have provided further examples of chromatin remodeling by mimicking these epigenetically altered genotypes via supplementation of fungal cultures with HDAC and DNA methyltransferase inhibitors.
Applications
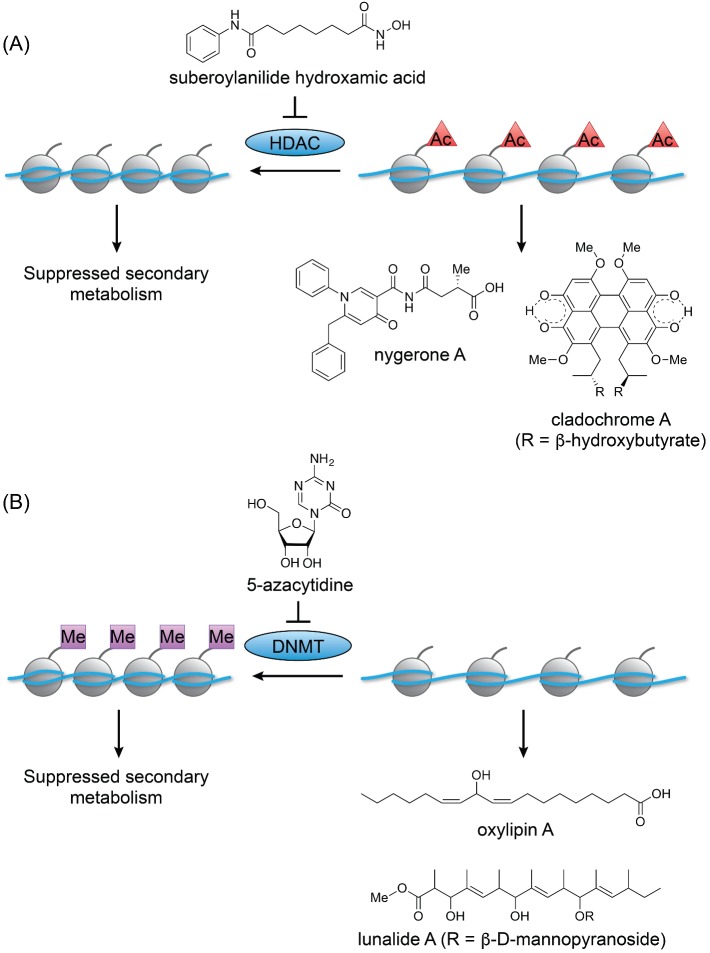
In the study demonstrating the importance of HdaA in regulating secondary metabolism, Keller and colleagues included a small set of proof-of-concept experiments that laid the groundwork for chromatin remodeling. Trichostatin A, a class 1/2 HDAC inhibitor, was added to Alternaria alternata and Penicillium expansum. This treatment caused an increase in the production of multiple unidentified small molecules (Shwab et al. 2007). Cichewicz expanded on these studies, subjecting a set of 12 fungi to a library of DNA methyltransferase (DNMT) and HDAC inhibitors at various concentrations. A total of 11 of the 12 strains could be induced by at least one member of the library to increase their levels or diversity of secondary metabolism. Two of the fungi were studied further. Cladosporium cladosporioides was stimulated by 5-azacytidine (5-AC, Fig. 1, 9), a DNMT inhibitor, to produce several cryptic oxylipins, or by suberoylanilide hydroxamic acid (SAHA, Fig. 1, 10), an HDAC inhibitor, to produce a series of cryptic perylenequinones, including both known and novel cladochrome analogs (Fig. 4). The second fungus, a Diatrype species, was induced by 5-AC to produce two new cryptic polyketides, lunalides A and B (Fig. 4B) (Williams et al. 2008).
Figure 4.
Mechanisms underlying chromatin remodeling. (A) Treatment with the HDAC inhibitor SAHA results in increased histone acetylation and consequently increased transcription of biosynthetic gene clusters. (B) Treatment with the DNMT inhibitor 5-AC results in decreased DNA methylation, altering the transcription of various biosynthetic gene clusters.
This approach was also successful in A. niger, which could be induced by SAHA to produce the new cryptic metabolite nygerone A (Fig. 4A) (Henrikson et al. 2008). Similarly, 5-AC or SAHA treatment of Alternaria sp. led to the production of several cryptic mycotoxins including alternariol, altenusin and tenuazonic acid, and increased production of altertoxin II (Sun et al. 2012). A study of the global effects of 5-AC and SAHA on the transcription of 55 polyketide synthase (PKS), non-ribosomal peptide synthetase (NRPS) or PKS/NRPS gene clusters was consistent with the results above (Fisch et al. 2009). It showed that ∼70% of these biosynthetic gene clusters were inactive when the fungus was grown using standard laboratory culture conditions, and that addition of 5-AC or SAHA induced transcription of all but seven of these (Fisch et al. 2009). Oberlies and coworkers took a slightly different approach to this method, using the proteasome inhibitor bortezomib to induce a filamentous fungus to produce cryptic metabolite 11 (Fig. 1) (VanderMolen et al. 2014). While mode-of-action restrictions largely limit this method's application to fungi, the McArthur group was able to apply it in Streptomyces coelicolor (Moore et al. 2012). Using qPCR, the group showed that the HDAC inhibitor sodium butyrate induced five cryptic pathways in Streptomycetes. They also performed activity-based assays on Streptomyces sp. KY5 and Pseudonocardia sp. P1 and saw induced activity against Candida albicans when the bacteria were cultured with sodium butyrate (Moore et al. 2012).
Mechanism
Fungal DNA, like that of higher eukaryotes, is organized onto histones. Histones compact the DNA into chromatin and regulate replication and transcription. There are two states of chromatin: a ‘closed’ state, called heterochromatin, is more densely packed and therefore transcriptionally silent. In contrast, the ‘open’ state, called euchromatin, is more loosely packed and transcriptionally active (Brosch, Loidl and Graessle 2008; Gacek and Strauss 2012).
Fungi can transition portions of chromatin between these two states through various post-translational modifications, the most well studied of which is histone acetylation. Unmodified lysine is predominately cationic, allowing it to bind to DNA; acetylation abrogates the positive charge on lysine, occluding DNA binding. Accordingly, more highly acetylated histones are generally more loosely packed and transcriptionally accessible (Fig. 4A) (Cichewicz 2010). Further, the DNA base cytosine can be methylated to form 5-methylcytosine. This modification can have different effects on transcription depending on specific contexts, but often results in gene silencing (Fig. 4B) (Suzuki and Bird 2008). Fungi are primed for chromatin remodeling because of the organization of their genome: most fungal secondary metabolite genes are grouped together in locations that tend to be near the telomeres of their chromosomes (Keller, Turner and Bennet 2005; Yu and Keller 2005). As a result, histone acetylation and DNA methylation have a large impact on the transcription of these loci (Bok and Keller 2004; Shwab et al. 2007; Palmer and Keller 2010; Gacek and Strauss 2012). Targeting the enzymes that regulate these markers is a straightforward way to probe the effects of different epigenetic conditions. Thus, DNMT and HDAC inhibitors serve as effective elicitors of fungal secondary metabolite. The use of proteasome inhibitors takes advantage of the same underlying mechanism in an indirect manner: proteasomes degrade many proteins, including several transcriptional regulators (VanderMolen et al. 2014).
Because bacterial DNA is not organized onto histones, it is somewhat surprising that McArthur and coworkers had success using this method in Streptomycetes (Moore et al. 2012). The authors explained their results by citing a parallel to chromatin organization: the bacterial genome is compacted by nucleoid-associated proteins, RNAs and differential supercoiling, which could lead to differential compaction for certain genes. In addition, bacteria have their own versions of HDAC proteins (Lombardi et al. 2011). The mechanism for how these bacterial HDAC-like enzymes function is still unclear, so the pathway for the epigenetic approach in bacteria ultimately awaits further clarification (Moore et al. 2012). They might indeed act on nucleoid proteins, though other proposed targets include polyamines, acetoin and regulators of primary metabolism (Leipe and Landsman 1997).
Chromatin remodeling has proven to be a straightforward, low-cost, effective means of activating cryptic gene clusters in fungi (Gacek and Strauss 2012). Inherent limitations result from its mechanism of action, which largely (theoretically) confine it to fungi and to telomere proximal biosynthetic gene clusters. Its recent application in Streptomycetes indicates that it may be a more general method than previously thought, but is currently the only example of chromatin remodeling outside of fungi. We eagerly await further assessments of this method's scope, as well as investigation of its mechanism in bacteria.
HiTES
Motivation
The preceding sections show that antibiotics are often elicitors of secondary metabolism in fungi and bacteria. However, which antibiotic, or more generally small molecule, serves as an inducer for a specific strain or biosynthetic gene cluster, needs to be determined experimentally. Consider the example of goadsporin (Onaka et al. 2001). It was shown to be a Streptomyces-specific antibiotic at high concentrations but to elicit secondary metabolism at subinhibitory levels. However, its mode of induction was likely independent of its mode of growth inhibition, as a number of other toxins tested—including streptomycin, kanamycin, thiostrepton, bacitracin and gramicidin D—did not stimulate secondary metabolism. In this regard, bioinformatic tools are of little help: while we are increasingly adept at providing an in silico analysis of a biosynthetic gene cluster and predicting the class of molecule that might result, it is extremely difficult, if not impossible, to predict which small molecule signal, if any, will activate it. To address this shortcoming and identify small molecule elicitors for a chosen gene cluster, high-throughput methods have been developed, an approach that we refer to as HiTES (high-throughput elicitor screens).
Applications
There are three key components involved in this approach: the choice of microorganism, read-out and small molecule library to be screened. The first rendition of HiTES was carried out by Nodwell and coworkers (Craney et al. 2012). They used Streptomyces coelicolor based on the historical importance and fruitfulness of this strain and because it synthesizes the blue polyketide actinorhodin and the red prodiginines, the production of which can be rapidly monitored. The Canadian Compound Collection, which contains 30 569 small molecules, was screened for compounds that increased actinorhodin production in S. coelicolor. From this screen, 112 compounds were identified as hits, and a subset of these containing four related compounds (named the ‘ARC2 series’) were used for further studies. In addition to upregulating actinorhodin synthesis 2- to 5-fold, ARC2 (Fig. 1, 12) was able to stimulate 3-fold overproduction of the germicidins, while decreasing yields of the daptomycin-like CDA and prodiginines approximately 2-fold. Thus, ARC2 elicitors displayed pleiotropic effects on secondary metabolism. ARC2 is structurally similar to triclosan (Fig. 1, 13), which provided some clues regarding the mode of induction by this elicitor (see below). Strikingly, ARC2 and triclosan, which also induces actinorhodin synthesis, both displayed hormetic properties, and modulated secondary metabolism only at subinhibitory concentrations.
Spurred by the results with ARC2, the authors also investigated its effects on other actinomycetes and found it to serve as a potent general elicitor: it altered the secondary metabolome of Kutzneria sp., S. pristinaespiralis and S. peucetius, where it induced production of cryptic metabolites. A derivative of ARC2, called Cl-ARC (Fig. 1, 14), has recently been applied to 50 additional Streptomyces, resulting in at least one induced compound in a majority of the strains for a total of 216 cryptic metabolites (Pimentel-Elardo et al. 2015).
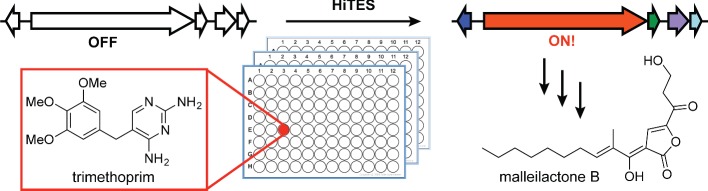
The other example of HiTES took a more general approach that can be applied to non-pigmented secondary metabolites. We demonstrated the effectiveness of HiTES in Burkholderia thailandensis, which was chosen for its importance as a model strain for the Pseudomallei group pathogens, genetic tractability and wealth of silent biosynthetic gene clusters (Liu and Cheng 2014; Seyedsayamdost 2014). In place of a phenotypic screen, a lacZ translational fusion to an essential gene within a selected biosynthetic gene cluster was employed as a reporter system. We chose the mal gene cluster, which synthesizes the cryptic virulence factor malleilactone, a potent inhibitor of Caenorhabditis elegans growth (Fig. 5). A 640-member library of bioactive small molecules was screened by monitoring the lacZ activity induced by each member of the compound library. From 640 compounds, nine elicitors were identified, which upregulated expression of the mal cluster (Fig. 5). Intriguingly, all nine elicitors were clinical antibiotics: trimethoprim, piperacilllin, ceftazidime, cefotaxime and five members of the fluoroquinolone family. At high concentrations, all of these antibiotics killed B. thailandensis, whereas subinhibitory levels served as potent inducers of the mal cluster. The top two elicitors, trimethoprim and piperacillin, showed a 5- to 45-fold upregulation of selected genes in the mal gene cluster, as determined by RT-qPCR. At optimal concentration, a ∼150-fold induction of malleilactone was observed with trimethoprim.
Figure 5.
Workflow for HiTES. A library of small molecules is screened to identify elicitors that induce expression of a cryptic gene cluster. The read-out can be pigment production or a genetic reporter via transcriptional or translational reporter fusion constructs. In B. thailandensis, trimethoprim was identified as an elicitor of malleilactone production using this approach.
Further investigations showed that trimethoprim served as a global activator of secondary metabolism in B. thailandensis (Okada et al. 2016). Production of at least five secondary metabolite families was induced as demonstrated by qPCR and/or metabolic profiling studies. These include thailandamide (8- to 36-fold overproduction depending on the analog), burkholdac (3-to 4-fold), 4-hydroxy-3-methyl-2-alkylquinolines (∼7-fold) and capistruin lasso peptides (not produced under normal growth), as well as a family of new cryptic metabolites called acybolins that has recently been structurally elucidated. Additional effects of trimethoprim on the secondary metabolic output of B. thailandensis have also been characterized.
Mechanism
A great strength and weakness of HiTES is that it is mechanism agnostic. It does not rely on a specific hypothesis for the mode of action of the elicitors identified. As a consequence, it is impossible to generalize its mode of induction. While this makes assigning a specific mechanism to a given elicitor more difficult, it also allows for the discovery of elicitors that operate through unforeseen or unprecedented modes of induction, thus broadening the scope of gene clusters that can be activated.
In the case of the ARC2 compound series, a molecular target was proposed based on the series’ structural similarity to triclosan, a known inhibitor of fatty acid synthesis that targets the FabI enoyl reductase (Craney et al. 2012). When tested against S. coelicolor, triclosan recapitulated the pigmentation induction of the ARC2 compounds. In addition, an S. coelicolor strain expressing a triclosan-resistant FabI paralog from Pseudomonas aeruginosa showed significantly reduced pigmentation compared to the control strain. Finally, biochemical assays of FabI with and without the ARC2 compounds confirmed that it could be inhibited by the series. Based on these results, the authors hypothesized that inhibition of FabI might redirect precursors, such as acetyl-CoA and malonyl-CoA, that are shared between fatty acid and polyketide synthesis, increasing the yield of the polyketide natural products. This link between fatty acid and polyketide biosynthesis, the authors proposed, helps set the upper limit on polyketide yields. Recent results show that the regulatory mechanism by ARC2 might be more complex and future studies will likely shed more light on this issue (Ahmed et al. 2013).
A mechanism for trimethoprim and other inducers found for B. thailandensis still remains to be determined. Based on the known antibiotic activity of the identified elicitors, it seems likely that their effects are mediated through stress response mechanisms. Trimethoprim has been shown to induce stress response in Escherichia coli (Khan and Yamazaki 1972). In addition, beta-lactam resistance in Staphylococcus aureus is mediated via stress response-related pathways (Aedo and Tomasz 2016). As discussed earlier in the context of ribosome engineering, cellular stress can alter the activity of the ribosome and thus induce secondary metabolism at the start of stationary phase. It is likely that the subinhibitory concentrations of antibiotics used in our study initiated stress response in B. thailandensis, resulting in the pleiotropic stimulation of secondary metabolism. Nonetheless, an alternative mechanism cannot be excluded at this point. For example, results by Davies (Goh et al. 2002) have shown that erythromycin-mediated transcriptional modulation occurred to the same extent in both wt and mutants defective in a variety of stress response pathways. Hu et al. reached a similar conclusion when examining the transcriptional changes induced by the antibiotic cecropin in E. coli O157 (Hong et al. 2003).
HiTES is a versatile, theoretically general method for activation of cryptic gene clusters. While relatively new, it promises to uncover new natural products and identify elicitors that operate through unprecedented mechanisms of activation, thus providing insights into the still poorly-understood regulatory pathways of secondary metabolite production.
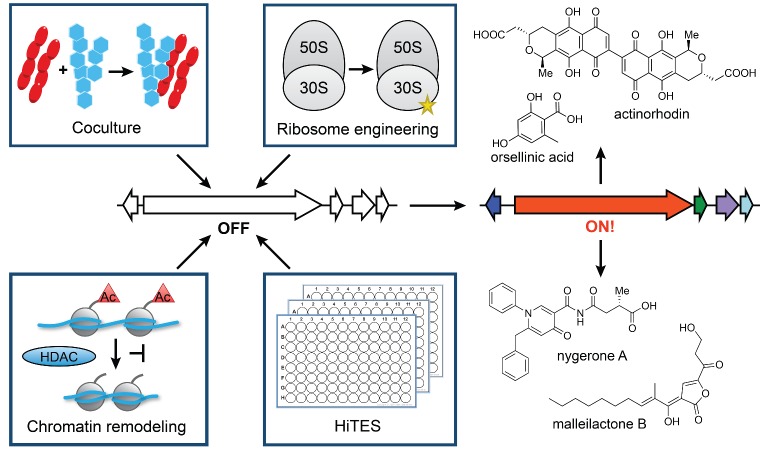
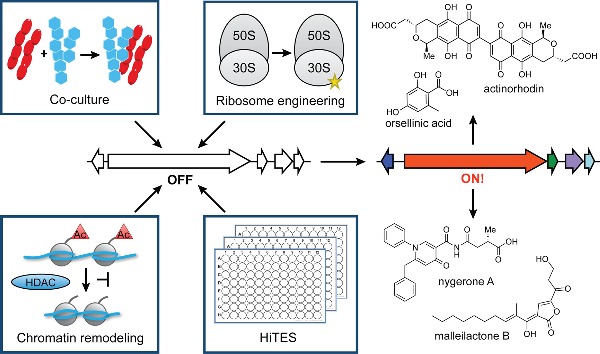
CONCLUSIONS
Recent technological advances have rejuvenated the field of natural products discovery, unearthing a vast untapped cache of silent biosynthetic gene clusters. Current efforts to take advantage of this new knowledge are limited by a lack of understanding of how secondary metabolism is regulated and therefore how the clusters might be activated. In this review, we have summarized the methods that have been developed to induce secondary metabolism using chemical approaches as well as the underlying biological mechanisms (Fig. 6). While the methods described in the preceding sections have distinct rationales, two underlying commonalities can be identified. The first key theme is the existence of hormesis—an inherent concentration dependence in the induced response to growth-inhibitory molecules (Davies 2006). Nearly 65 years ago, Hessayon provided perhaps the first report of hormesis by showing that the antibiotic trichothecin, produced in very small quantities by Trichothecium roseum, produced stimulatory effects on the growth of the plant pathogen Fusarium oxysporum, whereas higher concentrations had toxic effects (Hessayon 1953). This phenomenon, recently emphasized and shown to be prevalent by Davies and colleagues, was observed in numerous examples above. We have highlighted these instances of hormesis in the preceding sections, where subtoxic concentrations of antibiotics elicited sporulation, development or secondary metabolite biosynthesis, while higher titers caused cell death.
Figure 6.
Summary of approaches discussed in this review for activation of silent biosynthetic gene clusters using small molecules.
The second common theme is that growth inhibition is in many cases key to stimulation of secondary metabolism. Numerous examples demonstrate that microorganisms often respond to growth-inhibitory molecules by producing their own growth-inhibitory molecules. This observation has important repercussions. First, it suggests that old antibiotics may be used to discover new ones. In strong support of this proposition are a number of results described above, including (a) multiple examples of antibiotic synthesis in response to growth-inhibitory molecules in coculture assays; (b) the discovery of piperidamycin using streptomycin and rifampicin in ribosome engineering; (c) chromatin remodeling studies, where SAHA and other chromatin modulators display growth-inhibitory properties and elicit bioactive metabolite production; and (d) early results with HiTES, where antibiotics have been identified as inducers of cryptic virulence factors. Second, the observation above provides hints regarding how bacteria perceive antibiotics; that is, they give us clues about antibiotics’ natural roles. That bacteria in many cases respond to antibiotics with antibiotics could point to the existence of chemical warfare among neighboring colonies. However, Waksman, Davies and others have pointed out that naturally occurring concentrations of antibiotics are not high enough to serve as biocides in the environment (Waksman 1961; Davies 2006; Yim, Wang and Davies 2007). As such, the exchange of antibiotics should in most cases be interpreted as an interaction or communication. That is, in the wild, antibiotics usually serve as mediators of microbial interactions and shape multispecies communities in their microenvironment. What would be the role of antibiotic resistance in this context? In an anthropomorphic sense, resistance may be akin to putting on headphones and not participating in a conversation.
Additional insights regarding the roles of some antibiotics may be gained by considering the following: studies by Ochi and a number of coculture experiments show that the response of Streptomycetes to nutrient limitation is often similar to their response to antibiotic treatment. This may bring another role for antibiotics to the fore, namely that of deterrents or warning signals. A nutrient-poor environment is an undesirable one, and the similar responses of some bacteria to antibiotics would lead one to conclude that antibiotics can act as deterrents. There are parallels from a molecular point of view as well: nutrient limitation triggers the stringent response, which leads to an altered RNAP specificity via the molecule ppGpp. Similarly, antibiotic treatment or resistance can lead to activation of the stringent response or even an RNAP that acts as if ‘stringent’ even in the absence of ppGpp. The idea of inducible defense is well accepted in the plant research community. Phytoalexins are inducible defense metabolites produced by plants in response to certain threats. Might some bacterially produced antibiotics serve as an inducible defense mechanism—say, as bacterioalexins? Note that this does not imply that they are necessarily bactericidal, but simply that they may be inducible responses to undesired conditions.
Ultimately, it is difficult to ascribe a specific role to antibiotics simply because there is no single answer. The role of an antibiotic depends on the specific antibiotic and the nature of the recipient cell. To the recently discovered strain Amycolatopsis sp. AA4, which is resistant to 15 antibiotics, including glycopeptides, vancomycin plays no role at all (D'Costa et al. 2006). To organisms that do not have the requisite resistance genes, vancomycin may serve as a threat or a signal and therefore elicit complex changes in gene expression. However, some antibiotics, which kill at concentrations as low as a few molecules per cell, such as enediynes or bleomycins, should be considered chemical warfare agents (Povirk 1996). Thus, depending on the antibiotic and the recipient, the roles may vary from no function at all, to signals that elicit changes in gene expression patterns, to inducers of stress and to cell death-causing toxins. It is especially important to consider the role of the recipient cell. Because there is a concentration gradient of an antibiotic emanating from the producing strain, the neighboring cell will always initially sense subinhibitory concentrations of most antibiotics. Transcriptional regulation in bacteria is rapid and thus the recipient could activate antibiotic production or sporulation pathways to redirect or entirely evade the conversation.
The foregoing discussion shows that this research field, referred to as small molecule biology by Davies, is a fascinating one. Small molecules can certainly be used to probe and activate the store of silent biosynthetic gene clusters that have been found in microbial genomes. It is clear that bacteria have evolved complex response mechanisms to antibiotics. Much like quorum-sensing molecules, subinhibitory concentrations of antibiotics affect the expression of a set of particular genes, an antibiotic-specific regulon. Further investigations of this regulon will deepen our understanding of small molecule biology and, by stimulating secondary metabolism, provide additional bioactive molecules for use in human health and beyond.
Acknowledgments
The authors thank Clarissa Forneris and Paul Rosen for comments on the manuscript, and Marcus Gibson for assistance with figures.
FUNDING
The authors thank the National Institutes of Health (DP2-124786-01 to MRS) and the Searle Scholars Program of the Kinship Foundation (to MRS) for generous support of our work.
Conflict of interest. None declared.
REFERENCES
- Aedo S, Tomasz A. Role of the stringent stress response in the antibiotic resistance phenotype of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Ch. 2016;60:2311–7. doi: 10.1128/AAC.02697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Craney A, Pimentel-Elardo SM, et al. A synthetic, species-specific activator of secondary metabolism and sporulation in Streptomyces coelicolor. Chembiochem. 2013;14:83–91. doi: 10.1002/cbic.201200619. [DOI] [PubMed] [Google Scholar]
- Amano S, Morota T, Kano Y, et al. Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J Antibiot. 2010;63:486–91. doi: 10.1038/ja.2010.68. [DOI] [PubMed] [Google Scholar]
- Amano S, Sakurai T, Endo K, et al. A cryptic antibiotic triggered by monensin. J Antibiot. 2011;64:703. doi: 10.1038/ja.2011.69. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Patlan V, Sekine S, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeño-Tárraga A-M, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–7. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Berdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Bode HB, Bethe B, Höfs S, et al. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 2002;3:619–27. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a Regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–35. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch G, Loidl P, Graessle S. Histone modifications and chromatin dynamics: a focus on filamentous fungi. FEMS Microbiol Rev. 2008;32:409–39. doi: 10.1111/j.1574-6976.2007.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchko RA, Adams TH, Keller NP. Aspergillus nidulans mutants defective in stc gene cluster regulation. Genetics. 1999;153:715–20. doi: 10.1093/genetics/153.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzini P. Batch and fed-batch carotenoid production by Rhodotorula glutinis-Debaryomyces castellii co-cultures in corn syrup. J Appl Microbiol. 2001;90:843–7. doi: 10.1046/j.1365-2672.2001.01319.x. [DOI] [PubMed] [Google Scholar]
- Chai YJ, Cui CB, Li CW, et al. Activation of the dormant secondary metabolite production by introducing gentamicin-resistance in a marine-derived Penicillium purpurogenum G59. Mar Drugs. 2012;10:559–82. doi: 10.3390/md10030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YM, Chang SL, Oakley BR, et al. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol. 2011;15:137–43. doi: 10.1016/j.cbpa.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichewicz RH. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat Prod Rep. 2010;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–8. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- Cragg GM, Newman DJ. Natural products: a continuing source of novel drug leads. Biochim Biophys Acta. 2013;1830:3670–95. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craney A, Ozimok C, Pimentel-Elardo SM, et al. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem Biol. 2012;19:1020–7. doi: 10.1016/j.chembiol.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Cueto M, Jensen PR, Kauffman C, et al. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J Nat Prod. 2001;64:1444–6. doi: 10.1021/np0102713. [DOI] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. P Natl Acad Sci USA. 2002;99:10494–9. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Costa VM, McGrann KM, Hughes DW, et al. Sampling the antibiotic resistome. Science. 2006;311:374–7. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- Davies J. Are antibiotics naturally antibiotics? J Ind Microbiol Biot. 2006;33:496–9. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–53. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Fourati-Ben Fguira L, Smaoui S, Karray-Rebai I, et al. The antifungal activity of the terrestrial Streptomyces US80 strain is induced by heat-killed fungi. Biotechnol J. 2008;3:1058–66. doi: 10.1002/biot.200700155. [DOI] [PubMed] [Google Scholar]
- Fisch KM, Gillaspy AF, Gipson M, et al. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J Ind Microbiol Biot. 2009;36:1199–213. doi: 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–93. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol. 2015;197:1146–56. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacek A, Strauss J. The chromatin code of fungal secondary metabolite gene clusters. Appl Microbiol Biot. 2012;95:1389–404. doi: 10.1007/s00253-012-4208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudêncio SP, Pereira F. Dereplication: racing to speed up the natural products discovery process. Nat Prod Rep. 2015;32:779–810. doi: 10.1039/c4np00134f. [DOI] [PubMed] [Google Scholar]
- Goh EB, Yim G, Tsui W, et al. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. P Natl Acad Sci USA. 2002;99:17025–30. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrikson JC, Hoover AR, Joyner PM, et al. A chemical epigenetics approach for engineering the in situ biosynthesis of a cryptic natural product from Aspergillus niger. Org Biomol Chem. 2008;7:435–8. doi: 10.1039/b819208a. [DOI] [PubMed] [Google Scholar]
- Hessayon DG. Fungitoxins in the soil: I. Historical. Soil Sci. 1953;75:317–28. [Google Scholar]
- Hong RW, Shchepetov M, Weiser JN, et al. Transcriptional profile of the Escherichia coli response to the antimicrobial peptide cecropin A. Antimicrob Agents Chemother. 2003;47:1–6. doi: 10.1128/AAC.47.1.1-6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka T, Ohnishi-Kameyama M, Muramatsu H, et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol. 2009;27:462–4. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Xu J, Ochi K. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol Microbiol. 2006;61:883–97. doi: 10.1111/j.1365-2958.2006.05285.x. [DOI] [PubMed] [Google Scholar]
- Hosoya Y, Okamoto S, Muramatsu H, et al. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob Agents Ch. 1998;42:2041–7. doi: 10.1128/aac.42.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Zhang Q, Ochi K. Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase beta subunit) of Streptomyces lividans. J Bacteriol. 2002;184:3984–91. doi: 10.1128/JB.184.14.3984-3991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakovleva EP, Sokolova EN. Dissociation of a Candida tropicalis culture and its capacity to stimulate levorin synthesis when cultured together with Actinomyces levoris. Antibiotiki. 1978;23:199–203. [PubMed] [Google Scholar]
- Ikeda H, Ishikawa J, Hanamoto A, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–31. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- Imai Y, Fujiwara T, Ochi K, et al. Development of the ability to produce secondary metabolites in Streptomyces through the acquisition of erythromycin resistance. J Antibiot. 2012;65:323–6. doi: 10.1038/ja.2012.16. [DOI] [PubMed] [Google Scholar]
- Imai Y, Sato S, Tanaka Y, et al. Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl Environ Microb. 2015;81:3869–79. doi: 10.1128/AEM.04214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennet JW. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3B:937–47. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Khan SR, Yamazaki H. Trimethoprim-induced accumulation of guanosine tetraphosphate (ppGpp) in Escherichia coli. Biochem Bioph Res Co. 1972;48:169–74. doi: 10.1016/0006-291x(72)90358-0. [DOI] [PubMed] [Google Scholar]
- Kolter R, van Wezel GP. Goodbye to brute force in antibiotic discovery? Nat Microbiol. 2016;1:15020. doi: 10.1038/nmicrobiol.2015.20. [DOI] [PubMed] [Google Scholar]
- Lai C, Xu J, Tozawa Y, et al. Genetic and physiological characterization of rpoB mutations that activate antibiotic production in Streptomyces lividans. Microbiology. 2002;148:3365–73. doi: 10.1099/00221287-148-11-3365. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Landsman D. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res. 1997;25:3693–7. doi: 10.1093/nar/25.18.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi PM, Angell HD, Whittington DA, et al. Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases. Biochemistry. 2011;50:1808–17. doi: 10.1021/bi101859k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RA, Azam F. Antagonistic interactions among marine pelagic bacteria. Appl Environ Microb. 2001;67:4975–83. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cheng YQ. Genome-guided discovery of diverse natural products from Burkholderia sp. J Ind Microbiol Biot. 2014;41:275–84. doi: 10.1007/s10295-013-1376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmann A, Aly AH, Lin W, et al. Co-cultivation—a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar Drugs. 2014;12:1043–65. doi: 10.3390/md12021043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mearns-Spragg A, Bregu M, Boyd KG, et al. Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett Appl Microbiol. 1998;27:142–6. doi: 10.1046/j.1472-765x.1998.00416.x. [DOI] [PubMed] [Google Scholar]
- Meng L, Li M, Yang SH, et al. Intracellular ATP levels affect secondary metabolite production in Streptomyces spp. Biosci Biotech Bioch. 2011;75:1576–81. doi: 10.1271/bbb.110277. [DOI] [PubMed] [Google Scholar]
- Miller SJ, Clardy J. Natural products: Beyond grind and find. Nat Chem. 2009;1:261–3. doi: 10.1038/nchem.269. [DOI] [PubMed] [Google Scholar]
- Mohamed A, Nguyen CH, Mamitsuka H. Current status and prospects of computational resources for natural product dereplication: a review. Brief Bioinform. 2016;17:309–21. doi: 10.1093/bib/bbv042. [DOI] [PubMed] [Google Scholar]
- Moore JM, Bradshaw E, Seipke RF, et al. Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in streptomyces bacteria. Methods Enzymol. 2012;517:367–85. doi: 10.1016/B978-0-12-404634-4.00018-8. [DOI] [PubMed] [Google Scholar]
- Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–84. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural product scaffolds as leads to drugs. Future Med Chem. 2009;1:1415–27. doi: 10.4155/fmc.09.113. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–61. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- Nützmann HW, Reyes-Dominguez Y, Scherlach K, et al. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. P Natl Acad Sci USA. 2011;108:14282–7. doi: 10.1073/pnas.1103523108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K. From microbial differentiation to ribosome engineering. Biosci Biotech Bioch. 2007;71:1373–86. doi: 10.1271/bbb.70007. [DOI] [PubMed] [Google Scholar]
- Ochi K, Hosaka T. New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biot. 2013;97:87–98. doi: 10.1007/s00253-012-4551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K, Okamoto S, Tozawa Y, et al. Ribosome engineering and secondary metabolite production. Adv Appl Microbiol. 2004;56:155–84. doi: 10.1016/S0065-2164(04)56005-7. [DOI] [PubMed] [Google Scholar]
- Oh D-C, Jensen PR, Kauffman CA, et al. Libertellenones A–D: induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg Med Chem. 2005;13:5267–73. doi: 10.1016/j.bmc.2005.05.068. [DOI] [PubMed] [Google Scholar]
- Okada BK, Wu Y, Mao D, et al. Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chem Biol. 2016;11:2124–30. doi: 10.1021/acschembio.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ola ARB, Thomy D, Lai D, et al. Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J Nat Prod. 2013;76:2094–9. doi: 10.1021/np400589h. [DOI] [PubMed] [Google Scholar]
- Oliynyk M, Samborskyy M, Lester JB, et al. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol. 2007;25:447–53. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- Onaka H, Tabata H, Igarashi Y, et al. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and characterization. J Antibiot. 2001;54:1036–44. doi: 10.7164/antibiotics.54.1036. [DOI] [PubMed] [Google Scholar]
- Palmer JM, Keller NP. Secondary metabolism in fungi: does chromosomal location matter? Curr Op Microbiol. 2010;13:431–6. doi: 10.1016/j.mib.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GML, Bolis CM. Fungal cell-wall polysaccharides elicit an antifungal secondary metabolite (Phytoalexin) in the cyanobacterium Scytonema ocellatum. J Phycol. 1997;33:54–60. [Google Scholar]
- Pettit RK. Mixed fermentation for natural product drug discovery. Appl Microbiol Biot. 2009;83:19–25. doi: 10.1007/s00253-009-1916-9. [DOI] [PubMed] [Google Scholar]
- Pimentel-Elardo SM, Sørensen D, Ho L, et al. Activity-independent discovery of secondary metabolites using chemical elicitation and cheminformatic inference. ACS Chem Biol. 2015;10:2616–23. doi: 10.1021/acschembio.5b00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- Projan SJ. Why is big Pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 2003;6:427–30. doi: 10.1016/j.mib.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Rigali S, Titgemeyer F, Barends S, et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–5. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–23. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- Santos CN, Stephanopoulos G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol. 2008;12:168–76. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Schroeckh V, Scherlach K, Nützmann H-W, et al. Intimate bacterial–fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. P Natl Acad Sci USA. 2009;106:14558–63. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. P Natl Acad Sci USA. 2014;111:7266–71. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Case RJ, Kolter R, et al. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem. 2011a;3:331–5. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Traxler MF, Zheng SL, et al. Structure and biosynthesis of amychelin, an unusual mixed-ligand siderophore from Amycolatopsis sp. AA4. J Am Chem Soc. 2011b;3:11434–7. doi: 10.1021/ja203577e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima J, Hesketh A, Okamoto S, et al. Induction of actinorhodin production by rpsL (Encoding Ribosomal Protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2) J Bacteriol. 1996;178:7276–84. doi: 10.1128/jb.178.24.7276-7284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CS, Kim HJ, Kim MJ, et al. Morphological change and enhanced pigment production of Monascus when cocultured with Saccharomyces cerevisiae or Aspergillus oryzae. Biotechnol Bioeng. 1998;59:576–81. doi: 10.1002/(sici)1097-0290(19980905)59:5<576::aid-bit7>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Shwab EK, Bok JW, Tribus M, et al. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–64. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Rajbhandari I, Wesson K. Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb Ecol. 2001;41:90–6. doi: 10.1007/s002480000084. [DOI] [PubMed] [Google Scholar]
- Sonnenbichler J, Dietrich J, Peipp H. Secondary fungal metabolites and their biological activities, V. Investigations concerning the induction of the biosynthesis of toxic secondary metabolites in basidiomycetes. Biol Chem H-S. 1994;375:71–9. doi: 10.1515/bchm3.1994.375.1.71. [DOI] [PubMed] [Google Scholar]
- Starosta AL, Lassak J, Jung K, et al. The bacterial translation stress response. FEMS Microbiol Rev. 2014;38:1172–201. doi: 10.1111/1574-6976.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Awakawa T, Noguchi H, et al. Induced production of mycotoxins in an endophytic fungus from the medicinal plant Datura stramonium L. Bioorg Med Chem Lett. 2012;22:6397–400. doi: 10.1016/j.bmcl.2012.08.063. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Seyedsayamdost MR, Clardy J, et al. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol Microbiol. 2012;86:628–44. doi: 10.1111/mmi.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Kawai S, Ogawa H, et al. Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J Antibiot. 2000;53:979–82. doi: 10.7164/antibiotics.53.979. [DOI] [PubMed] [Google Scholar]
- VanderMolen KM, Darveaux BA, Chun W-L, et al. Epigenetic manipulation of a filamentous fungus by the proteasome-inhibitor bortezomib induces the production of an additional secondary metabolite. RSC Adv. 2014;4:18329–35. doi: 10.1039/C4RA00274A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman SA. The role of antibiotics in nature. Perspect Biol Med. 1961;4:271–87. [Google Scholar]
- Williams RB, Henrikson JC, Hoover AR, et al. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008;6:1895–7. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- Xu J, Tozawa Y, Lai C, et al. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2) Mol Genet Genomics. 2002;268:179–89. doi: 10.1007/s00438-002-0730-1. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Oikawa H, Ogawa HO, et al. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology. 2005;151:2899–905. doi: 10.1099/mic.0.28139-0. [DOI] [PubMed] [Google Scholar]
- Yim G, Wang HH, Davies J. Antibiotics as signaling molecules. Philis T R Soc B. 2007;362:1195–200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, Keller N. Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol. 2005;43:437–58. doi: 10.1146/annurev.phyto.43.040204.140214. [DOI] [PubMed] [Google Scholar]