Abstract
To better understand genetic regulation of differential growth of plant organs, a dominant and semidwarf mutant, constitutive differential growth 1-Dominant (cdg1-D), was isolated utilizing the technique of activation tagging. cdg1-D showed pleiotropic phenotype including dwarfism, exaggerated leaf epinasty, and twisted or spiral growth in hypocotyl, inflorescence stem, and petiole. Hypocotyls of cdg1-D were longer than those of wild type under light conditions. The phenotype was caused by activation tagging of CDG1 gene that encodes a receptor-like cytoplasmic kinase of RLCKVII subfamily. When treated with high concentrations of brassinolide, light-grown wild-type seedlings showed long hypocotyls and strong leaf epinasty as observed in cdg1-D seedlings. Treatment of cdg1-D with brassinazole, a specific inhibitor of brassinosteroid (BR) biosynthesis, did not rescue the mutant phenotype. Gene expression of CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM involved in BR biosynthesis and phyB ACTIVATION-TAGGED SUPPRESSOR1 that inactivates BR was repressed and induced, respectively, in cdg1-D plants, suggesting constitutive activation of BR signaling in the mutant. CDG1 was expressed at a very low level in all the organs of the wild type tested. We isolated two independent intragenic suppressors of cdg1-D. However, they showed normal morphology and responded to BR in a similar manner to wild type. Taken together, CDG1 gene may interfere with signal transduction of BR when overexpressed, but is not an essential factor for it in the wild type.
After differentiating in the apical meristem or embryo, plant organs continue to grow to reach their adult size, accompanying cell division and enlargement (Howell, 1998). Plant cells in an organ do not always grow uniformly, resulting in differential growth. Since plant cells are surrounded by cell walls, they do not change their position relative to neighbor cells. Thus, differential growth is one of the major means for plant organs to change their shape after cell division ceases. Plants change their form depending on their developmental stage and in response to environmental conditions. Consequently, plants often exhibit differential growth throughout their life. The process of differential growth is important for plant development and survival and represents a fundamental difference in how plants and animals react to environmental change.
Phototropism and gravitropism are the best characterized differential-growth responses induced by environmental conditions. Auxin plays a central role in these processes (Iino, 2001; Tatematsu et al., 2004). Hypocotyl cells at the apex grow differentially under dark conditions, so that a hook-like structure is formed. Hooks disappear when irradiated with light or as seedlings get old. Formation and disappearance of the hook structure is an example of differential growth caused by a developmental cue (Raz and Ecker, 1999). Ethylene has long been considered as a major factor controlling hook formation, inhibiting cell growth at the inner side of hook (Peck et al., 1998). Epinasty is another example of differential growth. Ethylene, high concentrations of auxin (Abeles et al., 1992), and brassinosteroid (BR; Schlagnhaufer and Arteca, 1985) induce leaf epinasty, which occurs as a result of differential growth of leaf blade and petiole, in many plants. Thus, several plant hormones appear to be involved in epinasty.
Activation tagging with the enhancer from the cauliflower mosaic virus (CaMV) 35S transcript promoter is a technique in plant functional genomics that can create transgenic plants in which the T-DNA carrying 35S enhancer at its right border is spliced into the plant genome at random sites (Hayashi et al., 1992; Weigel et al., 2000). Analysis of gain-of-function mutants created through activation tagging can provide insight to a gene's function. In fact, study on activation-tagged mutants has played a pivotal role in dissecting cytokinin (Kakimoto, 1996), BR (Neff et al., 1999), and light (Nakazawa et al., 2001) signaling.
In an attempt to determine factors involved in genetic regulation of differential growth of plant organs, we have carried out screening of gain-of-function mutants by use of activation-tagging methods in this study. From 10,000 activation-tagged lines, a novel, dominant, and semidwarf mutant, constitutive differential growth 1-Dominant (cdg1-D), was isolated. Although adult cdg1-D mutants have a semidwarf phenotype, cdg1-D hypocotyls are longer than wild-type hypocotyls under white light. In cdg1-D, the hypocotyl, stem, petiole, and fruit grow in a spiral or twisted shape and the leaves are small and show severe epinasty. Here we show that these defects were induced by activation of a gene for a Ser/Thr protein kinase and that the kinase may interfere with BR signaling.
RESULTS
Mutant Screening of Activation-Tagging Lines
We generated approximately 10,000 activation-tagged primary lines of Arabidopsis ecotype Columbia by using pPCVICEn4HPT (Hayashi et al., 1992; Nakazawa et al., 2001). For mutant screening, 11 T2 seeds of each transformed line were grown in the dark for 3 d on vertically oriented plates after germination. The length and shape of hypocotyls and the hook structure of the etiolated seedlings were monitored.
As a result, we isolated a dominant mutant that showed a twisted or spiral hypocotyl and an open-hook structure in the dark and excessively epinastic leaves when grown in the light condition. Three-quarters of the T2 progeny of this line showed the mutant phenotype and hygromycin resistance. F1 plants of the mutant allowed to backcross to Columbia also showed the mutant phenotype. These traits of the mutant are consistent with it having a single dominant allele. This dominant mutation was named cdg1-D after its leaf epinasty and twisted growth in the hypocotyl.
Mutant Phenotype
Defects of the cdg1-D mutant were pleiotropic. cdg1-D plants were dwarf and had small and remarkably epinastic leaves (Fig. 1, a–c). Mature leaves of the mutant were smaller than those of the wild type. Rosettes of the mutant plants showed a pineal shape because of its small, curly leaves and shorter petioles (Fig. 1c). Mutant plants bore only a small number of seeds. These phenotypes were more severe in homozygous cdg1-D plants (Fig. 1c) than heterozygous plants (Fig. 1b), indicating that cdg1-D is a semidominant mutation.
Figure 1.
Morphologies of wild-type and cdg1-D plants. a to c, Five-week-old light-grown wild type (a), heterozygous (b), and homozygous (c) cdg1-D. d, Eight-week-old light-grown cdg1-D. e and f, Three-day-old etiolated seedlings (e) and 1-week-old light-grown seedlings (f) of wild type (left) and cdg1-D (right). g, Two-week-old light-grown cdg1-D. Plants were grown on soil (a–d and g) or on agar plates (e and f). Arrowhead in g shows spiral growth in petiole. Bars, 5 mm in all sections.
cdg1-D hypocotyls grew in a twisted manner (Fig. 1e) or sometimes spirally in the dark with a vertical screw axis showing normal gravitropic responses. The hypocotyls were as long as those of the wild type. Twisted growth was also seen in various organs of adult cdg1-D plants such as inflorescence stem, petiole, and fruit (Fig. 1d). Petioles of cdg1-D also sometimes showed spiral growth (Fig. 1g, arrowhead). All leaves of cdg1-D, including cotyledons, rosette, and cauline leaves, showed exaggerated epinasty (Fig. 1, c and f). cdg1-D leaves were bent at the junction of the blade and petiole in a longitudinal direction before the leaf blade began to expand (Fig. 1g). Then they started to expand and curl in both longitudinal and latitudinal directions.
Microscopic observation was performed to examine cellular basis of abnormal differential growth such as leaf epinasty and twisted growth of hypocotyl and inflorescence stem (Fig. 2). The overall anatomy of the mutant organs appeared unaltered, suggesting that the mutation did not affect organ differentiation. But tissue development in each organ was disordered in the mutant. Cortex and epidermal cells of cdg1-D hypocotyl expanded randomly (Fig. 2, d and h). They became uneven in shape and size. cdg1-D tissue had extensive intercellular spaces in comparison with wild type. Consequently, the surface of cdg1-D hypocotyls was not as smooth as that of the wild type. Longitudinal observations of cdg1-D hypocotyls after immersion in India ink also showed a distortion of epidermal cell files and rough surface, which was dented and bulged from place to place (Fig. 2, b and f).
Figure 2.
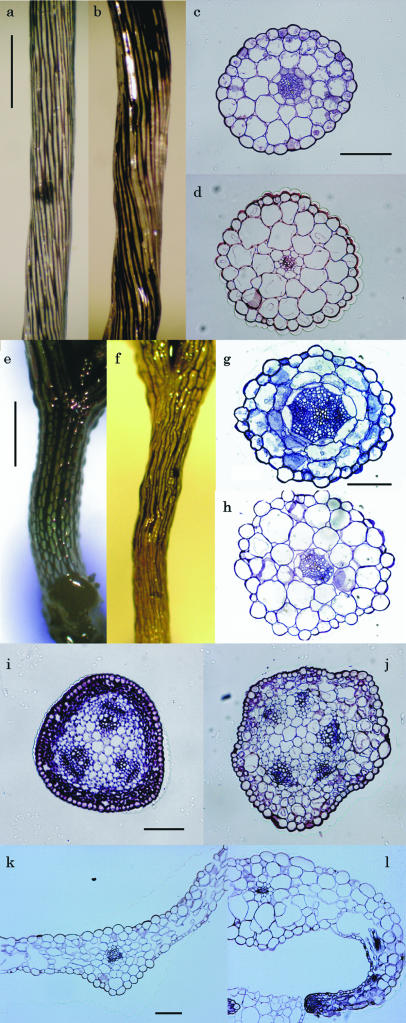
Structure of cdg1-D organs. a to h, Three-day-old etiolated (a–d) or 1-week-old light-grown (e–h) hypocotyls of wild-type (a, c, e, and g) and cdg1-D (b, d, f, and h) seedlings. Photographs were taken after immersion in India ink (a, b, e, and f) or cross-sectioning (c, d, g, and h). i and j, Cross-sections of inflorescence stems of 5-week-old wild-type (i) and cdg1-D (j) plants. k and l, Cross-sections of leaves of 3-week-old wild-type (k) and cdg1-D (l) plants. Bars, 500 (a and e) and 100 (c, g, i, and k) μm.
Epidermal and cortex cells of cdg1-D were also enlarged randomly in inflorescence stems, while those of the wild type were small and organized in order (Fig. 2, i and j). In contrast, the central stele of the mutant was relatively normal, with a smooth and round shape (Fig. 2j). These observations suggest that abnormal radial growth of the cortex and epidermal cells in cdg1-D disrupts epidermal cell files and causes twisted or spiral growth in both hypocotyls and inflorescence stems.
Leaf epidermal cells on the adaxial side were generally larger than those on the abaxial side in wild-type plants (Fig. 2k). In cdg1-D, epidermal cells on the adaxial side were larger than those of wild type, while those on the abaxial side were much smaller than those of wild type, especially in the region of the leaf margin (Fig. 2l). Namely, the difference in cell size between the adaxial and abaxial epidermal cells was expanded by the cdg1-D mutation. The amplified differential expansion of leaf cells appeared to be the cause of leaf epinasty in the latitudinal direction displayed in the mutant. Leaf parenchyma cells of cdg1-D, like the adaxial epidermal cells, also swelled. Epinasty in the longitudinal direction was also increased in a similar manner (data not shown). Underdevelopment of vascular tissues, another prominent characteristic of cdg1-D, was observed in all organs examined (Fig. 2, d, h, j, and l).
Responses to Growth Regulators in Light-Grown Seedlings
Hypocotyls of cdg1-D were about three times longer than those of the wild type under white-light condition (Fig. 1f). cdg1-D hypocotyls had longer epidermal cells than those of wild type (Fig. 2, e and f). Therefore, the longer cdg1-D hypocotyls probably resulted from an increase in length rather than an increase in the number of epidermal cells.
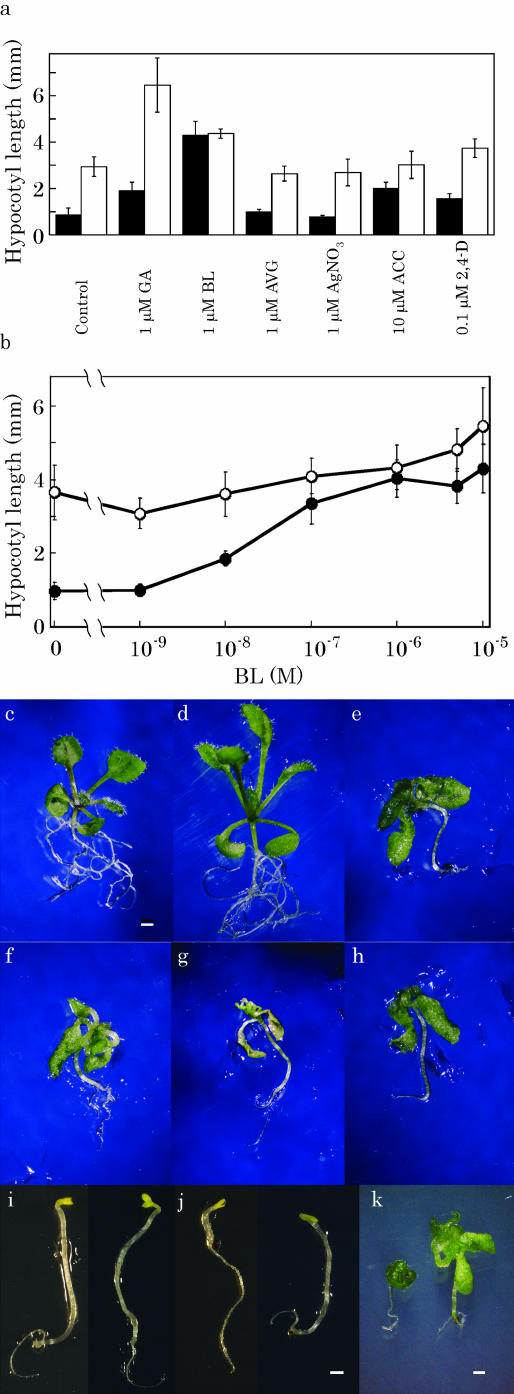
Since plant hormones are often involved in the long-hypocotyl phenotype, effects of growth regulators were examined on the growth of hypocotyls under white-light conditions. Seedlings were grown for 1 week on agar plates that contained various substances, including gibberellin (1 μm GA3), BR (1 μm brassinolide [BL]), inhibitors of ethylene synthesis (1 μm aminoethoxyvinylglycine) and perception (1 μm AgNO3), ethylene precursor (10 μm 1-aminocyclopropane-1-carboxylic acid [ACC]), and a synthetic auxin (0.1 μm 2,4-dichlorophenoxyacetic acid; Fig. 3a). GA treatment increased hypocotyl length about 2-fold in both wild-type and cdg1-D seedlings, showing that GA exhibited an additive interaction with the cdg1-D mutation. GA treatment did not affect leaf morphology in either the wild type or cdg1-D (Fig. 3, d and g).
Figure 3.
Effects of growth regulators on elongation of hypocotyls and epinasty of leaves. After seedlings were grown under continuous white light for 1 (a and c–h) or 2 (b) weeks on agar plates, length of hypocotyls of wild type (black bars and circles in a and b, respectively) and cdg1-D (white bars and circles in a and b, respectively) was measured. Wild-type (c–e) and cdg1-D (f–h) plants were grown in the absence (c and f) or presence (d and g) of 1 μm GA or BL (e and h). Wild-type (left) and cdg1-D (right) plants were grown in the absence (i) or presence (j and k) of 2 μm brassinazole for 4 d in the dark (i and j) or for 10 d under continuous white light (k). Bars, 1 mm.
BR also promoted hypocotyl elongation in both wild type and cdg1-D (Fig. 3a). Dose response of the BR effects in Figure 3b showed that BL started to increase hypocotyl length of the wild type at 10 nm and that the promotive effects reached a plateau at 1 μm. In contrast, growth of cdg1-D hypocotyls was not affected as much as the wild type. Consequently, cdg1-D hypocotyls were as long as those of the wild type at BL concentrations higher than 1 μm. Since BL treatment also induced leaf epinasty in wild type (Fig. 3e), wild-type plants grown in the presence of 1 μm BL looked very similar to cdg1-D grown in the same condition (Fig. 3, e versus h) and somewhat resembled cdg1-D grown in the absence of the hormone (Fig. 3, e versus f). However, treatment of cdg1-D with 1 μm brassinazole, a specific inhibitor of BR biosynthesis (Asami and Yoshida, 1999; Min et al., 1999), did not restore the aberrant phenotype of cdg1-D (Fig. 3, i–k). Thus, BL treatment appeared to mimic the cdg1-D phenotype in wild-type seedlings under white-light conditions, and the mutant phenotype was not likely due to overproduction of BR in it.
Neither aminoethoxyvinylglycine nor AgNO3 had any effects on hypocotyl elongation (Fig. 3a) or leaf epinasty (data not shown) in the wild type or cdg1-D. On the other hand, ACC induced hypocotyl elongation in the wild type. In the presence of ACC, hypocotyls of the wild type were more than twice as long as those of the control. They were, however, still shorter than those of cdg1-D (Fig. 3a). ACC also induced leaf epinasty in the wild type (data not shown), but its effects were smaller in extent than those observed by either the cdg1-D mutation (Fig. 3f) or BR treatment (Fig. 3e). These results suggest that ethylene is not a major factor for the expression of the cdg1-D phenotype, although sensitivity to ethylene was reduced in cdg1-D. Similarly, 2,4-dichlorophenoxyacetic acid had only a slight promotive effect on hypocotyl elongation in both the wild type and cdg1-D (Fig. 3a).
Cloning of CDG1 Gene
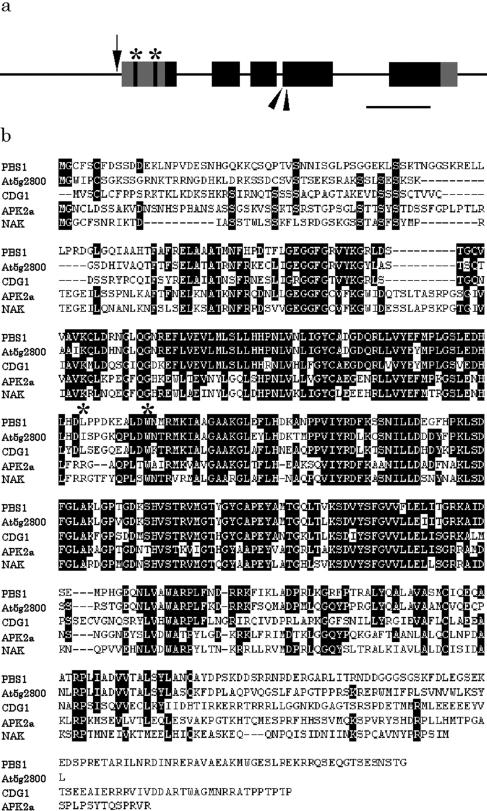
The CDG1 gene was cloned by plasmid rescue. Nucleotide sequences adjacent to both ends of T-DNA perfectly matched sequences in a P1 genomic clone, MOJ10, which is mapped on chromosome 3. The tetramerized CaMV 35S enhancer sequences were perfectly conserved in the fragment. A hypothetical gene (GI, 9279618, At3g26940) with similarity to Ser/Thr protein kinases was downstream of the enhancer sequences. Since no expressed sequence tags were available for the predicted gene, cDNA was isolated by the PCR after reverse transcription of RNA (RT-PCR) using primers designed to amplify the fragment from the putative start to stop codons of this gene. Results of 3′ and 5′ RACE showed that polyadenylation occurred 146 bp downstream of the putative stop codon and that the 5′ initiation site was located 332 bp upstream of the putative start codon. These results indicate that the CDG1 gene is transcribed into a sequence of 1,754 nucleotides and consists of 5 exons (Fig. 4a). T-DNA was inserted at 47 bp upstream of the determined 5′ end of the CDG1 transcript. No canonical TATA box was found upstream of the initiation site. The CDG1 transcript contained two open reading frames (ORFs) in addition to the protein kinase ORF at the 5′ end (Fig. 4a, indicated by asterisks). Start codons of these upstream ORFs (uORFs) were 242 and 79 bp upstream of the start codon of the protein kinase ORF, respectively. The uORFs can be translated into short peptides, 11 and 8 amino acid residues, respectively, that exhibit no similarity to any of the protein sequences in the GenBank database.
Figure 4.
Structure of CDG1 gene. a, Boxes show exons. Black and gray boxes indicate coding and noncoding regions, respectively. Arrow indicates the T-DNA insertion site. Arrowheads indicate suppressor mutation sites. Asterisks indicate upstream ORFs. Bar, 500 bp. b, Alignment of amino acid sequences of CDG1, PBS1 (Swiderski and Innes, 2001), APK2a (Ito et al., 1997), NAK (Moran and Walker, 1993), and At5g02800. Asterisks indicate suppressor mutation sites in CDG1. The alignment was made with CLUSTALW (http://www.ddbj.nig.ac.jp/E-mail/clustalw-j.html).
The longest ORF of the CDG1 gene can be translated into a sequence of 431 amino acid residues (Fig. 4b) that contains all the 11 conserved subdomains of eukaryotic protein kinases. All the invariant amino acid residues are conserved in their proper positions (Hanks and Quinn, 1988). The putative kinase domain is flanked by short nonkinase domains on both sides. Since this protein contains no membrane-spanning regions or extra membrane domains, it is likely to be either a cytoplasmic or nuclear kinase.
Expression of CDG1 Gene
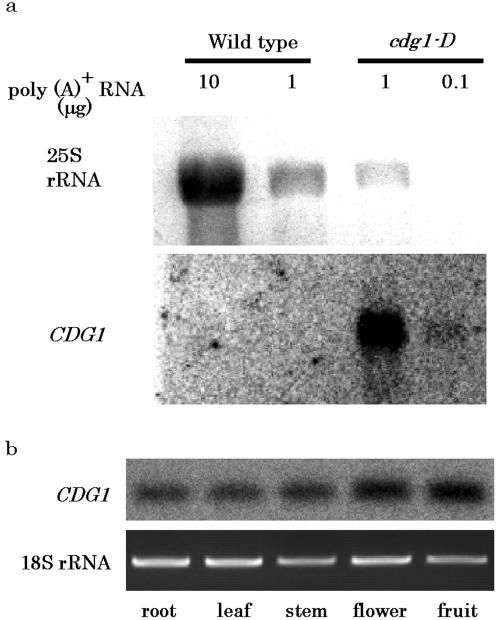
An RNA gel-blot analysis using the CDG1 kinase ORF as a probe revealed overexpression of this gene in the cdg1-D mutant (Fig. 5a). In the wild type, no signal of CDG1 was detectable in 10 μg poly(A)+ RNA, while a signal was observed in cdg1-D using 0.1 μg poly(A)+ RNA. Therefore, CDG1 mRNA appeared to accumulate more than 100-fold in cdg1-D. This result suggests that the abnormalities of cdg1-D plants result from activation tagging of the CDG1 protein kinase gene. Expression of the CDG1 gene was higher in homozygous cdg1-D than in heterozygous cdg1-D (data not shown), suggesting that the severity of the cdg1-D phenotype depends on the dose of the CDG1 gene. Expression of the CDG1 gene was not detected in the wild type by RNA gel-blot analysis, but it was observed by RT-PCR in various organs of the wild type, such as root, stem, leaf, flower, and fruit (Fig. 5b). The amounts of CDG1 messages seem to be slightly higher in the flower and fruit. These results show that CDG1 mRNA accumulates at a very low level in all organs of wild-type plants.
Figure 5.
mRNA accumulation of CDG1 gene. a, RNA gel-blot analysis of CDG1 gene in the wild type and cdg1-D. Poly(A)+ RNA was prepared from aerial tissue of wild-type and cdg1-D adult plants. The kinase ORF of CDG1 was used as probe. b, GDG1 gene expression in different organs of the wild type examined with RT-PCR. RT-PCR for amplification of CDG1 and 18S ribosomal RNA was carried out using 20 ng of total RNA prepared from each organ of wild type as templates. The numbers of cycles were 25 for CDG1 and 13 for 18S ribosomal RNA. RT-PCR product of CDG1 was detected by DNA gel-blot analysis using 32P-labeled probes because of its low expression level, while that of 18S ribosomal RNA was detected by staining with ethidium bromide.
Under control of the CaMV 35S promoter in the sense orientation, kinase ORF of the CDG1 mRNA was introduced into the genome of the wild type by the use of Agrobacterium tumefaciens. Of the nine independent T1 lines we generated, five lines showed dwarfism and epinastic leaves as observed in cdg1-D mutants. Their phenotype was heritable in T2 generation, linking with selection markers of T-DNA. Twelve-day-old plants of these transgenic lines were almost identical to cdg1-D plants with respect to their morphology (Fig. 6). They showed all the phenotypes seen in cdg1-D including dwarfism, twisted stem and fruit, and frizzy growth in dark condition and long hypocotyls under white-light condition (data not shown). Therefore, all the phenotypes seen in cdg1-D were reproduced in the CDG1 overexpressors. These results indicate that the cdg1-D phenotype is caused by overexpression of the protein kinase encoded by the CDG1 gene.
Figure 6.
Overexpressors of the CDG1 ORF for kinase. a, Wild type. b, cdg1-D. c and d, T2 generation of transformant lines ovD2 (c) and ovD4 (d). Plants were grown for 12 d on agar plates. Bar, 3 mm.
On the other hand, one of the nine transgenic lines showed the wild-type phenotype, and the other three lines exhibited weaker phenotype. Rosette leaves of the latter lines were as large as those of wild type; they displayed epinastic and twisted growth that was often seen in BR-treated plants, but their petioles were not as elongated as BR-treated ones (data not shown). These results clearly indicate that overexpression of CDG1 activates signaling pathway that is overlapping, but not identical with BR signaling.
Control of Gene Expression Involved in BR Metabolism
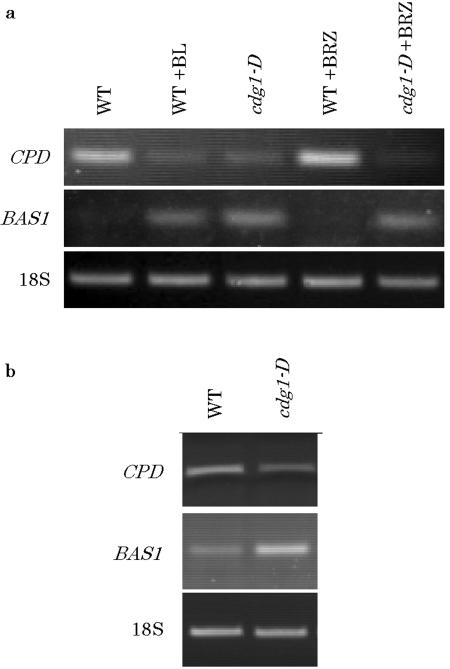
Recently BR biosynthesis has been shown to be controlled in a feedback fashion (Fujioka and Yokota, 2003). Genes involved in BR biosynthesis are repressed and those in BR catabolism are activated in the presence of higher levels of BR. If cdg1-D phenotype is related to BR action, such gene regulation should be observed in cdg1-D. This argument prompted us to examine gene expression of CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD; Szekeres et al., 1996) and phyB ACTIVATION-TAGGED SUPPRESSOR1 (BAS1; Neff et al., 1999; Turk et al., 2003), which encode cytochrome P450 enzymes involved in biosynthesis and inactivation of BL, respectively. RT-PCR in Figure 7a shows that mRNA level of CPD and BAS1 was decreased and increased in cdg1-D, respectively, compared to that of wild type. The change was very similar to that induced by treatment of wild type with 1 μm BL and was not affected by the treatment of the mutant with 1 μm brassinazole. These results suggest that cdg1-D phenotype is not caused by overproduction of BR, but that BR signaling is constitutively activated in cdg1-D seedlings. The repression of CPD and activation of BAS1 were also observed in mature cdg1-D plants (Fig. 7b).
Figure 7.
Expression of CPD and BAS1 genes determined by RT-PCR. RT-PCR for amplification of CPD, BAS1, and 18 S ribosomal RNA was carried out using 20 ng of total RNA prepared from 10-d-old seedlings grown in the absence or presence of 1 μm BL or 2 μm brassinazole under continuous white light (a) or from aerial tissue of 5-week-old plants (b). Numbers of PCR cycles were 27 for CPD, 29 for BAS1, and 13 for 18S ribosomal RNA. RT-PCR products were detected by staining with ethidium bromide.
Isolation and Characterization of cdg1, a Loss-of-Function Mutation of CDG1
By screening 10,000 M2 seeds, we isolated two independent suppressor mutants of cdg1-D that showed hygromycin resistance and suppressed all the phenotypes of cdg1-D. F1 plants of both mutants that were allowed to backcross with Columbia also showed the wild-type phenotype and hygromycin resistance. These traits of the suppressors suggest two possibilities: the mutations occurred at dominant loci, or they were intragenic suppressors that had mutations within the CDG1 gene downstream of the enhancer sequences in the T-DNA. Genomic sequencing of each line revealed a single-base substitution in CDG1 gene that results in a premature stop codon in exon 4 or disruption of a splice site junction between intron 3 and exon 4 (Fig. 4a). These findings mean that the mutated and overexpressed CDG1 mRNA in each allele should give rise to a nonfunctional protein that contains only subdomains I to V (Fig. 4b). Therefore, homozygotes of these suppressors must be loss-of-function mutants of CDG1 and were named cdg1-1 and 2, respectively. In contrast to the pleiotropic effects of overexpression of the CDG1 gene, cdg1-1 and 2 showed normal morphology in all organs (data not shown). We also examined effects of BR and GA on growth of these mutant alleles and found that they responded to the hormones essentially in the same fashion as the wild type (data not shown). These effects suggest that CDG1 gene possibly interferes with signal transduction of BR when overexpressed, but is not an essential factor for it in the wild type.
DISCUSSION
BR Signaling May Be Interfered within cdg1-D at the Seedling Stage
In this study we showed that activation of the Arabidopsis CDG1 gene by the use of CaMV 35S enhancers produced a dominant mutant, cdg1-D, which displayed pleiotropic defects in organ growth regulation after organogenesis. These defects included dwarfism, exaggerated differential growth in multiple organs such as hypocotyls, leaves, and inflorescence stems, and long-hypocotyl phenotype under white-light conditions. Treatment of wild-type seedlings with only BL phenocopied the long-hypocotyl phenotype of cdg1-D. Application of BL also induces epinasty of cotyledons, resulting in a cdg1-D-like morphology. It has been well known that BR biosynthesis is controlled by an elaborate feedback regulation (Fujioka and Yokota, 2003). BL treatment of Arabidopsis seedlings decreases mRNA level of several cytochrome P450 enzymes, including CPD (Mathur et al., 1998), DWF4, and ROT3 (Bancoş et al., 2002; Goda et al., 2002; Müssig et al., 2002), which function in BR biosynthesis. On the other hand, expression of BAS1/CYP72B1 gene, whose product inactivates BL through hydroxylation (Neff et al., 1999; Turk et al., 2003), is induced by exogenous BRs (Goda et al., 2002), suggesting that BAS1 acts to maintain steady-state levels of endogenous BRs. Decrease in CPD mRNA level and increase in BAS1 mRNA level were observed in cdg1-D, which were very similar to those observed in the wild-type seedlings treated with 1 μm BL. Furthermore, neither the change in gene expression level nor cdg1-D phenotype was influenced by treatment of cdg1-D with an inhibitor of BR biosynthesis, brassinazole (Asami and Yoshida, 1999; Min et al., 1999; Fujioka and Yokota, 2003). These findings strongly suggest that cdg1-D phenotype at the seedling stage results from constitutive activation of BR signaling.
Interestingly, CDG1 encodes a receptor-like kinase (RLK), and a few kinases have been identified that constitute the signal transduction pathway of BR (Thummel and Chory, 2002). A putative receptor of BR is a plasma membrane-localized Leu-rich repeat (LRR)-RLK, BRI1 (Li and Chory, 1997; Wang et al., 2001). Another LRR-RLK, BAK1, may form a receptor complex with BRI1 on the plasma membrane (Li et al., 2002; Nam and Li, 2002). BIN2, a negative regulator of BR signaling, is a glycogen synthase kinase-3 (Li and Nam, 2002; Pérez-Pérez et al., 2002). BIN2, together with BRI1 and BAK1, may constitute a phosphorylation cascade. The presumed constitutive activation of BR signaling in cdg1-D may thus result from interference of the phosphorylation cascade by the overexpressed CDG1 kinase.
Although cdg1-D seedlings look like BL-treated wild-type seedlings, mature cdg1-D plants are dwarf. In contrast, transgenic plants overexpressing BRI1-green fluorescent protein fusion protein (Wang et al., 2001) and wild-type plants grown with exogenously added BR (Arteca and Arteca, 2001) appear larger than the wild type because of their longer petioles, which is probably caused by activation of BR signaling. On the other hand, the mRNA level of CPD and BAS1 was decreased and increased, respectively, even in mature cdg1-D plants (Fig. 7b). Thus, the presumed interference of BR signaling by the overexpressed CDG1 may occur in mature plants of cdg1-D, although they are dwarf. A few overexpression lines of CDG1 kinase ORF showed weaker phenotype than cdg1-D and were not dwarf. In this case, however, their leaf petioles were not as elongated as those of the BRI1-overexpressing plants. At present, therefore, it is unknown what causes dwarfism in mature cdg1-D. There may exist another mechanism of dwarfism distinct from BR signaling that is also activated by CDG1.
CDG1 Is a Member of a Poorly Understood Subfamily of Receptor-Like Protein Kinases
Based on its amino acid sequence similarity, GDG1 was judged to encode a Ser/Thr kinase of the RLK subfamily. CDG1 kinase is closely related to PBS1 of Arabidopsis (Swiderski and Innes, 2001), with 66.8% identity within the kinase domain (Fig. 4b), and is also similar to several Arabidopsis protein kinases, NAK (Moran and Walker, 1993), ARSK1 (Hwang and Goodman, 1995), APK1 (Hirayama and Oka, 1992), and APK2 (Ito et al., 1997). It is also similar to Esi47 of wild wheatgrass (Lophophyrum elongatum; Shen et al., 2001). All of these proteins belong to NAK subfamily of plant protein kinases (Hardie, 1999). Like the product of CDG1, PBS1 and all the members of the NAK subfamily have a central kinase domain flanked by short nonkinase domains on both sides.
Based on the kinase domain phylogeny, Shiu and Bleeker (2001) have recently defined 44 subfamilies of the 610 RLKs in Arabidopsis that have a monophyletic origin. They include 11 subfamilies, which are named receptor-like cytoplasmic kinases (RLCKs), with no apparent signal sequence or transmembrane domain. CDG1 and PBS1 as well as members of the NAK subfamily are classified into the RLCKVII subfamily, which consists of 47 members. The subfamily forms a monophyletic group separated from the other RLCKs. Though CDG1 is most closely related to At5g02800 of Arabidopsis with 67.2% amino acid identity within the kinase domains, At5g02800 shows higher similarity to PBS1 and At3g20530 with 81.5% and 73.5% identities, respectively. In addition, the highest similarity between CDG1 and the other Arabidopsis RLCKVII genes is just as high as that observed between a monocot RLCKVII subfamily gene, Esi47, and Arabidopsis At3g09830 (69.3% identity). These observations indicate that CDG1 is a rather isolated gene in the RLCKVII subfamily of Arabidopsis.
Though several genes of the subfamily have been described in the literature, physiological significance is well characterized for only one of them, PBS1. Limited information is available on three others: ARSK1 may be involved in osmotic response of root (Hwang and Goodman, 1995), APK2a may be involved in flower development (Ito et al., 1997), and Esi47 of wild wheatgrass may play a role in abscisic acid or GA signaling of root (Shen et al., 2001). PBS1, one of the most closely related genes to CDG1 in the RLCKVII subfamily, is necessary for recognition of a bacterial pathogen, Pseudomonas syringae pv phaseolicola, by Arabidopsis (Swiderski and Innes, 2001). Pathogens secrete products of the avirulence (avr) genes into host plant cells. Specific recognition of the Avr protein of P. syringae pv phaseolicola, AvrPphB, requires at least two resistance genes in Arabidopsis, PBS1 and RPS5. Recently AvrPphB has been shown to be a Cys protease (Shao et al., 2002). It is proposed that RPS5, which is a nucleotide binding site-LRR protein, recognizes pathogens by binding to a peptide released from proteolysis of PBS1 kinase by the AvrPphB protease (Schneider, 2002; Shao et al., 2002). According to this model the kinase activity of PBS1 may not be involved in recognition of pathogens.
CDG1 Expresses Ubiquitously at a Low Level in Wild-Type Plants
The CDG1 transcript, in addition to a major ORF for protein kinase, contains two uORFs in the 5′ leader sequence (Fig. 4a). uORFs occur in about 7% to 10% of plant genes and may have a role in reducing the translation efficiency of the downstream major ORFs (for review, see Futterer and Hohn, 1996; Gallie, 1996; Geballe and Sachs, 2000). A uORF positioned upstream of the Lc gene of maize (Zea mays) reduces gene expression about 30-fold (Damiani and Wessler, 1993). A missense mutation that causes premature termination of uORF of Myb-like transcription factor, ATR1, results in a dominant mutant of Arabidopsis, in which ATR1 expression is increased 2.5-fold (Bender and Fink, 1998). Interestingly, the 5′ leader of Esi47 of wild wheatgrass also contains a 17-codon, small uORF, and the DNA sequence around the ATG start codon of this uORF has been shown to mediate the repression of basal-level expression of Esi47 in maize callus (Shen et al., 2001). It is suggested that such repression may be relieved by abscisic acid. At3g09830, which is the gene most closely related to Esi47 in Arabidopsis, also contains a uORF in the 5′ leader sequence (Shen et al., 2001). In contrast, PBS1 does not include any uORFs.
The presence of two uORFs in CDG1 gene raises the possibility that its expression is also regulated translationally. Although an RNA gel-blot analysis revealed more than a 100-fold accumulation of CDG1 gene in cdg1-D plants (Fig. 5a), CDG1 expression may be modified by translational repression in cdg1-D plants because transcripts of CDG1 contain intact uORFs in cdg1-D (Fig. 4a). On the other hand, overexpressors of CDG1 created in this study by the use of 35S promoter, contained only the kinase ORF of CDG1 mRNA without uORFs but still had the cdg1-D phenotype (Fig. 6). Thus, it is apparent that the uORFs do not contribute to abnormal growth in transgenic plants, and this is probably also the case in cdg1-D. In wild-type plants, however, the CDG1 mRNA level is quite low, and translation of the downstream ORF may be repressed by uORF. This suggests that activity of CDG1 is tightly repressed by both transcriptional and translational control in the wild type. This is probably the reason why CDG1 overexpression causes severe neomorphological abnormalities in cdg1-D mutants.
To better understand the function of CDG1 gene in BR signaling, we have isolated loss-of-function mutants of CDG1. However, we could not detect any alterations of phenotype, including responses to BR, indicating that CDG1 is not an essential factor for BR in Arabidopsis. Redundancy of RLCKVII kinases in Arabidopsis might explain no aberrant phenotype of loss-of-function mutants of CDG1, though it is rather an isolated gene in the subfamily as described above. cdg1-D, at least, could be a useful tool for dissecting cellular processes involved in differential and elongation growth caused by BR.
MATERIALS AND METHODS
Plant Materials
Transformation of Arabidopsis ecotype Columbia with activation-tagging vector, pPCVICEn4HPT (Hayashi et al., 1992), was performed using vacuum infiltration methods (Bechtold and Pelletier, 1998). T2 seeds of each hygromycin-resistant plant were used for mutant screening.
Seeds were surface sterilized with 1.5% (v/v) sodium hypochlorite and 0.02% Triton X-100 for 5 min with vigorous shaking, washed several times with sterile water, chilled in water at 4°C for 2 to 4 d, and plated onto half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with half-strength Gamborg B5 vitamins (Gamborg et al., 1968), 1% Suc, and 1% agar (Wako, Osaka). The plates were illuminated with continuous white light for 24 h at 23°C to induce germination. In some experiments, plants were grown on a 1:1 (v/v) mixture of vermiculite and Metromix 350 (Scotts-Sierra Horticultural Products, Marysville, OH) in a pot and grown at 23°C under continuous light.
DNA Preparation and Plasmid Rescue
Plants that had been stored frozen were ground with mortar and pestle with liquid N2. Ten milliliters of cetyltrimethylammonium bromide buffer that consisted of 140 mm sorbitol, 220 mm Tris-HCl, pH 8.0, 22 mm EDTA, 800 mm NaCl, 1% sarkosyl, and 0.8% hexadecyltrimethylammonium bromide were added to the powdered tissue. The homogenate was heated at 65°C for 15 min, and the same volume of chloroform was added. The mixture was mixed gently by a rotator for 15 min and centrifuged at 3,000 rpm for 10 min. The upper aqueous phase was taken, and the same volume of chloroform was added to it again. The sample was mixed and centrifuged as described above. After centrifugation, DNA was recovered by ethanol precipitation. Then DNA was dissolved in 10 mm Tris-HCl, pH 7.6, containing 1 mm EDTA (Tris-EDTA) buffer, and centrifuged at 90,000 rpm for 16 h in 1 mg/mL CsCl (CS100GX; Hitachi, Tokyo). After centrifugation, DNA was recovered and purified and finally dissolved in Tris-EDTA buffer.
For plasmid rescue, 2 μg of DNA were digested overnight with EcoRI (Takara, Kusatsu, Japan). After phenol-chloroform extraction, digested DNA was ligated at 16°C overnight with T4 DNA ligase (Takara), precipitated with ethanol, and was used for transformation of Escherichia coli DH5α competent cells (Toyobo, Tokyo) by heat shock at 42°C. Plasmid DNA extracted from ampicillin-resistant clones was sequenced using a Thermo Sequencing Primer Cycle sequencing kit 7-deaza dGTP (Amersham, Buckinghamshire, UK) with the primer 5′-TGGACGTGAATGTAGACACGTCGA-3′, which was designed from the nucleotide sequence of the T-DNA left border. Denaturing gel electrophoresis was run by a DNA sequencer (4000 L; LI-COR, Lincoln, NE).
RT-PCR
RT-PCR was performed with the Access RT-PCR system (Promega, Madison, WI), according to the manufacturer's directions. The primers for amplification of CDG1 cDNA were 5′-TTTACTTCACTTTCATGGAGTCGGAGT-3′ and 5′-ATGGTTAGTTGCTTGTGTTTTCGT-3′. For amplification of CPD, BAS1, and 18S ribosomal RNA, primers were designed according to Shimada et al. (2003). The numbers of amplification cycles were optimized to be quantitative in each gene. For CPD and BAS1, PCR was carried out between 25 and 29 cycles, and PCR with 27 and 29 cycles was found optimal, respectively. We conducted PCR for CDG1 with 20 and 25 cycles, and found the latter optimal. For 18S rRNA, we compared PCR with 13 and 15 cycles, and found the former condition optimal. RT-PCR products were detected by staining with ethidium bromide after electrophoresis or DNA gel-blot analysis using 32P-labeled probes. Each experiment was repeated independently three times.
Hormone Treatment
Seeds were imbibed in water in the dark at 4°C for 4 d, surface sterilized as described above, incubated for 1 d at 23°C under continuous white light in liquid medium that contained half-strength Murashige and Skoog salt, half-strength B5 vitamins, and 1% Suc, transferred onto 1% agar medium that consisted of the above nutrients supplemented with various growth regulators, and grown at 23°C under continuous white light thereafter. Hypocotyl length was measured 1 week after the transfer.
Transgenic Plants
RT-PCR products of the CDG1 kinase ORF were cloned into a binary vector pBI121 (CLONTECH Laboratories, Palo Alto, CA) under the control of the CaMV 35S promoter in the sense orientation. Then, the plasmid was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Arabidopsis wild-type plants, ecotype Columbia, were transformed with Agrobacterium by the floral dip method (Clough and Bent, 1998). T1 plants were selected by kanamycin resistance at 20 μg/mL.
Isolation of Suppressors
About 500 mg of seeds were harvested from heterozygous cdg1-D plants, treated with 0.1% ethyl methanesulfonate for 12 h at room temperature, extensively washed with water, vernalized at 4°C for 2 d, and sown in the soil. Plants showing the wild-type phenotype were discarded from the M1 population. The M1 plants that showed heterozygous or homozygous cdg1-D phenotype were self-pollinated, and M2 seeds were collected in five pools. About 10,000 M2 seeds from each pool were plated onto agar medium containing 40 μg/mL hygromycin and grown under white light for 2 weeks. Plants that showed suppression of cdg1-D phenotype were selected and grown in the soil.
Microscopic Analysis
Light microscopic observations were carried out according to the method of Kurata and Yamamoto (1998). Plants were fixed overnight in formaldehyde-acetic acid and dehydrated in a graded-ethanol series at room temperature. Completely dehydrated samples were embedded in Technovit 7100 (Kulzer, Wehrheim, Germany) as described by Tsukaya et al. (1993). Sections of 5-μm thickness were cut with Histoknives (Kulzer) on a microtome (RM2135; Leica, Nubloh, Germany), affixed to glass slides, and stained with 0.1% (w/v) toluidine blue at room temperature for 1 min. Specimens were examined with a microscope (Axioplan; Zeiss, Jena, Germany), and their images were recorded by a digital camera (DXM1200; Nikon, Tokyo). Intact seedlings were examined with a dissection microscope (Stemi 2000-C; Zeiss) after immersion in India ink.
Sequence data from this article have been deposited with the DDBJ/EMBL/GenBank data libraries under accession number AB099698.
Acknowledgments
We are grateful to Ms. N. Nishizaki for her technical assistance. This study was partly carried out in the Laboratory of Genetic Research, Center for Advanced Science and Technology, Hokkaido University.
This work was supported in part by a Grant-in-Aid for Scientific Research (B) (to K.T.Y. and N.Y.), and by Grants-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, and from the National Space Development Agency of Japan and the Japan Space Forum (Ground Research for Space Utilization; to K.T.Y.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046805.
References
- Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in Plant Biology, Ed 2. Academic Press, New York
- Arteca JM, Arteca RN (2001) Brassinosteroid-induced exaggerated growth in hydroponically grown Arabidopsis plants. Physiol Plant 112: 104–112 [DOI] [PubMed] [Google Scholar]
- Asami T, Yoshida S (1999) Brassinosteroid biosynthesis inhibitors. Trends Plant Sci 4: 348–353 [DOI] [PubMed] [Google Scholar]
- Bancoş S, Nomura T, Sato T, Molnar G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR (1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95: 5655–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Damiani RD Jr, Wessler SR (1993) An upstream open reading frame represses expression of Lc, a member of the R/B family of maize transcriptional activators. Proc Natl Acad Sci USA 90: 8244–8248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Futterer J, Hohn T (1996) Translation in plants: rules and exceptions. Plant Mol Biol 32: 159–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR (1996) Translational control of cellular and viral mRNAs. Plant Mol Biol 32: 145–158 [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Geballe AP, Sachs MS (2000) Translational control by upstream open reading frames. In N Sonenberg, JWB Hershey, MB Mathews, eds, Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 595–614
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM (1988) The protein kinase family: conserved features and deduced phylogeny of catalytic domains. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- Hardie DG (1999) Plant protein serine/threonine kinases: classification and functions. Annu Rev Plant Physiol Plant Mol Biol 50: 97–131 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Czaja I, Lubenow H, Schell J, Walden R (1992) Activation of a plant gene by T-DNA tagging: auxin-independent growth in vitro. Science 258: 1350–1353 [DOI] [PubMed] [Google Scholar]
- Hirayama T, Oka A (1992) Novel protein kinase of Arabidopsis thaliana (APK1) that phosphorylates tyrosine, serine and threonine. Plant Mol Biol 20: 653–662 [DOI] [PubMed] [Google Scholar]
- Howell SH (1998) Molecular Genetics of Plant Development. Cambridge University Press, Cambridge, UK
- Hwang I, Goodman HM (1995) An Arabidopsis thaliana root specific kinase homolog is induced by dehydration, ABA, and NaCl. Plant J 8: 37–43 [DOI] [PubMed] [Google Scholar]
- Iino M (2001) Phototropism in higher plants. In DP Hader, M Lebert, eds, Photomovement. Elsevier Science, Amsterdam, pp 657–810
- Ito T, Takahashi N, Shimura Y, Okada K (1997) A serine/threonine protein kinase gene isolated by an in vivo binding procedure using Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol 38: 248–258 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (1996) CKI1, a histidin kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985 [DOI] [PubMed] [Google Scholar]
- Kurata T, Yamamoto KT (1998) petit1, a conditional growth mutant of Arabidopsis defective in sucrose-dependent elongation growth. Plant Physiol 118: 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnár G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593–602 [DOI] [PubMed] [Google Scholar]
- Min YK, Asami T, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (1999) New lead compounds for brassinosteroid biosynthesis inhibitors. Bioorg Med Chem Lett 9: 425–430 [DOI] [PubMed] [Google Scholar]
- Moran TV, Walker JC (1993) Molecular cloning of two novel protein kinase genes from Arabidopsis thaliana. Biochim Biophys Acta 1216: 9–14 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assay with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Müssig C, Fisher S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25: 213–221 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a precptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubaki M, Honda T, Takatsuto S, Yoshida S, et al (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck SC, Pawlowski K, Kende H (1998) Asymmetric responsiveness to ethylene mediates cell elongation in the apical hook of peas. Plant Cell 10: 713–720 [Google Scholar]
- Pérez-Pérez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242: 161–173 [DOI] [PubMed] [Google Scholar]
- Raz V, Ecker JR (1999) Regulation of differential growth in the apical hook of Arabidopsis. Development 126: 3660–3668 [DOI] [PubMed] [Google Scholar]
- Schlagnhaufer CD, Arteca RN (1985) Brassinosteroid-induced epinasty in tomato plants. Plant Physiol 78: 300–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DS (2002) Plant immunity and film noir: what gumshoe detectives can teach us about plant-pathogen interactions. Cell 109: 537–540 [DOI] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE (2002) A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109: 575–588 [DOI] [PubMed] [Google Scholar]
- Shen W, Gomez-Cadenas A, Routly EL, Ho THD, Simmonds JA, Gulick PJ (2001) The salt stress-inducible protein kinase gene, Esi47, from the salt-tolerant wheatgrass Lophopyrum elongatum is involved in plant hormone signaling. Plant Physiol 125: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleeker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinase. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski MR, Innes RW (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J 26: 101–112 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and deetiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS, Chory J (2002) Steroid signaling in plants and insects: common themes, different pathways. Genes Dev 16: 3113–3129 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Naito S, Redei GP, Komeda Y (1993) A new class of mutation in Arabidopsis thaliana, acaulis1, affecting the development of both inflorescences and leaves. Development 118: 751–764 [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM (2003) CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol 133: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]