Abstract
We have identified a T-DNA insertion mutation of Arabidopsis (ecotype C24), named sto1 (salt tolerant), that results in enhanced germination on both ionic (NaCl) and nonionic (sorbitol) hyperosmotic media. sto1 plants were more tolerant in vitro than wild type to Na+ and K+ both for germination and subsequent growth but were hypersensitive to Li+. Postgermination growth of the sto1 plants on sorbitol was not improved. Analysis of the amino acid sequence revealed that STO1 encodes a 9-cis-epoxicarotenoid dioxygenase (similar to 9-cis-epoxicarotenoid dioxygenase GB:AAF26356 [Phaseolus vulgaris] and to NCED3 GB:AB020817 [Arabidopsis]), a key enzyme in the abscisic acid (ABA) biosynthetic pathway. STO1 transcript abundance was substantially reduced in mutant plants. Mutant sto1 plants were unable to accumulate ABA following a hyperosmotic stress, although their basal ABA level was only moderately altered. Either complementation of the sto1 with the native gene from the wild-type genome or supplementation of ABA to the growth medium restored the wild-type phenotype. Improved growth of sto1 mutant plants on NaCl, but not sorbitol, medium was associated with a reduction in both NaCl-induced expression of the ICK1 gene and ethylene accumulation. Osmotic adjustment of sto1 plants was substantially reduced compared to wild-type plants under conditions where sto1 plants grew faster. The sto1 mutation has revealed that reduced ABA can lead to more rapid growth during hyperionic stress by a signal pathway that apparently is at least partially independent of signals that mediate nonionic osmotic responses.
An important aspect of the adaptive responses of plants to osmotic stresses is that it includes both abscisic acid (ABA)-dependent and -independent mechanisms, as reported by Grillo et al. (1995) and in further studies (Ishitani et al., 1997; Bray, 2002a). Many studies involving increased tissue ABA levels by exogenous application, and reduced ABA levels by use of pharmacological agents or ABA-deficient mutants, have clearly established the importance of the role of ABA in response to osmotic stresses (Himmelbach et al., 1998). Understanding the manner by which ABA levels increase in response to osmotic stress has been greatly assisted by knowledge of the biosynthetic pathway for ABA and the use of various mutants impaired in specific steps in the pathway (Giraudat, 1995; Giraudat and Schroeder, 2001).
In plants, ABA biosynthesis occurs mainly via an indirect pathway from the C40 carotenoid (Rock and Zeevaart, 1991; Schwartz et al., 1997a; Iuchi et al., 2000). Genes encoding three important enzymes involved in ABA biosynthesis have been identified and cloned. The first encodes zeaxanthin epoxidase (ZEP), which converts zeaxanthin to epoxycarotenoid and is defective in the Arabidopsis mutant aba1 (Marin et al., 1996). The cleavage enzyme 9-cis-epoxycarotenoid dioxygenase (NCED) catalyzes the conversion of epoxycarotenoids to xanthoxin and is homologous to the VP14 protein from maize (Zea mays; Marin et al., 1996; Schwartz et al., 1997b). Abscisic acid aldehyde oxidase (AAO) carries out the final enzymatic step converting ABA aldehyde to ABA and is defective in the Arabidopsis aao3 mutant (Seo et al., 2000). This final step also requires a molybdenum-containing cofactor (MoCo). MoCo requires sulfuration for activation that is catalyzed by a MoCo sulfurase, which is encoded by the ABA3/LOS5 locus in Arabidopsis (Xiong et al., 2001). Expression of all four of these genes, ZEP, NCED, AAO3, and ABA3/LOS5, is increased by osmotic stress (Xiong et al., 2001). Furthermore, mutations in all four of the genes encoding these proteins result in plants with impaired stomatal function. All exhibit wilty phenotypes compared to wild-type plants when challenged with osmotic stress.
In a screen of T-DNA insertion-tagged Arabidopsis lines for mutants with increased ability to germinate and grow in the presence of 145 mm NaCl, we identified a particularly fast-growing mutant. Cosegregation with the insertion marker, identification of the disrupted locus by thermal asymmetric interlaced (TAIL)-PCR, and confirmation by genetic complementation identified the responsible gene to be NCED3 (Iuchi et al., 2000). ABA measurements confirmed the inability of this mutant to accumulate ABA during osmotic stress. The nced3 mutant also exhibited increased water loss and sensitivity to osmotic challenge, as reported for other nced3 mutants (Iuchi et al., 2001; Tan et al., 2003). However, under conditions of saturated humidity, where stomatal function is minimally important, nced3 mutants displayed unexpected characteristics of increased growth during conditions of salt-induced osmotic stress. Furthermore, this enhanced growth was not correlated with increased osmotic adjustment that would be expected for growth resumption after osmotic challenge (Morgan, 1984). Complementation of the mutant with a wild-type NCED3 gene or treatment with ABA resulted in normal growth inhibition by NaCl.
The nced3 mutation has been reported to affect stomatal function by increasing the rate of water loss from plants in drying soil. Changes in transpiration rate of these mutants have been typically measured as bulk loss of water over time or as static time points using various porometer/photosynthesis apparatus (Iuchi et al., 2001). By examining dynamic water flux rates during complete diurnal cycles, we have found that gradual soil desiccation imposes increasingly narrower limits on diurnal cycles of water flux out of the plant. The nced3 mutation prevents the restriction of such limits during gradual soil desiccation.
RESULTS
Identification of the sto1 Mutant from a T-DNA Mutagenized Population of Arabidopsis
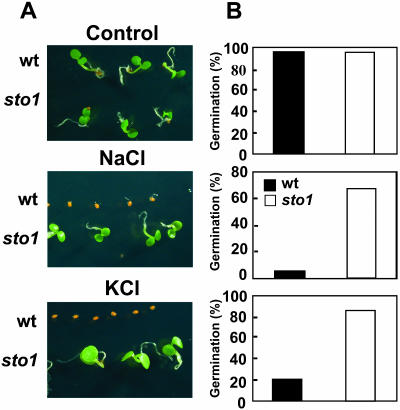
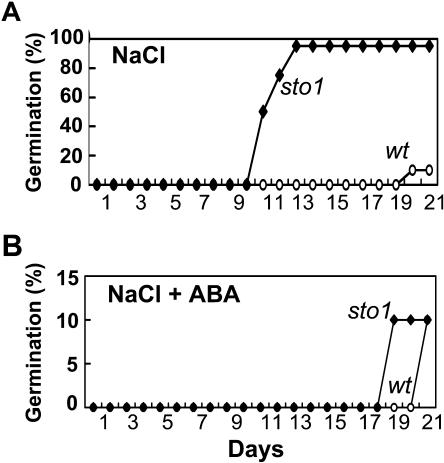
A population of over 300,000 T-DNA-tagged Arabidopsis (ecotype C24) mutants was generated as described in Koiwa et al. (2002). Approximately 11,000 T2 lines were screened based on their ability to germinate and grow on Murashige and Skoog (MS) media containing 145 mm NaCl. Two stable genetic mutants able to germinate and grow faster than wild type were isolated and confirmed. One of these two mutants, designated sto1 (for salt tolerant), was selected for further characterization (Fig. 1). Germination assays on MS media plus or minus salt revealed a normal phenotype of the sto1 seeds in the absence of salt and enhanced germination in the presence of both KCl and NaCl (Fig. 1A). Fourteen days after sowing, 80% and 60% of sto1 seeds germinated on 160 mm KCl- and NaCl-containing media, respectively, whereas wild-type seeds showed 95% (NaCl) or 80% (KCl) inhibition compared to germination on MS control media (Fig. 1B). Within the first 7 d after germination, sto1 seedlings developed normal cotyledons and true leaves. Experiments both in vitro (Fig. 2A) and in soil under high atmospheric humidity (Fig. 2B) using increasing NaCl concentrations confirmed the enhanced ability of sto1 seeds to germinate and sto1 plants to grow faster in the presence of high salt.
Figure 1.
The sto1 mutation enhances germination on media supplemented with NaCl or KCl. A, Photographs are representative of wild-type and sto1 mutant plants. Seeds were placed on MS medium or MS medium supplemented with 160 mm NaCl or KCl and allowed to germinate and grow for 14 d. B, Percentage of germinated seeds after 14 d (significant difference at 99% confidence level). Germination was assessed on 60 seeds of wild-type or sto1 mutant plants distributed in three replica plates per each treatment (20 seeds per each genotype).
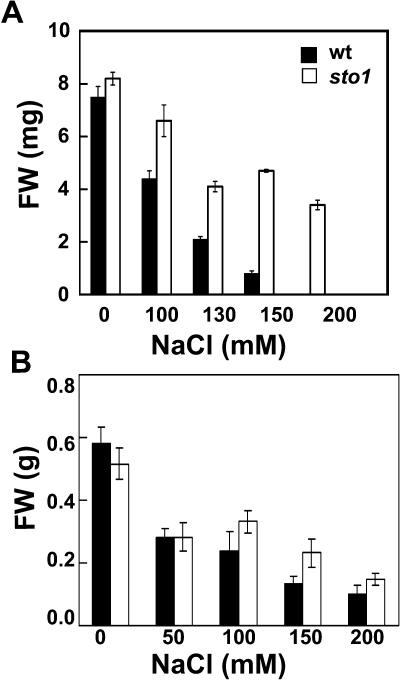
Figure 2.
Growth response of wild-type and sto1 seedlings and plants at increasing NaCl concentrations. A, Seeds were placed in petri plates on MS medium or MS medium supplemented with increasing concentrations of NaCl and allowed to germinate and grow. Plant fresh weight was measured after 21 d. Values are means of 60 plants ± se. B, Seeds were germinated and grown in soil at saturated atmospheric humidity (see “Materials and Methods” for details) and irrigated every day with saline water with different NaCl concentrations. After 28 d, shoot fresh weight was measured. Values are means of 20 plants ± se.
Ionic Specificity of the Enhanced Growth Response of sto1 Plants
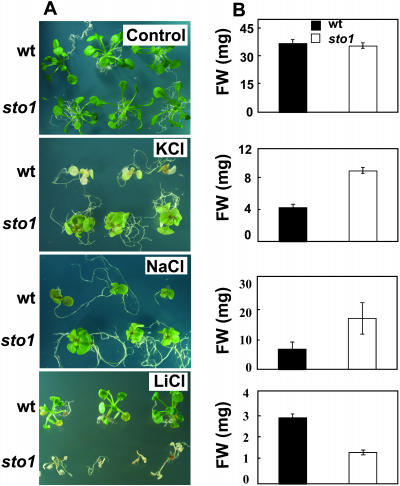
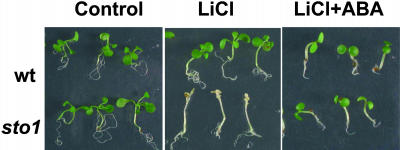
Because germination and growth in the presence of NaCl may involve distinct physiological adaptive responses to stress, sto1 seedlings were allowed first to germinate on nonsaline media and then to be transferred (at the stage of fully expanded cotyledons) to hyperosmotic media to examine their growth response independent of germination. In the presence of both elevated KCl and NaCl, sto1 plantlets were able to withstand the abrupt hyperosmotic stress and grow until flowering (data not shown), whereas wild-type seedlings became chlorotic upon exposure to high salinity and never flowered (Fig. 3). However, in the presence of LiCl, a salt commonly used at low concentrations to discriminate between ionic toxicity and osmotic effects of nutrient media (Rus et al., 2001), sto1 was remarkably impaired in its normal growth (Fig. 3), indicating that the sto1 seedlings are hypersensitive to Li+ ions, possibly by the altered activity of an ion transport system specific to Li+ or to an alteration in the expression of a Li+-sensitive component such as HAL2 (Serrano and Rodriguez-Navarro, 2001). Hypersensitivity of sto1 seed germination to Li+ was also observed (data not shown).
Figure 3.
sto1 mutant plants are tolerant to KCl and NaCl and hypersensitive to LiCl stresses. A, Seeds were germinated in petri plates on standard MS medium and 7-d-old seedlings were subsequently transferred to MS medium supplemented with 160 mm NaCl, KCl, or 20 mm LiCl and allowed to grow for an additional 20 d. Photographs are representative of wild-type and sto1 mutant plants after 20 d from transferring onto saline medium. B, Plant fresh weights were measured after 20 d of growth on indicated medium. Values are means of 60 plants ± se.
Nonionic Osmotic Stress Tolerance of sto1 Plants
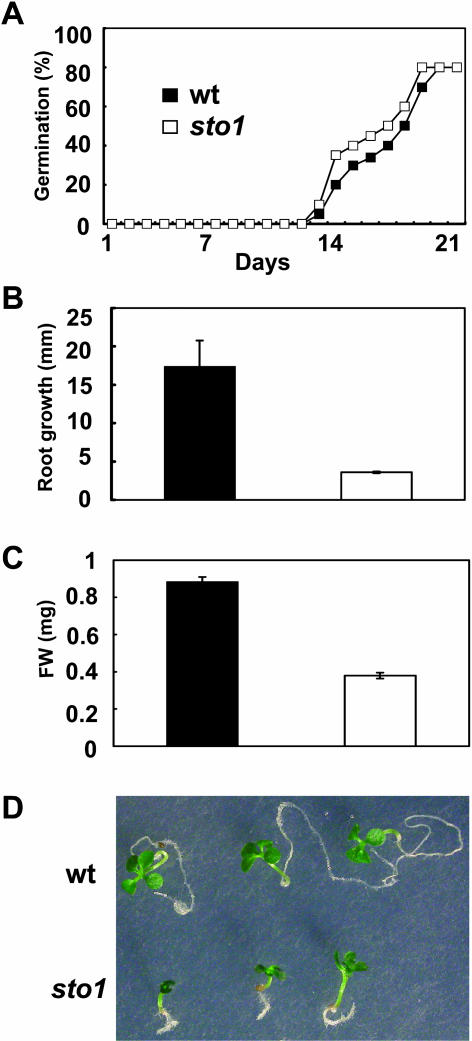
The osmotic stress tolerance of sto1 mutants was assessed upon exposure to 300 mm sorbitol (Fig. 4). Seeds of sto1 plants were able to germinate slightly earlier than wild type at 300 mm sorbitol (Fig. 4A). In contrast to salt treatment (NaCl and KCl), growth of sto1 plants on sorbitol medium, after germination on medium without sorbitol, was inhibited more than the wild type (Fig. 4, B–D). These results indicate that osmotic stress-induced growth inhibition is reduced in sto1 plants only in salt stress medium.
Figure 4.
The sto1/nced3 mutation inhibits growth on media supplemented with sorbitol. A, Percentage of germinated wild-type and sto1/nced3 seeds on MS medium supplemented with 300 mm sorbitol over a 21-d time period. Germination was assessed on 60 seeds of wild-type and sto1/nced3 plants distributed on three replica plates (20 seeds per each genotype). B, Root length of 14-d-old seedlings growing on sorbitol-containing medium. Values are means of 60 plants ± se. C, Fresh weight of 14-d-old seedlings growing on sorbitol-containing medium. Values are means of 60 plants ± se. D, Seeds were germinated in petri plates on standard MS medium and 3-d-old seedlings were subsequently transferred on MS medium (not shown) or MS medium supplemented with 300 mm sorbitol and allowed to grow for an additional 20 d and then photographed.
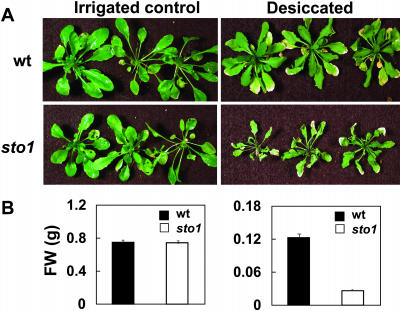
Desiccation and Salt Stress Tolerance of sto1 Plants Grown in Soil
The tolerance of sto1 plants to desiccation stress was also evaluated on soil-grown plants. Irrigation was interrupted at the stage of six to seven fully expanded leaves, and plant response to gradual dehydration of the soil was assessed visually and quantified in terms of plant fresh weight. Mutant sto1 plants were much more sensitive than wild type to soil desiccation. After 1 week from interruption of irrigation, sto1 plants were wilted and weighed approximately 30% of the wild-type plants (Fig. 5B). These results are in sharp contrast to those observed during growth of sto1 plants in petri plates with saturated humidity and exposed to ion stress (Fig. 2A). In fact, when sto1 plants are grown in soil and exposed to salt stress under high atmospheric humidity (>95%; Fig. 2B), they grow faster than wild-type plants exposed to the same stress conditions.
Figure 5.
sto1 mutant plants are sensitive to soil desiccation. A, Representative photograph of wild-type and sto1 mutant plants exposed to desiccation. Plants were grown in soil under a standard irrigation regime until four to five fully expanded leaves were formed, at which stage irrigation was stopped. After 15 d, in coincidence with the appearance of clear symptoms of leaf desiccation, plants were rewatered and left to recover for 48 h, at which time pictures were taken. B, Shoot fresh weights of desiccation-stressed wild-type and sto1 mutant plants after rewatering. Values are means of 20 plants ± se.
Genetic Analysis of sto1 Mutant Plants and Identification of the STO1 Locus
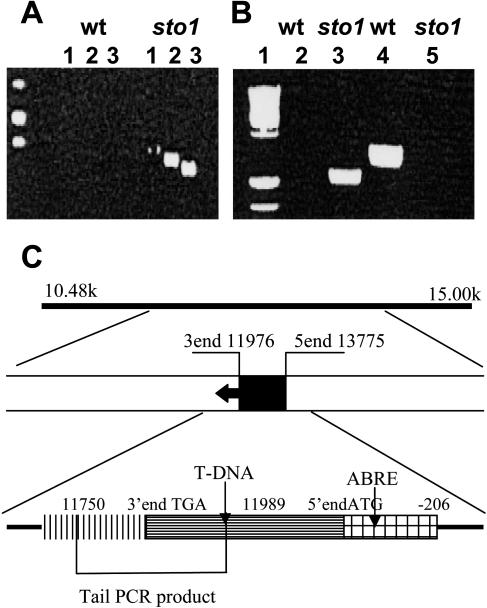
The sto1 mutants were crossed to C24 wild type, and the resulting F1 progeny all presented the wild-type salt-sensitive phenotype and were bialaphos resistant (bialaphos herbicide resistance was the selection marker of the activation-tagging vector), indicating that the mutation is recessive (Table I). F2 seedlings from selfed F1 plants revealed a segregation ratio of approximately 3:1 for NaCl sensitivity (C24)/NaCl tolerance (sto1) phenotypes (χ2 test; P > 0.05), which cosegregated with the herbicide-resistant phenotype. It was concluded that the sto1 phenotype was caused by a single recessive nuclear mutation. TAIL-PCR analysis of DNA from sto1 plants (Koiwa et al., 2002; Fig. 6A) located a single T-DNA insertion in an open reading frame of chromosome III (position 11,989 of P1 clone MOA2.4; GenBank accession no. AB028617). Examination of the predicted cDNA nucleotide sequence revealed that the T-DNA was located inside the coding region (−13 bp from the 3′ end) of a gene (NCED3) encoding a 9-cis-epoxicarotenoid dioxygenase (similar to 9-cis-epoxicarotenoid dioxygenase GB: AAF26356 [Phaseolus vulgaris]), a key enzyme in the ABA biosynthetic pathway (Fig. 6C; Table II).
Table I.
sto1 genetic analysis
| F1 | Bialophos Tolerance | F2 | Bialophos Tolerance | |
|---|---|---|---|---|
| % | % | |||
| Total seedlings | 400 | 100 | 400 | 75 |
| Salt tolerant | 0 | 0 | 98 | 100 |
| Seedlings | ||||
| Salt sensitive | 400 | 100 | 302 | 66 |
| Seedlings | ||||
| χ2 | 0.05 |
Figure 6.
PCR analysis and genome location of T-DNA insertion in sto1/nced3 mutants. A, Secondary TAIL-PCR product (lane 2) and shift of the tertiary PCR product (lane 3) in sto1. B, Correct genomic integration of the T-DNA insertion was verified by diagnostic PCR; lanes 1 (marker); 2 (DNA template, wild type; primers, T-DNA LB [3′] and STO1-specific primer [5′]); 3 (DNA template, sto1; primers, T-DNA LB [3′] and STO1-specific primer [5′]); 4 (DNA template, wild type; primers, sto1-specific primer [3′] and STO1-specific primer [5′]); 5 (DNA template, sto1; primers, STO1-specific primer [3′] and STO1-specific primer [5′]). Oligonucleotide sequences are reported in Table V. C, Physical map of the sto1 locus and insertion site of the T-DNA. Solid line represents fragment of the bacterial artificial chromosome clone MOA2.4. Black box indicates the coding region of the gene; arrow indicates the predicted transcription direction.
Table II.
STO1/AtNCED gene family
Molecular and Functional Evidence for Inactivation of the NCED3 Gene in sto1/nced3 Mutants
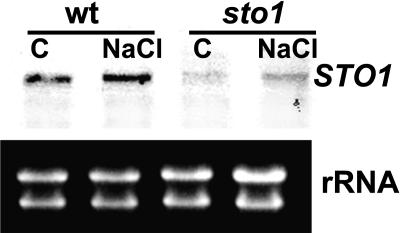
The STO1/NCED3 transcript is expressed in both leaf and root (data not shown) tissue of unstressed wild-type plants, and its level is increased moderately upon 145 mm NaCl treatment (Fig. 7), as previously reported (Iuchi et al., 2001). The STO1/NCED3 transcript abundance was substantially reduced in unstressed sto1/nced plants compared to wild-type plants, possibly due to instability of the transcript. Upon 145 mm NaCl treatment, there was an increase of the STO1/NCED3 transcript abundance in the mutant plants, but the level remained well below that observed in stressed wild-type plants (Fig. 7). Consistent with the fact that the STO1/NCED3 gene encodes a key enzyme in ABA biosynthesis, the level of this hormone was significantly affected in sto1/nced3 plants (Table III). Specifically, the ABA content was moderately lower in sto1/nced3 compared to wild-type unstressed plants, yet, more importantly, it did not accumulate as it did in wild-type plants following exposure to hyperosmotic stress (Table III). These results indicate that deficiency in the NCED3 enzyme blocked the stress-induced increase in ABA while having a much smaller effect on the ABA content of unstressed leaves.
Figure 7.
NCED3 transcript abundance is increased by NaCl treatment and is reduced in sto1/nced3 mutant plants. Ten micrograms of total RNA were isolated from 21-d-old sto1/nced3 mutant and wild-type plants that were germinated and grown on 145 mm NaCl, separated on a denaturing formaldehyde-agarose gel, and blotted onto nylon membrane. The membrane-bound RNA was hybridized with DIG-labeled DNA probe (Roche, Indianapolis). The probe was produced by PCR reaction using the primers listed in Table V.
Table III.
ABA content in wild-type and sto1 plants (ng g−1 fresh weight)a
| Treatment | WT | sto1 |
|---|---|---|
| Control | 396.37 ± 9.2 | 311.3 ± 9.4 |
| Sorbitol | 951.8 ± 4.0 | 342.0 ± 4.4 |
Values are means of 3 plants ± se.
Genetic and Pharmacological Complementation of the sto1/nced3 Mutant Phenotype Confirm That Inactivation of the STO1/NCED3 Gene Is Responsible for Enhanced Germination on Salt and Hypersensitivity to Desiccation and LiCl
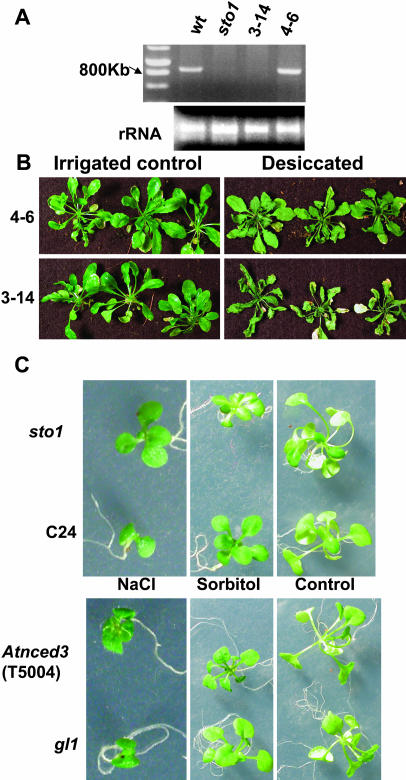
To determine whether the mutated STO1/NCED3 gene was responsible for the sto1/nced3 mutant phenotype in terms of both enhanced seed germination on hyperosmotic medium and soil desiccation tolerance, a 4.5-kb Sma1 (5′)/Kpn1 (3′) genomic fragment, including the full-length STO1/NCED3 gene and its promoter, was cloned from wild-type C24 plants and introduced via Agrobacterium transformation (pBIB vector) into sto1/nced3 mutant plants. Ten independent hygromycin-resistant T1 transformants were confirmed by reverse transcription (RT)-PCR analysis and one was selected for further study. Phenotypic and molecular evaluation of lines 4 to 6 revealed that expression of the wild-type STO1/NCED3 gene in mutant plants (Fig. 8A) eliminated their enhanced ability to germinate on salt (data not shown) and their hypersensitivity to desiccation (Fig. 8B). Pharmacological complementation of sto1/nced3 plants by addition of ABA to the medium also reverted both their enhanced germination on NaCl medium (Fig. 9) and growth sensitivity in LiCl (Fig. 10). A separate allelic mutation of the STO1/NCED3 locus was reported by Iuchi et al. (2001). Although this nced3 mutant exhibited increased sensitivity to desiccation stress, the growth of these plants during salt and osmotic stress under conditions of saturated humidity was not examined. We obtained the nced3 mutant reported by Iuchi et al. (2000, 2001) and found that it also exhibited similar faster growth than wild-type plants (gl1 Columbia) in NaCl stress but not during stress induced by nonionic osmotic stress (sorbitol; Fig. 8C). Both the nced3 (Iuchi et al., 2001) and our sto1/nced3 mutants fail to accumulate ABA to normal levels with stress, and thus the observed increased growth during NaCl treatment is apparently affected by the inability to increase ABA during NaCl treatment and does not require normal levels of ABA to be reduced substantially.
Figure 8.
Complementation with the STO1/NCED3 gene reverts the soil desiccation-sensitive phenotype of the sto1/nced3 mutant. A, STO1/NCED3 transcript abundance detected by RT-PCR in wild type, sto1/nced3, lines 3 to 14 (pBI vector control), and lines 4 to 6 (expressing STO1/NCED3). One microliter of cDNA was used as a template for the first PCR amplification (20 cycles). B, Representative photograph of lines 3 to 14 (pBI vector control) and lines 4 to 6 (pBI::STO1/NCED3). Top, sto1/nced3 plants complemented with STO1/NCED3 (lines 4–6) grown under standard irrigation regime (left) and exposed to desiccation (right) showing the reverted desiccation-sensitive phenotype; Bottom, sto1/nced3 plants complemented with vector only (lines 3–14) grown under standard irrigation regime (left) and exposed to desiccation (right). C, Responses of C24, sto1/nced3, gl1, and Atnced3 (T5004) mutant seedlings to NaCl and sorbitol treatments. Seeds were germinated and grown in petri plates on standard MS medium or supplemented with 160 mm NaCl or 300 mm sorbitol. Plant fresh weights (mg) after 21 d were as follows: MS, 36.2 ± 1.7 (C24); 37.5 ± 1.6 (sto1); 35.5 ± 1.5 (gl1); 36.5 ± 1.3 (Atnced3). NaCl, 4.3 ± 0.34 (C24); 8.5 ± 0.3 (sto1); 3.9 ± 0.32 (gl1); 7.7 ± 0.24 (A-nced3). Sorbitol, 9.35 ± 0.15 (C24); 3.92 ± 0.68 (sto1); 7.2 ± 0.08 (gl1); 5.2 ± 0.31 (Atnced3). (Values are means ± se, n = 20.)
Figure 9.
ABA treatment abolishes the enhanced germination of sto1/nced3 seeds on NaCl medium. Seeds of wild type and sto1/nced3 mutant plants were surface sterilized and placed on MS medium supplemented with 145 mm NaCl (A) or with 145 mm NaCl + 20 μm ABA (B). Germination on standard MS medium was also included (not shown). For each genotype (wild type and sto1/nced3), the number of germinated seeds (out of 60) was assessed over 21 d and expressed as a percentage.
Figure 10.
ABA reverts the LiCl sensitivity of sto1/nced3 seedlings to wild type. Wild-type and sto1 seeds were surface sterilized and germinated on standard MS medium or MS medium supplemented with 20 mm LiCl or 20 mm LiCl + 20 μm ABA. A representative photograph of 15-d-old seedlings (from three replica plates per treatment) is displayed.
Enhanced Growth of sto1/nced3 Plants on Hyperosmotic Medium Is Associated with Blockage of ABA-Mediated Growth Inhibition Independent of the Degree of Osmotic Adjustment
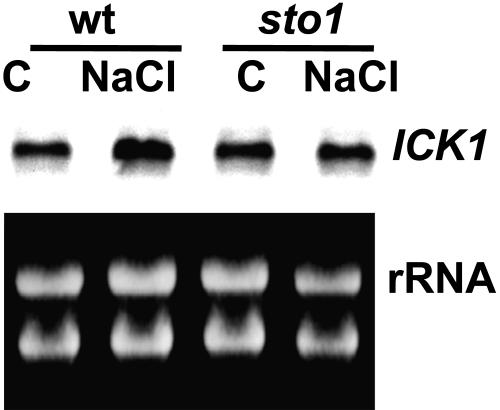
To verify whether the enhanced growth of sto1/nced3 plants was correlated with an increased accumulation of solutes, osmotic potentials in the absence or presence of stress were measured. Surprisingly, cellular saps from sto1/nced3 plants had less negative osmotic potential under stress compared to wild-type plants (Table IV). Since the growth of sto1/nced3 plants was not associated with an increased osmotic adjustment, it appears that ABA-mediated stress-induced growth reduction can be uncoupled from osmotic adjustment. It follows that insufficient osmotic adjustment is not the cause of stress-induced growth reduction. Rather, NaCl stress appears to directly affect the growth of sto1/nced3 plants through an ABA-mediated mechanism that is independent from osmotic adjustment. In fact, growth-related genes, such as cyclin kinase inhibitors (ICK1), have been reported to be activated under stress (Wang et al., 1998) and, interestingly, we also detected an increase of ICK1 transcript level in NaCl-stressed wild-type plants, which in contrast did not change in sto1/nced3 plants (Fig. 11).
Table IV.
Osmotic potential (MPa)a
| Control (MS) | NaCl | Sorbitol | |
|---|---|---|---|
| Germinationb | |||
| Wild type | −2.2 ± 0.1 | −5.3 ± 0.2 | −3.4 ± 0.2 |
| sto1 | −2.2 ± 0.15 | −1.6 ± 0.7 | −2.4 ± 0.1 |
| Growthc | |||
| Wild type | −2.3 ± 0.13 | −3.1 ± 0.8 | −3.2 ± 0.1 |
| sto1 | −2.1 ± 0.09 | −1.9 ± 0.1 | −2.2 ± 0.1 |
Values are means of three seedling preparation ± se.
Values refer to seeds germinated and grown on nonsaline (control), NaCl, or sorbitol media.
Values refer to seeds germinated on nonsaline medium and subsequently transferred and grown on nonsaline. NaCl, or sorbitol media (see “Materials and Methods” for details).
Figure 11.
Expression of the ICK1 gene in sto1/nced3 and wild-type plants. Total RNA was extracted from whole seedlings that were grown for 7 d on MS medium or MS medium supplemented with 145 mm NaCl. Twenty micrograms of total RNA were loaded in each lane. The probe was produced by PCR reaction using the primers listed in Table V. Bottom, ethidium bromide-stained total RNA gel image as a loading control.
Ethylene Treatment of Wild-Type Plants Phenocopies the sto1/nced3 Mutation
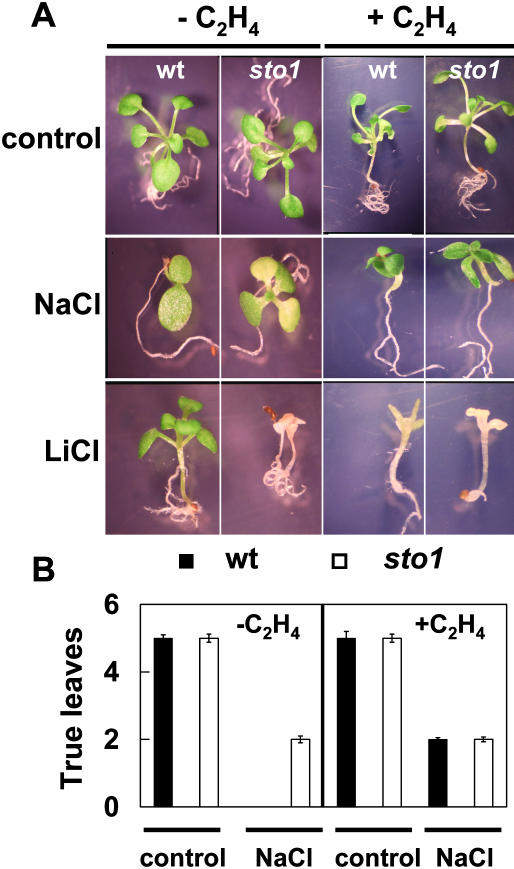
Since the restriction of ethylene production is a well-established function of ABA and, moreover, considering that ethylene also affects seed germination and growth (Beaudoin et al., 2000; Sharp and Le Noble, 2002), we examined the possibility of a link between ABA and ethylene levels in sto1/nced3 plants. Consistent with other observations (Ghassemian et al., 2000), the ethylene concentration in sto1/nced3 was slightly higher compared to the wild type (data not shown). The ethylene level was also moderately decreased in wild-type salt-stressed plants, as previously reported, yet it was unaffected in stressed sto1/nced3 mutant plants. Ethylene treatment of wild-type seeds allowed rapid germination similar to that observed with seeds of the sto1/nced3 mutant. Exogenous applications of ethylene also phenocopied the sto1/nced3 mutation by suppressing the NaCl-induced growth inhibition and LiCl hypersensitivity of wild-type plants (Fig. 12). Since the ethylene level of sto1/nced3 mutant plants was only marginally increased, the reduced ABA of this mutant may have also resulted in increased sensitivity to ethylene.
Figure 12.
Ethylene-treated wild-type plants mimic the sto1/nced3 phenotype. A, Representative photographs of 15-d-old wild-type and sto1/nced3 seedlings germinated and grown in the presence or absence of 20 ppm of C2H4 on petri plates containing standard MS medium, or MS medium supplemented with 145 mm NaCl or 20 mm LiCl. B, Number of new leaves per plant formed after 15-d treatment. Values are means of 100 plants ± se.
Effects of sto1/nced3 Mutation on Stomatal Function Do Not Override the Diurnal Effects of the Day/Night Cycle
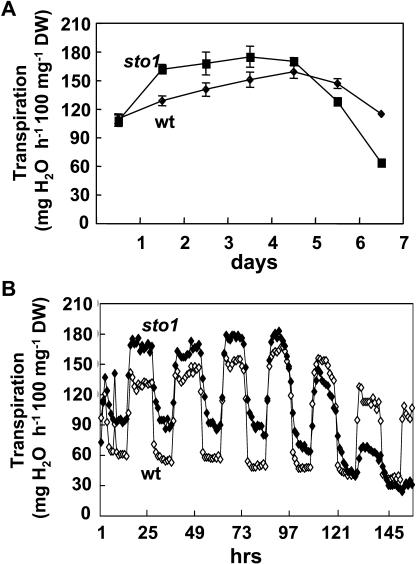
Because ABA is also directly involved in the regulation of stomatal aperture, we examined water loss characteristics of sto1/nced3 plants. Wild-type and mutant plants were grown in the soil for 2 weeks under a normal irrigation regime. At the stage of six to eight fully expanded leaves, irrigation was interrupted and plant water loss via transpiration was monitored over a 7-d time period (Fig. 13A). Mutant sto1/nced3 plants always transpired more than wild-type plants until day 5, after which they began to lose turgor and to exhibit a wilty appearance. This result is consistent with an impaired stomatal response to dehydration and to the previously observed desiccation sensitivity of plants with decreased NCED3 expression (Fig. 5; Iuchi et al., 2001). Because we found that the osmotic stress-induced ABA level was reduced in sto1/nced3 mutant plants, stomatal malfunction was expected to occur also during mid-day water stress. Measurement of daily fluctuations of transpiration fluxes over a 7-d time period confirmed higher transpiration of sto1/nced3 plants during the midday photoperiod. However, to our surprise, our results also revealed higher transpiration fluxes of these plants during night hours (Fig. 13B). The upper and lower limits of stomatal aperture, but not the amplitude (max-min) of the daily transpiration flux, changed in the mutant plants (Fig. 13B). We cannot fully explain the reduced night stomatal closure of sto1/nced3 plants. However, this may be caused by a residual effect of an altered diurnal activity of NCED3 enzyme (Wilkinson and Davies, 2002) affecting ABA levels that subsequently impair stomatal closure at night. Changes in guard cell ABA levels may result from a complex interaction of ABA synthesis, catabolism storage, and transport (Wilkinson and Davies, 2002). Nevertheless, the amount of ABA accumulation (Table III) in the leaves that is controlled by NCED3 appears not to be able to override the diurnal cycle behavior of stomatal aperture, which is apparently determined by the composite effects of several simultaneous signals (Shinozaki and Yamaguchi-Shinozaki, 1996; Giraudat and Schroeder, 2001; Bray, 2002a; Zhu, 2002).
Figure 13.
Transpiration of wild-type and sto1/nced3 mutant plants. A, Transpiration of wild-type and sto1/nced3 mutant plants was measured as daily water loss over a 7-d time interval. Each value represents the average daily water loss of three plants ± se. B, Daily fluctuations of rate of water loss in wild-type and sto1/nced3 mutant plants. The diurnal patterns displayed are representative of three independent experiments.
DISCUSSION
Complementation of the altered growth and desiccation tolerance phenotypes of sto1/nced3 plants by transformation with a wild-type NCED3 gene confirms the identity of the locus responsible for the phenotypic syndrome of the sto1/nced3 mutation. Also, application of exogenous ABA to sto1/nced3 plants is able to induce a phenotype that mimics wild type, and a second mutant allele of nced3, reported by Iuchi et al. (2001), exhibited a similar stress response phenotype.
Because abiotic stress-induced increase in ABA level has been implicated in the mediation of many stress responses (Bray, 2002b), ablation of the ABA response in plants with a mutated nced3 gene should lead to many changes in gene expression. At this time, however, only a few have been examined. Antisense expression of the NCED3 gene leads to suppression of desiccation induction of the RD29B gene, but not the RAB18 or KIN1 genes (Iuchi et al., 2001). We have found that in sto1/nced3 plants the transcript of the ABA-induced ICK1 gene also fails to accumulate as it does in wild-type plants. The effects of the nced3 mutation on other gene expression and physiological responses may depend on the specific effects of this mutation on tissue and cell-specific perturbations in ABA content (Wilkinson and Davies, 2002). It has recently been reported that the NCED3 gene is expressed in both leaf and root tissues (Tan et al., 2003) and, as shown here and by Iuchi et al. (2001), can affect both constitutive and stress-induced ABA levels in shoot tissues, implying a complex role for NCED3 genes in the regulation of tissue ABA levels.
Disruption of the NCED3 gene has revealed that in Arabidopsis this leads to a syndrome of phenotypic changes that indicates that there may be a number of ABA-associated responses to stress that are not always necessarily coupled in previously assumed ways. For instance, it has been generally believed that when plants are exposed to decreased water potential, cell growth ceases or slows due to the inadequate solute potential needed to compensate for the water potential imbalance with the environment (Morgan, 1984; Bressan et al., 1990). The resulting low-tissue water potential and turgor loss leads to increased ABA or some other signal response, which in turn affects osmotic adjustment that compensates for the reduced turgor, allowing resumption of water uptake and regrowth (Bressan et al., 1990; Wilkinson and Davies, 2002). Indeed, many studies have explored this relationship between osmotic adjustment and growth of plants during desiccation or other osmotic stresses (Sharp et al., 1994). In fact, even with plants grown in the field, osmotic adjustment has been positively correlated with greater growth and productivity in a water-limited environment (Morgan, 1984). Certainly an important role for ABA in coordinating growth and osmotic adjustment responses to environments with decreased availability of water has been well established (Sharp et al., 1994; Giraudat, 1995). However, the primary effects of ABA in this coordination are complex and never have been clearly defined. Some general principles, such as increased ABA accumulation in plants resulting from osmotic stress often facilitates root growth and restricts shoot growth, have emerged from the observations of many studies using hormone and other pharmacological treatments (Spollen et al., 2000). Surprisingly few studies have attempted to clarify this issue using recently available ABA biosynthesis mutants that are available in Arabidopsis and other species (Sharp et al., 2000; Sharp, 2002; Sharp and LeNoble, 2002).
Identification of the sto1/nced3 mutant and the nced3 mutant reported by Iuchi et al. (2001) as mutants that exhibit enhanced germination and increased root and shoot growth at low water potentials (Figs. 1–3) indicates that ABA increase during osmotic stress in wild-type plants reduces growth. In fact, not only do roots and shoots of sto1/nced3 mutant seedlings grow faster than wild-type plants at low water potentials caused by salinized medium, but also they have significantly more positive osmotic potentials (less osmotic adjustment). These results indicate that the effect of ABA on growth can be direct and not always dependent on ABA effects on osmotic adjustment. In fact, the greater osmotic adjustment in wild-type plants may largely be facilitated by ABA-mediated reduction of growth rate, allowing a higher concentration of solutes without increased solute production. Nuccio et al. (1999) calculated that reduced cell growth rate after adaptation to salt may account for 60% of the observed accumulation of the solute Pro. Even though results indicate that, in response to osmotic stress, ABA-mediated signaling affecting osmotic adjustment is at least partially uncoupled from growth, the mechanism of uncoupling remains unclear. Cell growth rate is dependent on and thus appears to be tightly linked to osmotic adjustment (Bressan et al., 1990), perhaps as in yeast (Saccharomyces cerevisiae), where the same receptor protein mediates both cell growth and osmotic adjustment pathways (Maggio et al., 2002a, 2002b).
Although abi5 mutant seedlings were also reported to have exhibited postgermination osmosensitivity, only survival and not growth was assessed in that study (Lopez-Molina et al., 2001). The more rapid germination of sto1/nced3 seeds exposed to osmotic stress is also consistent with a recent report describing a similar germination phenotype of aba2 mutants of Arabidopsis that also fail to increase ABA levels after osmotic stress (Gonzáles-Gúzman et al., 2002). It is interesting that these authors reported that growth after germination was, in contrast to our results with nced3 mutants, more inhibited by NaCl compared to wild-type plants in closed petri plates. This difference could be explained by some variation in the environment (the aba2 mutants were grown in the absence of Suc). Alternatively, different lesions in the ABA biosynthesis pathway, although all resulting in reduced gross tissue ABA levels, may not result in the same phenotypic changes.
Both NaCl- and sorbitol-treated sto1/nced3 plants have sufficient osmotic adjustment to sustain the growth observed in wild-type plants, since their apparent turgor level differences with wild-type seedlings (seedlings are at or very near water potential equilibrium with the medium inside the closed petri plates) are similar to those of unstressed plants. However, only the NaCl-treated sto1/nced3 plants do not exhibit normal stress-induced growth inhibition. Sorbitol-treated plants, however, actually exhibit somewhat more growth inhibition (compared to wild-type plants). Apparently, under these conditions, osmotic stress-induced growth inhibition caused by salt is largely dependent on ABA, and ablation of the stress-induced increase in ABA accumulation in sto1/nced3 mutant plants allows faster growth. However, nonionic osmotic stress mediated by sorbitol can apparently inhibit growth by an ABA-independent pathway, since sto1/nced3 plants exhibit osmotic stress-induced growth reduction without the normal increased ABA accumulation associated with osmotic stress (Fig. 4). In fact, increased ABA appears to be required for growth maintenance during nonionic osmotic stress, as reported previously (Sharp, 2002). Importantly, this strongly implies that plants are able to sense ionic and nonionic osmotic stresses differently.
The role of ABA in nonturgor-dependent growth inhibition has recently received more experimental attention (Sharp et al., 2000). Recent studies have revealed that there is a very close connection between ABA and C2H4, and that the interaction of these two hormones may often affect plant tissue growth rates at low water potentials (Sharp, 2002; Sharp and LeNoble, 2002). Spollen et al. (2000) have reported that, in maize, ABA is required to maintain root growth at low water potentials and that this effect can be related to dependence of growth on ABA-restricted ethylene production. In addition, Arabidopsis seed germination and growth can be stimulated by ethylene (Ghassemian et al., 2000). However, ethylene treatment of sto1/nced3 mutant seeds did not further stimulate germination and growth as it does in wild-type seeds and plants (Fig. 12). Furthermore, consistent with the maintenance of a basal level of ABA, ethylene levels in the sto1/nced3 mutant were modestly higher than wild-type seedlings (data not shown), suggesting that impaired ABA production during osmotic stress in sto1/nced3 plants may also affect their sensitivity to ethylene, allowing faster germination and growth (Ghassemian et al., 2000).
Whatever the involvement of ABA and ethylene in controlling stress-induced growth reduction, the mechanism by which ABA level exerts its influence on growth is unknown. This mechanism should, however, involve control of cell division, since the rate of leaf formation and not just fresh-weight gain of sto1/nced3 plants during stress was also accelerated compared to controls (Fig. 12B). This indicates that there is a clear increase in rate of leaf cell division and not just cell enlargement in sto1/nced3 plants during salt exposure compared to wild-type plants. Considerable progress in elucidating important regulators of plant cell division has been made in recent years (Wang et al., 1998; De Veylder et al., 2001), and the ICK1 gene encoding a cyclin-dependent kinase inhibitor appears to play a crucial role in cell cycle regulation in Arabidopsis. Furthermore, this gene is regulated by ABA and has been proposed to mediate ABA effects on cell growth and development (Wang et al., 1998, 2000). Plants with a defective NCED3 gene fail to increase ABA accumulation in response to salt stress and also fail to increase levels of ICK1 transcript as much as wild-type plants under the same conditions. This is further evidence that the almost universally observed decrease in plant growth after exposure to osmotic stress is mediated by signal transduction events directly leading to growth through altered activities of cell cycle progression and cell division and other growth processes, rather than indirectly affecting only the physical-chemical changes controlling water flux (Wang et al., 1998).
Iuchi et al. (2001) have reported that overexpression of the NCED3 gene results in plants with increased ability to survive severe desiccation stress. However, they did not compare growth responses of either nced3 mutant plants or plants overexpressing nced3 on salinized versus desiccated soil. In addition, our results imply that NCED3-overexpressing plants may display reduced growth even in the absence of stress that is independent of the consequences of increased ABA on plant water status.
Although we have clearly shown through specific characteristics of the sto1/nced3 mutant that there exists a role for ABA in directly controlling stress-induced growth and development that is also independent from ABA effects on stomatal function, the diminished capacity of sto1/nced3 plants to accumulate ABA at low water potentials dramatically alters stomatal behavior as well (Fig. 13). Interestingly, studies on stomatal function where the effects of exogenously applied ABA and mutations that impair ABA synthesis (Iuchi et al., 2000) or sensitivity (Rodriguez et al., 1998) have been utilized, there has not been any examination of the relationship between these alterations in ABA physiology and the diurnal cycle of stomatal movement that is largely controlled by light and CO2 levels (Hedrich et al., 2001). In fact, only recently have any effects of mutation on stomatal diurnal cycle behavior been reported (Klein et al., 2003). This is partly the result of most studies employing only epidermal peels (Raschke et al., 1979), which are, of course, disconnected from mesophyll, root, and other tissues. Our observations reveal that a failure to increase ABA levels after osmotic stress in sto1/nced3 plants leads to very normal (without stress) diurnal stomatal movements with only altered upper and lower limits of stomatal aperture being imposed by the level of ABA. As soil desiccation is increased, diurnal opening is suppressed and diurnal closing is enhanced by the normal, wild-type accumulation of ABA (Fig. 13B). Somewhat surprising to us was the small degree to which the diurnal range of stomatal aperture was affected by reduced ABA levels in sto1/nced3 plants. Even under continually desiccating conditions of the soil that eventually lead to plant death, the degree of water loss rate in sto1/nced3 plants was increased only about 20%. Klein et al. (2003) also observed similar moderate changes in the diurnal cycle of stomatal movements that lead to clear changes in wilting phenotype. This indicates that relatively small changes in the dynamics of water absorption and loss can result in dramatic consequences for plant growth and survival.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis C24 line homozygous for the chimeric RD29A::LUC reporter gene (Ishitani et al., 1997) was provided by Professor J.K. Zhu (University of Arizona). A T-DNA population was obtained after Agrobacterium tumefaciens floral transformation with GV3101 (pMP90RK) carrying the binary vector pSK1015 and selection based on Liberty (bialaphos; 30 mg/L) resistance (Koiwa et al., 2002). Herbicide-resistant plants were combined into 10-plant pools and T2 seeds were collected for screening. Plants from T2 seeds were grown either in a controlled environment room with 16 h of light at 22°C and 8 h of darkness at 18°C or in a greenhouse. The screening was performed in vitro for the ability of the seeds to germinate in a medium containing 145 mm NaCl. Seeds were surface sterilized (2% sodium hypochloride for 15 min), sowed onto medium containing MS basal salt mixture, 2% Suc, 145 mm NaCl, solidified with 1% agar (pH 5.7). After sowing, the seeds were stratified for 2 to 4 d at 4°C. For the mutant characterization in vitro, MS either contained no supplement or was supplemented with 100, 130, 145, 150, 200, or 250 mm NaCl ± ABA, 20 mm LiCl ± ABA, or 160 mm KCl. For hygromicin selection, seeds were plated onto medium containing 1/4× MS salt, 30 μg mL−1 hygromicin B, 100 μg mL−1 timentin, and 1% agar.
Germination Assay
Wild-type and sto1 seeds were sterilized and sowed on petri plates containing either basic MS agar medium or MS medium supplemented with 100, 130, 150, 160, 200, or 250 mm NaCl, 160 mm KCl, or 20 mm LiCl, stratified at 4°C for 4 d and transferred in a growth chamber with 16 h of light at 22°C and 8 h of darkness at 18°C. Fourteen days after sowing, the number of germinated seeds was assessed. After 7 additional days of growth on saline media, plant fresh weight was measured.
Growth on Saline Medium after Germination on Nonsaline Medium
Wild-type and sto1 mutant plants were germinated in vitro on MS medium. Seven-day-old seedlings were transferred onto MS medium or MS medium supplemented with 160 mm NaCl, 160 mm KCl, or 20 mm LiCl. After 20 d from transferring the plants, they were collected for fresh-weight measurements.
Germination and Growth on Nonionic Osmotic Stress Medium
Wild-type and sto1 seeds were sterilized and sowed on petri plates containing either basic MS agar medium or MS medium supplemented with 300 mm sorbitol, stratified at 4°C for 4 d, and transferred in a growth chamber with 16 h of light at 22°C and 8 h of darkness at 18°C. Twenty-one days after sowing, the number of germinated seeds was assessed. Wild-type and sto1 mutant plants were germinated in vitro on MS medium, and 3-d-old seedlings were transferred onto MS medium or MS medium supplemented with 300 mm sorbitol. After 14 d from transferring the plants, they were collected for fresh-weight and root length measurements.
Desiccation Stress and Salt Stress Tolerance in Soil
Seeds of wild-type and sto1 mutant plants were directly sowed in Turface (MPV, Bancroft, LA). Fifty uniform plants from the wild-type and sto1/nced3 populations were selected and grown in the greenhouse using a standard irrigation regime. After 15 d, when plantlets had reached the stage of five to six fully expanded leaves, irrigation was stopped for one-half of the plants (25 for each genotype), whereas the other half were grown under a standard irrigation regime as a control. After 15 d, water-stressed plants were rewatered and for 2 additional days were allowed to recover before measuring plant fresh weight. To assess salt stress tolerance in soil under saturated atmospheric humidity, Arabidopsis seeds were directly sowed in Turface and allowed to grow for 1 week under nonsaline irrigation. At this stage, five groups of 20 plants each were placed under PVC domes and irrigated daily with saline water (50, 100, 150, and 200 mm NaCl plus a nonsalinized control). After 28 d, plants were collected for fresh-weight measurements.
Identification of the T-DNA Insertion Region, Northern-Blot Analyses, and RT-PCR
Genomic sequence flanking the T-DNA insertion was determined by using the TAIL-PCR procedure of Liu et al. (1995) with primers corresponding to the nested regions internal to the left border and degenerate primers as listed in Table V.
Table V.
Oligonucleotides used in this study
| Name | Experiment | Sequence |
|---|---|---|
| 1 LB1 | TAIL-PCR | 5-atacgacggatcgtaatttgtc-3 |
| 2 LB2 | TAIL-PCR | 5-taataacgctgcggacatctac-3 |
| 3 LB3 | TAIL-PCR diagnostic PCR | 5-ttgaccatcatactcattgctg-3 |
| 4 DEG1 | TAIL-PCR | 5-wgcnagtnagwanaag-3 |
| 5 DEG2 | TAIL-PCR | 5-awgcangncwganata-3 |
| 6 NCED F | Gene cloning | 5-gggctctgttatgaggcaaagacgc-3 |
| 7 NCED R | Gene cloning | 5-ggtggatcccatcagtgtgg-3 |
| 8 NCED F | Gene cloning | 5-ccacactgatgggatccacc-3 |
| 9 NCED R | Gene cloning | 5-agctgagctcgaacggctag-3 |
| 10 NCED F | T-DNA detection diagnostic PCR | 5-tggtctctccactgcgttacc-3 |
| 11 NCED R | T-DNA detection diagnostic PCR | 5-cagatgaagtcgtcgtgatga-3 |
| Primers for Northern Probes | ||
| 12 NCED F | 5-cgtgaaatccgtacggaacc-3 | |
| 13 NCED R | 5-ccggaatccggtgaactctt-3 | |
| 14 ICK1 F | 5-ccgtcgtcggtgataatgga-3 | |
| 15 ICK1 R | 5-ctaatggcttctccttctcg-3 | |
Total RNA was isolated from wild-type and sto1/nced3 plants germinated in control medium or in medium containing 145 mm NaCl. Using the RNAeasy total RNA isolation kit (Qiagen, Valencia, CA), 10 μg of total RNA were isolated and electrophoretically separated on denaturing formaldehyde-agarose gels and blotted onto nylon membrane (Schleicher & Schuell, Keene, NH). RNA was cross-linked to the membrane and the membrane was hybridized with DIG-labeled DNA probe (Roche, Indianapolis). The probe was produced by PCR reaction using the primers listed in Table V. The blots were washed twice in 2× SSC and 0.1% (w/v) SDS at 25°C, and twice in 0.5× SSC and 0.1 (w/v) SDS at 65°C.
Total RNA for RT-PCR was extracted as described. First-strand cDNA was synthesized using the SuperscriptII kit (Gibco BRL, Rockville, MD). First-strand cDNA of total RNA (4 μg) from shoots of 3-week-old plants was used for PCR amplification. PCR was carried out using ExTaq DNA polymerase (TaKaRa, Shiga, Japan) and gene-specific primers for sto1/nced3 as described in Table V.
Genetic Analysis and Complementation
Stable F3 sto1 mutant plants were back-crossed with the parental C24 wild-type plants and cosegregation for the sto1 salt tolerance (germination assay) and herbicide-resistant phenotypes was determined in F1 and F2 generations.
Three separate fragments of 2.2, 0.8, and 2.3 kb, respectively, were amplified from the bacterial artificial chromosome clone MOA2.4 and subcloned into pBluescript. The resulting 4.5-kb DNA fragment was digested with Sma1 (5′) and Kpn1 (3′) and ligated into the pBIB vector (Hygromycin+). The plasmid was transferred into Agrobacterium strain GV3101. The Agrobacterium-transformed colonies were selected with 50 mg/L kanamycin (binary vector marker), 30 mg/L rifampicin (strain marker), and 30 mg/L gentamycin (Ti-plasmid marker). Single-transformed colonies were isolated and grown in liquid medium, confirmed for their insert size by PCR, and stored frozen at −80°C. A 5-μL aliquot from the −80°C stock was used to inoculate 250 mL of yeast extract phosphate medium (DIFCO, Becton-Dickinson, Sparks, MD) plus appropriate antibiotics and incubated on a shaker in the dark at 28°C and allowed to grow to an OD600 of >1.5 to 2.0 (16–20 h). The bacteria were then centrifuged for 10 min at 4,000 rpm and the resulting pellet was resuspended in 500 mL of Agrobacterium infiltration medium (2.3 g L−1 MS salt, 50 g L−1 Suc, 0.01 mg/L N6-benzilaminopurine, and 200 μL L−1 Silwet L-77, pH 5.7. Flower buds of 30-d-old sto1 plants were sprayed with the infiltration solution and kept at high humidity for 24 h. Plants were then transferred to the greenhouse and grown under standard conditions. F1 seeds were collected and selected for hygromycin resistance on MS medium. F2 plants from five Hygromycin+ lines that were confirmed homozygous for the insertion in the NCED3 gene were grown and analyzed by RT-PCR for the expression of the NCDE3 gene. One of the five transgenic lines (4–6) was tested for salt tolerance using the germination assay and for soil desiccation tolerance.
ABA Extraction and Quantification
ABA was extracted as described by Xiong et al. (2001) from 3-week-old wild-type and sto1/nced3 plants grown in petri plates on standard MS medium or MS medium supplemented with 145 mm sorbitol. ABA was quantified using an immunoassay ELISA kit according to the manufacturer's instructions (Phytodetek ABA, AGDIA, Elkhart, IN).
Osmotic Potential Measurements
The sap osmotic potential of wild-type and sto1/nced3 plants was measured after germination and growth on 145 mm NaCl- and 150 mm sorbitol-containing media. Fifteen days after germination, plantlets were collected, frozen in liquid nitrogen, and centrifuged for 20 min at 4,000 rpm in microcentrifuge tubes. Further separation of the cellular fluid from plant debris was obtained by centrifugation at 10,000 rpm for 10 min, and osmotic potential was measured using 10-μL samples with a Wescor 5500 vapor pressure osmometer (Wescor, Logan, UT). The same procedure was followed for plants sown and grown for 7 d on basic MS medium and then transferred and grown for an additional 15 d on NaCl or sorbitol medium before measuring the osmotic potential.
Transpiration Measurements
Wild-type and sto1/nced3 mutant plants were grown in Turface in 100-mL pots. At day 21 after sowing, each pot was covered with a plastic bag with the sealed shoot protruding outside the bag. This system was used to avoid water loss from the soil surface. Each plant was then placed on an electronic balance under a light intensity of 140 μmol m−2 s−1 at 25°C, and the weight loss was automatically measured every hour for 24 h using PC software. Water loss values were normalized for plant dry weights taken at the end of the experiment.
Pharmacological Complementation of the sto1 Salt-Tolerant Phenotype
Wild-type and sto1/nced3 mutant seeds were sterilized and sown in vitro on MS medium or MS medium supplemented with 160 mm NaCl ± 20 μm ABA, stratified at 4°C for 4 d, and placed in a growth chamber with 16 h of light at 22°C and 8 h of darkness at 18°C. The number of germinated seeds was determined periodically over a 21-d time interval.
Pharmacological Complementation of LiCl Sensitivity of sto1 Plants
Wild-type and sto1/nced3 mutant plants were germinated in vitro on MS medium. Seven-day-old seedlings were transferred onto MS medium or MS medium supplemented with 20 mm LiCl ± 20 μm ABA. Seedlings were allowed to grow for an additional 7 d before visual assessment of LiCl sensitivity.
Ethylene Treatment and Measurements
Wild-type and sto1/nced3 mutant plants were germinated on MS medium. Seven-day-old seedlings were transferred onto MS medium or MS medium supplemented with 160 mm NaCl, 160 mm KCl, or 20 mm LiCl. Ten plates (each containing 20 seedlings per each genotype) were placed in two sealed Plexiglas chambers (five plates per chamber). Ethylene was added in one of the two chambers to a final concentration of 20 ppm. After 10 d, plant growth and development were quantified by counting the number of new leaves.
Ethylene production in wild-type and sto1/nced3 mutant plants was assayed on 7-d-old seedlings germinated on basal MS medium and then transferred in liquid medium. Seedlings were allowed to grow for 7 d in 3 mL of a 6-mL plastic syringe filled with liquid MS medium or MS medium supplemented with 160 mm NaCl. Ethylene accumulated during this time in the remaining 3-mL volume of the syringe and was collected by injecting the nonliquid volume (air) of the syringe in sealed vials. The ethylene concentration was subsequently quantified using standard gas chromatography analysis and normalized per 0.5 g of plant fresh weight.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB028617.
Acknowledgments
We thank Professor Kazuo Shinozaki for providing Atnced3 seeds.
This work was supported, in part, by a National Science Foundation Plant Genome Award (no. DBI–9813360).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046169.
References
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA (2002. a) Classification of genes differentially expressed during water-deficit stress in Arabidopsis thaliana: an analysis using microarray and differential expression data. Ann Bot (Lond) 89: 803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA (2002. b) Abscisic acid regulation of gene expression during-water deficit stress in the era of the Arabidopsis genome. Plant Cell Environ 25: 153–161 [DOI] [PubMed] [Google Scholar]
- Bressan RA, Nelson DE, Iraki NM, LaRosa PC, Singh NK, Hasegawa PM, Carpita NC (1990) Reduced cell expansion and changes in cell walls of plant cells adapted to NaCl. In F Katterman, ed, Environmental Injury to Plants. Academic Press, San Diego, pp 137–171
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J (1995) Abscisic acid signaling. Curr Opin Cell Biol 7: 232–238 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Schroeder JI (2001) Cell signaling and gene regulation—Plant signal transduction pathways: graying of the black boxes—Editorial overview. Curr Opin Plant Biol 4: 379–381 [Google Scholar]
- Gonzáles-Gúzman M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodriguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo S, Leone A, Xu Y, Tucci M, Francione R, Hasegawa PM, Monti L, Bressan RA (1995) Control of osmotin gene expression by ABA and osmotic stress in vegetative tissues of wild type- and ABA-deficient mutants of tomato. Physiol Plant 93: 498–504 [Google Scholar]
- Hedrich R, Neimanis S, Savchenko G, Felle HH, Kaiser WM, Heber U (2001) Changes in apoplastic pH and membrane potential in leaves in relation to stomatal responses to CO2, malate, abscisic acid or interruption of water supply. Planta 213: 594–601 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Iten M, Grill E (1998) Signalling of abscisic acid to regulate plant growth. Philos Trans R Soc Lond B Biol Sci 353: 1439–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2000) A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol 123: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Perfus-Barbeoch L, Frelet A, Gaedeke N, Reinhardt D, Mueller-Roeber B, Martinoia E, Forestier C (2003) The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. Plant J 33: 119–129 [DOI] [PubMed] [Google Scholar]
- Koiwa H, Barb AW, Xiong L, Li F, McCully MG, Lee BH, Sokolchik I, Zhu J, Gong Z, Reddy M, et al (2002) C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci USA 99: 10893–10898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua N-H (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the AB15 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio A, Matsumoto T, Hasegawa PM, Pardo JM, Bressan RA (2002. b) The long and winding road to halotolerance genes. In A Lauchli, U Luttge, eds, Salinity: Environment—Plants—Molecules. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 505–533
- Maggio A, Miyazaki S, Veronese P, Fujita T, Ibeas JI, Damsz B, Narasimhan ML, Hasegawa PM, Joly RJ, Bressan RA (2002. a) Does proline accumulation play an active role in stress-induced growth reduction? Plant J 31: 699–712 [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15: 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- Morgan JM (1984) Osmoregulation and water stress in higher plants. Annu Rev Plant Physiol 35: 299–319 [Google Scholar]
- Nuccio ML, Rhodes D, McNeil SD, Hanson AD (1999) Metabolic engineering of plants for osmotic stress resistance. Curr Opin Plant Biol 2: 128–134 [DOI] [PubMed] [Google Scholar]
- Raschke K, Haupt W, Feinleb ME (1979) Movement of stomata. In Encyclopedia of Plant Physiology. Springer, Berlin, pp 385–441
- Rock CD, Zeevaart JA (1991) The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc Natl Acad Sci USA 88: 7496–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 24: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Leon-Kloosterziel KM, Koornneef M, Zeevaart JA (1997. a) Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol 114: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR (1997. b) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276: 1872–1874 [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T (2000) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R, Rodriguez-Navarro A (2001) Ion homeostasis during salt stress in plants. Curr Opin Cell Biol 13: 399–404 [DOI] [PubMed] [Google Scholar]
- Sharp RE (2002) Interaction with ethylene: changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ 25: 211–222 [DOI] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME (2002) ABA, ethylene and the control of shoot and root growth under water stress. J Exp Bot 53: 33–37 [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME, Else MA, Thorne ET, Gherardi F (2000) Endogenous ABA maintains shoot growth in tomato independently of effects on plant water balance: evidence for an interaction with ethylene. J Exp Bot 51: 1575–1584 [DOI] [PubMed] [Google Scholar]
- Sharp RE, Wu Y, Voetberg GS, Saab IN, LeNoble ME (1994) Confirmation that abscisic acid accumulation is required for maize primary root elongation at low water potentials. J Exp Bot 45: 1743–1751 [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. Curr Opin Biotechnol 7: 161–167 [DOI] [PubMed] [Google Scholar]
- Spollen WG, LeNoble ME, Samuels TD, Bernstein N, Sharp RE (2000) Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol 122: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Joseph LM, Deng WT, Liu L, Li QB, Cline K, McCarty DR (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou Y, Gilmer S, Whitwill S, Fowke LC (2000) Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J 24: 613–623 [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Davies WJ (2002) ABA-based chemical signalling: the co-ordination of responses to stress in plants. Plant Cell Environ 25: 195–210 [DOI] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]