Abstract
In this study, we describe a large-scale expression-profiling approach to identify genes differentially regulated during the symbiotic interaction between the model legume Medicago truncatula and the nitrogen-fixing bacterium Sinorhizobium meliloti. Macro- and microarrays containing about 6,000 probes were generated on the basis of three cDNA libraries dedicated to the study of root symbiotic interactions. The experiments performed on wild-type and symbiotic mutant material led us to identify a set of 756 genes either up- or down-regulated at different stages of the nodulation process. Among these, 41 known nodulation marker genes were up-regulated as expected, suggesting that we have identified hundreds of new nodulation marker genes. We discuss the possible involvement of this wide range of genes in various aspects of the symbiotic interaction, such as bacterial infection, nodule formation and functioning, and defense responses. Importantly, we found at least 13 genes that are good candidates to play a role in the regulation of the symbiotic program. This represents substantial progress toward a better understanding of this complex developmental program.
Legume plants have the unique capacity to enter a nitrogen-fixing endosymbiosis with prokaryotes of the genera Rhizobium, Sinorhizobium, Mesorhizobium, and Bradyrhizobium (collectively termed rhizobia). In exchange for plant photosynthates, the endosymbiotic rhizobia convert dinitrogen to ammonia that is supplied to the plant for incorporation into amino acids and ultimately proteins. Symbiotic nitrogen fixation thus allows legumes to grow and produce protein-rich seeds even on nitrogen-depleted soil.
Endosymbiotic interactions represent a particular case of biotrophic interactions (Parniske, 2000) where the microorganism is enclosed in a host-derived membrane within transient organelles, termed symbiosomes. These are harbored in a specific organ that differentiates from root tissues, the root nodule. Nodule formation and bacterial infection are strictly controlled by the plant (Schultze and Kondorosi, 1998; Stougaard, 2000). First of all, in wild-type legumes, nodulation is possible only when alternative sources of assimilable nitrogen (nitrate or ammonium) are not available. Second, legumes allow invasion of a very limited range of bacteria species producing highly specific signals, the Nod factors (chitolipooligosaccharidic molecules whose perception is essential to trigger the plant symbiotic program), and proper cell wall components (notably exopolysaccharides and lipopolysaccharides). Finally, nodules and infection threads (tubular structures of plant origin) develop in defined places and limited numbers. This is regulated by the plant via a locally operating mechanism that involves the plant hormone ethylene and a systemically operating mechanism, with a mobile signal of as yet unknown nature (Penmetsa et al., 2003). In our experimental system, the differentiation of a nitrogen-fixing nodule takes about 1 week. Such a functional nodule consists of central tissues (the distal meristematic zone I, the prefixing zone II [infection zone], the amyloplast-rich interzone II–III, and the nitrogen-fixing zone III), surrounded by peripheral tissues (the nodule parenchyma, vascular bundles, endodermis, and cortex; Vasse et al., 1990). Young nodules (e.g. 4 d old in our conditions) comprise essentially dividing (or newly divided) cells and a network of infection threads.
In the past decades, one key goal in studying endosymbioses with Rhizobium was the identification of plant genes involved in root nodule formation and in the nitrogen fixation process. Differential or subtractive hybridization techniques resulted in the identification of several dozen nodule-specific or nodule-enhanced genes in different legumes, termed nodulin genes. The recent development of ambitious genomics programs (Udvardi, 2002; VandenBosch and Stacey, 2003) on two model legumes, Medicago truncatula (indeterminate nodules; nodulated by Sinorhizobium meliloti) and Lotus japonicus (determinate nodules; nodulated by Mesorhizobium loti), and on soybean (Shoemaker et al., 2002) opens new perspectives on transcriptome studies. Indeed, it becomes possible now to have as realistic goals the identification of (1) extensive sets of genes accompanying the symbiotic interaction; (2) regulation networks associated with different developmental stages (signal perception, organogenesis, infection, functioning); and (3) global regulatory genes controlling these networks. Indeed, although major breakthroughs have recently been achieved with the identification of genetic determinants of Nod factor perception and transduction (see Cullimore and Denarie, 2003), much remains to be discovered regarding later stages of nodulation. In particular, it can be noted that only a few transcriptional regulators have been described so far (Heard and Dunn, 1995; Schauser et al., 1999; Frugier et al., 2000), which cannot account for the complexity of the nodulation/infection processes.
Several expressed sequence tag (EST) sequencing projects resulted in the deposition of approximately 187,000 M. truncatula, 111,000 L. japonicus, and 300,000 soybean ESTs in public databases from a variety of cDNA libraries. Genes up-regulated in M. truncatula root nodules could thus be predicted by in silico approaches (Fedorova et al., 2002; Journet et al., 2002; Lamblin et al., 2003). Macroarrays were used with L. japonicus by Colebatch et al. (2002), who found 83 significantly nodule-induced genes from 2,304 cDNA clones, and with M. truncatula by Liu et al. (2003), who identified genes accompanying arbuscular mycorrhiza (AM) symbiosis from 2,268 clone arrays. We present here a larger-scale study, based upon three M. truncatula cDNA libraries dedicated to the study of root symbiotic interactions (Journet et al., 2002). Macro- and microarrays were constructed from 6,048 cluster-representative cDNAs (Küster et al., 2004), and expression-profiling experiments led us to identify a large number of genes differentially regulated at different nodulation stages and in different genetic backgrounds. These can be put in the perspective of the complex set of events taking place during nodulation and opens interesting perspectives for functional studies, particularly regarding the genes encoding possible regulators of the symbiotic program.
RESULTS AND DISCUSSION
Experimental Design and cDNA Arrays
For our global transcriptome studies, both wild-type and mutant plant material were used to enable the identification of genes that are controlled by the Nod factor signaling pathway and/or require infection for their activation. Young and mature (nitrogen-fixing) S. meliloti-induced root nodules were taken from wild-type material (Jemalong A17 line) 4 and 10 d postinoculation (dpi). Whole root systems from the supernodulating M. truncatula mutant TR122 (Sagan et al., 1995), 3 and 6 dpi, were also used. This mutation, allelic to sunn (Penmetsa et al., 2003; E.-P. Journet, personal communication), results in a 5- to 10-fold increase in root infections and nodule number. Compared to wild-type roots, those of TR122 become infected earlier and show continuous infections and nodule initiations during several days, while mature nodules take longer to form so that 6-d-old nodules of TR122 do not fix nitrogen. Two nodulation-defective mutants of M. truncatula were studied 3 dpi: an nfp (Nod factor perception) mutant that does not respond to Nod factors and shows no preparation for infection and an hcl (hair curling) mutant that still responds to Nod factors and prepares for infection, which is blocked just prior to root hair curling (Catoira et al., 2001; Ben Amor et al., 2003). Finally, both A17 and TR122 were also inoculated with an S. meliloti nodA mutant that is defective in Nod factor production and consequently unable to infect and nodulate.
For cDNA arrays, we exploited three cDNA libraries representing young root nodules, mycorrhiza, and noninoculated roots (Journet et al., 2002). Inserts from approximately 6,000 cDNA clones were PCR amplified and spotted to generate macro- and microarrays (Küster et al., 2004). Most of them corresponded to nonredundant genes, and relatively short inserts were chosen whenever possible to give more relative importance to the 3′-untranslated region. Both types of arrays were analyzed in parallel, with the same RNA samples being used for hybridizations. A minimum ratio of 2 with a P value ≤0.05 was chosen to define differentially expressed genes (except for Supplemental Table IV), with nodules or inoculated roots being compared to roots that were harvested just before inoculation.
The correlation observed between expression ratios determined on the basis of macro- and microarrays was often good, although a larger range of induction or repression ratios was generally observed on microarrays. However, a subset of genes was scored as differentially regulated with one type of array only (corresponding to 286 spots for microarrays and 47 for macroarrays), possibly because of (1) different elimination of data due to local experimental artifacts; (2) the fact that the induction or repression of genes concerned was below the chosen threshold; and (3) the use of different normalization procedures found to be most suitable for either array tool. Still, it is important to note that only one serious inconsistency (i.e. up-regulation on one type of array versus down-regulation on the other) was found, which is extremely low, considering the number of values examined and the fact that micro- and macroarray data were generated in two different array platforms.
Since neither micro- nor macroarray results alone gave a complete picture of differential gene expression, we decided to compile data from both series of experiments. In order to be more stringent for genes identified from one type of array only, we kept those showing a consistent expression pattern in at least two of the four symbiotic samples (supernodulated roots 3 and 6 dpi, isolated nodules 4 and 10 dpi), except for Supplemental Table IV (identification on macroarrays of early and sometimes transiently expressed genes). It is likely that such a stringent procedure eliminates some valid candidates, as we could see in one case of particular interest to us (MtC00457; see Table V), but also it helps to limit the rate of false positives. Among genes identified from microarrays only, five (MtC10811, MtC40082, MtC45259, MtC50559, and MtC90927; Supplemental Table I, available at www.plantphysiol.org) were validated by quantitative reverse transcription (qRT)-PCR (for one not confirmed, MtC00405) and five by independent probes representing the same cDNAs and/or previously published expression data (MtGSIb, MtN14, MtN28, MtC10310, and MtSucS1). Similarly, six genes found from macroarrays only were validated by qRT-PCR (MtC00340, MtC00663, MtC10733, MtC91107, and MtC91406, Supplemental Table I; MtC50408, Fig. 3) for one not confirmed (MtC91072), whereas seven more were validated by independent spots and published data (MtAnn1, MtN12, MtN20, ENOD2, MtENOD12, MtENOD20, and MtENOD40).
Table V.
Examples of differentially regulated M. truncatula genes potentially involved in regulatory mechanisms
| Cluster | Description | TW3/T0
|
N4/N0
|
TW6/T0
|
N10/N0
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mic | Mac | Mic | Mac | Mic | Mac | Mic | Mac | ||
| Transcription Factors | |||||||||
| MtC00457 | No apical meristem protein-like | −0.71 | 1.18 | −0.88 | |||||
| MtC00553 | Putative transcription factor | 0.14 | 1.76 | 2.22 | 1.94 | 0.91 | 1.27 | ||
| MtC10310 | Kruppel-like zinc finger protein | 0.76 | 1.21 | ||||||
| MtC10372 | WRKY type transcription factor | −0.42 | −0.99 | −0.50 | −1.41 | −1.04 | |||
| MtC10582 | CCAAT-binding transcription factor | 1.76 | 1.05 | 2.98 | 2.18 | 4.12 | 1.69 | 0.74 | |
| MtC10648 | Basic-helix-loop-helix DNA-binding protein | 0.56 | 0.46 | 1.53 | |||||
| MtC20187 | MADS box transcription factor | 0.50 | 0.45 | 0.72 | 1.17 | 1.38 | 0.95 | 0.85 | |
| MtC30157 | Homeobox-Leu zipper protein | −0.12 | −0.76 | −0.50 | −0.26 | −0.43 | −1.03 | −1.01 | |
| MtC50408 | AP2 domain protein | 0.09 | 1.31 | 0.99 | 0.38 | ||||
| MtC10711 | Homeodomain-Leu zipper protein | −0.39 | −1.17 | −1.23 | −0.22 | −0.50 | −0.83 | −1.20 | |
| MtC50689 | Homeodomain-Leu zipper protein | −1.21 | −0.92 | ||||||
| MtC90017 | Homeodomain protein JUBEL1-like protein | 0.43 | 1.06 | ||||||
| MtC90852 | bZIP transcription factor | 0.17 | 0.29 | −0.59 | −0.51 | −1.68 | −1.79 | ||
| MtC91049 | Basic-helix-loop-helix DNA-binding protein | 0.67 | 0.67 | 1.03 | 0.84 | ||||
| Signal Transduction | |||||||||
| MtC00791.1 | 14-3-3 protein Autointerpro: 14-3-3 protein | 0.20 | −0.67 | −0.39 | −1.20 | ||||
| MtC10022 | 14-3-3 protein | −0.27 | −0.81 | −0.3 | −1.00 | ||||
| MtC10811 | Remorin | 2.71 | 2.77 | 3.46 | |||||
| MtC10950 | Zn finger Ran-binding protein | −0.31 | −0.26 | −1.39 | |||||
| MtC20129 | Annexin | 0.72 | 0.00 | 1.92 | 0.98 | 1.38 | 0.75 | 1.44 | 1.19 |
| MtC20218 | [NODULIN] MtAnn1 annexin | 1.30 | 1.36 | 1.51 | 1.06 | ||||
| MtC30108 | IQ calmodulin binding | −0.5 | −0.25 | −1.18 | −1.08 | −0.78 | −0.88 | −0.85 | |
| MtC30551 | G-protein β WD-40 repeat gene | 1.33 | 1.17 | ||||||
| MtC40043 | Shaggy-related protein kinase | 0.10 | 0.41 | 0.59 | 0.02 | 1.35 | |||
| MtC40161 | Ser/Thr protein kinase | 0.24 | 1.21 | 1.12 | 3.05 | 1.77 | 0.25 | ||
| MtC45594 | Calcium-binding GTP-binding protein | 0.36 | −1.91 | −1.87 | −0.97 | −1.85 | |||
| MtC90003 | Ser/Thr protein kinase | −0.39 | 0.33 | −0.91 | −0.98 | −1.05 | −0.86 | −1.22 | −2.22 |
| MtC90276 | Dual specificity protein phosphatase | 0.28 | 0.43 | 1.11 | |||||
| MtC90498.1 | crinkly4/clavata 1 receptor kinase-like | −0.66 | −1.20 | −0.92 | −2.21 | ||||
| MtC91314 | Calcium-binding protein | 0.83 | 0.16 | 1.12 | |||||
| MtC91827 | Phospholipase A2 precursor | 0.47 | 1.60 | ||||||
All ratios are expressed as log2 values (legend as in Table II).
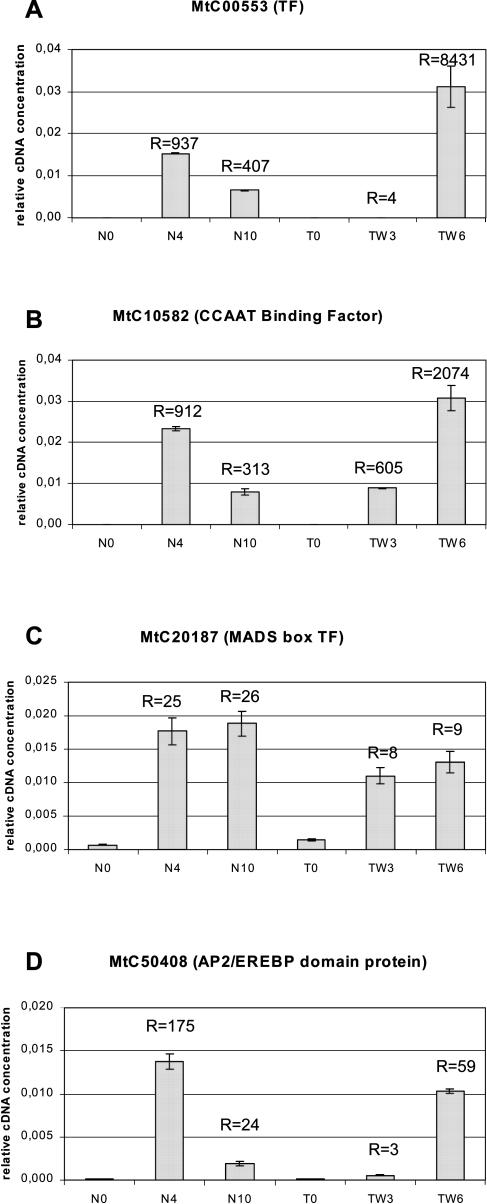
Figure 3.
Expression analysis by qRT-PCR of four putative TF genes predicted by macro- and microarrays to be up-regulated in nodules. Corresponding cDNA concentrations were estimated according to quantified DNA standards in the following samples: roots before inoculation with S. meliloti (N0), isolated nodules at 4 and 10 dpi (N4 and N10), TR122 roots before inoculation (T0), and at 3 or 6 dpi (TW3 and TW6). Concentrations were normalized using their respective EF1-α cDNA concentration, measured by qRT-PCR. Means and sds of two to three repeated experiments are reported in the graphs. Induction ratios to control noninoculated samples (N0 and T0) are indicated above bars (R=).
More Than 750 M. truncatula Genes Are Differentially Regulated during Symbiosis with S. meliloti
An overview of the genes scored as being differentially regulated is presented in Table I, and a complete list is provided in Supplemental Table II. In total, we found that 366 probes (323 genes) were activated at least 2-fold with a P value ≤0.05 in one or more symbiotic condition, whereas 474 probes (460 genes) were repressed. The expression of about 14% of the collection of genes tested was therefore identified as being affected. Thirty of these were also up- or down-regulated to a roughly similar level in response to the nonnodulating nodA S. meliloti mutant (Supplemental Table III; Fig. 1, cluster IV), possibly as a consequence of stress responses, older root systems, or longer nitrogen starvation. These genes were not further taken into consideration, resulting in a set of 756 genes either up- or down-regulated (313 and 443, respectively) at different stages of the nodulation process.
Table I.
Overview of the results obtained from macro- and microarray analyses
| Samples | R ≥ 2 | Maximum Induction Ratio | R ≤ 0.5 | Maximum Repression Ratio |
|---|---|---|---|---|
| Micro N10 | 235 | 294 | 269 | 32 |
| Macro N10 | 158 | 501 | 247 | 11 |
| Total different clones | 273 | 385 | ||
| Micro N4 | 156 | 96 | 162 | 9 |
| Macro N4 | 147 | 45 | 156 | 10 |
| Total different clones | 191 | 230 | ||
| Micro TR122 6 dpi | 143 | 100 | 140 | 11 |
| Macro TR122 6 dpi | 127 | 62 | 109 | 7 |
| Total different clones | 177 | 187 | ||
| Micro TR122 3 dpi | 66 | 14 | 50 | 3 |
| Macro TR122 3 dpi | 37 | 6 | 9 | 3 |
| Total different clones | 81 | 51 | ||
| Macro A17 3 dpi | 10 | 3.5 | 0 | |
| Total different clones | 5 |
Indicated are the number of spotted cDNAs exhibiting an activation or inhibition ratio (R) of at least 2-fold in comparison to the noninoculated reference sample, with a P value ≤0.05, as determined from micro- and macroarray hybridizations (labeled Micro and Macro, respectively). All samples were inoculated with wild-type S. meliloti, 4-d and 10-d nodules (labeled N4 and N10, respectively); TR122 supernodulated whole root systems, 3 or 6 dpi; wild-type A17 whole root systems, 3 dpi.
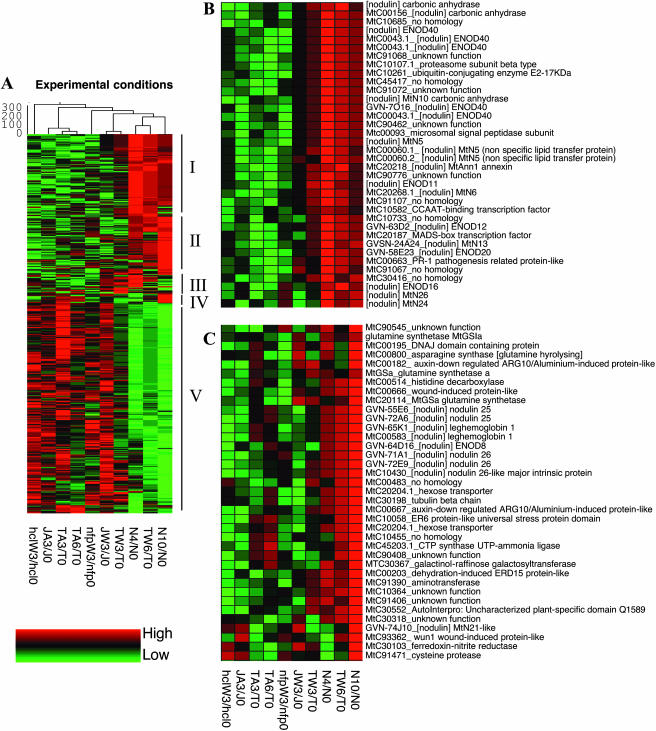
In support of the robustness of these results, it should be underlined that in all 45 cases where genes were represented on arrays by several cDNAs, patterns obtained for the corresponding spots were very similar (see examples in Fig. 1, B and C). In addition, a number of genes identified from these macro- and microarray results are validated by other approaches. Thus, all known nodulation marker genes represented on our arrays (83 cDNAs, corresponding to 41 genes) gave hybridization signals consistent with their published expression patterns (e.g. Gamas et al., 1996). Finally, hierarchical clustering analyses showed in many cases a consistent expression pattern in different samples (e.g. nodules 4 and 10 dpi and supernodulated roots).
Figure 1.
Hierarchical clustering of M. truncatula genes differentially regulated during symbiotic interactions with S. meliloti. The expression patterns of 539 genes found as differentially regulated in Mt6k-RIT macroarray hybridizations were analyzed under 10 symbiotic conditions, and ratios were calculated between the inoculated and the corresponding noninoculated samples. All genes exhibit a minimum 2-fold expression ratio with a P value ≤0.05 in at least one condition. Whole root systems from wild-type Jemalong A17 (J), supernodulating mutant TR122 (T), and nfp and hcl mutants were harvested at 3 or 6 dpi with either wild-type (W3, W6) or nodA (A3, A6) S. meliloti strains and compared to noninoculated material (0). Isolated nodules were collected at 4 (N4) and 10 dpi (N10) and compared to noninoculated roots (N0). Hierarchical clustering was carried out using average linkage values and Pearson correlations. The global clustering analysis is presented in (A), where five clusters (designated I–V) can be distinguished. Close-ups of clusters I and II, respectively, are shown in B and C, with corresponding MtC EST cluster numbers and annotations.
Figure 1 shows such a hierarchical clustering analysis comparing 10 conditions (nodules, TR122, A17, nfp, and hcl plant material inoculated with S. meliloti wild type and a nodA mutant). The clustering of conditions (columns) makes good biological sense from (1) nonnodulated material on the left (with the three S. meliloti nodA-treated root samples well clustered) to hypernodulating roots and isolated nodules on the right and (2) from young to mature nodules. In addition, five clusters of expression profiles (lines) can be distinguished (Fig. 1A): genes up-regulated at all nodulation stages with maximum relative transcript abundance in young nodules (cluster I; e.g. MtENOD11 or MtN5); genes activated mainly in mature nodules (cluster II; e.g. leghemoglobin genes); genes transiently induced at 3 to 4 dpi (cluster III); genes induced in response both to wild type and nodA S. meliloti (cluster IV); and genes down-regulated during nodulation (cluster V). The distribution of nodulin genes for which the pattern of expression is known gives credence to this division (Fig. 1, B and C).
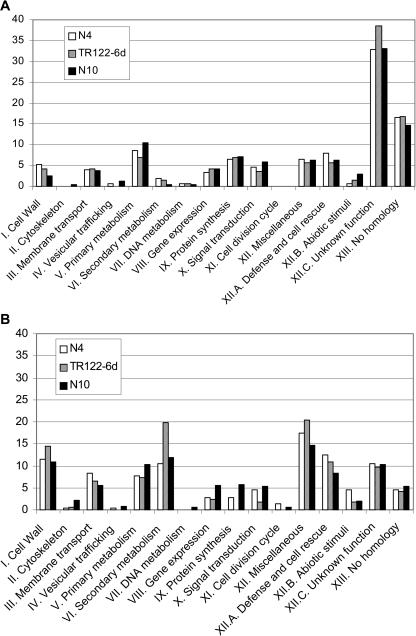
Genes identified as being differentially regulated encode proteins belonging to 16 different functional classes, as defined in the MENS database (Fig. 2). The overall distribution is similar between the three symbiotic reference samples (N4, N10, and TR122, 6 dpi). Almost one-half of the up-regulated genes belong to the unknown function (including a number of early nodulin genes) and the no-homology classes, in contrast to the down-regulated genes (Fig. 2B), which are distributed more homogeneously.
Figure 2.
Functional class distribution of M. truncatula genes identified as differentially regulated during nodulation. The up-regulated (A) and down-regulated (B) genes were sorted into the 16 broad functional categories used for classification in the MENS database. This diagram shows relative distributions expressed as percentages of the total number of up- or down-regulated genes in each sample: 4-d nodules (N4), TR122 nodulated roots, 6 dpi (TR122-6d), and 10-d nodules (N10). Bars represent the relative abundance of each class expressed as a percentage of the up- or down-regulated genes.
M. truncatula Gene Activation during Early Nodulation Stages
We examined on macroarrays the transcriptome expressed at 3 dpi with S. meliloti, using wild-type Jemalong A17 and supernodulating TR122 plants, on the one hand, and two early symbiotic plant mutants, nfp and hcl, on the other hand, to discriminate some of the activated genes. We decided to use a low threshold for induction ratios because three early nodulin genes (MtENOD40, MtN3, and MtN13) were up-regulated 3 dpi with an induction ratio of below 2 in wild-type Jemalong, probably because of a dilution problem, i.e. a low proportion of cells responding to symbiotic signals. The use of the TR122 supernodulating mutant substantially increased the numbers of genes observed as being differentially regulated upon inoculation with S. meliloti, in accordance with the higher number of infections and nodule primordia formed.
Forty-eight genes showed an induction ratio of at least 1.5 in either wild-type or TR122 plants, with a P value ≤0.05 and no induction by an S. meliloti nodA mutant (Supplemental Table IV). These included 30 genes up-regulated in nodules, among which were eight known nodulin genes (MtENOD11, 12, 40; MtN3, 5, 6, 13; and a carbonic anhydrase gene). The activation of 27 genes could be detected in wild-type Jemalong, thus allowing for a direct comparison with nfp and hcl mutants. Interestingly, seven proteins potentially involved in signal transduction or gene regulation were among them (DNA-binding protein S1Fa, bZIP transcription factor (TF)- and Myb-like proteins, a protein kinase, a seven transmembrane domain protein, and two proteasome-related proteins).
None of the 27 genes was found to be induced in the nfp mutant. This confirms the importance of the NFP-dependent Nod factor signaling pathway and suggests that no other signaling pathway plays a significant role in its absence. In the hcl mutant, three genes (MtN5, MtPR1-1, and MtENOD40) were identified as being induced at a statistically significant level. These genes are thus early markers of events preceding infection, probably involving Nod factor signaling. hcl mutants still prepare for infection and exhibit cortical cell divisions with which the observed MtENOD40 induction is almost certainly associated (Catoira et al., 2001). Intriguingly, MtN5 belongs to the family of nonspecific lipid transfer proteins that includes DIR1, a protein recently shown to play a key role in long-distance signaling for systemic acquired resistance, possibly as a lipid sensor (Maldonado et al., 2002). PR1 is another gene that is induced both in systemic acquired resistance establishment and during early responses to S. meliloti. A lipolytic enzyme gene (MtC00057), which might generate a lipid signal, is also transiently induced in wild-type plants. It is thus conceivable that a signaling process similar to that operating during pathogenic interactions takes place during early interactions with S. meliloti.
It is interesting to note that, in contrast to most genes, six genes were significantly less induced in TR122 than in wild-type A17 at 3 dpi, among which several potentially related to defense reactions (encoding a germin-like protein, an endo-β-glucanase, and a wound-induced protein), as well as a putative bZIP TF gene. This latter gene was transiently up-regulated in wild-type plants and could thus be related to a transient defense response or the nodule/infection regulation process inhibited in TR122.
Genes Associated with Nodule Formation
Secondary and Hormone Metabolism
Cytokinins and auxins are thought to play an important role during nodule initiation, and both hormones are able to induce an expression of early nodulin genes (Hirsch et al., 1997; Jimenez-Zurdo et al., 2000; Mathesius et al., 2000). In young nodules, we found that a UDP-glucuronosyltransferase gene is induced (MtC90141, Table II), which is highly homologous to a gene, PsUGT1, strongly induced in dividing cells (Woo et al., 1999). This gene is essential for growth and development in pea (Pisum sativum) and alfalfa (Medicago sativa), and it is postulated that it regulates the activity of a ligand needed for cell division. Interestingly, MtC90141 is also homologous to a zeatin O-xylolyltransferase (Martin et al., 1999), which raises the question of cytokinins as possible ligands for this UDP-glucuronosyltransferase. It is also worth noting that a gene for adenine phosphoribosyltransferase (MtC00578), potentially involved in cytokinin interconversion (Moffatt et al., 2000), is detected as being induced in young nodules. Concerning auxin, no definite picture emerges from our data, since in mature nodules a concomitant activation of small auxin-up RNAs (MtC10140) and two homologs of auxin-down-regulated genes (MtC00667 and MtC10128) was observed (Table II).
Table II.
Examples of differentially regulated M. truncatula genes related to nodule formation
| Cluster | Description | TW3/T0
|
N4/N0
|
TW6/T0
|
N10/N0
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mic | Mac | Mic | Mac | Mic | Mac | Mic | Mac | ||
| Secondary and Hormone Metabolism | |||||||||
| MtC00343.2 | Gibberellin biosynthesis enzyme ent-kaurene synthase | 0.54 | −0.82 | −2.38 | −2.98 | −2.47 | |||
| MtC00442 | Squalene monooxygenase | −0.76 | −0.88 | −0.49 | −1.09 | −0.57 | −1.58 | −1.12 | |
| MtC00578 | [NODULIN]MtN30 adenine phosphoribosyltransferase | 0.41 | 0.69 | 0.85 | 1.32 | −0.65 | 0.05 | ||
| MtC00650 | Diphosphomevalonate decarboxylase | −0.26 | −1.17 | −0.15 | −0.68 | ||||
| MtC00667 | Auxin-down regulated ARG10/Aluminum-included protein-like | 0.37 | 0.39 | 0.51 | 0.52 | 0.73 | 1.58 | 3.17 | |
| MtC10128 | Auxin-down regulated /Aluminum-induced protein-like | −0.18 | 0.25 | 0.35 | 0.27 | 1.38 | 1.47 | ||
| MtC10140 | Auxin responsive SAUR protein-like | 1.21 | 1.74 | ||||||
| MtC10438 | Auxin-induced protein-like | −0.90 | −2.29 | −1.39 | −1.38 | −2.31 | −1.26 | ||
| MtC10905 | Squalene synthase | −0.75 | −0.63 | 0.15 | −0.69 | −0.68 | −1.04 | −1.07 | |
| MtC20178 | Gibberellin induced protein-like | −1.80 | −1.07 | −2.11 | −1.65 | −2.52 | −1.45 | −1.64 | |
| MtC30281.1 | β-amyrin synthase | 0.94 | −0.10 | −0.64 | −0.11 | −1.64 | −0.73 | −2.32 | −1.12 |
| MtC90141 | UDP-glucuronosyltransferase | 0.23 | 1.26 | 0.59 | 1.13 | 2.17 | 0.29 | ||
| MtC90482 | 1-Deoxy-d-xylulose 5-phosphate synthase 2 | −0.39 | 0.47 | −1.24 | −0.59 | −1.85 | −2.21 | −2.68 | −3.33 |
| MtC91838 | β-Amyrin synthase | −1.17 | −0.67 | −0.42 | −0.67 | −1.98 | −1.01 | −1.07 | |
| Cell Wall | |||||||||
| GVSN-9F12 | [NODULIN] nodulin 75 (ENOD2) | −0.29 | 1.13 | 2.79 | 0.50 | 5.49 | 0.66 | 5.73 | |
| JVC-PG | [NODULIN]ENOD11 | 2.01 | 2.33 | 3.88 | 4.33 | 1.83 | 2.32 | 2.51 | 1.49 |
| JVC-PG | [NODULIN]ENOD16 | 1.08 | −0.12 | 2.69 | 2.48 | 2.56 | 2.56 | 2.88 | 2.32 |
| MtC00071 | [NODULIN]ENOD20 | 0.23 | 3.31 | 2.81 | 1.37 | ||||
| MtC00240 | [NODULIN]ENOD12 | 0.80 | 0.84 | 2.28 | 3.51 | 1.35 | 2.31 | 2.21 | 1.14 |
| MtC00351 | Arabinogalactan-protein (AGP4)-like Pro-rich protein | −1.08 | −0.54 | −1.07 | −0.88 | −1.15 | −1.30 | −1.65 | −1.48 |
| MtC00548.2 | Endoxyloglucan transferase | −0.92 | −0.64 | −1.20 | −1.26 | ||||
| MtC00582 | Endo-β-1,4-glucanase | 0.40 | 1.10 | −0.16 | −0.82 | ||||
| MtC00611.2 | Xyloglucan endo-transglycosylase | −0.51 | −0.66 | −1.06 | |||||
| MtC10168 | Pectin methyl-esterase | 0.32 | 0.98 | 1.28 | 1.74 | 0.49 | 0.70 | 1.15 | |
| MtC10353 | Arabinogalactan protein | −1.00 | −0.44 | −1.47 | −0.68 | −1.13 | −0.73 | −2.30 | −1.25 |
| MtC10404 | Pro-rich protein | −0.08 | −1.61 | −2.12 | −0.26 | −0.84 | −1.97 | −1.42 | |
| MtC10798 | Expansin | −0.47 | 0.10 | −1.58 | −1.18 | −1.08 | −0.81 | −1.31 | −1.03 |
| MtC10880 | UDP-glucose 6-dehydrogenase | −0.34 | −1.19 | −0.97 | −1.48 | −0.96 | −2.00 | −1.29 | |
| MtC20373 | Endo-β-1,4-glucanase | 2.66 | 1.66 | 0.69 | 0.55 | 0.22 | |||
| MtC30160 | Xyloglucan endo-transglycosylase | −0.95 | −0.06 | −1.92 | −1.21 | −2.07 | −1.01 | −3.17 | −1.39 |
| MtC30178.2 | Extensin | 0.33 | −1.33 | −1.33 | −0.73 | −0.95 | −1.58 | −1.53 | |
| MtC30264 | Cellulose synthase catalytic subunit | −0.31 | −1.25 | −0.59 | −1.08 | ||||
| MtC90424 | Expansin | −0.41 | −1.31 | −1.27 | −1.41 | ||||
| MtC90807 | Pectate lyase | 1.10 | 1.07 | −0.36 | 0.18 | 0.14 | −0.66 | ||
| Protein Synthesis and Processing | |||||||||
| MtC00093 | Microsomal signal peptidase 25 kD subunit | 0.39 | 0.45 | 1.38 | 1.90 | 0.85 | 1.81 | 1.26 | 1.02 |
| MtC00153 | Microsomal signal peptidase 18 kD subunit | 0.40 | −0.04 | 1.64 | 1.12 | 1.59 | 1.14 | 0.59 | 0.43 |
| MtC00546 | Microsomal signal peptidase 12 kD subunit | 0.34 | 2.24 | 1.09 | 1.17 | 0.72 | 0.80 | 0.27 | |
| MtC10107.1 | Proteasome subunit β type | 0.66 | 1.20 | 1.55 | 3.16 | 1.09 | 2.27 | 1.16 | 1.77 |
| MtC10260 | AutoInterpro: Cyclin-like F-box | −0.24 | 0.58 | −0.27 | 1.49 | 1.02 | |||
| MtC10261 | Ubiquitin-conjugating enzyme E2-17 kD | 0.38 | 1.31 | 1.21 | 2.56 | 0.55 | 1.91 | 0.97 | 1.51 |
| MtC10489 | 30S ribosomal protein S20 | 0.85 | 0.65 | 1.86 | 3.58 | 1.51 | 2.34 | 1.39 | 1.25 |
| MtC20366.1 | Cys proteinase | −0.76 | −0.13 | −0.83 | −1.00 | −1.11 | −0.55 | −1.63 | −1.45 |
| MtC30111 | 26S protease regulatory subunit 4 | −0.67 | −0.15 | −1.77 | −1.03 | −0.95 | −0.52 | −1.51 | −0.71 |
| MtC30415 | Microsomal signal peptidase 23 kD subunit | 0.34 | 0.50 | 1.49 | 2.04 | 1.48 | 1.81 | 0.65 | 0.60 |
| MtC90940 | [NODULIN]MtN7 AutoInterpro: Cyclin-like F-box | 0.36 | 0.26 | 1.38 | 1.29 | 1.26 | 1.82 | 1.38 | 0.66 |
| MtC91471 | Cys proteinase | 2.10 | 2.23 | ||||||
All ratios are expressed as log2 values between normalized signals obtained by microarray (Mic) or macroarray (Mac) analysis for the following samples and corresponding noninoculated roots: isolated 4- and 10-d A17 nodules (N4 and N10); supernodulated TR122 whole root systems, 3 and 6 dpi (TW3 and TW6). All nodules were induced by wild-type S. meliloti inoculation. AutoInterpro corresponds to automatic annotation generated by Interproscan analysis on proteins predicted from the EST cluster sequences.
Regarding gibberellins, it is worth noting that the gene encoding the gibberellin biosynthesis enzyme ent-kaurene synthase (MtC00343.2, Table II) is down-regulated in nodules. Down-regulation was also observed for a homolog (MtC20178) of the GASA family of gibberellin-regulated proteins, which play an important role in plant development, e.g. during lateral root initiation (Taylor and Scheuring, 1994). Switching off this gene could be important to redirect the root developmental program toward nodule organogenesis. The root cap protein gene (Matsuyama et al., 1999), which is also turned down during nodulation (MtC00237), is probably a good marker for this fundamental developmental switch.
Generally speaking, many more secondary metabolism genes were found to be down-regulated rather than activated in nodules in comparison to roots. The biological interpretation is delicate, since many of them can be involved in various biological processes. This is the case for the terpenoid pathway (e.g. diphosphomevalonate decarboxylase, MtC00650; squalene synthase, MtC10905), from which a wide range of metabolites can be synthesized, including gibberellins and brassinosteroids, phytosterol, or defense molecules.
Cell Wall Proteins
Cell wall Pro-rich proteins (PRPs) are among the first and best-characterized markers of different symbiotic stages, from response to Nod factors to infection and nodule formation. The expression pattern established by macro- and microarrays (Table II; Supplemental Table II) for the following genes was quite consistent with previous studies: MtENOD11 and MtENOD12, encoding PRPs proposed to facilitate cell wall porosity and plasticity, and consequently the infection process and exchange of metabolites (Journet et al., 1994, 2001); MtENOD16 and MtENOD20, encoding proteins structurally related to phycocyanins, postulated to play a role in cell wall assembly and reorganization (Greene et al., 1998); ENOD2, shown to accumulate in the nodule parenchyma (van de Wiel et al., 1990); and MtN12, encoding a protein highly homologous to a pea extensin-like protein found in the infection thread matrix (Rathbun et al., 2002). Interestingly, MtN12 was found to be up-regulated in 4-d nodules but also, to a lower level, in roots inoculated with nodA mutants of S. meliloti, which could relate to a localized defense reaction (see also subsection “Nodulation and Defense Reactions”). By contrast, several root PRP genes are repressed in nodules (e.g. MtC30178.2, MtC10404, and MtC00351), which is consistent with a profound modification of cell wall structures accompanying the infection and nodulation process.
In terms of cell wall-related enzymes, the up-regulation of a pectinesterase gene was observed (MtC10168), notably in young nodules, which could be related to cell wall softening to facilitate the infection process. This could also be the case for two endo-β-1,4-glucanases induced at 3 and 4 dpi (MtC20373 and MtC00582), and of a pectate lyase (MtC90807). Interestingly, the latter is homologous to a protein potentially involved in the development of transmitting tissues of the style and/or the facilitation of pollen tube growth (Budelier et al., 1990), both processes being somewhat reminiscent of infection thread growth.
A range of genes encoding enzymes involved in cell wall growth (e.g. encoding an endoxyloglucan transferase, MtC00548.2; a xyloglucan endotransglycosylase, MtC00611.2; a UDP-Glc/GDP-Man dehydrogenase involved in the synthesis of hemicellulose and pectin, MtC10880; and a cellulose synthase, MtC30264) is poorly expressed in nodules in comparison to roots, which could correspond in some or all cases to root-specific members of gene families, since, for example, two expansin genes (MtC10798 and MtC90424) are turned down in nodules, whereas another is activated (MtC50648, Supplemental Table II).
Protein Synthesis and Maturation/Degradation
The protein synthesis machinery can play an important role for the organogenic and infectious processes, accompanied by the coexpression of a number of specific proteins. Therefore, it is interesting to observe the coordinated activation, particularly in young nodules, of genes encoding four subunits of a microsomal signal peptidase (MtC00546, MtC00153, MtC30415, and MtC00093, Table II). This might be related to the production of abundant and specific secreted proteins, such as PRPs, the large family of nodule-specific Cys-rich proteins (Fedorova et al., 2002; Mergaert et al., 2003), or PA1b/leginsulin-like proteins (see subsection “Nodulation and Defense Reactions”). Several proteins within these families share a highly homologous signal peptide (Journet et al., 2001; Mergaert et al., 2003), which may thus be cleaved by a specific signal peptidase such as the one found here.
Potentially also very interesting is the fact that several genes involved in protein degradation (ubiquitin pathway, a range of proteases, and heat shock protein genes) appear to be activated or repressed, again with different behaviors within the same gene family. Protein degradation can be part of the normal cellular protein turnover process but can also play an important role in the control of plant development and plant-microbe interactions (Hellmann and Estelle, 2002; Serino and Deng, 2003). Thus, up-regulated cyclin-like F-box proteins (MtN7 early nodulin/MtC90940 and MtC10260) could be part of a ubiquitin ligase complex involved in signaling via protein degradation.
Unknown Function/No-Homology Proteins
As already mentioned, it is not possible to propose a function, and, in some cases, even to find homologs in public databases for a large number of genes activated during nodulation. It is worth noting that 40 of them correspond to ESTs exclusively found in nodule libraries (see Table III) and may thus be highly specific for nodulation and legumes in general. These include many nodulin genes already described in M. truncatula or other legumes, which provide a useful internal validation of our data.
Table III.
M. truncatula genes scored by macro- and micro-array analyses as up-regulated in nodules and for which corresponding ESTs are exclusively found in nodule libraries
| Cluster | Description | TW3/T0
|
N4/N0
|
TW6/T0
|
N10/N0
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mic | Mac | Mic | Mac | Mic | Mac | Mic | Mac | ||
| MtC30497.1 | [NODULIN] PsN466/PsENOD3-related protein | −0.11 | 2.1 | 1.97 | 2.09 | 1.21 | 1.11 | 0.6 | |
| MtC90572 | Late nodulin PsNOD6-related | −0.11 | 1.45 | 0.59 | 2.21 | 1.75 | 1.13 | 1.12 | |
| MtC90604 | Unknown function | 0.48 | 0.78 | 1.27 | 0.12 | 1.09 | 1.01 | 0.17 | |
| MtC90621 | Late nodulin NOD-CCP3-like | 2.02 | 1.54 | 0.75 | |||||
| MtC90752 | Distantly related to Vicia faba late nodulin NOD-CCP5 | 0.21 | 0.72 | 1.13 | 1.58 | 1.09 | 0.62 | ||
| MtC90842 | Unknown function | −0.27 | 2.36 | 2.12 | 3.48 | 2.57 | 0.63 | 1.06 | |
| MtC90940 | [NODULIN] MtN7 AutoInterpro: Cyclin-like F-box | 0.36 | 0.26 | 1.38 | 1.29 | 1.26 | 1.82 | 1.38 | 0.66 |
| MtC90944 | Nodule-specific Gly-rich family protein-like | −0.36 | 0.77 | 1.67 | 0.73 | 0.52 | 1.41 | ||
| MtC91045 | [NODULIN] MtN17 MtN16 MtN11 nodulin-like | 0.01 | 1.45 | 2.42 | 0.47 | 1.27 | 0.5 | ||
| MtC00436 | Pea PsN466 nodule specific protein-like | 0.31 | 0.16 | 1.39 | 2.16 | 0.93 | 1.81 | 1.29 | 0.81 |
| MtC40167 | [NODULIN] MtN9-like protein | 1.65 | 0.04 | 5.16 | 1.62 | 5.28 | 1.35 | 3.08 | 0.87 |
| MtC90571 | [NODULIN] MtN19 | 1.23 | 0.07 | 4.08 | 2.57 | 3.4 | 2.22 | 2.4 | 0.96 |
| MtC00290 | Embryo-specific protein 3 (ATS3)-like | 0.23 | 1.78 | 1.59 | 2.16 | 1.36 | 1.93 | 1.72 | |
| MtC00746 | [NODULIN] MtN11 | 1.39 | 0 | 6.59 | 4.78 | 4.51 | 3.17 | 2.99 | 1.33 |
| MtC20268.1 | [NODULIN] MtN6 | 2.25 | 1.16 | 4.43 | 2.66 | 3.41 | 1.57 | 0.75 | |
| MtC00515 | NOD-CCP5 late nodulin-like | 0.11 | 1.74 | 0.42 | 3.6 | 1.59 | 1.83 | 0.9 | |
| MtC00186 | [NODULIN] MtN16 | 0.84 | 0.18 | 4.18 | 3.46 | 2.36 | 2.92 | 2.98 | 1.06 |
| MtC20267.1 | [NODULIN] MtN26 | 0.37 | 1.88 | 2.86 | 6.14 | 3 | 2.34 | 2.83 | |
| MtC90643 | [NODULIN] MtN28 | 0.89 | 1.43 | ||||||
| MtC00043.1 | [NODULIN] ENOD40 | 1.77 | 3.71 | 2.82 | 2.34 | ||||
| MtC10358 | [NODULIN] ENOD3 distantly related protein | 0.39 | −0.33 | 0.92 | 0.62 | 0.75 | 1.7 | 1.87 | 1.67 |
| MtC00120 | [NODULIN] MtN29 | 1.97 | 0.18 | 4.2 | 3.95 | 4.11 | 3.87 | 3.2 | 2.69 |
| MtC00588 | [NODULIN] MtN25 | −0.21 | 2.31 | 2.81 | 3.04 | 3 | 1.44 | 1.8 | |
| MtC00643 | [NODULIN] MtN15 related protein | 0.25 | 1.61 | 1.27 | 1.03 | 1 | 1.76 | 1.78 | |
| MtC91028 | No homology | 0.62 | 0.38 | 1 | |||||
| MtC91055 | No homology | 0.05 | 3.03 | 1.06 | 4.33 | 0.93 | 1.02 | 0.58 | |
| MtC91067 | No homology | 0.82 | 2 | 1.65 | 0.83 | ||||
| MtC91074 | No homology | 0.68 | 0.26 | 2.52 | 2.43 | 2.28 | 1.66 | 1.82 | 0.92 |
| MtC91107 | No homology | 1.02 | 3.37 | 1.96 | 0.87 | ||||
| MtC91127 | No homology | 0.79 | 0.75 | 1.99 | |||||
| MtC91157 | No homology | 0.47 | 2.03 | 1.97 | 1.16 | 0.66 | 0.43 | ||
| MtC00697 | No homology | 0.85 | 1.26 | ||||||
| MtC10766 | No homology | 2.99 | 0.14 | 3.62 | 1.33 | 5.58 | 3.04 | 4.52 | 2.15 |
| MtC30515 | No homology | 0.49 | −0.16 | 1.27 | 1.3 | 1 | 1.07 | 1.58 | 0.85 |
| MtC30530 | No homology | 0.19 | 1.41 | 2.24 | 1.46 | 1.01 | 0.85 | 0.6 | |
| MtC30542.1 | No homology | 0.17 | 2.45 | 1.32 | 1.52 | 0.74 | 0.47 | 0.19 | |
| MtC45417 | No homology | 0.5 | 1.19 | 0.93 | 0.75 | ||||
| MtC10733 | No homology | 0.99 | 1.93 | 1.35 | 1.04 | ||||
| MtC00483 | No homology | 0.43 | 0.48 | 0.07 | 0.4 | 1.73 | 1.38 | 3.11 | |
All ratios are expressed as log2 values (legend as in Table II).
Several of the genes listed in Table III and Supplemental Table II (MtC00436, MtC00515, MtC30497.1, and MtC91023) probably belong to the large family of nodule Cys-rich proteins recently described (Fedorova et al., 2002; Mergaert et al., 2003), postulated to be involved in cell-to-cell signaling or in the formation of a defense system.
Genes Involved in Nodule Functioning
Carbon Metabolism
Nodule development and functioning involves the reallocation of plant photosynthates to this new organ. This creation of an additional sink for carbon takes place quite early and is accompanied by changes in the expression of a series of genes related to carbon metabolism (Supplemental Table II; Table IV; Fig. 1).
Thus, a transcriptional activation of the nodule-enhanced (Hohnjec et al., 1999, 2003) Suc synthase MtSucS1 (e.g. MtC00042) and of cell wall or cytoplasmic invertase genes (MtC00187.1 and MtC90461, respectively) is probably involved in catabolism of the Suc unloaded from the phloem. A β-amylase-encoding gene (MtC30105) is also up-regulated, possibly allowing the starch found in nodule primordia and the nodule interzone II-III to be degraded. The induction of a Fru-bisphosphate aldolase gene (MtC92047) is likely to be linked to the activation of glycolysis. Glycolysis leads to the production of phosphoenolpyruvate (PEP), itself providing oxaloacetate by the action of PEP carboxylase (PEPC), important for providing both organic acids to bacteroids and substrates for nitrogen assimilation. PEPC was previously described to be induced in nodules (Vance and Gantt, 1992), and a corresponding gene is found here to be transcriptionally activated (MsPEPC). PEPC uses HCO3− as a substrate to produce oxaloacetate from PEP. A carbonic anhydrase gene (MtC00156), the product of which catalyzes the CO2 hydration to form HCO3−, is highly up-regulated at early and late symbiotic stages, as already documented (Coba de la Pena et al., 1997; Galvez et al., 2000), which could relate to the control of osmolarity and oxygen diffusion.
Several genes involved in different metabolic processes are also clearly up-regulated during early- or late-nodulation stages, notably related to lipid degradation (MtC00057, MtC50550), amino acid metabolism (MtC91090, a phenylpyruvate dioxygenase, and MtC00514, a His decarboxylase), and riboflavin biosynthesis (MtC45259). Two thioredoxin genes (MtC00388 and MtC00198) are also activated, which could affect the activities of various proteins by controlling their redox status (e.g. the malate dehydrogenase as described for the M-type, represented here by MtC00388).
Nitrogen Fixation
As expected, a strong induction of two nitrogen assimilation genes is detectable in mature nodules (Table IV; Fig. 1). Those encode the Gln synthetase Ia (e.g. MtC20114) and an Asn synthase (MtC00800), consistent with previous reports (Küster et al., 1997; Shi et al., 1997; Carvalho et al., 2000). An l-asparaginase gene (MtC20062) is found to be up-regulated only in immature nodules and should not consequently be linked to ammonium assimilation.
Table IV.
Examples of differentially regulated genes related to M. truncatula nodule functioning
| Cluster | Description | TW3/T0
|
N4/N0
|
TW/T0
|
N10/N0
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mic | Mac | Mic | Mac | Mic | Mac | Mic | Mac | ||
| Primary Metabolism | |||||||||
| JVC-PG | Glutamine synthetase MtGSlb | −0.48 | −0.82 | −1.21 | −0.66 | −0.69 | −1.25 | −0.24 | −1.22 |
| MsPEPC | PEP carboxylase | 0.64 | 1.50 | 1.40 | 1.12 | ||||
| MtC00042 | Sucrose synthase | 2.29 | −0.03 | 3.77 | 0.95 | 3.26 | 0.50 | 2.94 | 1.55 |
| MtC00057 | Esterase | 0.42 | −0.05 | −1.34 | −0.98 | ||||
| MtC00100 | Fructose-bisphosphate aldolase | −0.42 | −0.33 | −0.52 | −0.16 | −0.75 | −0.72 | −1.43 | −1.05 |
| MtC00156 | [NODULIN] Carbonic anhydrase | 3.26 | 2.25 | 4.44 | 4.52 | 5.11 | 3.84 | 4.26 | 2.74 |
| MtC00187.1 | Cell wall invertase β-fructofuranosidase | −0.38 | 1.46 | 1.02 | −0.22 | 0.30 | |||
| MtC00198 | Thioredoxin M-type chloroplast precursor | −0.01 | 2.63 | 1.09 | 1.97 | 0.80 | 0.25 | ||
| MtC00241 | Asp synthase | −0.86 | −2.48 | −1.96 | 0.49 | ||||
| MtC00388 | Thioredoxin M-type | 0.07 | 2.26 | 2.48 | 1.67 | 1.11 | 0.56 | ||
| MtC00514 | His decarboxylase | 0.21 | 0.38 | 0.73 | 0.60 | 0.96 | 1.23 | 2.13 | |
| MtC00800 | Asp synthase [Glu-hydrolysing] | −1.00 | −0.48 | 0.70 | 0.12 | −0.54 | −0.43 | 4.06 | 3.82 |
| MtC10059 | Carbonic anhydrase | −0.20 | −1.07 | −0.62 | −0.44 | ||||
| MtC20039 | E2 component of pyruvate dehydrogenase complex | −0.48 | −0.16 | −1.50 | |||||
| MtC20062 | l-Asp | 1.42 | 1.33 | 0.96 | 0.70 | 0.96 | 1.07 | −0.34 | |
| MtC20114 | MtGSa Glu synthase | 0.57 | 0.17 | 0.50 | 1.85 | 2.36 | |||
| MtC20236 | Alkaline/neutral invertase | 0.22 | 0.18 | 0.62 | 0.49 | 0.43 | 0.04 | 1.19 | 1.06 |
| MtC30105 | β-Amylase | 0.87 | 0.06 | 1.83 | 0.62 | 2.81 | 1.01 | 3.53 | 1.63 |
| MtC30278 | Pyruvate dehydrogenase E1 component | −0.51 | −0.22 | −1.25 | |||||
| MtC45259 | 3,4-Dihydroxy-2-butanone 4-phosphate synthase | 1.16 | 1.90 | ||||||
| MtC50550 | Lipase | 0.65 | −0.17 | 1.55 | 1.47 | 1.10 | 0.29 | ||
| MtC90461 | Acid β-fructofuranosidase precursor | −0.73 | −1.59 | −0.87 | −1.53 | ||||
| MtC91090 | 4-Hydroxyphenylpyruvate dioxygenase | 0.03 | 1.18 | 0.10 | 0.84 | 0.28 | 2.14 | 1.05 | |
| MtC92047 | Fructose-bisphosphate aldolase | 0.33 | 0.88 | 0.55 | 1.01 | ||||
| MtGSa | Glu synthetase a | 0.25 | 0.61 | 1.02 | 0.55 | 0.33 | 0.50 | 3.77 | 2.84 |
| Membrane Transport | |||||||||
| MtC00001 | Aquaporin | −1.23 | −1.10 | −1.51 | −0.65 | −1.39 | −1.42 | −2.70 | −1.94 |
| MtC10430 | [NODULIN] nodulin 26-like | 0.61 | 0.21 | 2.71 | 1.23 | 2.90 | 1.88 | 5.25 | 3.84 |
| MtC20134.1 | MtPT2 Phosphate transporter | −0.12 | −1.09 | −0.53 | −1.02 | ||||
| MtC20134.2 | MtPT1 Phosphate transporter | −0.65 | −1.91 | −1.06 | −1.68 | ||||
| MtC20204.1 | Hexose transporter | 0.09 | 1.67 | 0.56 | 2.13 | 0.74 | 3.88 | 2.08 | |
| MtC45479 | High-affinity nitrate transporter | −0.81 | −0.12 | −2.67 | −1.82 | −1.21 | −0.98 | −3.11 | −2.03 |
| MtC90689 | Cationic amino acid transporter | 0.00 | 1.56 | 1.05 | 1.69 | 0.74 | 1.99 | 1.73 | |
| MtC91857 | CHL1-like nitrate/chlorate transporter | −0.38 | −1.41 | −0.55 | −1.14 | ||||
| MtC93235 | Plasma membrane H+ATPase | −0.45 | 0.01 | −1.14 | −1.29 | −0.53 | −0.44 | −1.26 | −1.13 |
| Miscellaneous | |||||||||
| MtC00583 | [NODULIN] leghemoglobin 1 | 3.37 | 5.27 | 2.74 | 5.12 | 5.08 | 6.99 | 8.97 | |
| MtC10279 | Heme oxygenase | 0.26 | −0.19 | 0.19 | 0.31 | 1.21 | 2.02 | ||
| MtC20131 | Ferritin | 1.09 | 0.42 | 3.07 | 2.10 | 2.58 | 1.44 | 2.43 | 1.73 |
All ratios are expressed as log2 values (legend as in Table II).
Several leghemoglobin genes, providing an efficient system both to decrease the concentration of free oxygen molecules and deliver sufficient oxygen to the infected cells, are also strongly induced. An interesting correlation can be observed with the expression of a ferritin gene (MtC20131), encoding a protein that is likely to provide iron for incorporation into the oxygen-binding haem cofactor of leghemoglobin proteins. The concomitant induction of a haem oxygenase gene (MtC10279) in mature nodules is consistent with a recently published study that proposes a role for the encoded protein in the metabolism of haem cofactors (Baudouin et al., 2004).
Down-Regulated Metabolic Genes
A number of primary metabolism genes appear to be much less expressed in nodules than in roots. It can be noted in several instances that some members of multigene families are repressed concomitantly to the activation of other members: A well-documented example is the MtGSIb gene (Table IV), but this seems also to be the case for genes encoding a carbonic anhydrase (MtC10059), an invertase (MtC90461), an Asn synthase (MtC00241), or a Fru-bisphosphate aldolase (MtC00100). Such differential regulation within multigene families has often been described in plants, and several other examples are observed during nodulation in other functional classes (see below).
The large number of down-regulated genes may also reflect substantial differences in primary metabolism of roots and nodules, the latter being highly specialized for nitrogen fixation. Thus, dicarboxylic acids are a major carbon source for bacteroids. These are derived from PEP being first converted to oxaloacetate and subsequently to malate via the successive action of PEPC and malate dehydrogenase. This might explain some nodule-enhanced pathways, such as the fueling of the citric acid cycle via PEPC rather than pyruvate dehydrogenase (MtC20039, MtC30278), which, in contrast to PEPC, is more expressed in roots.
Membrane Transport
Membrane transporters are obviously important players for metabolic exchanges within the root nodule. The observed transcriptional activation of a hexose transporter gene (MtC20204.1) is certainly related to the root nodule functioning as a carbon sink. An increased expression of an amino acid transporter gene (MtC90689) can also be noted in young and mature nodules, which could reflect possible roles for amino acids, either as an alternative carbon source for the bacteroids or as the major compound of reduced nitrogen exported into the root phloem tissue. The most striking activation concerns the Nodulin 26 aquaporin-like gene (MtC10430), observed from early to late-nodulation stages, i.e. already before the peribacteroid membrane formation. At the same time, it can be observed that the expression of a series of 10 other aquaporin-encoding genes is turned down (e.g. MtC00001, Table IV), suggesting, as discussed above, that a symbiosis-specific gene takes over the function of nonsymbiotic members of the same family. The observed repression of a plasma membrane H+ ATPase (MtC93235) could correspond to a similar mechanism.
The two phosphate transporter genes, MtPT1 and MtPT2 (Liu et al., 1998), appear to be turned down in nodules (MtC20134.1, MtC20134.2), possibly reflecting the fact that nodules play no role in mineral uptake from the soil, in contrast to roots. Two further prominent examples of down-regulated genes are those encoding high- and low-affinity nitrate transporters (MtC45479 and MtC91857, respectively). This down-regulation is observed both in mature and immature nodules, suggesting a signaling mechanism that does not involve the reduced nitrogen resulting from nodule activity.
Nodulation and Defense Reactions
It is generally believed that a successful symbiotic interaction involves an inhibition of defense mechanisms to permit the establishment of a foreign organism within the plant host (Mithöfer, 2002). Part of our data is consistent with this point of view. A large collection of genes coding for pathogenesis and defense-related proteins, including many members of the PR10/Betv1, peroxidases, nonspecific lipid transfer proteins, and Kunitz-type trypsin inhibitor gene families (e.g. MtC00168, MtC10567.1, MtC10860/MtC10297, and MtC10756, respectively), as well as two disease resistance-related proteins (EDS1, MtC40039; HSR201, MtC50593), were found to be down-regulated (Supplemental Tables II and V). In addition, several genes found to be induced in M. truncatula leaves during the compatible interaction with the pathogenic fungus Colletotrichum trifolii (Torregrosa et al., 2004) were also found to be down-regulated. This includes several genes coding for lipoxygenases (MtC00480.1, MtC10441, MtC10070, MtC40012), particularly in the supernodulating background, a gene coding for a germin (MtC00172), as well as eight genes coding for PRPs (MtC10757, MtC00455, MtC10404, MtC10118, MtC93202, MtC10169, MtC00393, and MtC00014).
However, it should also be noted that several strongly up-regulated genes have clear homologies to pathogenesis-related proteins: knottins and thionins (MtN1/MtC00068, MtN15/MtC00380), PR1 (MtC00259/MtPR1-1), PR10 (MtN13/MtC10690), nonspecific lipid transfer proteins (MtN5/MtC00060.1), chitinases (MtC10312), thaumatin (MtC00235), proteinase inhibitors (MtC00300, MtC10968, MtC93406), and proteins induced by fungal elicitors or during syringolide-induced hypersensitive cell death (MtC90971, MtC91319; Hagihara et al., 2004). A gene coding for an isoflavone 7-O-methyltransferase highly homologous to an isoflavone 7-O-methyltransferase involved in the production of the phytoalexin medicarpin (He et al., 1998) is also induced in wild-type nodules (MtC45591). Finally, several genes coding for proteins involved in general stress responses are also found to be up-regulated during nodulation. These include a universal stress protein (MtC10058) and a 1-aminocyclopropane-1 carboxylic acid oxidase (MtC10108.2), the last enzyme of the ethylene biosynthetic pathway, often associated with biotic or abiotic stress (Kim et al., 1998), which we find induced at 3 dpi in the supernodulating background, possibly as a result of the high number of infections.
As already discussed (Gamas et al., 1998), this apparent contradiction could be explained by the necessity both to allow for Rhizobium infection and to control the extent of this infection, or to protect the nodules (carbon and nitrogen-rich organs) from pathogen and pest attacks. It should be mentioned here that some insect larvae feed exclusively on root nodules (Gerard, 2001). Many of the above-mentioned proteins are toxic for insects (Rudiger and Gabius, 2001), including α-amylase inhibitors, proteinase inhibitors, and knottin-like and thaumatin-like proteins. Lectins can also play such a role, which could explain the accumulation of MtC00344 transcripts in nodules. Small Cys-rich nodulins of the A1b/leginsulin family are strongly expressed both in seeds and in nodules, where they include MtN11, 16, and 17 (MtN11/MtC00746, MtN16/MtC00186, MtN17/MtC91045). PA1b, a 37-amino acid peptide found in pea seeds, was shown to be toxic against insect larvae (Gressent et al., 2003; Jouvensal et al., 2003) and could thus play a protective role. It could be the same for nodule-specific related proteins, which is supported by the fact that insect toxicity was detected in M. truncatula roots and even more in nodulated roots (B. Delobel and Y. Rahbé, personal communication).
Genes Encoding Regulatory Proteins
One of the important goals of our study was to identify regulatory genes controlling the developmental program associated with S. meliloti infection and nodule organogenesis and functioning. We found 79 differentially regulated genes (34 and 45 up- and down-regulated, respectively) potentially involved in regulatory mechanisms (see Supplemental Table II and Table V, classes VIII and X).
Signal Transduction Genes
Regarding early-nodulation stages, it is worth underlining the up-regulation of a gene coding for a β-subunit G-protein containing WD repeats (MtC30551), which is reminiscent of the potential involvement of G-proteins in Nod factor signal transduction (Pingret et al., 1998). A strongly induced gene, MtC10811, encodes a remorin, an intriguing protein able to bind complex galacturonides and for which a role in cell-to-cell communication has been proposed (Reymond et al., 1996). One additional gene coding for an annexin differing from the already characterized MtAnn1 (De Carvalho-Niebel et al., 2002) was also identified (MtC20129). These proteins could transduce a calcium signal, shown to play a key role in Nod factor signaling to cell cycle, cytoskeleton rearrangements, and/or vesicular trafficking.
In nodules, a variety of signaling genes were found to be induced, notably encoding an EF-hand calcium-binding protein (MtC91314), a protein phosphatase (MtC90276), protein kinases (MtC40161, MtC40043, MtC50559), or a phospholipase A (MtC91827).
Proteins encoded by repressed genes include a calcium-GTP binding protein (MtC45594), an IQ calmodulin-binding protein (MtC30108), two 14-3-3 proteins (MtC10022 and MtC00791.1), a Zn-finger protein (MtC10950), a protein kinase (MtC90003), and a putative receptor kinase (MtC90498.1) homologous to CLAVATA1, and Lj HAR or GmNARK, which are involved in autoregulation of nodulation (Krusell et al., 2002; Searle et al., 2003).
Transcription Factors
We have a special interest in TF genes because they represent potential master regulators able to control sets of genes associated with defined stages of the symbiotic developmental program. Thirteen and 11 putative TF genes were found to be up- or down-regulated, respectively, in nodules. Hierarchical clustering analysis (Fig. 1) clearly suggested a coregulation of several of them with nodulin genes, the expression of which they might thus control. qRT-PCR experiments were conducted on four TFs up-regulated in young nodules and belonging to different classes (Fig. 3). Array and RT-PCR data were consistent, confirming the robustness of our data, but the observed range of induction was clearly much larger with qRT-PCR. This reduced range on arrays could be due to cross-hybridization with member(s) of the same gene families having a different expression pattern and/or to the difficulty of quantifying very low expression levels (e.g. in the case of MtC00553 and MtC10582, these being very poorly expressed in roots).
One of the striking features of the symbiosis with Rhizobium is the development of a meristem leading to the formation of a new organ, the nodule. It is thus very interesting to find among the genes induced in young nodules several homologs of homeotic genes, known to play a role, among other things, in organ identity. The closest homolog of a MADS box TF gene activated in young and mature nodules (MtC20187, Fig. 3C) is the so-called SVP (short vegetative phase) gene of the jointless family that functions as a negative regulator of the vegetative-to-floral meristem transition in Arabidopsis (Hartmann et al., 2000). MtC00457, the induction of which was confirmed in nodules by qRT-PCR (data not shown), is a member of the no-apical meristem family, which includes genes like CUC3 required for boundary and shoot meristem formation (Vroemen et al., 2003). MtC50408, strongly induced in developing nodules (Fig. 3D), encodes a protein belonging to the large family of AP2/EREBP (APETALA2/ethylene-responsive element-binding protein) TFs, which play key roles in development or stress responses (see Riechmann and Meyerowitz, 1998). Ethylene is an important factor for the control of nodule development, possibly by cross-talk with the Nod factor signal transduction pathway (Oldroyd et al., 2001); it will be interesting to test whether MtC50408 is involved in that regulation process.
MtC10582 is one of the few TF genes strongly activated at 3 dpi (Fig. 3B). It encodes a B-subunit of the heterotrimeric CBF (CCAAT-binding factor) protein complex, which binds the CCAAT box found in many eukaryotic promoters. In plants, the CBF subunits are encoded by small gene families bearing specialized members that can play key roles in development (Lotan et al., 1998; Kwong et al., 2003).
Several other putative TF genes were more strongly induced in mature nodules and are likely to participate in later steps of nodule development or functioning. These encode a homeobox protein (MtC90017), basic helix-loop-helix proteins (MtC91049, MtC10648), and a homolog of the Krüppel-like Mszpt2-1 TF (MtC10310) involved in the differentiation of the nitrogen-fixing zone of nodules (Frugier et al., 2000).
Finally, several putative TFs were down-regulated in nodules and could thus correspond to regulatory elements specifically controlling root development. These include a homeobox Leu zipper protein (MtC30157) and a DNA-binding WRKY protein (MtC10372).
CONCLUSION
The current study, added to other large-scale transcriptome studies carried out on L. japonicus and on M. truncatula (Colebatch et al., 2002; Fedorova et al., 2002; Journet et al., 2002), confirms the power of global expression analyses and how complementary they can be to genetic approaches to identify symbiotic genes. Here, the use of arrays based upon root, nodule, and mycorrhiza cDNA libraries of biological samples highly enriched in symbiotic tissues and of various controls allowed us to identify with a good level of confidence a large collection of genes differentially regulated during nodulation. The fact that a series of well-characterized symbiotic gene markers exhibits expression patterns on micro- and macroarrays consistent with previous studies gives credence to the numerous new nodulin genes found here. A modest but inevitable level of false positives is likely to remain in our list of genes scored as differentially regulated, considering statistical arguments (we chose a P value ≤0.05) and the fact that cDNA clone resequencing allowed us to detect a global error rate of about 9% (see “Materials and Methods”).
The switch from a root-specific to a nodule-specific gene expression program is accompanied by the concomitant up- and down-regulation of many genes, including paralogs within gene families (observed here in at least 30 cases). Whether legume/nodule-specific genes can be found is a commonly raised question that is not easy to answer. Indeed the results of automatic analyses obviously depend on the thresholds chosen to define homologies at the DNA or protein levels and have to be completed by manual detailed studies to reach robust conclusions. As shown in the MENS database, a small group of genes were predicted to be legume specific on the basis of automatic protein analyses (WU-TBLASTN of MtC cluster predicted proteins against legume and nonlegume DNA databases, with similarity ≥60% and E value ≤1e−6). A few of them were found on macro- and microarrays to be up-regulated during nodulation (e.g. MtC10073, MtC90450, MtC30530, MtC10455). It would be interesting to investigate with finer studies whether or not other genes, notably those from the no-homology class exhibiting a strong induction during nodulation, have homologs restricted to legume species, and test their importance for symbiosis with Rhizobium.
A comparison to gene activation in AM is of obvious interest since symbiosis with Rhizobium and mycorrhizal fungi share some important genetic components (Gualtieri and Bisseling, 2000). RT-PCR experiments, as well as the analysis of promoter-gus fusions, have already indicated that several genes (e.g. MtENOD11, 12, 40, Nodulin 26, MtSucS1) are activated in both symbioses (van Rhijn et al., 1997; Journet et al., 2001; Hohnjec et al., 2003; Brechenmacher et al., 2004). Nevertheless, comparisons to recent results from Liu et al. (2003) and Küster et al. (2004) on AM-induced M. truncatula genes indicate that the overlap between nodule- and AM-related genes identified solely on the basis of DNA arrays is probably limited. As an example, 13 genes identified as being Sinorhizobium- or nodule-induced in this study were found to be activated at least 2-fold in AM with Glomus intraradices (Küster et al., 2004). These EST clusters represent the Nodulin 26 gene (MtC10430 and two additional cDNAs), homologs of auxin-down-regulated genes (MtC00667, MtC10128, MtC00182), a gene specifying a Cys proteinase (MtC91471), and a dehydration-induced protein (MtC00203). In addition, six EST clusters encode proteins of either no homology or unknown function (MtC00397, MtC10364, MtC30370, MtC45510, MtC91415, MtC10987, MtC92032). The defense gene PR1 was also found as transiently induced in mycorrhizae (Gianinazzi-Pearson et al., 1996). Although more comprehensive experiments need to be performed to draw a final conclusion, the finding that only a low number of genes was found to be coinduced during these two endosymbioses is not totally surprising since (1) these symbioses show important differences in terms of organs and functions involved; (2) the number of infected cells is significantly lower in AM as compared to root nodules, leading to significant dilution effects; and (3) possible common genes involved in signal perception and transduction during the initiation of both symbioses are probably expressed below a level that would allow their detection in pooled tissue samples.
It is difficult to make precise comparisons between studies based upon different references (e.g. MENS clusters versus The Institute for Genomic Research [TIGR] tentative consensus clusters for M. truncatula). Next generations of M. truncatula microarrays based upon oligonucleotides, hopefully shared by a large community, should be more powerful both to examine differential regulation within gene families and to compare the transcriptome expressed during various developmental responses, e.g. pathogenic and symbiotic interactions or nodule versus flower/seed development (two sinks for carbon).
Another important development of M. truncatula transcriptomics studies consists of various tools now available to assess the functional importance of genes identified from global expression analyses, e.g. based upon RNAi or TILLING approaches. We are now undertaking such functional studies for genes of interest, particularly those susceptible to play a regulatory role during symbiotic interactions, and the next challenge will be to define corresponding regulation networks.
MATERIALS AND METHODS
Bacterial Strains, Media, and Growth Conditions
Sinorhizobium meliloti RCR2011 pXLGD4 (GMI 6526; Ardourel et al., 1994) and S. meliloti RCR2011 nodA::Tn5 pXLGD4 (GMI 6702; Debelle et al., 1986) were grown in TY medium (Sambrook et al., 1989), supplemented with 6 mm calcium chloride at 30°C using tetracycline at 10 μg mL−1. For plant inoculation, 10 mL of this culture diluted to an OD600 of 1 in nitrogen-free plant culture medium were added to 10 L of seedling medium in aeroponic caissons, as described below.
Cultivation of Plants, Harvesting of Tissues, and Isolation of Total RNA
Plant growth chamber conditions were the following: temperature, 22°C; 75% hygrometry; light intensity, 200 μE m−2 s−1; light/dark photoperiod, 16 h/8 h. Plants of wild-type Medicago truncatula Gaertn cv Jemalong genotype A17 and of supernodulating M. truncatula TR122 mutant (Sagan et al., 1995) and nfp and hcl A17 mutants (Catoira et al., 2001; Ben Amor et al., 2003) were grown aeroponically during 11 d using a nitrogen-rich medium, as in Journet et al. (2001), with around 90 plants per caisson. Plants were then nitrogen starved for 4 d before being inoculated with S. meliloti. Roots were harvested just before inoculation as reference material, taking the whole root system from about 2 cm below the crown. Whole root systems (A17, TR122, nfp and hcl plants) were harvested at 3 and 6 dpi for TR122, whereas isolated A17 nodules were harvested at 4 and 10 dpi (from about 30 plants). All material was frozen in liquid nitrogen after no more than 5 min on ice. Total RNA was then extracted with Trizol (Invitrogen, Cergy Pontoise cedex, France).
Printing and Layout of Mt6k-RIT Macro- and Microarrays
The generation of Mt6k-RIT PCR-based micro- and macroarrays was described in detail by Küster et al. (2004).
Control Sequencing of Selected PCR Products and Reannotation of Arrayed PCR Products
A technical validation of the macro- and microarray printing process was carried out as described (Küster et al., 2004). To assess the error rate across the arrayed PCR products, 165 randomly distributed clones were sequenced from the DNA used for gridding macroarrays. Fifteen of them (i.e. 9%) were found to differ from the expected sequences found in the MENS database (http://medicago.toulouse.inra.fr/MENS). Probes where the control sequencing confirmed the array of wrong or multiple PCR products were not considered further. The annotation of all EST clusters showing a differential regulation on macro- or microarrays was updated using tools provided in the MENS database.
Cy Labeling, Microarray Hybridization, Image Acquisition, and Data Analysis
Twenty micrograms of total RNA were used to synthesize Cy3- or Cy5-labeled first-strand cDNA targets, as described in Küster et al. (2004). Spot detection, image segmentation, and image quantification were performed using ImaGene 5.0 software (BioDiscovery, Los Angeles). ImaGene output files were imported into the EMMA 1.0 microarray analysis software (Dondrup et al., 2003). During import, spots flagged as empty or poor were removed. After local background subtraction and after applying a floor value of 20, the resulting signal intensities were used for data normalization using a local regression (Lowess) procedure. Subsequently, M values (expression ratios) and A values (average signal intensities) were calculated according to Dudoit et al. (2002). Genes significantly up- or down-regulated were identified by applying a t test using EMMA 1.0. Genes were regarded as being differentially expressed if P ≤ 0.05 and M ≥ 1 or ≤ −1. Technical and biological variation was addressed by investigating two different biological samples in two (TR122 samples) or three (4 and 10 dpi A17 nodules and noninoculated samples) technical replicates. In each case, dye-specific variations were accounted for by applying a dye swap between the two different biological samples.
33P Labeling of Hybridization Targets, Hybridization, and Image Acquisition
Twenty micrograms of total RNA were reverse transcribed in the presence of 50 μCi [α-33P]dCTP with SuperscriptII RTase (Invitrogen), using anchored oligo(dT)17 primer and 0.5 mm d[A, G, T]TP. A cold chase (1 h with 2 mm dCTP) was carried out after 1 h incubation at 42°C. Reaction was stopped and RNA degraded in 0.3 m NaOH (30 min at 68°C). After neutralization in 0.4 m Tris-HCl, pH 7.5, cDNAs were purified on MicroSpin S-200 columns (Amersham Biosciences Europe, Orsay, France). The labeling efficiency was quantified by liquid scintillation counting. Membranes were prehybridized during 3 to 4 h at 65°C in 15 mL of Church buffer (Church and Gilbert, 1984). The hybridization was performed at 65°C for 16 h in 10 mL of fresh hybridization solution containing the denaturated labeled target (25−50 × 106 cpm). Membranes were rapidly rinsed with solution A (2× SSC, 0.1% [w/v] SDS), washed at 65°C for 15 min in 100 mL of solution A, then 100 mL of solution B (0.2× SSC, 0.1% [w/v] SDS). Membranes were exposed for 4 d to a low-energy screen (Kodak Molecular Dynamics, Uppsala), which was then scanned with a storm 840 PhosphoImager (Molecular Dynamics; Amersham Biosciences Europe).
Analysis of Image Data from Mt6k-RIT Macroarray Hybridizations
Signals were quantified by ImaGene 5.0 and the resulting data analyzed with GeneSight 3.5 software (both BioDiscovery). The total signal of each spot was corrected by subtracting the median background of the corresponding subgrid (16 neighboring spots). Negative values were floored to a value of 20. The resulting total net signal values were log2 transformed, and log2 ratios between experiments and noninoculated controls were calculated. For membrane-to-membrane comparisons, results were normalized by using the mean of all spot signals. Replicates (two to four biological replicates, as well as duplicate spots) were combined and averaged. Any values showing more than 2 sds from the mean were considered as outliers and discarded. The significance of our results was then assessed on the basis of a t test (provided by the GeneSight 3.5 package), combined with an examination of induction and repression ratios.
All data files for Mt6k-RIT macro- and microarrays are available through the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress; array accession nos. A-MEXP-80 and A-MEXP-81) under experiment accession number E-MEXP-129.
qRT-PCR
Analyzed RNA samples were previously used for macro- and microarray studies. Total RNA was treated with Dnase I (Roche Diagnostics, Penzberg, Germany), and 5 μg were reverse transcribed following the recommended protocol (SuperscriptII; Invitrogen). The qRT-PCR reaction was performed in a LightCycler (Roche) with 1 μL of 50-fold diluted cDNA, 6.75 μL of water, 1 μL of 25 mm MgCl2, 0.25 μL of mixed primers (25 μm each), and 1 μL of LightCycler FastStart DNA Master SYBR green I (Roche). Primers (Supplemental Table VI) were designed using Vector NTI (InforMax, Oxford). Samples were preincubated at 95°C for 8 min, which was followed by 45 cycles (95°C, 5 s; 60°C, 7 s; 72°C, 7 s) with a temperature transition rate of 20°C s−1. The melting/fusion curves were set up between 65°C and 95°C, with a transition rate of 0.1°C s−1. To get the crossing point (cycle threshold), we used the second derivative maximum method and the arithmetic signal baseline adjustment. The cDNA abundance was assessed from a standard curve established from a serial dilution (from 10−2 to 10−8 ng μL−1) of an appropriate template (plasmid or PCR product) containing the corresponding insert. The values were normalized with an internal control, EF1-α, verified to be expressed at a similar level in the tested conditions.
Supplementary Material
Acknowledgments
We thank Jean Dénarié for his very valuable comments on the manuscript. Eva Schulte-Berndt and Silvia Rüberg (Institute of Genome Research and Department of Genetics, Bielefeld University, Germany) are acknowledged for their expert support in the initial phase of array construction. We thank J. Garcia for his help on plant growth and harvesting. We are grateful to Yvan Rahbé (INRA-INSA, Villeurbanne, France) for sharing unpublished results with us and to Gabriella Endre and Kate VandenBosch (University of Minnesota, St. Paul, MN), Julie Cullimore (INRA-CNRS, Toulouse, France), and Jeff Landgraf (Plant Research Laboratory, Michigan State University, East Lansing) for sharing cDNA clones for array construction.
This work was supported by the European Union (project MEDICAGO; QLG2–CT–2000–00676) and the Deutsche Forschungsgemeinschaft (grant no. BIZ 7). N.H. and H.K. acknowledge financial support from the International NRW Graduate School in Bioinformatics and Genome Research. F.E.Y. was supported by grants from the FP5 MEDICAGO program and INRA. B.B.A. was financed by a grant from the French government in the spirit of French-Tunisian cooperation.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.043612.
References
- Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Prome JC, Denarie J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin E, Frendo P, Le Gleuher M, Puppo A (2004) A Medicago sativa haem oxygenase gene is preferentially expressed in root nodules. J Exp Bot 55: 43–47 [DOI] [PubMed] [Google Scholar]
- Ben Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Denarie J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Brechenmacher L, Weidmann S, Van Tuinen D, Chatagnier O, Gianinazzi S, Franken P, Gianinazzi-Pearson V (2004) Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza 14: 253–262 [DOI] [PubMed] [Google Scholar]
- Budelier KA, Smith AG, Gasser CS (1990) Regulation of a stylar transmitting tissue-specific gene in wild-type and transgenic tomato and tobacco. Mol Gen Genet 224: 183–192 [DOI] [PubMed] [Google Scholar]
- Carvalho H, Lescure N, de Billy F, Chabaud M, Lima L, Salema R, Cullimore J (2000) Cellular expression and regulation of the Medicago truncatula cytosolic glutamine synthetase genes in root nodules. Plant Mol Biol 42: 741–756 [DOI] [PubMed] [Google Scholar]
- Catoira R, Timmers AC, Maillet F, Galera C, Penmetsa RV, Cook D, Denarie J, Gough C (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coba de la Pena T, Frugier F, McKhann HI, Bauer P, Brown S, Kondorosi A, Crespi M (1997) A carbonic anhydrase gene is induced in the nodule primordium and its cell-specific expression is controlled by the presence of Rhizobium during development. Plant J 11: 407–420 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15: 411–420 [DOI] [PubMed] [Google Scholar]
- Cullimore J, Denarie J (2003) Plant sciences. How legumes select their sweet talking symbionts. Science 302: 575–578 [DOI] [PubMed] [Google Scholar]
- Debelle F, Rosenberg C, Vasse J, Maillet F, Martinez E, Denarie J, Truchet G (1986) Assignment of symbiotic developmental phenotypes to common and specific nodulation (nod) genetic loci of Rhizobium meliloti. J Bacteriol 168: 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho-Niebel F, Timmers AC, Chabaud M, Defaux-Petras A, Barker DG (2002) The Nod factor-elicited annexin MtAnn1 is preferentially localised at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. Plant J 32: 343–352 [DOI] [PubMed] [Google Scholar]
- Dondrup M, Goesmann A, Bartels D, Kalinowski J, Krause L, Linke B, Rupp O, Pühler A, Meyer F (2003) EMMA: a platform for consistent storage and efficient analysis of microarray data. J Biotechnol 106: 135–146 [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP (2002) Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica 12: 111–139 [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugier F, Poirier S, Satiat-Jeunemaitre B, Kondorosi A, Crespi M (2000) A Kruppel-like zinc finger protein is involved in nitrogen-fixing root nodule organogenesis. Genes Dev 14: 475–482 [PMC free article] [PubMed] [Google Scholar]
- Galvez S, Hirsch AM, Wycoff KL, Hunt S, Layzell DB, Kondorosi A, Crespi M (2000) Oxygen regulation of a nodule-located carbonic anhydrase in alfalfa. Plant Physiol 124: 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, de Billy F, Truchet G (1998) Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol Plant Microbe Interact 11: 393–403 [DOI] [PubMed] [Google Scholar]
- Gamas P, de Carvalho-Niebel F, Lescure N, Cullimore J (1996) Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact 9: 233–242 [DOI] [PubMed] [Google Scholar]
- Gerard PJ (2001) Dependence of Sitona lepidus (Coleoptera: Curculionidae) larvae on abundance of white clover Rhizobium nodules. Bull Entomol Res 91: 149–152 [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Dumas-Gaudot E, Golotte A, Tahiri-Alaoui A, Gianinazzi S (1996) Cellular and molecular defence-related responses to invasion by arbuscular mycorrhizal fungi. New Phytol 133: 45–57 [Google Scholar]
- Greene EA, Erard M, Dedieu A, Barker DG (1998) MtENOD16 and 20 are members of a family of phytocyanin-related early nodulins. Plant Mol Biol 36: 775–783 [DOI] [PubMed] [Google Scholar]
- Gressent F, Rahioui I, Rahbé Y (2003) Characterization of a high affinity binding site for the pea albumin 1b (PA1b) entomotoxin in the weevil Sitophilus. Eur J Biochem 270: 2429–2435 [DOI] [PubMed] [Google Scholar]
- Gualtieri G, Bisseling T (2000) The evolution of nodulation. Plant Mol Biol 42: 181–194 [PubMed] [Google Scholar]
- Hagihara T, Hashi M, Takeuchi Y, Yamaoka N (2004) Cloning of soybean genes induced during hypersensitive cell death caused by syringolide elicitor. Planta 218: 606–614 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- He XZ, Reddy JT, Dixon RA (1998) Stress responses in alfalfa (Medicago sativa L). XXII. cDNA cloning and characterization of an elicitor-inducible isoflavone 7-O-methyltransferase. Plant Mol Biol 36: 43–54 [DOI] [PubMed] [Google Scholar]
- Heard J, Dunn K (1995) Symbiotic induction of a MADS-box gene during development of alfalfa root nodules. Proc Natl Acad Sci USA 92: 5273–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297: 793–797 [DOI] [PubMed] [Google Scholar]
- Hirsch AM, Fang Y, Asad S, Kapulnik Y (1997) The role of phytohormones in plant-microbe symbioses. Plant Soil 194: 171–184 [Google Scholar]
- Hohnjec N, Becker JD, Pühler A, Perlick AM, Küster H (1999) Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Mol Gen Genet 261: 514–522 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Pühler A, Küster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant Microbe Interact 16: 903–915 [DOI] [PubMed] [Google Scholar]
- Jimenez-Zurdo JI, Frugier F, Crespi MD, Kondorosi A (2000) Expression profiles of 22 novel molecular markers for organogenetic pathways acting in alfalfa nodule development. Mol Plant Microbe Interact 13: 96–106 [DOI] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Arnould C, Morandi D, Barker DG, Gianinazzi-Pearson V (2001) Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol Plant Microbe Interact 14: 737–748 [DOI] [PubMed] [Google Scholar]
- Journet EP, Pichon M, Dedieu A, de Billy F, Truchet G, Barker DG (1994) Rhizobium meliloti Nod factors elicit cell-specific transcription of the ENOD12 gene in transgenic alfalfa. Plant J 6: 241–249 [DOI] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvensal L, Quillien L, Ferrasson E, Rahbe Y, Gueguen J, Vovelle F (2003) PA1b, an insecticidal protein extracted from pea seeds (Pisum sativum): 1H-2-D NMR study and molecular modeling. Biochemistry 42: 11915–11923 [DOI] [PubMed] [Google Scholar]
- Kim YS, Choi D, Lee MM, Lee SH, Kim WT (1998) Biotic and abiotic stress-related expression of 1-aminocyclopropane-1-carboxylate oxidase gene family in Nicotiana glutinosa L. Plant Cell Physiol 39: 565–573 [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426 [DOI] [PubMed] [Google Scholar]
- Küster H, Albus U, Frühling M, Tchetkova S, Tikhonovich IA, Pühler A, Perlick AM (1997) The asparagine synthetase gene VfAS1 is strongly expressed in the nitrogen-fixing zone of broad bean (Vicia faba L.) root nodules. Plant Sci 124: 89–95 [Google Scholar]
- Küster H, Hohnjec N, Krajinski F, El Yahyaoui F, Manthey K, Gouzy J, Dondrup M, Meyer F, Kalinowski J, Brechenmacher L, et al (2004) Construction and validation of comprehensive cDNA-based macro- and microarrays to explore root endosymbioses in the model legume Medicago truncatula. J Biotechnol 108: 95–113 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamblin AF, Crow JA, Johnson JE, Silverstein KA, Kunau TM, Kilian A, Benz D, Stromvik M, Endre G, VandenBosch KA, et al (2003) MtDB: a database for personalized data mining of the model legume Medicago truncatula transcriptome. Nucleic Acids Res 31: 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ (1998) Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant Microbe Interact 11: 14–22 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Mok DW (1999) A gene encoding the cytokinin enzyme zeatin O-xylosyltransferase of Phaseolus vulgaris. Plant Physiol 120: 553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Charon C, Rolfe BG, Kondorosi A, Crespi M (2000) Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol Plant Microbe Interact 13: 617–628 [DOI] [PubMed] [Google Scholar]
- Matsuyama T, Yasumura N, Funakoshi M, Yamada Y, Hashimoto T (1999) Maize genes specifically expressed in the outermost cells of root cap. Plant Cell Physiol 40: 469–476 [DOI] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A (2002) Suppression of plant defence in rhizobia-legume symbiosis. Trends Plant Sci 7: 446–450 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Wang L, Allen MS, Stevens YY, Qin W, Snider J, von Schwartzenberg K (2000) Adenosine kinase of Arabidopsis. Kinetic properties and gene expression. Plant Physiol 124: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Engstrom EM, Long SR (2001) Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13: 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M (2000) Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr. Opin Plant Biol 3: 320–328 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR (2003) Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingret J-L, Journet EP, Barker DG (1998) Rhizobium Nod factor signalling: evidence for a G protein-mediated transduction mechanism. Plant Cell 10: 659–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbun EA, Naldrett MJ, Brewin NJ (2002) Identification of a family of extensin-like glycoproteins in the lumen of rhizobium-induced infection threads in pea root nodules. Mol Plant Microbe Interact 15: 350–359 [DOI] [PubMed] [Google Scholar]
- Reymond P, Kunz B, Paul-Pletzer K, Grimm R, Eckerskorn C, Farmer EE (1996) Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell 8: 2265–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Rudiger H, Gabius HJ (2001) Plant lectins: occurrence, biochemistry, functions and applications. Glycoconj J 18: 589–613 [DOI] [PubMed] [Google Scholar]
- Sagan M, Morandi D, Tarenghi E, Duc G (1995) Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after g-ray mutagenesis. Plant Sci 111: 63–71 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Schultze M, Kondorosi A (1998) Regulation of symbiotic root nodule development. Annu Rev Genet 32: 33–57 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112 [DOI] [PubMed] [Google Scholar]
- Serino G, Deng XW (2003) The COP9 signalosome: regulating plant development through the control of proteolysis. Annu Rev Plant Biol 54: 165–182 [DOI] [PubMed] [Google Scholar]
- Shi L, Twary SN, Yoshioka H, Gregerson RG, Miller SS, Samac DA, Gantt JS, Unkefer PJ, Vance CP (1997) Nitrogen assimilation in alfalfa: isolation and characterization of an asparagine synthetase gene showing enhanced expression in root nodules and dark-adapted leaves. Plant Cell 9: 1339–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R, Keim P, Vodkin L, Retzel E, Clifton SW, Waterston R, Smoller D, Coryell V, Khanna A, Erpelding J, et al (2002) A compilation of soybean ESTs: generation and analysis. Genome 45: 329–338 [DOI] [PubMed] [Google Scholar]
- Stougaard J (2000) Regulators and regulation of legume root nodule development. Plant Physiol 124: 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BH, Scheuring CF (1994) A molecular marker for lateral root initiation: The RSI-1 gene of tomato (Lycopersicon esculentum Mill) is activated in early lateral root primordia. Mol Gen Genet 243: 148–157 [DOI] [PubMed] [Google Scholar]
- Torregrosa C, Cluzet S, Fournier J, Huguet T, Gamas P, Prosperi JM, Esquerré-Tugayé M-T, Dumas B, Jacquet C (2004) Cytological, genetic and molecular analysis to characterise compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii. Mol Plant Microbe Interact 17: 909–920 [DOI] [PubMed] [Google Scholar]
- Udvardi MK (2002) Legume genomes and discoveries in symbiosis research. Genome Biol 3: reports4028 [DOI] [PMC free article] [PubMed]
- van de Wiel C, Scheres B, Franssen H, van Lierop MJ, van Lammeren A, van Kammen A, Bisseling T (1990) The early nodulin transcript ENOD2 is located in the nodule parenchyma (inner cortex) of pea and soybean root nodules. EMBO J 9: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rhijn P, Fang Y, Galili S, Shaul O, Atzmon N, Wininger S, Eshed Y, Lum M, Li Y, To VV, et al (1997) Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium-induced nodules may be conserved. Proc Natl Acad Sci USA 94: 5467–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Gantt JS (1992) Control of nitrogen and carbon metabolism in root nodules. Physiol Plant 85: 266–274 [Google Scholar]
- VandenBosch K, Stacey G (2003) Summaries of legume genomics projects from around the globe. Community resources for crops and models. Plant Physiol 131: 840–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Camut S, Truchet G (1990) Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol 172: 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HH, Orbach MJ, Hirsch AM, Hawes MC (1999) Meristem-localized inducible expression of a UDP-glycosyltransferase gene is essential for growth and development in pea and alfalfa. Plant Cell 11: 2303–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.