Abstract
The ancestral shared synteny concept has been advocated as an approach to positionally clone genes from complex genomes. However, the unified grass genome model and the study of grasses as a single syntenic genome is a topic of considerable controversy. Hence, more quantitative studies of cereal colinearity at the sequence level are required. This study compared a contiguous 300-kb sequence of the barley (Hordeum vulgare) genome with the colinear region in rice (Oryza sativa). The barley sequence harbors genes involved in endosperm texture, which may be the subject of distinctive evolutionary forces and is located at the extreme telomeric end of the short arm of chromosome 5H. Comparative sequence analysis revealed the presence of five orthologous genes and a complex, postspeciation evolutionary history involving small chromosomal rearrangements, a translocation, numerous gene duplications, and extensive transposon insertion. Discrepancies in gene content and microcolinearity indicate that caution should be exercised in the use of rice as a surrogate for map-based cloning of genes from large genome cereals such as barley.
Gene content among higher eukaryotes appears to be relatively constant, ranging from 25,000 to 43,000 genes even though the genome size varies by 600-fold among angiosperms alone (Bennett et al., 1982; Bennett and Leitch, 1995, 1997; Miklos and Rubin, 1996). In the Gramineae, the allohexaploid genome of bread wheat (Triticum aestivum; 17,000 Mb) is approximately 3, 6, and 35 times larger than the barley (Hordeum vulgare; 5,300 Mb), maize (Zea mays; 2,500 Mb), and rice (Oryza sativa; 440 Mb) genomes, respectively (Arumuganathan and Earle, 1991; Shields, 1993). Comparative mapping studies have shown that, despite substantial variation in genome size and chromosome number, grass species have maintained significant conservation of gene and marker order (colinearity) and have sustained a minimal number of large chromosomal rearrangements since their divergence 50 to 80 million years ago (Wolfe et al., 1989; Crepet and Feldman, 1991; Ahn and Tanksley, 1993; Clark et al., 1995; Moore et al., 1995; Devos and Gale, 1997; Gale and Devos, 1998; Keller and Feuillet, 2000). The high degree of observed colinearity, coupled with the assumption that the essential components for growth and development are conserved among plants, led to the use of model organisms with small genome sizes, namely Arabidopsis and rice, as tools for plant genomics studies.
Despite the apparent conservation of gene order and content on a full genome scale, at the local level various small chromosomal rearrangements, such as segmental inversions, translocations, insertions, and deletions, have been reported to disrupt the degree of microcolinearity (for review, see Bennetzen, 2000; Bennetzen and Ramakrishna, 2002; Feuillet and Keller, 2002; Bennetzen and Ma, 2003). Even in instances where gene order was found to be conserved, the presence of large expanses of nested transposable sequence in plants of large genome size, including maize, barley, and wheat, were found to have a notable impact on the distribution of genes relative to closely related species of smaller genome size, such as rice and sorghum (Sorghum bicolor; Chen et al., 1998; Dubcovsky et al., 2001). To date, 11 large contiguous barley genomic sequences have been reported in the literature representing 1.35 Mb of sequence (Panstruga et al., 1998; Shirasu et al., 2000; Dubcovsky et al., 2001; Rostoks et al., 2002; Wei et al., 2002; Yan et al., 2002; Gu et al., 2003). Coupled with descriptions of large contiguous regions of the wheat and maize genomes, this information provides invaluable insight into the genome organization of large genome crop species (SanMiguel et al., 1996; Wicker et al., 2001).
Although several studies have described the levels of microcolinearity between Triticeae species and rice (Kilian et al., 1997; Han et al., 1998, 1999; Druka et al., 2000; Li and Gill, 2002), only two previous studies have compared large orthologous regions from rice and barley at the sequence level (Dubcovsky et al., 2001; Brunner et al., 2003). Such comparisons are vital for predicting the extent to which comparative genomics approaches based on the fully sequenced rice genomes (Goff et al., 2002; Sasaki et al., 2002; Yu et al., 2002; Wu et al., 2004) will be applicable for the transfer of information between organisms for molecular breeding, association mapping, and positional cloning strategies in related organisms of large genome size.
This paper describes the generation and sequencing of a contiguous genomic region of the barley genome represented by three bacterial artificial chromosomes (BACs) that cover the Ha locus and comparison with the colinear genomic region in rice. Our results indicate that comparative genomics can be an invaluable resource for the identification and determination of gene structure and provide new insights in the processes of genome evolution. However, the extensive number of small chromosomal rearrangements, including the absence of the entire grain (endosperm) texture gene family in rice, could complicate shuttle mapping and cloning approaches (Delseny, 2004) based exclusively on model genomes such as rice.
RESULTS
Identification and Sequencing of Barley BAC Clones
Fluorescent-based fingerprinting of 14 BACs believed to harbor the grain softness protein (GSP) gene identified BAC122.a5 as the clone that exhibited the most extensive coverage of the genomic region flanking the GSP locus. To extend this physical region to include the genetically linked hordoindoline genes (Rouves et al., 1996; Beecher et al., 2001), additional screens of the Morex BAC library were performed using gene-specific probes designed from the orthologous wheat sequences (puroindolines; GenBank accession nos. AJ249929 and AJ249928). Size determination and BAC end sequencing enabled the selection of two clones (BACs 519.k7 and 799.c8) that would provide minimal overlap and maximum coverage of the target region. The three contiguous BACs were sequenced and assembled using a shotgun sequence approach (6,912 clones with an average length of 600 quality bp) to obtain approximately 14 times coverage of the overall physical contig (303 kb). Two problematic regions prevented the completion of a continuous sequence. The first difficult region was composed of an approximately 340-bp AT-rich tandemly repeated segment located within the truncated Caspar_AY643842_1 transposon at the extreme 5′ region of the contig (BAC517.k9; Fig. 1A). PCR amplification confirmed the subcontig assembly and the estimated gap length indicated that three to four copies of the tandem duplication are missing from the sequence. The second problematic region also involved an AT-rich tandem duplication (42 bp) located approximately 3 kb downstream of the chalcone synthase (HvCHS) gene (BAC799.c8; Fig. 1A).
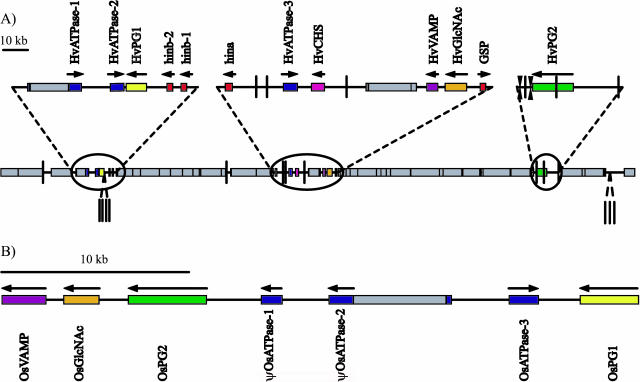
Figure 1.
A linear representation of the gene content and organization of the (A) region containing the barley Ha locus and its (B) colinear rice region. Coding sequence is represented by colored boxes, and arrows designate gene orientation. Repetitive sequence is represented by shaded boxes. MITEs are indicted by a vertical bar. tRNA are indicated by double arrowheads.
Gene Density of the Barley Genomic Region
The gene density of this region was determined through the integration of several different gene prediction applications and homology to previously characterized genes and expressed sequence tags (ESTs) available in the public databases. In total, 12 putative protein-coding and two duplicated tRNAARG genes (Fig. 1A) were identified within the 303-kb contiguous sequence. All exon:intron splice junctions contained the conserved GT and AG intron borders and a minimum of five of the nine (5′-CAG:GTAAGT-3′) and three of the five (5′-GCAG:G-3′) consensus nucleotides for the respective exon:intron and intron:exon splice sites in plants, with one exception. The border between exon 1 and intron 1 of the putative synaptobrevin (vesicle associated membrane protein, HvVAMP) gene contained only four of the nine exon:intron consensus nucleotides. However, both the presence of the mandatory GT intron border and splice agreement with more than one EST provided further support that this is a functional splice site.
Three of the four candidate grain texture genes, hinb-1, hinb-2, and hina, were found in the same orientation. However, HvGSP was in the opposite orientation (Fig. 1A). Homology at the protein level suggests that all four are members of the same gene family and may have resulted from duplications of a single ancestral gene. Based on nucleotide sequence homology, the original duplication resulted in HvGSP and one of the hordoindoline genes. Subsequent duplications generated templates for the gradual divergence of hina and hinb and an additional hinb copy.
Three of the putative genes belong to the ATPase associated activities superfamily characterized by one or two conserved domains (ATPase associated activities modules) responsible for ATP binding (Patel and Latterich, 1998). This family of genes is ubiquitous for all organisms and is involved in numerous cellular activities including membrane fusion, proteolysis, and DNA replication (Ogura and Wilkinson, 2001). HvATPase-2 and HvATPase-3 code for 518 and 516 amino acid proteins that are 84% and 80% identical at the nucleotide and protein level, respectively. The ψHvATPase-1 pseudogene has maintained 84% and 91% nucleotide homology to HvATPase-2 and HvATPase-3, respectively, despite the insertion of the HORPIA-2_AY643843 retrotransposon and several insertion and deletion events causing shifts in the reading frame. Remnants of an additional ATPase gene (ψHvATPase-4) were detected immediately downstream to HvATPase-3, demonstrating 81% homology to HvATPase-3. This copy has been severely truncated by a deletion of over 1 kb from the internal portion of the coding sequence. Evidence of yet another degenerate ATPase (ψHvATPase-5) gene exists in the region flanked by ψHvATPase-1 and HvATPase-2. A stretch of approximately 500 bp exhibits 88% homology to the immediate 5′ flanking sequence of HvATPase-2. This precedes a shorter segment with 88% homology. The full-length ATPase genes have maintained considerable identity across the entire coding region. However, little homology was detected among the flanking sequences. This hinders resolution of the history of duplication of this gene family cluster. Based on coding sequence homology alone, the original duplication probably resulted in HvATPase-2 and one of the other two full-length copies with a second duplication generating the third copy (Fig. 3). Additional duplications of both HvATPase-2 and HvATPase-3 resulted in ψHvATPase-4 and ψHvATPase-5, respectively. Genomic sequences of other barley lines or close barley relatives are needed to discern the exact series of events.
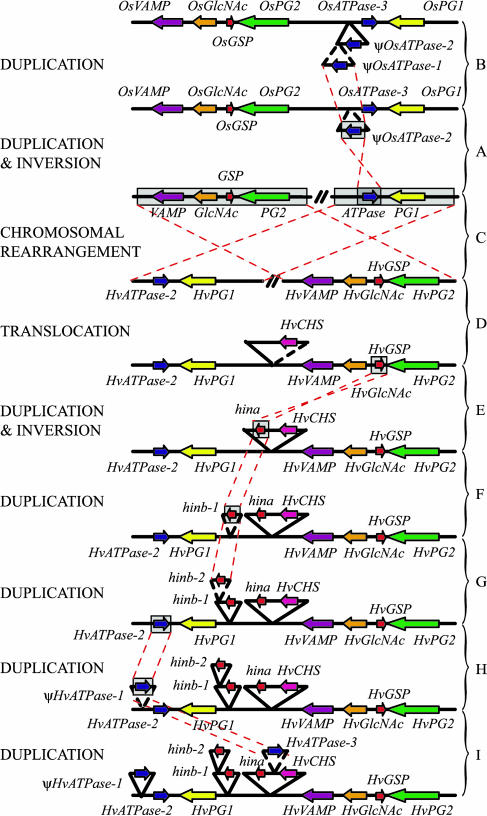
Figure 3.
A visual representation of one possible evolutionary scheme between the rice and barley colinear sequences. Evolutionary events move upwards toward present day rice (A and B) and downward toward present day barley (C–I) from the presumed last common ancestor. C, An intra-chromosomal rearrangement results in the repositioning of two conserved gene clusters. D, Translocation involves the relocation of CHS. E to G, Subsequent duplications and a gene inversion generate the individual grain texture genes. A and B, H to I, Independent gene duplications and inversions generate numerous copies of ATPase in both species. The two severely degenerate copies of barley ATPase are not present in this scheme.
Three out of the five remaining genes showed significant homology to previously described proteins: naringenin-chalcone synthase (HvCHS), N-acetylglucosaminyltransferase (HvGlcNAc), and synaptobrevin (HvVAMP), a vesicle associated membrane protein. CHS is a member of the chalcone synthase gene family. Chalcone is a key compound in the phenylpropenoid pathways involved in various cellular functions, including flower pigmentation (anthocyanin) and microbial defense (phytoalexins; Dixon et al., 1995, 1996; Dixon and Paiva, 1995; Shirley, 1996). Synaptobrevin is involved in a complex of SNARE proteins that control the regulation of vesicle docking and fusion during transport (Trimble et al., 1988; Baurnert et al., 1989; Sollner et al., 1993; Weber et al., 1998; Chen and Scheller, 2001). GlcNAc is a member of the large enzymatic superfamily of UDP glycosyltransferases. UDP glycosyltransferases regulate the transfer of sugar molecules (glycosyl residues) between different chemical R-groups (aglycones), thus indirectly regulating the biochemical properties of aglycones, i.e. secondary metabolites involved in abiotic stress and defense responses, hormones, and foreign chemical substances (xenobiotics, such as pesticides and herbicides; Li et al., 2001; Ross et al., 2001). Two additional putative genes (HvPG1 and HvPG2) whose functions remain to be determined are also present in the contig. Although EST homology is low (pLog > E-6) for both genes and is limited to members of the grass family, HvPG2 shows significant protein homology (pLog ≥ E-44) to several predicted proteins from mammalian species, including Rattus norvegicus, Homo sapiens, and Mus musculus (GI accession nos. 34867764, 13376072, and 21313472, respectively).
Composition of the Barley Intergenic Space
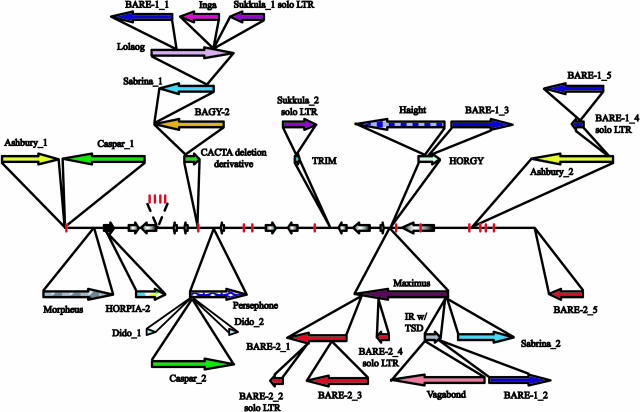
Over 75% of the contiguous barley sequence was composed of repetitive elements (Table I). The position, orientation, and order of insertion of the different transposable elements are depicted in Figure 2. The major portion of insertional activity has been directed to the intergenic space between hinb-1 and hina and between HvGSP and HvPG2. Approximately 93% of the 78-kb region separating hinb-1 and hina is composed of two separate nested element clusters. The Sukkula_AY643843_1 solo long terminal repeat (LTR), the BARE-1_AY643843_1 retrotransposon, and the truncated Inga_AY643843 retrotransposon represent the last of a series of insertions forming the largest of the two clusters involving the now degenerate CACTA transposon and BAGY-2_AY643843, Sabrina_AY643843_1, and Lolaog_AY643843 retrotransposons. The smaller cluster is composed of the full-length Caspar_AY643843_2 transposon immediately flanked by two identical putative short interspersed nuclear elements (SINEs; Dido_AY643843_1 and Dido_AY643843_2). All three elements are inserted into the extreme 5′ end of the novel long interspersed nuclear element (LINE) Persephone_AY643843.
Table I.
Summary of the transposable elements found within the 300-kb barley sequence
| Name | Element Type | Element Subgroup | Size | TSD | Reference Sequence |
|---|---|---|---|---|---|
| bp | |||||
| Ashbury_AY643842_1 | LTR retrotransposon | Ty3/gypsy | 8,278 | N/A | Novel |
| Ashbury_AY643844_2 | LTR retrotransposon | Ty3/gypsy | 12,131 | GTGAG | Novel |
| BAGY-2_AY643843 | LTR retrotransposon | Ty3/gypsy | 10,260 | CTAAA | TREP206; AF254799 |
| BARE-1_AY643843_1 | LTR retrotransposon | Ty1/copia | 8,917 | GTTGA | TREP725; AF227791 |
| BARE-1_AY643844_2 | LTR retrotransposon | Ty1/copia | 8,932 | GCGTG | TREP725; AF227791 |
| BARE-1_AY643844_3 | LTR retrotransposon | Ty1/copia | 8,957 | CATGT | TREP725; AF227791 |
| BARE-1_AY643844_5 | LTR retrotransposon | Ty1/copia | 8,503 | CAAGA | TREP725; AF227791 |
| BARE-1_AY643844_4 solo LTR | LTR retrotransposon | Ty1/copia | 1,818 | GGAAG | TREP725; AF227791 |
| BARE-2_AY643844_1 | LTR retrotransposon | Ty1/copia | 9,203 | ACACC | AJ279072 |
| BARE-2_AY643844_2 | LTR retrotransposon | Ty1/copia | 8,619 | GTGAC/G | AJ279072 |
| BARE-2_AY643844_5 | LTR retrotransposon | Ty1/copia | 5,021 | N/A | AJ279072 |
| BARE-2_AY643844_3 solo LTR | LTR retrotransposon | Ty1/copia | 1,807 | GTTAC | AJ279072 |
| BARE-2_AY643844_4 solo LTR | LTR retrotransposon | Ty1/copia | 1,813 | AT/GGCT | AJ279072 |
| CACTA_AY643843 | Transposon | CACTA | 2,140 | TAT | Novel |
| Caspar_AY643842_1 | Transposon | CACTA | 7,646 | N/A | TREP788 |
| Caspar_AY643844_2 | Transposon | CACTA | 12,085 | TTA | TREP788 |
| Dido_AY643843_1 | Non-LTR retrotransposon | SINE | 256 | N/A | Novel |
| Dido_AY643843_2 | Non-LTR retrotransposon | SINE | 256 | N/A | Novel |
| Haight_AY643844 | LTR retrotransposon | Ty3/gypsy | 13,050 | CCCGC | Novel |
| HORGY_AY643844 | LTR retrotransposon | Ty3/gypsy | 3,077 | TCCTC | TREP728; AF427791 |
| HORPIA-2_AY643843 | LTR retrotransposon | Ty1/copia | 4,285 | CGCGC | TREP730;AF427791 |
| Inga_AY643843 | LTR retrotransposon | Ty1/copia | 5,650 | N/A | TREP704; AF474982 |
| IR with TSD | Unclassified | N/A | 2,244 | ATAGG | Novel |
| Lolaog_AY643843 | LTR retrotransposon | Ty3/gypsy | 10,698 | GCATA | AY268139 |
| Maximus_AY643844 | LTR retrotransposon | Ty1/copia | 13,775 | CCAAC | Novel |
| Morpheus_AY643843 | Non-LTR retrotransposon | LINE | 7,966 | ATGCCG | Novel |
| Persphone_AY643843 | Non-LTR retrotransposon | LINE | 7,889 | ATGTCTGCCCAACGG | Novel |
| Sabrina_AY643843_1 | LTR retrotransposon | Ty3/gypsy | 8,000 | GTCAT | TREP710; AF474071 |
| Sabrina_AY643844_2 | LTR retrotransposon | Ty3/gypsy | 8,183 | GAC/ACC | TREP710; AF474071 |
| Sukkula_AY643843_1 solo LTR | LTR retrotransposon | Ty3/gypsy | 4,961 | CAAGC/CG | TREP715; AF474072 |
| Sukkula_AY643843_2 solo LTR | LTR retrotransposon | Ty3/gypsy | 4,844 | ACTGG | TREP715; AF474072 |
| TRIM_AY643843 | Non-LTR retrotransposon | TRIM | 725 | GCCGG | AY164585 |
| Vagabond_AY643844 | LTR retrotransposon | Ty3/gypsy | 13,918 | GGTCAA | Novela |
Similarity to TREP253; AF459639 was to internal region only.
Figure 2.
Stacked representation of the genome organization of the region containing the Ha locus in barley. Arrows directly on the “base” sequence represent putative genes; designation can be seen in Figure 1. Arrows above and below the base sequence represent the position, orientation, and order of insertion of various transposable elements. Vertical bars illustrate MITES.
The 97-kb intergenic space between HvGSP and HvPG2 is also primarily composed (97%) of two independent transposable element clusters. The insertion of the novel copia-like element Maximus_AY643844 provided a platform for eight additional independent insertions including a highly degenerate element with identifiable inverted repeats, an intact 5-bp target site duplication (TSD), and remnants of ancient coding capacity and seven retrotransposons: Sabrina_AY643844_2, BARE-1_AY643844_2, the novel Latidu-like Vagabond_AY643844, and four BARE-2 (BARE-2_AY643844_1–4; two remain only as solo LTRs). Likewise, the degenerate HORGY_AY643844 retrotransposon acted as the receptor for the insertion of the novel gypsy-like element Haight_AY643844 and an additional BARE-1 copy (BARE-1_AY643844_3).
Two other examples of nested transposable elements are found within the contiguous barley sequence. A second Sukkula solo LTR (Sukkula_AY643843_2) was found inserted into the only terminal-repeat retrotransposons in miniature (TRIM) within the region (TRIM_AY643843). This small cluster is located between the HvCHS and HvVAMP genes. Similarly, the full-length BARE-1_AY643844_4 and the BARE-1_AY643844_5 solo LTR were found sequentially inserted into the novel gypsy element Ashbury_AY643844_2. This cluster is located downstream of HvPG2. In addition, a second copy of Ashbury (Ashbury_AY643842_1) appears to have inserted into a second Caspar (Caspar_AY643842_1) transposon. However, the entire sequence of both elements could not be obtained as they extend beyond the extreme 5′ end of the contig. Likewise only the partial sequence of an additional BARE-2 (BARE-2_AY643844_5) element was found as a consequence of its location at the extreme 3′ end of the contig. A second novel LINE, Morpheus_AY643843, was found located just upstream of ψHvATPase-1. The internally truncated HORPIA-2_AY643843 retrotransposon inserted into ψHvATPase-1 represents the only gene interruption by a large repetitive insertion.
In total, 15 different miniinverted transposable element (MITE) insertions were found composing less than 1% of the total genomic region. The majority of these were members of the Stowaway and Tourist families contributing seven and four respective copies. One full-length and one partial copy of the XI element were also located in the region. This element, previously described as a potential novel element (Brunner et al., 2003), demonstrates high homology to intron five of an Aegilops tauschii isoamylase gene (GenBank accession no. AF548379). The isoamylase copy maintains 36/41-bp imperfect miniinverted repeats, suggesting that this element originated in the Triticeae as a MITE. However, only two of the six copies located within the barley contig 211252 (GenBank accession no. AF521177) remain as intact full-length copies, and both sets of miniinverted repeats have degenerated to less than 75% identity, suggesting that additional mechanisms such as nonreciprocal recombination could account for the high accumulation of this element in this region. This is further supported by lack of intact TSDs and the tandem nature of several copies. The presence of all known copies near or within (TA)n microsatellites suggests a strong insertion bias.
Characterization of the Colinear Region in Rice
To facilitate a comparison between rice and barley sequences, all repetitive elements were removed from the barley genomic sequence and flanking segments were merged at the site of target duplication. The resulting 69-kb barley sequence was used as a template for additional searches of the nonredundant database (nrdb) and EST database (dbEST) at the National Center of Biotechnology Information (NCBI). Several regions of considerable homology were identified across a 34-kb unannotated segment of rice chromosome 12 (GenBank accession nos. AL928743 and AL732378). All seven conserved regions corresponded to the genic space of the barley contig, and no significant sequence identity longer than 25 bp was found beyond the coding regions of the genes.
Similar to the barley region, the rice region also contains three ATPase gene copies (Fig. 1B). However, a greater degree of sequence homology exists among paralogs within species than between orthologs of the different species. This indicates that gene duplication occurred independently post speciation (Fig. 3). OsATPase-3 is the only functional rice copy encoding a 524-amino acid protein with 68% and 72% identity (82% similarity) to ψHvATPase-2 and HvATPase-3, respectively. OsATPase-3 maintains a minimum of 81% nucleotide homology to both rice paralogs and was probably a product of the original duplication event. ψOsATPase-1 contains a premature stop codon resulting in the truncation of the C-terminal end of the protein. Although ψOsATPase-1 maintains 94% homology to the first two-thirds of ψOsATPase-2, no significant homology is observed after the truncation, suggesting that either ψOsATPase-1 resulted from a partial gene duplication event or the terminal end has been subsequently deleted. ψOsATPase-2 has been interrupted by the insertion of a novel 5-kb copia-like element between codons 57 and 58. This is the only retrotransposable element located within the rice region.
A TBLASTN comparison using the GSP protein identified a small stretch of 120 bp in the colinear rice sequence with high similarity (64%; E = 0.55) to the C-terminal end of the protein. This putative unannotated rice protein was previously identified through a similar comparison using the monococcum GSP gene, and further analysis revealed the presence of both a putative TATA-box and polyadenylation signal (Chantret et al., 2004). To determine the most closely related sequence to the barley grain texture genes in the rice genome, BLASTP and TBLASTN comparisons to the annotated rice proteins and the rice genomic sequence, respectively, were preformed. The highest protein similarity found was to a family of rice prolamin genes (51%–54% similarities; 2E-14–0.0025). This similarity is not surprising as puroindolines have previously been classified in the prolamin superfamily, albeit a different class than the prolamins themselves, characterized by the conserved number and spacing of Cys residues (Shewry et al., 2002). Although a higher E value was obtained in comparison with the prolamin genes than the unannotated protein described above, several lines of evidence exist that suggest prolamins are not orthologous to the grain texture gene ancestor. Similarity to GSP did not extend across the entire protein and was predominantly restricted to the conserved Cys backbone. Furthermore, prolamins show a higher similarity to other barley ESTs (59% similarity; 2e-26) that extends beyond the Cys and Gln residues.
Homologs to four of the five remaining barley genes were located within the colinear region of rice. However, the orientation and organization of these genes is not entirely conserved between the two grass species (Fig. 1). A chromosomal rearrangement has reversed the positions of two gene clusters (an ATPase and PG1 and VAMP, GlcNAc, GSP, and PG2) while maintaining gene order and orientation within clusters. Although a CHS homolog is not present within the colinear rice region, a homolog with 91% identity at the nucleotide level exists on rice chromosome 7 (GI number 34395291). This suggests a past translocation event involving either the relocation of CHS from chromosome 12 to chromosome 7 in rice or of CHS from another region of the barley genome to the region surrounding the Ha locus (Fig. 3).
Gene Structure of the Barley, Rice, and Arabidopsis Homologs
The gene structure of the barley and rice orthologs was compared to Arabidopsis using BLASTP and BLASTN searches at The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/Blast/; Table II). Grain texture homologs could not be detected through BLASTN searches. The highest protein similarities were 48% to the C-terminal end of a seed storage protein (At4g27140; score 39.5; E = 0.002) and 39% to the N-terminal end of a protease inhibitor/seed storage/lipid transfer protein (At3g42720; score 32.1; E = 0.26).
Table II.
BLASTP comparisons between the predicted barley protein, the predicted colinear rice protein or closest homolog, and the closest Arabidopsis homolog
| Hv Gene | Size (Amino Acids) | Predicted Os
|
Arabidopsis Gene | BLASTP
|
EST Accession | BLASTN
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Score | Expect | Score | Expect | Score | Expect | ||||
| HvATPase-2 | 518 | N/A | N/A | At5g40010 | 520 | e-147 | CD939530, wheat | 634 | 0 |
| BJ257579, wheat | 698 | 0 | |||||||
| BJ265958, wheat | 323 | e-85 | |||||||
| HvPG1 | 535 | 830 | 0 | At1g74780 | 536 | e-152 | CA731405, wheat | 959 | 0 |
| CA007346, barley | 1,235 | 0 | |||||||
| BU996747, barley | 1,132 | 0 | |||||||
| CA005797, barley | 825 | 0 | |||||||
| Hinb-2 | 147 | N/A | N/A | N/A | N/A | N/A | BE454227, barley | 874 | 0 |
| Hinb-1 | 147 | N/A | N/A | N/A | N/A | N/A | BG36753, barley | 874 | 0 |
| Hina | 149 | N/A | N/A | N/A | N/A | N/A | BQ65384, barley | 886 | 0 |
| HvATPase-3 | 516 | At5g40010 | 514 | e-156 | BI778940, barley | 971 | 0 | ||
| CA684810, wheat | 753 | 0 | |||||||
| HvCHS | 432 | 691 | 0 | At4g34850 | 527 | e-150 | BG343835, barley | 1,055 | 0 |
| CA600207, wheat | 825 | 0 | |||||||
| CA502438, wheat | 323 | e-85 | |||||||
| HvVAMP | 215 | 362 | e-99 | At1g04760 | 309 | e-83 | CB667109, rice | 224 | e-55 |
| CA667948, wheat | 490 | e-135 | |||||||
| HvGluNAc | 425 | 709 | 0 | At5g39990 | 559 | e-159 | BM368259, barley | 618 | e-174 |
| BG948458, sorghum | 507 | e-140 | |||||||
| CB861600, barley | 1,152 | 0 | |||||||
| BU983520, barley | 841 | 0 | |||||||
| HvGSP | 164 | N/A | N/A | N/A | N/A | N/A | BE454072, barley | 975 | 0 |
| HvPG2 | 723 | 1,076 | 0 | At1g74790 | 805 | 0 | BU997791, barley | 952 | 0 |
| BG369772, barley | 922 | 0 | |||||||
| BJ278101, wheat | 670 | 0 | |||||||
| CB631610, rice | 113 | e-21 | |||||||
| BU100503, wheat | 963 | 0 | |||||||
BLASTN comparisons between the predicted barley gene and the dbEST database. No significant homologs were found to the grain texture genes in either rice or Arabidopsis.
The putative barley ATPases were the only gene products within the contig to maintain a higher similarity to the Arabidopsis (At5g40010; 75% similarity; Table I) than the closest rice homolog (72%). The ATPases of all three species contained a single exon.
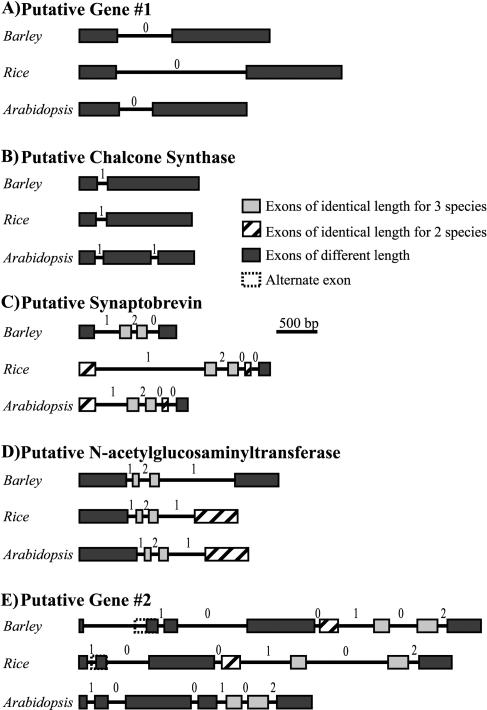
The predicted HvPG1 protein (535 amino acids) shows 87% and 67% similarity to the predicted OsPG1 protein (526aa) and that of the closest Arabidopsis homolog (At1g74780, 533 amino acids; Table I). All three genes contain two exons. However, neither exon is of similar length in any of the three species (Fig. 4A). The gene structure in barley and rice was confirmed by alignment of the genomic sequence with Triticeae and rice ESTs, respectively (Table I). Although the precise function of this gene has yet to be determined, the Arabidopsis homolog is annotated as containing similarity to a nodule-specific protein in Lotus japonicus (GI no. 3329366).
Figure 4.
Structure of the genes located within the barley contig, the colinear rice region, and their closest Arabidopsis homologs (also see Table I). Intron phase is indicated by the number above each intron.
The predicted HvCHS (432 amino acids) gene product showed a high level of similarity to its closest rice (GI no. 34395291; 405 amino acids; 87% similarity) and Arabidopsis (At4g3450; 392 amino acids; 78% similarity) homologs (Table I). The gene structure in barley was confirmed by alignment of the genomic sequence with wheat and barley ESTs (Table I). Both the rice and barley genes contain two exons and the Arabidopsis gene contains three exons (Fig. 4B). Exon 1 from all three species differs in length by only nine codons. The main difference between exon 2 from Arabidopsis and rice is the presence of a (GC)9 microsatellite just before the stop codon in rice. Interestingly, the translated amino acids of the rice microsatellite are conserved in the barley protein although the microsatellite structure is no longer present. Exon 2 of HvCHS contains an additional stretch of 23 codons not found in either of the other two species.
A good similarity exists between the HvVAMP protein (215 amino acids) and both the OsVAMP protein (219 amino acids; 90% similarity) and closest Arabidopsis homolog (At1g04760; 220 amino acids; 84% similarity; Table I). The gene structure in barley and rice was confirmed by alignment of the genomic sequence with wheat and rice ESTs (Table I). However, two rice ESTs (GenBank accession nos. CB667109 and CB667110) showed that the second intron of the rice transcript was not being spliced. Both the 5′ and 3′ splice sites show homology to the 5′-CAG:GTAAGT-3′ and 5′-GCAG:G-3′ plant consensus sites and the uracil/adenine content of the intron is within the expected range. However, intron 2 does not contain a strong branchpoint consensus and this could reduce splice efficiency (Simpson et al., 2002). In addition, both ESTs were from 3-week-old leaf tissue that had been inoculated with rice blast 24 h before harvest. It is, therefore, possible that improper splicing is either tissue specific or somehow induced by infection. Without ESTs from other tissue types, however, this is speculative. The elimination of this splice event introduces a premature stop codon immediately after the predicted splice site (codon 111). If the splice site is preserved, OsVAMP would maintain identical exon length and structure to the entire Arabidopsis gene with the exception of one less codon in the fourth exon (represented in Fig. 4C). The length of exons 2 and 3 is also conserved in HvVAMP. However, intron 4 has been removed in comparison to the coding sequences of the other species.
The predicted HvGlcNAc and OsGlcNAc proteins are almost identical in length (425 versus 426 amino acids, respectively) and demonstrate a high degree of similarity (87%; Table I). The slightly larger Arabidopsis homolog (At5g39990; 447 amino acids) is 76% similar to both the barley and rice proteins. The gene structure in barley and rice was confirmed by alignment of the genomic sequence with barley, sorghum, and rice ESTs (Table I). Exons 2 and 3 are identical in length in all three species, and exon 4 is identical in length in Arabidopsis and rice. In addition, the first and fourth exons of HvGlcNAc differ in length from those of OsGlcNAc by only three and two codons, respectively (Fig. 4D). Alternative splicing in Arabidopsis to conserve the length of the first exon is highly unlikely as six of the nine consensus bases are absent including the mandatory GT at the site of excision.
A high level of similarity exists between the HvPG2 protein (753 amino acids) and both the OsPG2 protein (683 amino acids; 84% similarity) and the closest Arabidopsis homolog (At1g74790; 695 amino acids; 72% similarity; Table I). The gene structure in barley and rice was confirmed by alignment of the genomic sequence with wheat and rice ESTs, respectively (Table I). However, no homologous ESTs were found for the extreme 5′ end of either gene. Therefore, two alternate structures for the barley protein were considered. The first, predicted by the rice genome automated annotation system (RiceGAAS; Sakata et al., 2002), introduced an additional exon and resulted in a 723-amino acid gene product (Fig. 4E). The second involved locating the first in-frame start codon upstream of the last confirmed gene region and encoded a 753-amino acid protein. This second gene structure is represented by a dashed region extending the length of exon 2 in Figure 4E. Neither alternative contained any identity in the extreme 5′ region of either OsPG2 or the Arabidopsis homolog at the protein or nucleotide level. However, the latter maintains the exon/intron structure of the Arabidopsis gene and EST homology extends 56 bp into what would otherwise be the intron region of the first alternative structure. Two alternate structures were also considered for the 5′ terminal end of OsPG2. The first, predicted by RiceGAAS, maintained the exon/intron structure of the Arabidopsis gene and resulted in a 683-amino acid protein (Fig. 4E). The second, determined by the first ATG start codon upstream of the last confirmed region with ESTs, eliminated an exon and introduced a premature stop codon nine amino acids into the protein. This second gene structure is represented by a dashed region extending the length of exon 2 in Figure 4E. Again, no identity to the 5′ terminal end of either barley or the Arabidopsis gene was observed at the protein or nucleotide level. Exons 5 and 6 are of identical length in all three species, and exon 4 is of identical length in barley and rice. The Arabidopsis homolog is annotated as containing similarity to a hedgehog interacting protein from M. musculus (GI no. 4868122).
DISCUSSION
Gene Islands and Intergenic Space
This study describes the sequencing and analysis of a region of the barley genome covering over 300 kb at 10 times coverage. The current gene content of higher plants is estimated to range from 25,000 to 43,000 genes (Miklos and Rubin, 1996). Therefore, an average gene density of one gene every 123 to 250 kb would be expected in barley (5,300 Mb) assuming even gene distribution. Furthermore, cytogentic studies have previously reported an increase in gene density along the chromosome arms moving away from the centromere toward the telomeres (Gill et al., 1996; Akhunov et al., 2003). Regardless, despite the location of the hardness locus at the extreme distal end of 5HS, the results reported here suggest a local concentration of genes with approximately one gene every 25 kb. This is in concordance with the pattern of genome organization found within other large contiguous regions of barley that demonstrate an average density of one gene every 20 kb (one gene every 12–103 kb; Panstruga et al., 1998; Shirasu et al., 2000; Dubcovsky et al., 2001; Rostoks et al., 2002; Wei et al., 2002; Yan et al., 2002; Gu et al., 2003). Moreover, the presence of “gene islands” appears to be widespread among several members of the grass family with large genome size, namely maize and wheat (SanMiguel et al., 1996; Feuillet and Keller, 1999; Tikhonov et al., 1999; Wicker et al., 2001). However, not all genes are located within clusters. A span of 96 kb separates HvPG2 from the nearest upstream gene (HvGSP) and a minimum 43-kb gene void exists downstream. In addition, only a single gene was found within the 103-kb barley BAC 745c13 (Rostoks et al., 2002) and on Triticum monococcum BAC111I4 the RGA-1 gene was separated from other genes by a minimum of 31 kb (Wicker et al., 2001).
The presence of different transposable elements within the barley contig was the primary contributor to the patterns of genome organization and the major factor responsible for the vast difference in length between the colinear rice and barley sequences. Although over 75% of the barley contiguous region reported here is composed of repetitive elements, only one element, a 5-kb Ty1/copia retrotransposon, was present within the orthologous rice sequence (Fig. 1B). One-third of the repetitive sequence in the barley region consists of the BARE retrotransposon family, with both BARE-1 and BARE-2 contributing equally. This is 3-fold higher than average genome BARE-1 levels estimated in cultivated barley. However, other members of the Hordeae were found to have as much as 40% of their genomes composed of BARE-1 alone (Vicient et al., 1999). Evidence for the disruption of microcolinearity among grass species by nested transposable element insertion has also been reported between the closely related species of sorghum and maize. At the sh2/a1 locus, with the exception of a single gene duplication in sorghum, gene number and orientation was completely conserved between the two species despite a 3-fold difference in the overall lengths of the orthologous sequences (Chen et al., 1998). Furthermore, only 15% of the adh locus in sorghum was found to be composed of nongenic sequence compared to over 74% in the orthologous locus in maize (Tikhonov et al., 1999).
It is interesting that the only retrotransposon insertion in the rice sequence occurred within the ψOsATPase-2 gene. ψHvATPase-1 was also disrupted by the insertion of a copia element of similar length within the same region of the gene in barley. The complete lack of nucleotide homology and the presence of target site footprints of different lengths indicate that these insertions were separate events involving different retrotransposons. Given that the insertion of retrotransposons into coding sequence is rare (SanMiguel et al., 1996), the independent insertion of different elements into the same gene in colinear regions of two different grass species is surprising, particularly as this is the only retroelement insertion within the rice region.
It has been suggested that differences in intron length could also account for a portion of the differences observed in genome size. A greater proportion of rice introns (64%) were longer than their barley counterparts. However, the total length of intron sequence within a given gene was equally as likely to be longer in barley as in rice (two versus three genes, respectively; Fig. 4). When the introns of rice and Arabidopsis were compared, all but one rice intron was longer, and the total intron length within a gene was always greater for the rice gene. Interestingly this was not the case when comparing the barley and Arabidopsis genes despite a considerably larger difference in genome size. Although a greater number of barley introns (69%) were longer than their Arabidopsis equivalents, only three of the five genes gained extra additive length (Fig. 4). In both cases, the longer total intron length in Arabidopsis was a result of an extra intron. Although longer intron size within the grass genes suggests either a greater frequency of large insertions or a better retention of such insertions, this may be compensated for by a greater number of smaller introns within Arabidopsis genes. Similar comparisons in intron length were reported in barley BAC 635P2 (Dubcovsky et al., 2001). However, the positional bias for introns located between codons (phase 0) noted in BAC 635P2 was contrary to the results obtained in this study. These results indicate a bias toward introns positioned within codons (64%) and an additional bias toward phase 1 (located between the first and second codon positions) introns over phase 2 (located between the second and third codon positions) introns. In every case, intron phase was conserved between all three species (Fig. 4).
Gene Discovery and Determination of Gene Structure
Despite the extensive collection of ESTs in the public database, sequences of full-length ESTs are still fairly rare. In addition, ESTs for a particular gene are often represented only from a single developmental stage or tissue type and, therefore, may represent only one of many alternative splicing events. The only two available rice ESTs for the synaptobrevin gene indicate failure to splice intron 2, resulting in a severely truncated protein. However, the highly conserved gene structure and protein similarity compared to the barley and Arabidopsis homologs indicates that either this gene is still properly spliced in other tissues or under other conditions in rice or the mutations leading to improper splicing have occurred so recently that homology has not yet been degraded. Gene prediction programs, which are reasonably accurate in locating genic regions, often fall short in discerning the intricacies of specific gene structure. The automated gene prediction of HvPG2 eliminated two entire exons, truncated a third, and generated a false start site. The automated prediction of the OsPG2 generated an additional exon and introduced a new intron, which altered the termination site of the gene. However, automated prediction was helpful in discerning the most probable start site in the absence of full-length ESTs with the Arabidopsis sequence as a guide. In both instances, predicted genes from the completely sequenced Arabidopsis and rice genomes proved a valuable tool for discerning gene structure.
Microcolinearity and Genome Evolution
Although some repetitive sequences are remnants of ancient insertion events, the vast majority of transposable element insertions occurred post speciation (SanMiguel and Bennetzen, 1998). The presence of these elements can often complicate the detection of orthologous loci for comparative genomics studies as critical regions of similarity could be missed within the sea of nonhomologous intergenic DNA. The removal of all repetitive elements from the barley sequence generated a template that facilitated the identification of the colinear rice sequence.
A wide variety of small chromosomal rearrangements have occurred between the region containing Ha locus in barley and its colinear rice sequence (Fig. 3). An interchromosomal event concluded in the translocation of the putative chalcone synthase gene. Although at least three copies of ATPase were present within the colinear region in both species, sequence homology revealed a greater conservation among paralogs within the same species than between orthologs of the different species. This indicated a total of six different independent duplications involving one gene inversion post speciation. Three further gene duplications involving a minimum of one inversion also arose from the ancestral grain texture gene in the barley genome. An intrachromosomal rearrangement resulted in the repositioning of two conserved gene clusters. One of these gene clusters, GC2 (VAMP, GlcNAc, and GSP), has also been conserved in T. monococcum (Chantret et al., 2004). The high level of conservation in this particular region was further demonstrated by the low level of transposon insertion. No transposable elements were present within GC2 in T. monococcum compared to other sequenced contiguous regions of the genome that are composed of 70% to 80% repetitive elements (Wicker et al., 2001, 2003; SanMiguel et al., 2002). Furthermore, the only element insertions within GC2 in the barley region occurred outside of the conserved region with T. monococcum between GSP and PG2.
Several additional breaks in colinearity existed between the wheat and barley genomes. The rice and wheat sequences contained a putative gene just upstream of GC2, which was not present in the barley sequence (Chantret et al., 2004). Neither genome contained the CHS gene located in this position in the barley sequence indicating this translocation event occurred in the barley genome relative to the ancestral grass sequence. Similarly, a putative gene was present in the rice and barley sequences downstream of GC2, which was not found in wheat (Chantret et al., 2004). Therefore, it is probable that the intrachromosomal rearrangement observed between rice and barley involved the relocation of the other gene cluster, GC1 (ATPase and PG1). Furthermore, the puroindoline genes were positioned downstream of GC2 and in the same orientation as GSP in wheat (Chantret et al., 2004), while the hordoindolines were located upstream and in the opposite orientation in barley. All three grain texture genes in wheat and barley demonstrated orthologous relationships indicating that this rearrangement occurred post gene duplication. Extended sequencing of the T. monococcum region and additional sequences from related grass species are necessary to discern the exact series of evolutionary events.
A low level of microcolinearity still exists between the two grass species and Arabidopsis. The closest homologs to the putative N-acetylglucosaminyltransferase and ATPase are under 14 kb apart on chromosome 5 in reverse orientation and separated by one additional gene. In addition, the closest Arabidopsis homologs to PG1 and PG2 are located only 1 kb apart in similar orientation on chromosome 1. Although the closest homolog to the putative synaptobrevin gene was also located on chromosome 1 it was widely separated from this gene cluster.
Only two other studies have compared large orthologous regions from rice and barley at the sequence level. At the Xwg644 locus, despite one gene inversion and a single gene duplication in barley as compared to rice, the gene order of all four orthologs was completely conserved (Dubcovsky et al., 2001). A single gene inversion and one gene duplication was also reported at the Rph7 locus (Brunner et al., 2003). However, a segment of 153 kb containing six additional genes not present in the colinear rice region was inserted within the conserved order of four gene family members (Brunner et al., 2003). A rice homolog for each additional barley gene was found located elsewhere within the rice genome suggesting at least one past translocation event. The comparison of the colinear barley and rice regions presented here represents the most complicated configuration of small chromosomal rearrangements to be reported between grass species thus far involving numerous small chromosomal rearrangements, a translocation, several gene duplications, and the insertion of numerous transposable elements. This may reflect historical evolutionary pressures and/or the telomeric location of these genes in barley. Well-conserved colinearity with rice has been frequently reported along proximal regions of the Triticeae chromosomes such as the Vrn1 (Yan et al., 2003), Ph1 (Roberts et al., 1999), and Gpc-B1 (Distelfeld et al., 2004) loci in wheat. However, colinearity has recently been reported to be less conserved at the telomeric regions of the chromosomes among the wheat genomes. Moreover, a breakdown of microcolinearity has repeatedly been shown in comparative studies involving rice and distal regions of the wheat and barley genomes (Kurata et al., 1994), including the Rpg1 (Kilian et al., 1997), LMW Glu-A3/SRLK/Lrk10/Tak/Lr10 (Feuillet and Keller, 1999; Guyot et al., 2004), and Sh2/X1/X2/A1 (Li and Gill, 2002) regions in wheat and barley. Our results demonstrate that the trend of colinearity breakdown within telomeric chromosomal regions extends beyond the genetic level to the sequence level. Despite this trend, a comparison of the locations of physically mapped wheat ESTs and the first draft of the rice genomic sequence revealed that within the wheat genome, regardless of chromosomal location, even the most conserved regions of colinearity contain homologous sequences from more than one region of the rice genome (Sorrells et al., 2003; La Rota and Sorrells, 2004). These results support the view that grass genomes are more fluid than first anticipated and that structural and functional relationships are complex. The resultant breakdown of microcolinearity exemplifies the limitations of rice as a model organism for the application of comparative genomics in association mapping and positional cloning. These findings stress the importance of implementing genomic studies directly in the species of interest.
The extent of the difference between rice and barley in the organization of this region could be related to the function of the grain texture genes. Selective pressure may have led to the maintenance of subsequent duplications of the ancestral copy and the gradual ascertainment of new functions within the gene family. It is unlikely that the currently accepted function of these genes, namely in controlling grain texture (for review, see Morris, 2002), is the source of selection for the structure and copy number of these genes in barley and wheat. Grain endosperm texture is a characteristic that would only have been relevant during or after domestication of these species, an event too recent to account for the complex structure of this region and also inconsistent with the presence of these genes in wheat, barley, and wild members of the Triticeae. It has been suggested that the products of the grain endosperm texture genes may also protect against pathogen attack (Blochet et al., 1993; Dubreil et al., 1998; Krishnamurthy et al., 2001). Such a role would be consistent with the observed genome organization of the region and would provide an explanation for the maintenance and duplication of the genes. It will now be important to investigate fully alternative functions of these genes.
MATERIALS AND METHODS
BAC Selection
A set of 14 BACs (barley [Hordeum vulgare] cv Morex; Yu et al., 2000) identified through positive hybridization with a wheat (Triticum aestivum) GSP-1 cDNA clone was obtained from Professor Andris Kleinhof's lab at Washington State University (http://barleygenomics.wsu.edu/db3/db3.html). These BACs were fingerprinted in Professor Michele Morgante's lab at DuPont Agriculture and Nutrition (Newark, DE), and BAC122.a5 was selected for construction of a subclone library and full-length sequencing. Primers for the amplification of hordoindoline-a (5′-GGTCTGCTTGCTTTGGTAGC-3′ and 5′-AATAGTGCTGGGGATGTTGC-3′) and -b (5′-CTCCTAGCCCTCCTTGCTCT-3′ and 5′-CTCCCATGTTGCACTTTGAG-3′) were designed from GenBank accessions HVU249929 and HVU249928, respectively, using Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) for the generation of gene-specific probes for additional BAC library screens. Primers for the amplification of GSP (5′-CAACATTGACAACATGAAGACC-3′ and 5′-TTTGGCACAACTAACATTGG-3′) were designed from the Morex BAC122.a5 sequence. Positive clones were analyzed by Southern hybridization to validate the presence of the GSP, hina, and/or hinb. Size determination and BAC end sequencing were employed to identify BACs that would allow minimal overlap and ensure maximum coverage of the region. BACs 519.k7 and 799.c8 were selected for further experimentation.
BAC Sequence and Assembly
Purified BAC DNA was obtained using the Qiagen Large Construct kit (Qiagen USA, Valencia, CA) and sheared by nebulization for 15 s at 10 pounds per square inch. The 2-kb and 5-kb fractions were blunt ended, dephosphorylated, and ligated into pUC18 cloning vector. Individual clones were sequenced in the forward and reverse direction using ABI big dye terminator chemistry and analyzed on an ABI 3700 automated capillary sequencer (ABI, Sunnyvale, CA). Preassembly and assembly analysis of the sequencing reads were performed by using PHRED version 0.020425.c and PHRAP version 0.990329 software (University of Washington, Seattle; Ewing and Green, 1998; Ewing et al., 1998). The combined information was viewed and edited through CONSED version 12.0 software (University of Washington; Gordon et al., 1998). Gaps were closed and weak consensus regions strengthened by either direct sequencing of subclones using nested primers or sequencing PCR amplicons spanning the region between contig ends.
Sequence Analysis
Preliminary characterization of the sequenced barley and rice (Oryza sativa) regions was preformed using standard nucleotide-nucleotide (BLASTN; Altschul et al. 1997) and nucleotide-protein (BLASTX) searches against the nrdb at the NCBI (http://ncbi.nlm.nih.gov/BLAST/) and the Triticeae Repeat Sequence Database (TREP, http://wheat/pw.usda.gov/ggpages/ITMI/Repeats/balstrepeats3.html; Wicker et al., 2002). Inverted and direct repeats of previously uncharacterized elements were detected through Bestfit analysis using WebANGIS (http://www.angis.org.au/WebANGIS/WebFM). SINEs were detected by scanning the genomic sequence for similarity to the conserved Arabidopsis A (TRKYNNARNGG) and B (RGTTCRANHYY) boxes spaced 25 to 50 bp apart. Initial gene prediction analysis was performed using RiceGAAS (http://ricegaas.dna.affrc.go.jp/; Sakata et al., 2002), which couples the integration of several programs for the prediction of open reading frames (GENSCAN, RiceHMM, FGENESH, MZEF) with homology search analysis programs (BLAST, HMMER, ProfileScan, MOTIF). Expression of putative genes was determined using BLASTN analysis against the dbEST at the NCBI. Exon:intron splice junctions were determined by genomic alignment with ESTs. Splice junctions were confirmed by the presence of the conserved GT and AG intron borders and a minimum of five of the nine (5′-CAG:GTAAGT-3′) and three of the five (5′-GCAG:G-3′) consensus nucleotides for the respective exon:intron and intron:exon splice sites in plants. Putative functions and conserved protein domains were determined using BLASTP analysis against the nrdb and swissprot database at NCBI. Identification of colinear and homologous Arabidopsis and rice sequences were performed at TAIR (http://www.arabidopsis.org/Blast/), The Institute for Genomic Research (http://tigrblast.tigr.org/euk-blast/index.cgi?project=osa1), and Gramene Web sites (http://www.gramene.org/) using BLASTN, BLASTP, and TBLASTN functions. The Dotter program (Sonnhammer and Durbin, 1995; word length 25, similarity 80) was used to identify conserved regions of sequence homology between the barley BAC contig and the rice colinear sequence (GenBank accession no. AL928743).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY643842 to AY643844.
Acknowledgments
We thank A. Kleinhofs and M. Morgante for preliminary work in BAC identification and fingerprinting, respectively.
This work was supported by the Scottish Executive Environment and Rural Affairs Department.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.044081.
References
- Ahn S, Tanksley SD (1993) Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA 90: 7980–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov ED, Goodyear AW, Geng S, Qi LL, Echalier B, Gill BS, Miftahudin, Gustafson JP, Lazo G, Chao SM, et al (2003) The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res 13: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9: 211–215 [Google Scholar]
- Baurnert M, Maycox PR, Navone F, DeCarnilli P, Jahn R (1989) Synaptobrevin: an integral membrane protein of 18,000 Daltons present in small synaptic vesicles of rat brain. EMBO J 8: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecher B, Smidansky ED, See D, Blake TK, Giroux MJ (2001) Mapping and sequence analysis of barley hordoindolines. Theor Appl Genet 102: 833–840 [Google Scholar]
- Bennett MD, Leitch IJ (1995) Nuclear DNA amounts in angiosperms. Ann Bot (Lond) 76: 113–176 [Google Scholar]
- Bennett MD, Leitch IJ (1997) Nuclear DNA amounts in angiosperms: 583 new estimates. Ann Bot (Lond) 80: 169–196 [Google Scholar]
- Bennett MD, Smith JB, Heslop-Harrison JS (1982) Nuclear DNA amounts in angiosperms. Proc R Soc Lond B Biol Sci 216: 179–199 [Google Scholar]
- Bennetzen JL (2000) Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell 12: 1021–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J (2003) The genetic colinearity of rice and other cereals on the basis of genomic sequence analysis. Curr Opin Plant Biol 6: 128–133 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Ramakrishna W (2002) Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol Biol 48: 821–827 [DOI] [PubMed] [Google Scholar]
- Blochet JE, Chevalier C, Forest E, Pebaypeyroula E, Gautier MF, Joudrier P, Pezolet M, Marion D (1993) Complete amino-acid-sequence of puroindoline: a new basic and cystine-rich protein with a unique tryptophan-rich domain, isolated from wheat endosperm by Triton X-114 phase partitioning. FEBS Lett 329: 336–340 [DOI] [PubMed] [Google Scholar]
- Brunner S, Keller B, Feuillet C (2003) A large rearrangement involving genes and low-copy DNA interrupts the microcollinearity between rice and barley at the Rph7 locus. Genetics 164: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantret N, Center A, Sabot F, Anderson O, Dubcovsky J (2004) Sequencing of the Triticum monococcum hardness locus reveals good microcolinearity with rice. Mol Gen Genet 271: 377–386 [DOI] [PubMed] [Google Scholar]
- Chen MS, SanMiguel P, Bennetzen JL (1998) Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics 148: 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Scheller RH (2001) SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol 2: 98–106 [DOI] [PubMed] [Google Scholar]
- Clark LG, Zhang WP, Wendel JF (1995) A phylogeny of the grass family (Poaceae) based on Ndhf -sequence data. Syst Bot 20: 436–460 [Google Scholar]
- Crepet WL, Feldman GD (1991) The earliest remains of grasses in the fossil record. Am J Bot 78: 1010–1014 [Google Scholar]
- Delseny M (2004) Re-evaluating the relevance of ancestral shared synteny as a tool for crop improvement. Curr Opin Plant Biol 7: 1–6 [DOI] [PubMed] [Google Scholar]
- Devos KM, Gale MD (1997) Comparative genetics in the grasses. Plant Mol Biol 35: 3–15 [PubMed] [Google Scholar]
- Distelfeld A, Uauy C, Olmos S, Schlatter AR, Dubcovsky J, Fahima T (2004) Microcolinearity between a 2-cM region encompassing the grain protein content locus Gpc-6B1 on wheat chromosome 6B and a 350-kb region on rice chromosome 2. Funct Integr Genomics 4: 59–66 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Harrison MJ, Paiva NL (1995) The isoflavonoid phytoalexin pathway; from enzymes to genes to transcription factors. Physiol Plant 93: 385–392 [Google Scholar]
- Dixon RA, Lamb CJ, Masoud S, Sewalt VJH, Paiva NL (1996) Metabolic engineering: prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses. A review. Gene 179: 61–71 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A, Kudrna D, Han F, Kilian A, Steffenson B, Frisch D, Tomkins J, Wing R, Kleinhofs A (2000) Physical mapping of the barley stem rust resistance gene rpg4. Mol Gen Genet 264: 283–290 [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Ramakrishna W, SanMiguel PJ, Busso CS, Yan LL, Shiloff BA, Bennetzen JL (2001) Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol 125: 1342–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreil L, Gaborit T, Bouchet B, Gallant DJ, Broekaert WF, Quillien L, Marion D (1998) Spatial and temporal distribution of the major isoforms of puroindolines (puroindoline-a and puroindoline-b) and non specific lipid transfer protein (ns-LTPle(1)) of Triticum aestivum seeds: relationships with their in vitro antifungal properties. Plant Sci 138: 121–135 [Google Scholar]
- Ewing B, Green P (1998) Base calling of automated sequencer traces using phred. II: error probabilities. Genome Res 8: 186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I: accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Feuillet C, Keller B (1999) High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA 96: 8265–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C, Keller B (2002) Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann Bot (Lond) 89: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95: 1971–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill KS, Gill BS, Endo TR, Boyko EV (1996) Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang RL, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Gu YQ, Anderson OD, Londeore CF, Kong X, Chibbar RN, Lazo GO (2003) Structural organization of the barley D-hordein locus in comparison with orhtologous region of wheat genomes. Genome 46: 1084–1097 [DOI] [PubMed] [Google Scholar]
- Guyot R, Yahiaoui N, Feuillet C, Keller B (2004) In silico comparative analysis reveals a mosaic conservation of genes within a novel colinear region in wheat chromosome 1AS and rice chromosome 5S. Funct Integr Genomics 4: 47–58 [DOI] [PubMed] [Google Scholar]
- Han F, Kilian A, Chen JP, Kudrna D, Steffenson B, Yamamoto K, Matsumoto T, Sasaki T, Kleinhofs A (1999) Sequence analysis of a rice BAC covering the syntenous barley Rpg1 region. Genome 42: 1071–1076 [DOI] [PubMed] [Google Scholar]
- Han F, Kleinhofs A, Ullrich SE, Kilian A, Yano M, Sasaki T (1998) Synteny with rice: analysis of barley malting quality QTLs and rpg4 chromosome regions. Genome 41: 373–380 [Google Scholar]
- Keller B, Feuillet C (2000) Colinearity and gene density in grass genomes. Trends Plant Sci 5: 246–251 [DOI] [PubMed] [Google Scholar]
- Kilian A, Chen J, Han F, Steffenson B, Kleinhofs A (1997) Towards map-based cloning of the barley stem rust resistance genes Rpgl and rpg4 using rice as an intergenomic cloning vehicle. Plant Mol Biol 35: 187–195 [PubMed] [Google Scholar]
- Krishnamurthy K, Balconi C, Sherwood JE, Giroux MJ (2001) Wheat puroindolines enhance fungal disease resistance in transgenic rice. Mol Plant Microbe Interact 14: 1255–1260 [DOI] [PubMed] [Google Scholar]
- Kurata N, Moore G, Nagamura Y, Foote T, Yano M, Minobe Y, Gale M (1994) Conservation of genome structure between rice and wheat. Biotechnology 12: 276–278 [Google Scholar]
- La Rota M, Sorrells M (2004) Comparative DNA sequence analysis of mapped wheat ESTs reveals the complexity of genome relationships between rice and wheat. Funct Integr Genomics 4: 34–46 [DOI] [PubMed] [Google Scholar]
- Li WL, Gill BS (2002) The colinearity of the Sh2/A1 orthologous region in rice, sorghum and maize is interrupted and accompanied by genome expansion in the Triticeae. Genetics 160: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Miklos GLG, Rubin GM (1996) The role of the genome project in determining gene function: Insights from model organisms. Cell 86: 521–529 [DOI] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD (1995) Cereal genome evolution: grasses, line up and form a circle. Curr Biol 5: 737–739 [DOI] [PubMed] [Google Scholar]
- Morris CF (2002) Puroindolines: the molecular genetic basis of wheat grain hardness. Plant Mol Biol 48: 633–647 [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ (2001) AAA(+) superfamily ATPases: common structure-diverse function. Genes Cells 6: 575–597 [DOI] [PubMed] [Google Scholar]
- Panstruga R, Buschges R, Piffanelli P, Schulze-Lefert P (1998) A contiguous 60-kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res 26: 1056–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Latterich M (1998) The AAA team: related ATPases with diverse functions. Trends Cell Biol 8: 65–71 [PubMed] [Google Scholar]
- Roberts MA, Reader SM, Dalgliesh C, Miller TE, Foote TN, Fish LJ, Snape JW, Moore G (1999) Induction and characterization of Ph1 wheat mutants. Genetics 153: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim EK, Bowles DJ (2001) Higher Plant Glycosyltransferases. Genome Biology 2: 3004.1–3004.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostoks N, Park Y-J, Ramakrishna W, Ma J, Druka A, Shiloff BA, SanMiguel PJ, Jiang Z, Brueggeman R, Sandhu D, et al (2002) Genomic sequencing reveals gene content, genomic organization, and recombination relationships in barley. Funct Integr Genomics 2: 51–59 [DOI] [PubMed] [Google Scholar]
- Rouves S, Boeuf C, Zwickert-Menteur S, Gautier MF, Joudrier P, Bernard M, Jestin L (1996) Locating supplementary RFLP markers on barley chromosome 7 and synteny with homoeologous wheat group 5. Plant Breed 115: 511–513 [Google Scholar]
- Sakata K, Nagamura Y, Numa H, Antonio BA, Nagasaki H, Idonuma A, Watanabe W, Shimizu Y, Horiuchi I, Matsumoto T, et al (2002) RiceGAAS: an automated annotation system and database for rice genome sequence. Nucleic Acids Res 30: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot (Lond) 82: 37–44 [Google Scholar]
- SanMiguel P, Ramakrishna W, Bennetzen JL, Busso C, Dubcovsky J (2002) Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5Am. Funct Integr Genomics 2: 70–80 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420: 312–316 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Beaudoin F, Jenkins J, Griffiths-Jones S, Mills ENC (2002) Plant protein families and their relationships to food allergy. Biochem Soc Trans 30: 906–910 [DOI] [PubMed] [Google Scholar]
- Shields R (1993) Plant genetics: pastoral synteny. Nature 365: 297–298 [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P (2000) A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res 10: 908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW (1996) Flavonoid biosynthesis: ‘new’ functions for an ‘old’ pathway. Trends Plant Sci 1: 377–382 [Google Scholar]
- Simpson CG, Thow G, Clark GP, Jennings SN, Watters JA, Brown JWS (2002) Mutational analysis of a plant branchpoint and polypyrimidine tract required for constitutive splicing of a mini-exon. RNA 8: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE (1993) A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75: 409–418 [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: 1–10 [DOI] [PubMed] [Google Scholar]
- Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, Miftahudin, Mahmoud A, Ma XF, Gustafson PJ, et al (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13: 1818–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein NM, Bennetzen JL, Avramova Z (1999) Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc Natl Acad Sci USA 96: 7409–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble WS, Cowan DM, Scheller RH (1988) VAMP-1: a synaptic vesicle-associated integral membrane protein. Proc Natl Acad Sci USA 85: 4538–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Suoniemi A, Anamthamat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH (1999) Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11: 1769–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, Mcnew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE (1998) SNAREpins: minimal machinery for membrane fusion. Cell 92: 759–772 [DOI] [PubMed] [Google Scholar]
- Wei FS, Wong RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14: 1903–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Matthews DE, Keller B (2002) TREP: a database for Triticeae repetitive element. Trends Plant Sci 7: 561–562 [Google Scholar]
- Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B (2001) Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J 26: 307–316 [DOI] [PubMed] [Google Scholar]
- Wicker T, Yahiaoui N, Guyot R, Schlagenhauf E, Liu ZD, Dubcovsky J, Keller B (2003) Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and A(m) genomes of wheat. Plant Cell 15: 1186–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Gouy ML, Yang YW, Sharp PM, Li WH (1989) Date of the monocot dicot divergence estimated from chloroplast DNA-sequence data. Proc Natl Acad Sci USA 86: 6201–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yamagata H, Hayashi-Tsugane M, Hijishita S, Fujisawa M, Shibata M, Ito Y, Nakamura M, Sakaguchi M, Yosihara R, et al (2004) Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Echenique V, Busso C, SanMiguel P, Ramakrishna W, Bennetzen JL, Harrington S, Dubcovsky J (2002) Cereal genes similar to Snf2 define a new subfamily that includes human and mouse genes. Mol Gen Genet 268: 488–499 [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu SN, Wang J, Wong GKS, Li SG, Liu B, Deng YJ, Dai L, Zhou Y, Zhang XQ, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA (2000) A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theor Appl Genet 101: 1093–1099 [Google Scholar]