Abstract
Two nitrogen-fixing root nodule symbioses between soil bacteria and higher plants have been described: the symbiosis between legume and rhizobia and actinorhizal symbioses between plants belonging to eight angiosperm families and the actinomycete Frankia. We have recently shown that the subtilisin-like Ser protease gene cg12 (isolated from the actinorhizal plant Casuarina glauca) is specifically expressed during plant cell infection by Frankia. Here we report on the study of C. glauca cg12 promoter activity in the transgenic legume Medicago truncatula. We found that cg12 promoter activation is associated with plant cell infection by Sinorhizobium meliloti. Furthermore, applications of purified Nod factors and mycorrhizal inoculation failed to trigger expression of the cg12-reporter gene construct. This indicates that at least part of the transcriptional environment in plant cells infected by endosymbiotic nitrogen-fixing bacteria is conserved between legume and actinorhizal plants. These results are discussed in view of recent data concerning molecular phylogeny that suggest a common evolutionary origin of all plants entering nitrogen-fixing root nodule symbioses.
Two groups of plants are able to form nitrogen-fixing root nodule symbioses with soil bacteria: legumes (plus Parasponia in the Ulmaceae family) associate with rhizobia, while the so-called actinorhizal plants belonging to eight angiosperm families interact with Frankia. Inside root nodules, bacteria protected and nourished by the plant find a favorable environment for nitrogen fixation and, in exchange, provide the plant with fixed nitrogen. Recent molecular phylogeny studies based on the chloroplast gene rbcL indicate that plants entering rhizobial or actinorhizal symbioses belong to the same clade (Rosid I; Soltis et al., 1995), suggesting that a predisposition to form nitrogen-fixing root nodule symbioses originated once in the history of flowering plants. However, the nature of this predisposition remains unknown.
Depending on the plant species, bacteria infect the root either by root hair infection or through cellular spaces between epidermal cells (crack entry). Root hair infection is characteristic of most temperate legumes and of several actinorhizal genera like Alnus and Casuarina. In this case, bacteria induce a localized degradation of the cell wall of the root hair; the plasma membrane then invaginates leading to the formation of a tubular structure called the infection thread (IT). ITs are filled with bacteria and surrounded by newly deposited cell wall material and spread bacteria by growing inside plant cells and from one cell to another. Whereas actinorhizal ITs never release the bacteria, in most legume species, the ITs that reach the nodule release bacterial cells that then differentiate into bacteroids and start fixing nitrogen (Pawlowski and Bisseling, 1996).
In legume-rhizobia symbioses, secreted bacterial Nod factors play an essential role by mediating specific recognition between the two partners and activating a series of responses involved in nodule formation (Lerouge et al., 1990; Denarié and Cullimore, 1993; Downie and Walker, 1999). Among these responses is the transcription of the so-called nodulin genes that are specifically transcribed in symbiotic tissues and may participate in the establishment of the symbiosis (Schultze and Kondorosi, 1998). Some of these nodulin genes are also activated in response to endomycorrhizal colonization of roots (Albrecht et al., 1998), and the analysis of plant mutants indicates that the signaling pathways involved in legume-rhizobia and mycorrhizal symbioses at least partially overlap (Duc et al., 1989; Wegel et al., 1998; Endre et al., 2002; Stracke et al., 2002). These results suggest that at least some of the molecular mechanisms of root nodule symbioses may have been recruited from the more ancient and widespread mycorrhizal symbiosis. As in legumes, several genes specifically induced during actinorhizal symbioses have been described, but little information concerning the signaling pathways involved in their regulation is available (Franche et al., 1998a).
Among actinorhizal nodulin genes, cg12 is one of the earliest induced after Frankia inoculation. This gene was isolated from the actinorhizal tree Casuarina glauca and encodes a subtilisin-like Ser protease (Laplaze et al., 2000). Using transgenic Casuarinaceae containing cg12 promoter reporter gene fusions, we recently showed that cg12 expression is specifically linked to the infection of root hairs and cortical cells by Frankia and that cg12 could therefore be used as a marker gene of the early events that occur during cell infection by bacteria in actinorhizal plants (Svistoonoff et al., 2003). Moreover, cg12 expression is not induced in endo- or ectomycorrhizae nor by Frankia root hair deforming factors (Svistoonoff et al., 2003). Here we show that the cg12 promoter directs reporter gene expression specifically in rhizobia-infected cells in transgenic Medicago truncatula plants. This promoter is not activated either by infection by an endomycorrhizal fungus or by Nod factors. This indicates that a plant cell infection-related transcriptional environment is conserved between actinorhizal and rhizobial symbioses.
RESULTS
The C. glauca cg12 Promoter Retains Its Cell-Specific Expression in Transgenic M. truncatula
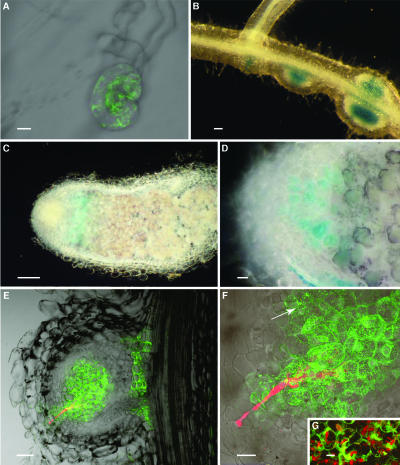
To test whether or not regulatory sequences in the cg12 promoter were recognized by legume transcription factors, transcriptional fusions between the cg12 promoter and gus- or gfp-reporter genes were introduced in M. truncatula. Reporter gene expression was similar in four independent transgenic lines. No gus or gfp expression was detected in shoots and leaves. In noninoculated roots, no green fluorescent protein (GFP) fluorescence was detected; however, slight β-glucuronidase (GUS) activity was present in the pericycle around the vascular tissues (data not shown). This activity was very slight as the blue color could be seen only after >6 h of incubation and then only in small amounts (<0.5 mm) of FeCN. After Sinorhizobium meliloti inoculation, root hair deformation occurred 24 h postinoculation, but the pattern of reporter gene expression did not change until 3 to 8 d postinoculation, when some deformed root hairs (1 out of 100–200) began to strongly express the reporter genes (Fig. 1A). Two to 3 d later, as a nodule primordium was formed beneath these root hairs, reporter gene expression was detected in the central tissues of the developing nodule (Fig. 1B). Strong reporter gene expression was then detected in bacteria-infected cells in the nodule primordia and in small nodules (Fig. 1B). In older nodules, reporter gene expression was detected in the infection zone (Fig. 1C) and decayed in the interzone II-III, as marked by starch accumulation, and in the fixation zone where cells begin to differentiate and fix nitrogen (Fig. 1, C and D). Infection with a S. meliloti strain that constitutively produces DsRed showed that all the cells containing ITs strongly express gfp (Fig. 1, E–G). GFP fluorescence was also detected in some uninfected cells close to the newly infected cells (Fig. 1, E and F). Consequently, the cg12 promoter is activated in cells infected or just about to be infected by the endosymbiotic bacteria. This pattern of expression is similar to the one observed with the same constructs in transgenic actinorhizal plants (Svistoonoff et al., 2003).
Figure 1.
Analysis of reporter gene expression in transgenic M. truncatula. A to D, Plants inoculated with wild-type S. meliloti. A, Composite image of a root hair 10 dai showing GFP fluorescence. B, Lateral root showing three early developmental stages of nodule formation. C and D, Longitudinal sections of mature nodules. To determine different nodule zones, starch granules that accumulate in the interzone II-III were stained with Lugol's solution. E to G, Composite images of longitudinal sections of young nodules obtained 10 to 12 dai with transgenic S. meliloti constitutively expressing DsRed. GFP fluorescence is seen in green; DsRed fluorescence is seen in red. F, Detail of E showing the progression of the IT inside the nodule and the consecutive GFP fluorescence in the infected cells. Note that some cells not yet invaded by bacteria already show weak levels of GFP fluorescence (arrow). G, Cells infected by ITs strongly expressing GFP. A, Bar = 10 μm; B, D, and E, bar = 50 μm; C, bar = 500 μm; F and G, bar = 20 μm.
To investigate whether the cg12 promoter is also activated during arbuscular mycorrhizal (AM) symbiosis, transgenic M. truncatula were inoculated with the endomycorrhizal fungi Glomus mossaeae, Glomus rosea, and Glomus deserticola. GUS activity was not detected in mycorrhized roots (data not shown), suggesting that the cg12 promoter is not activated in fungi-infected cells during AM formation.
Nod Factors Are Necessary But Not Sufficient for cg12 Promoter Activation in Transgenic M. truncatula
To determine whether Nod factor recognition is involved in cg12 promoter activation, roots from transgenic M. truncatula plants were treated either with wild-type or a nodA mutant S. meliloti strain or purified S. meliloti Nod factors or water (control plants). The nodA mutant used in this study carries a polar mutation that abolishes synthesis of the three enzymes encoded by the nodABC operon that are responsible for elaborating the Nod factor core structure and is therefore unable to produce Nod factors. Inoculated roots were removed 2 or 7 d after inoculation (dai). GUS activity was analyzed on one-half of the samples. The rest of the inoculated roots were used to test the endogenous MtENOD20 expression using a reverse transcription (RT)-PCR approach. MtENOD20 is rapidly activated by infection and purified Nod factors (Vernoud et al., 1999) and was therefore used as a positive control. The results of both tests are summarized in Table I.
Table I.
GUS activity and MtENOD20 expression in Pcg12-GUS transgenic plants following treatment with wild-type or nodA mutant S. meliloti strains or purified Nod factors
| Days After Inoculation
|
||||
|---|---|---|---|---|
| Treatment | 2
|
7
|
||
| GUSa | MtENOD20b | GUSa | MtENOD20b | |
| Noninoculated (water) | − | − | − | − |
| S. meliloti Rm2011 (wild type) | − | + | + | + |
| S. meliloti RM2011 nodA::Tn5 | − | − | − | − |
| S. meliloti Nod factors (10−7M) | − | + | − | + |
−, No GUS staining; +, GUS staining detected in some root hairs or cortical cells.
−, No MtENOD20 expression; +, MtENOD20 expression in the inoculated roots.
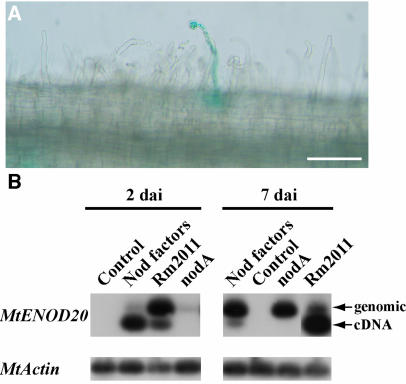
When plants were inoculated with wild-type S. meliloti, GUS activity was seen in some curled root hairs (Fig. 2A) and young nodule primordia 7 dai but not 2 dai. ENOD20 expression was detected at both 2 and 7 dai (Fig. 2B). Control plants (treated with water) showed neither GUS activity (data not shown) nor ENOD20 expression (Fig. 2B). No GUS activity nor ENOD20 expression was found in root hairs or cortical cells following treatment with the nodA mutant S. meliloti strain. This shows that a S. meliloti nodA mutant unable to produce Nod factors is unable to activate cg12 promoter. On the other hand, no GUS activity was found in transgenic plants treated with purified S. meliloti Nod factors (data not shown), whereas ENOD20 expression was detected (Fig. 2B), therefore showing that the plants reacted properly to Nod factors. Taken together, these results indicate that Nod factors are necessary but not sufficient for cg12 promoter activation in transgenic M. truncatula plants. Some GUS activity in the pericycle around the vascular tissues was seen for all treatments (data not shown).
Figure 2.
Analysis of Pcg12::GUS and endogenous MtENOD20 expression in transgenic M. truncatula plants inoculated with wild-type (Rm2011), mutant (nodA) S. meliloti, purified Nod factors, or water (control) 2 and 7 dai. A, Detail of a root 7 dai with S. meliloti Rm2011 showing GUS activity in a deformed root hair and the adjacent cortical cells. B, Transcriptional activation of MtENOD20 expression 2 and 7 dai. Inoculated roots were harvested, total RNA extracted, and MtENOD20 expression analyzed by RT-PCR. MtENOD20 primers are situated on both sides of the single MtENOD20 intron (Vernoud et al., 1999) so that cDNA and genomic amplification products can be separated. Amplification of a constitutive actin cDNA (MtActin; Cohn et al., 2001) was used as a positive control. A, Bar = 100 μm.
cg12 Promoter Is Induced after Inoculation with S. meliloti exoH Mutant Defective in Exopolysaccharide Synthesis
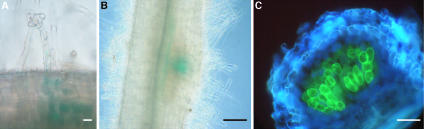
Since cg12 promoter activation is linked to plant cell infection by ITs, we addressed the question of whether an S. meliloti mutant that forms aborted ITs still elicits activation of the cg12 promoter. After inoculation of transgenic plants with the exoH mutant, root hair deformation occurred and was followed by cortical cell divisions as described (Yang et al., 1992). Six to 10 dai, small nodule-like structures devoid of bacteria appeared but did not evolve any further. Reporter gene activity was first seen in some deformed root hairs (Fig. 3A) then in cortical cells and in young nodule-like structures (Fig. 3, B and C). However, reporter gene expression was transient, and 2 weeks after inoculation reporter gene expression was no longer detected in the bacteria-free nodules.
Figure 3.
Analysis of reporter gene expression in transgenic M. truncatula plants inoculated with the Rm7225 exoH mutant. A, Detail of a lateral root showing GUS activity in a deformed root hair and in cortical cells beneath 10 dai. B, Nodule primordia 12 dai showing GUS activity in the pericycle and in primordium cells. C, Nodule-like structure obtained 2 weeks after inoculation showing GFP fluorescence in central parenchyma cells. A, Bar = 10 μm; B, bar = 60 μm; C, bar = 20 μm.
Putative cg12 Homologs Are Expressed in M. truncatula Nodules
To find homologs of cg12 in M. truncatula, we performed a BLAST search on M. truncatula cluster expressed sequence tag (EST) database (http://medicago.toulouse.inra.fr/Mt/EST). Many ESTs corresponding to subtilases and highly homologous to cg12 were found. The expression pattern of the 25 genes most similar to cg12 (Table II) was analyzed in silico using the iESTANT electronic northern facility (Journet et al., 2002). Four of those genes (MtC50058_GC, MtC10895_GC, MtD15912_GC, and MtC93070_GC) were predicted to be expressed in nodules or rhizobia-infected roots (data not shown) and might therefore represent orthologs of cg12.
Table II.
BLAST search results
| Rank | Name | BLAST Score | Expect Value | Identity | Positive | Gap |
|---|---|---|---|---|---|---|
| % | % | % | ||||
| 1 | MtC50058_GC | 1,424 | 1.6e-146 | 41 | 60 | 3 |
| 2 | MtC10895_GC | 654 | 1.2e-124 | 39 | 56 | 4 |
| 3 | MtC10895_GC | 584 | 1.2e-124 | 43 | 62 | 1 |
| 4 | MtC40177_GC | 1,188 | 1.6e-121 | 41 | 57 | 3 |
| 5 | MtC40177_GC | 639 | 2.4e-63 | 39 | 58 | 5 |
| 6 | MtD05536_GC | 646 | 4.2e-119 | 65 | 78 | 0 |
| 7 | MtD05536_GC | 543 | 4.2e-119 | 51 | 72 | 3 |
| 8 | MtC45210_GC | 513 | 5.6e-85 | 39 | 57 | 3 |
| 9 | MtC45210_GC | 394 | 5.6e-85 | 39 | 59 | 2 |
| 10 | MtD04916_GC | 463 | 3.6e-83 | 50 | 70 | 2 |
| 11 | MtD04916_GC | 398 | 3.6e-83 | 55 | 71 | 1 |
| 12 | MtC60376_GC | 444 | 3.1e-80 | 38 | 56 | 4 |
| 13 | MtC60376_GC | 424 | 3.1e-80 | 41 | 57 | 6 |
| 14 | MtD01919_GC | 769 | 9.5e-77 | 43 | 60 | 3 |
| 15 | MtD15912_GC | 654 | 1.2e-64 | 39 | 56 | 4 |
| 16 | MtC00585_GC | 636 | 1.0e-62 | 40 | 60 | 2 |
| 17 | MtD02656_GC | 634 | 1.5e-62 | 38 | 55 | 2 |
| 18 | MtC93070_GC | 585 | 3.0e-57 | 44 | 61 | 1 |
| 19 | MtD06873_GC | 459 | 8.6e-57 | 45 | 66 | 1 |
| 20 | MtD06873_GC | 152 | 8.6e-57 | 37 | 55 | 3 |
| 21 | MtD07861_GC | 579 | 1.5e-56 | 40 | 55 | 2 |
| 22 | MtC10182_GC | 570 | 9.8e-56 | 41 | 58 | 0 |
| 23 | MtC61392_GC | 569 | 1.2e-55 | 44 | 60 | 1 |
| 24 | MtD24960_GC | 539 | 3.8e-52 | 49 | 69 | 1 |
| 25 | MtC91611_GC | 524 | 1.6e-50 | 46 | 63 | 5 |
CG12 protein sequence was compared to cluster M. truncatula consensus EST database.
DISCUSSION
Infections of the legume M. truncatula by S. meliloti and the actinorhizal tree C. glauca by Frankia are morphologically very similar. The intracellular infection pathway shared by both plants begins with the penetration of bacteria through a deformed root hair. Invagination of the plasma membrane leads to the formation of an IT that progresses from one cell to another. In both symbiotic systems, the cells that are going to be infected by an IT undergo rearrangements leading to the movement of the nucleus toward the center of the cells and the formation of a phragmoplast-like structure, the preinfection thread (PIT), through which the IT can grow (Berg, 1999; Gage and Margolin, 2000). Our results suggest that activation of the cg12 promoter in M. truncatula is strongly associated with plant cell infection: reporter genes are expressed in cells containing ITs and in cells located near infected cells that presumably contain PITs. A similar expression pattern has been described for the legume early nodulin gene Enod5. Pea (Pisum sativum) Enod5 expression is detected in root 24 h after rhizobia inoculation (Albrecht et al., 1998) and in nodule cells recently invaded by rhizobia (Scheres et al., 1990). In contrast with other early-induced nodulin genes such as Enod12 or Enod40, Enod5 expression is mainly detected in cells that contain bacteria and is only slightly induced by Nod factor treatments (Heidstra et al., 1997). On the other hand, Enod5 expression is induced in AM roots (Albrecht et al., 1998), while cg12 is not. A factor produced in response to cell infection and acting over a short distance has been proposed to regulate Enod5 expression (Scheres et al., 1990). Such a factor could also be involved in the regulation of cg12 expression. In legumes, most early nodulin genes (such as MtEnod11, Enod12, Enod12A, and Enod20) respond to purified Nod factor treatments (Spaink, 1996; Niebel Fde et al., 1998). In this study, we showed that Nod factors are necessary but not sufficient to activate the cg12 promoter. Interestingly, the same is true for PIT formation in alfalfa (Medicago sativa), a very close relative of M. truncatula (Timmers et al., 1999). On the other hand, Nod factors induce PIT structures in pea (Yang et al., 1994).
In a recent study, we showed that in C. glauca, the activation of cg12 promoter is also strongly linked to plant cell infection by Frankia both in root hairs and in root and nodule cortical cells (Svistoonoff et al., 2003). This suggests considerable similarity between the transcriptional environments activated in response to plant cell infection by bacteria in both symbiotic systems. Similar results were obtained for hemoglobin genes whose expression is induced later in nodule development. The promoter of the soybean (Glycine max) lbc3 gene retains its cell specificity in transgenic Allocasuarina verticillata (Franche et al., 1998b) and so does the C. glauca promoter in transgenic Lotus japonicus (Jacobsen-Lyon et al., 1995).
In M. truncatula, cg12 promoter activity also occurred in pericycle cells located in front of a growing nodule primordium, whereas in C. glauca no expression in the pericycle cells was detected. The pericycle cells of the two systems behave differently in response to bacterial infection: in C. glauca the nodule primordia are formed in the pericycle, while in M. truncatula nodule primordia arise in the inner cortex. During the formation of legume nodule primordia, pericycle cells may be activated to enter the cell cycle (e.g. formation of nodule vasculature). cg12 activation may be related to this cell activation in a way similar to that in cells that contain PITs, which were also shown to reenter the cell cycle (Yang et al., 1994). Alternatively, it is possible that the very high levels of reporter gene expression in M. truncatula in comparison to C. glauca allowed the detection of slight activation in M. truncatula that was too low to be detected in C. glauca.
The exopolysaccharide succinoglycan (EPS) has an important signaling function during the infection of legumes by their rhizobial endosymbiont. exo mutants deficient in EPS synthesis are inefficient at initiating and extending ITs (Gage and Margolin, 2000; Cheng and Walker, 1998). The involvement of bacterial EPS in cg12 promoter activation was assessed using the exoH mutant that produces symbiotically nonfunctional high-molecular-weight EPS that lacks the succinyl modification. The exoH mutant is able to elicit root hair deformation, division of cortical cells, and even the formation of small nodule-like structures that are not colonized by bacteria (Cheng and Walker, 1998). ITs are initiated but fail to elongate and abort before reaching the middle of root hairs (Cheng and Walker, 1998). After inoculation with exoH mutant, the cg12 promoter is transiently activated in root hairs and nodule primordia in transgenic M. truncatula. This transient activation could be due to PIT formation in some cortical cells. This suggests that although the recognition of a proper EPS is not needed for cg12 promoter activation, it requires the maintenance and extension of ITs in infected cells and in cells preparing for infection (containing PITs).
Several nodulin genes are also activated in response to endomycorrhizal colonization of roots (Miklashevichs et al., 2001), and the analysis of plant mutants indicates that the signaling pathways involved in legume-rhizobia and mycorrhizal symbioses at least partially overlap (Duc et al., 1989; Wegel et al., 1998; Endre et al., 2002; Stracke et al., 2002). In this study, we showed that cg12 promoter is not activated by AM fungus infection in transgenic M. truncatula. Taken together, our results show that cg12 promoter contains regulatory motifs that are specifically recognized during plant cell infection by the endosymbiotic bacteria in both rhizobial and actinorhizal symbioses. This suggests that at least part of the molecular mechanisms leading to the transcription of infection-related symbiotic genes is conserved between and specific to the two systems.
cg12 codes for a subtilisin-like Ser protease secreted in the interface between the plant cell and Frankia (S. Svistoonoff, M. Nicole, and D. Bogusz, unpublished data). It may mature proteins and/or peptides involved in signal exchanges between the two partners or participate in cell wall loosening linked to IT growth. To our knowledge, no ortholog of cg12 has been studied in legumes. We found genes highly similar to cg12 among M. truncatula ESTs. Four of them are predicted to be expressed in nodules or rhizobia-infected roots and could therefore have a similar role in M. truncatula as cg12 in C. glauca. It will be interesting to know if a true homolog of cg12 exists in a model legume as this would allow the functional study of the role of this subtilase during the symbiotic infection process.
Actinorhizal plants are distributed among eight families that include symbiotic and nonsymbiotic plants. Phylogenetic studies have shown that together with legumes, all actinorhizal plants belong to the Rosid I clade (Soltis et al., 1995), thus suggesting that a predisposition to enter symbiosis appeared in the ancestor of this group. It is possible that key elements of this predisposition to enter symbioses with soil bacteria rely on their ability to recognize the symbiotic partner and to activate suitable responses to accommodate them. Our results suggest that actinorhizal plants and legumes share common mechanisms of transcriptional gene regulation activated specifically during bacterial infection. This conserved pathway may be part of the common heritage of legume and actinorhizal plants.
MATERIALS AND METHODS
Plant Material
Medicago truncatula cv Jemalong plants were grown in a growth chamber under a 16-h photoperiod and a day/night temperature of 24°C/22°C, respectively, with light intensity of about 100 μmol E−2 s−1 and relative humidity of 50%. Plants were cultivated on Murashige and Skoog medium with 3% Suc.
Transformation of M. truncatula
Transcriptional fusions between the cg12 promoter region and reporter genes gus and gfp generated as previously described (Svistoonoff et al., 2003) were transformed into Agrobacterium tumefaciens EHA105 strain (Trinh et al., 1998). Transgenic M. truncatula plants containing Pcg12-gus or Pcg12-gfp fusions were regenerated via somatic embryogenesis as described (Chabaud et al., 1996). Four transgenic lines coming from independent transformation events were selected for each construct. The transgenic nature of the plants was confirmed using kanamycin resistance, reporter gene expression, and PCR experiments performed with specific primers (data not shown).
Bacterial and Fungal Inoculation
The wild-type Sinorhizobium meliloti Rm2011 strain was used for nodulation experiments. A strain constitutively expressing DsRed was generated by introducing the pDG77 (ptrp-DsRed) plasmid (Gage, 2002) in the wild-type strain. The S. meliloti nodA mutant GMi5382 (Rm2011 nodA::Tn5 no.2208) that does not produce Nod factors was kindly provided by Dr J. Dénarié. The S. meliloti mutant defective in exopolysaccharide synthesis was the Rm7225 strain (exoH225; Leigh et al., 1987). Plants grown in petri dishes containing nitrogen-free Fahraeus medium (Fahraeus et al., 1957) were inoculated with S. meliloti strains resuspended in water at an OD600 = 0.003.
Inoculum of the mycorrhizal fungus Glomus mosseae, Glomus rosea, and Glomus deserticola propagated on Trifolium roots was kindly provided by L.G. Wall (National University of Quilmes, Argentina). Transgenic plants from in vitro culture were transferred to pots containing a sterilized mixture of soil-vermiculite mixed with the fungal inoculum and allowed to grow for 1 month. The roots were harvested at different times during the infection process, and reporter gene expression was assayed. The presence of mycorrhizal structures was checked by staining fungal hyphae with trypan blue in the same samples used to assay reporter gene expression as described (Svistoonoff et al., 2003).
Application of Nod Factors
M. truncatula plants were grown as described for S. meliloti inoculation. Twenty-two-week-old transgenic plants were used for Nod factor treatments. Solutions of purified Nod factors (10−6 or 10−7 m) from S. meliloti (kindly provided by J. Dénarié, Institut National de la Recherche Agronomique Toulouse, France) were directly applied to the roots. Reporter gene expression was monitored 24, 48, 36, and 64 h, 1 week, and 3 weeks after inoculation.
Histochemical GUS Assays and Microscopy
M. truncatula nodules were embedded in 3% agarose and sliced into 40- to 60-μm-thick sections on a vibratome (Leica VT1000E, Wetzlar, Germany). For the detection of GUS activity, explants from Pcg12-gus transformed plants were stained in a solution containing 1 mm X-gluc (5-bromo-4-chloro-3-indolyl β-d-glucuronide) and incubated for 0.5 to 12 h at 37°C. A total of 0.5 to 5 mm of K3Fe(CN)6 and K4Fe(CN)6 were added to limit the diffusion of the blue staining. Samples were fixed for 12 h in a solution containing 5% formaldehyde, 5% acetic acid, and 50% ethanol and washed several times in 70% ethanol. Whole root segments or sections were observed under a Leica DMRB microscope. For starch staining, samples were incubated for 5 min in Lugol's solution then washed in water. Samples were observed using a Leica DMRB microscope with appropriate filter sets for GFP fluorescence observation. Confocal microscopy imaging was performed on agar-embedded sections of living material as described (Svistoonoff et al., 2003).
RT-PCR Analysis of MtENOD20 Expression
Roots inoculated with S. meliloti 2011 (wild type) or GMi5283 (2011 nodA mutant) strain or treated with purified Nod factors or water (control plants) were harvested and frozen in liquid nitrogen. Total RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. First strand synthesis, amplification, and detection of the MtENOD20 cDNA fragment were done according to Vernoud et al. (1999). Parallel amplification of a constitutively expressed M. truncatula actin gene (Cohn et al., 2001) was used as an internal control.
BLAST Search and in Silico Analysis of Gene Expression
BLAST search was performed using the MtCD BLAST server (http://medicago.toulouse.inra.fr/Mt/EST). The predicted CG12 protein sequence was compared to MtCDJan2003 cluster DNA consensus EST database using TBLASTN. Gene expression was analyzed using the iESTANT electronic northern facility (http://medicago.toulouse.inra.fr/Mt/EST/; Journet et al., 2002).
Acknowledgments
We thank Dr. D. Barker (Laboratory of Plant-Microbe Interactions, Castanet-Tolosan, France) for critical reading of this manuscript, N. Lautredou (CRIC, Montpellier, France) for help in confocal microscopy, and Mariana Obertello (IRD, Montpellier, France) for help with analysis of endomycorrhizae.
This work was supported by IRD, FCT (project POCTI/BME/36191 and grant SFRH/BD/6493/2001 to J.L.), and by GRICES/French Embassy (bilateral project 616C2).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.048967.
References
- Albrecht C, Geurts R, Lapeyrie F, Bisseling T (1998) Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. Plant J 15: 605–614 [DOI] [PubMed] [Google Scholar]
- Berg RH (1999) Frankia forms infection threads. Can J Bot 77: 1327–1333 [Google Scholar]
- Chabaud M, Larsonneau C, Marmouget C, Huguet T (1996) Transformation of barrel medic (Medicago truncatula Gaertn.) by Agrobacterium tumefaciens and regeneration via somatic embryogenesis of transgenic plants with the MtENOD12 nodulin promoter fused to the gus reporter gene. Plant Cell Rep 15: 305–310 [DOI] [PubMed] [Google Scholar]
- Cheng HP, Walker GC (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180: 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn JR, Uhm T, Ramu S, Nam YW, Kim DJ, Penmetsa RV, Wood TC, Denny RL, Young ND, Cook R, et al (2001) Differential regulation of a family of apyrase genes from Medicago truncatula. Plant Physiol 125: 2104–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denarié J, Cullimore J (1993) Lipo-oligosaccharide nodulation factors: new class of signaling molecules mediating recognition and morphogenesis. Cell 74: 951–954 [DOI] [PubMed] [Google Scholar]
- Downie JA, Walker SA (1999) Plant responses to nodulation factors. Curr Opin Plant Biol 2: 483–489 [DOI] [PubMed] [Google Scholar]
- Duc G, Trouvelot A, Gianinazzi-Pearson V, Gianinazzi S (1989) First report of non-mycorrhizal plant mutants (myc-) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.). Plant Sci 60: 215–222 [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Fahraeus G (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Franche C, Diouf D, Laplaze L, Auguy F, Frutz T, Rio M, Duhoux E, Bogusz D (1998. b) Soybean (lbc3), Parasponia and Trema hemoglobin gene promoters retain symbiotic and nonsymbiotic specificity in transgenic Casuarinaceae: implications for hemoglobin gene evolution and root nodule symbioses. Mol Plant Microbe Interact 11: 887–894 [Google Scholar]
- Franche C, Laplaze L, Duhoux E, Bogusz D (1998. a) Actinorhizal symbioses: recent advances in plant molecular and genetic transformation studies. Crit Rev Plant Sci 17: 1–28 [Google Scholar]
- Gage DJ (2002) Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol 184: 7042–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ, Margolin W (2000) Hanging by a thread: invasion of legume plants by rhizobia. Curr Opin Microbiol 3: 613–617 [DOI] [PubMed] [Google Scholar]
- Heidstra R, Nilsen G, Martinez-Abarca F, van Kammen A, Bisseling T (1997) Nod factor-induced expression of leghemoglobin to study the mechanism of NH4NO3 inhibition of root hair deformation. Mol Plant Microbe Interact 10: 215–220 [DOI] [PubMed] [Google Scholar]
- Jacobsen-Lyon K, Jensen EO, Jorgensen JE, Marcker KA, Peacock WJ, Dennis ES (1995) Symbiotic and nonsymbiotic hemoglobin genes of Casuarina glauca. Plant Cell 7: 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring the root symbiotic programs of the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Ribeiro A, Franche C, Duhoux E, Auguy F, Bogusz D, Pawlowski K (2000) Characterization of a Casuarina glauca nodule-specific subtilisin-like protease gene, a homolog of Alnus glutinosa ag12. Mol Plant Microbe Interact 13: 113–117 [DOI] [PubMed] [Google Scholar]
- Leigh JA, Reed JW, Hanks JF, Hirsch AM, Walker GC (1987) Rhizobium meliloti mutants that fail to succinylate their calcofluor-binding exopolysaccharide are defective in nodule invasion. Cell 51: 579–587 [DOI] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Prome JC, Denarie J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Miklashevichs E, Rohring H, Schell J, Schmidt J (2001) Perception and signal transduction of rhizobial NOD factors. Crit Rev Plant Sci 20: 373–394 [Google Scholar]
- Niebel Fde C, Lescure N, Cullimore JV, Gamas P (1998) The Medicago truncatula MtAnn1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Mol Plant Microbe Interact 11: 504–513 [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Bisseling T (1996) Rhizobial and actinorhizal symbioses: what are the shared features? Plant Cell 8: 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, van Engelen F, van der Knaap E, van de Wiel C, van Kammen A, Bisseling T (1990) Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell 2: 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze M, Kondorosi A (1998) Regulation of symbiotic root nodule development. Annu Rev Genet 32: 33–57 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, Martin PG (1995) Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci USA 92: 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP (1996) Regulation of plant morphogenesis by lipochin oligosaccharides. Crit Rev Plant Sci 15: 559–582 [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Laplaze L, Auguy F, Runions J, Duponnois R, Haseloff J, Franche C, Bogusz D (2003) cg12 expression is specifically linked to infection of root hairs and cortical cells during Casuarina glauca and Allocasuarina verticillata actinorhizal nodule development. Mol Plant Microbe Interact 16: 600–607 [DOI] [PubMed] [Google Scholar]
- Timmers AC, Auriac MC, Truchet G (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628 [DOI] [PubMed] [Google Scholar]
- Trinh TH, Ratet P, Kondorosi E, Durand P, Kamaté K, Bauer P, Kondorosi A (1998) Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp. falcata lines improved in somatic embryogenesis. Plant Cell Rep 17: 345–355 [DOI] [PubMed] [Google Scholar]
- Vernoud V, Journet E-P, Barker DG (1999) MtENOD20, a Nod factor-inducible molecular marker for root cortical cell activation. Mol Plant Microbe Interact 12: 604–614 [Google Scholar]
- Wegel E, Schauser L, Sandal N, Stougaard J, Parniske M (1998) Mycorrhiza mutants of Lotus japonicus define genetically independent steps during symbiotic infection. Mol Plant Microbe Interact 11: 933–936 [Google Scholar]
- Yang C, Signer ER, Hirch AM (1992) Nodules initiated by Rhizobium meliloti exopolysaccharide mutants lack a discrete, persistent nodule meristem. Plant Physiol 98: 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, de Blank C, Meskiene I, Hirt H, Bakker J, van Kammen A, Franssen H, Bisseling T (1994) Rhizobium Nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6: 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]