Abstract
Plant roots can be highly colonized by fungal endophytes. This seems to be of particular importance for the survival of plants inhabiting stressful habitats. This study focused on the Identification of the fungal endophytic community associated with the roots of quinoa plants (Chenopodium quinoa) growing near the salt lakes of the Atacama Desert, Chile. One hundred endophytic fungi were isolated from healthy quinoa roots, and the internal transcribed spacer (ITS) region was sequenced for phylogenetic and taxonomic analysis. The isolates were classified into eleven genera and 21 distinct operational taxonomic units (OTUs). Despite a relatively high diversity of root endophytic fungi associated with quinoa plants, the fungal community was dominated by only the Ascomycota phyla. In addition, the most abundant genera were Penicillium, Phoma and Fusarium, which are common endophytes reported in plant roots. This study shows that roots of C. quinoa harbor a diverse group of endophytic fungi. Potential roles of these fungi in plant host tolerance to stressful conditions are discussed.
Keywords: Fungal endophytes, Quinoa, Internal transcribed spacer, Phylogenetic analysis, Atacama Desert
1. Introduction
Plant roots can be associated with a variety of endosymbiotic microbes including mycorrhizal fungi, rhizosphere bacteria and endophyte microorganisms [1]. Root-associated endophytic fungi commonly occur in angiosperms and a high rate of colonization has been described for different plants and environments [2], [3], [4], [5], [6]. Non-clavicipitaceous endophytes associated with plant roots involve different groups of fungi, including the ‘class II endophytes’ and the ‘dark septate endophytes’ (DSE) [7]. These groups differ in ecological aspects, such as host colonization patterns, mechanism of transmission and biodiversity levels [7], but both are capable of extensive tissue colonization and appear to be of particular relevance for plant survival in stress habitats [8], [9], [5], [10]. Although the study of endophytic fungal communities in plants has gained attention in recent years, little is known about the diversity and composition of endophytic fungi associated with agricultural crops [11], [12].
Desert habitats represent one of the most challenging environments for the growth of plants [13], whose distribution and diversity are restricted by environmental stresses, including low water availability and high temperatures, salinity and irradiance. The Atacama Desert of Northern Chile is considered to be one of the driest regions of the world [14]. It is a severe environment for plant growth due to its high salinity, low temperatures and drought. The pseudo-cereal Chenopodium quinoa is a native crop well-adapted to the harsh climatic conditions of the Atacama Desert. Quinoa is an important food source in the Andean region since 3000 BCE [15]. Recently, this crop plant has gained attention due to its high nutritional value, being considered an important crop with the potential of contributing to food security worldwide [16].
The goal of this study was to investigate the fungal endophyte community associated with the roots of C. quinoa growing near the Salt Lake of the Atacama Desert, Chile. We isolated and identified root-associated endophytic fungi of C. quinoa plants by the amplification of the internal transcribed spacer (ITS) region of fungus genomic DNA. This study is the first research performed in Chile that provides data on the relationship between C. quinoa and its endosymbiotic microbes.
2. Methods
2.1. Studied species
C. quinoa Willd. (Amaranthaceae) is a gynomonoecious annual plant with an erect stem. It bears alternate leaves that are variously coloured due to the presence of betacyanins. The inflorescence is a panicle, 15–70 cm in length, rising from the top of the plant and from the axils of lower leaves [17]. Quinoa is mainly cultivated in the Andean highlands and lowlands of Bolivia, Peru, Chile and Argentina. Root samples were collected from the plants of C. quinoa growing close to the Village of Socaire (23°36′00′′ S and 67°50′60′′ W), situated 3.500 m above sea level, 50 km East of the Salt Lake of the Atacama Desert. The climate is characterized as extreme arid [13]. Daily temperatures range from a maximal average of 24.5 °C to a minimum average of 7.1 °C and the mean annual precipitation is 18 mm [18].
2.2. Isolation of fungal endophytes
Six plants of C. quinoa were randomly selected for root collection in March 2015. Plant roots without visible damage were collected. Primary and secondary roots were washed under running tap water to remove soil debris and then surface-sterilized with ethanol (70%) for 3 min, sodium hypochlorite (1%) for 1 min, followed by three rinses in sterile distilled water for 3 min each. The success of surface sterilization was confirmed by the absence of any microbial growth from the water-wash on PDA plates (potato-dextrose-agar, Phyto Technology Laboratories). Small sections of sterilized roots (0.5–1.0 cm) were subsequently cultivated on PDA petri dishes plates. Plates were then incubated at room temperature for 3–4 weeks. After that time, emerging colonies were subcultured to obtain pure isolates. Fungal isolates were first grouped according to similar morphological characteristics. From those fungi that belonged to the same genus, only three pure isolates were considered for DNA extraction. Pure isolates were grown on PDA plates at room temperature for one month before DNA extraction and molecular identification.
2.3. Molecular characterization of endophytes
Genomic DNA was extracted from the mycelial mat of 45 pure isolates using the method described by Nicholson [19] with slight modifications. Species identification of endophytic fungi was performed using the primers ITS1-F-KYO1 (CTHGGTCATTTAGAGGAASTAA) and ITS4 (TCCTCCGCTTATTGATATGC). Amplification of the ITS regions (around 680 kbp) was conducted with 50 mL of PCR reaction mixtures, each containing 7 μL of total fungal genomic DNA, 1 μL of each primer (10 μM), 27.5 μL of SapphireAmp Fast PCR Master Mix (Takara) and 13.5 μL of sterilized water. PCR was performed in a Techne TC-5000 Thermal Cycler (Fisher Scientific) with the following program: 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 54 °C for 30 s and primer extension at 72 °C for 1 min, completed with a final extension at 72 °C for 7 min. PCR products were sent to Macrogen (South Korea) for purification and sequencing. Sequences were assembled using SeqTrace software. Consensus sequences were used for BLAST search at the NCBI (http://www.ncbi.nlm.nih.gov).
Sequences alignments and phylogenetic tree were constructed by using MEGA software, version 7.0 [20]. Alignments were performed with ClustalW [21], DNA weight matrix ClustaW 1.6 and default parameters. Phylogenetic reconstruction was performed by using neighbor-joining method [22], with p-distance substitution model and bootstrapping of 1000. Additional 18S rRNA sequences for the different genera were retrieved from NCBI.
3. Results
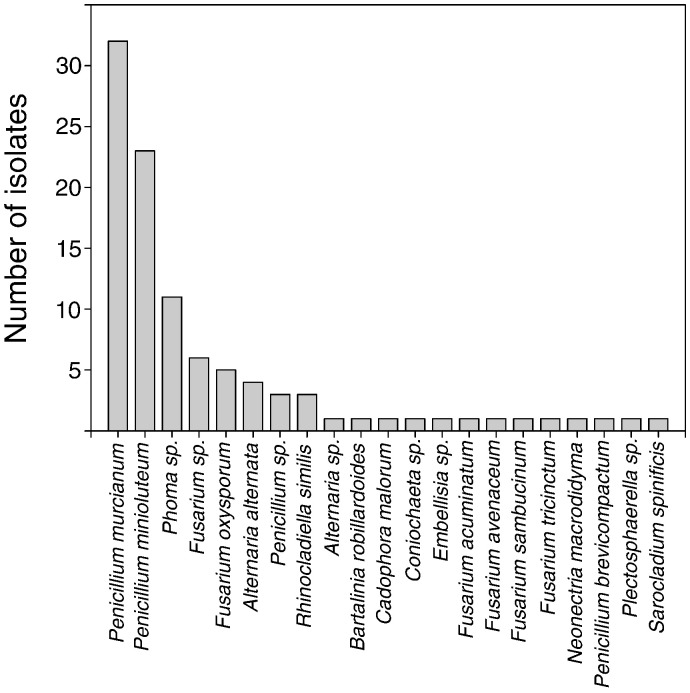
All six plants of C. quinoa screened in this study were associated with endophytic fungi in their roots. Molecular data revealed that C. quinoa roots were colonized by a diverse fungal community (Fig. 1). A total of 100 fungal isolates were purified into individual cultures, with a total species richness of 21 OTUs (Table 1).
Fig. 1.
Frequency of endophytes taxa isolated from healthy roots of plants of Chenopodium quinoa growing in the Atacama Desert.
Table 1.
Best BLAST matches for isolated fungal endophyte OTUs collected from roots of Chenopodium quinoa in the Atacama Desert.
| Phylum | Order | Description | Accession | Identity |
|---|---|---|---|---|
| Ascomycota | Pleosporales | Alternaria alternata | KX355190.1 | 99 |
| Ascomycota | Pleosporales | Alternaria sp. | KR094462.1 | 100 |
| Ascomycota | Xylariales | Bartalinia robillardoides | NR_126145.2 | 99 |
| Ascomycota | Undefined | Cadophora malorum | JQ796752.1 | 99 |
| Ascomycota | Coniochaetales | Coniochaeta sp. | KF367565.1 | 99 |
| Ascomycota | Pleosporales | Embellisia sp. | JN859373.1 | 99 |
| Ascomycota | Hypocreales | Fusarium acuminatum | KR051403.1 | 100 |
| Ascomycota | Hypocreales | Fusarium avenaceum | KT963799.1 | 100 |
| Ascomycota | Hypocreales | Fusarium oxysporum | KU059956.1 | 99 |
| Ascomicota | Hypocreales | Fusarium sambucinum | KM231813.1 | 98 |
| Ascomycota | Hypocreales | Fusarium sp. | KF727426.1 | 99 |
| Ascomicota | Hypocreales | Fusarium tricinctum | KF913341.1 | 99 |
| Ascomycota | Hypocreales | Neonectria macrodidyma | FR877539.1 | 99 |
| Ascomycota | Eurotiales | Penicillium brevicompactum | LT558911.1 | 99 |
| Ascomycota | Eurotiales | Penicillium minioluteum | JF910284.1 | 94 |
| Ascomycota | Eurotiales | Penicillium murcianum | NR_138358.1 | 99 |
| Ascomycota | Eurotiales | Penicillium sp. | HQ631007.1 | 99 |
| Ascomycota | Pleosporales | Phoma sp. | KF367493.1 | 99 |
| Ascomycota | Glomerellales | Plectosphaerella sp. | KU555990.1 | 85 |
| Ascomycota | Chaetothyriales | Rhinocladiella similis | KP132562.1 | 98 |
| Ascomycota | Hypocreales | Sarocladium spinificis | KF269096.1 | 98 |
The Simpson diversity index (range from 0 to 1, with 1 representing a higher sample diversity) was 0.83, suggesting a relatively high diversity of endophytic fungi associated with roots of C. quinoa. The fungal root-endophyte community was mainly dominated by the genus Penicillium that accounted for 59% of the total culturable community. Fusarium and Phoma occurred at frequencies lower than 15%; whereas other genera were found in rare instances, with frequencies between 1% and 5% (Fig. 1).
The phylogenetic tree (Fig. 2) showed eleven clades representing different genera as follows: Phoma, Alternaria, Rhinocladiella, Cadophora, Penicillium, Bartalinia, Coniochaeta, Neonectria, Fusarium, Sarocladium, and Plectosphaerella. The first clade contained ten sequences, which cluster with sequences from Phoma genera obtained from NCBI (ID: KF367492, KF367493, and KT989566) at 94% of bootstrap. The second clade was comprised by the genus Alternaria, which contains seven endophyte sequences, grouped with 93% of bootstrap to sequences from A. alternata and Alternaria sp. obtained from NCBI (ID: LC131449, LC131448, and GU584948). The third clade contained one endophyte sequence that grouped with 100% bootstrap to Rhinoclaediella sequences obtained from NCBI (ID: KF811429, HE608796, and KC254071). Following, we observed one endophyte that grouped with 100% bootstrapping to Cadophora malorum sequences retrieved form NCBI (ID: HM589226, KF646089, and JQ796752). In the Penicillium clade, we clearly observed two sub-clades. The first sub-clade contained sequences with 100% bootstrapping to the NCBI P. murcianum sequence (ID: NR138358). The second sub-clade showed 96% bootstrapping to the P. minioluteum sequence (ID: EU833222). Another clade contained one endophyte sequence that grouped with Bartalinea robillardoides sequences obtained from NCBI with 100% bootstrapping (ID: FR822822, KT269525, and NR126145). The next clade comprises sequences belonging to the Coniochaeta genera obtained from NCBI (ID: KF367558, KF367565, and KF367562) and one endophyte, with a 100% of bootstrapping. The next clade contained one endophyte that grouped with 99% bootstrapping to Neonectria sequences obtained from NCBI (ID: GQ131876, GQ131875, and FR877539). The following clade was divided in two sub-clades, one of them containing four endophyte sequences that grouped to Fusarium oxysporum sequences retrieved from NCBI (ID: EU839369, KU872840) with 95% of bootstrapping, which allows to classify the endophyte in this species. The second sub-clade comprised seven endophyte sequences, which were grouped with 98% bootstrapping to Fusarium sp. sequences obtained from NCBI (ID: KF727426, KU516466, and KT268939). The last two clades showed 100% of bootstrapping. One of them contained one endophyte sequence grouped to Sarocladium sp. sequences retrieved from NCBI (ID: KF269096, KP968451, and KP968441). The last clade comprised one endophyte sequence, and Plectosphaerella cucumerina sequences obtained from NCBI (ID: KU555990, KU350731, and KU350698), allowing classification of the endophyte to species level.
Fig. 2.
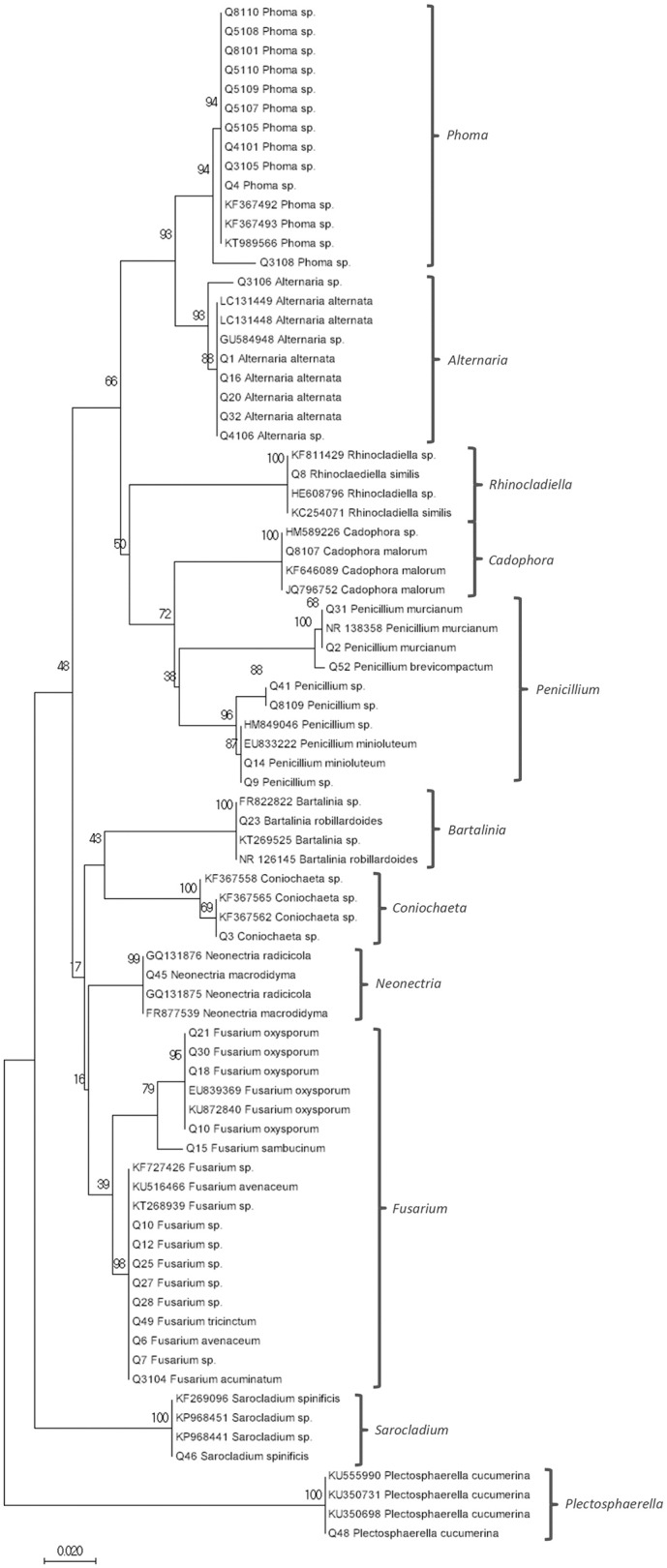
Neighbor Joining (NJ) tree showed phylogenetic relationship between 45 sequences of endophytic fungi from Ascomycota phylum, based on the ITS rDNA sequences. NCBI sequences were added for showing clades. Bootstrap 1000, values are shown at the branch nodes.
4. Discussion
All OTUs described in this study belong to the Ascomycota, which usually is the predominant root-colonizing fungal group [23], [24]. These data support earlier findings showing that root-associated fungal community is diverse for many plant species [25], [26], [5], nevertheless, only a few species dominate the community [27], [5]. Even though the taxonomical classification of DSE has not been well defined yet [7], Penicillium, Phoma and Fusarium, which are the most abundant fungi occurring in C. quinoa roots, have been described as class II endophytes [7]. Further microscopic analysis is required to thoroughly investigate the presence of DSE in roots of C. quinoa.
In desert environments fungal endophyte communities usually appear to be diverse [5], [24]. It has been proposed that heterogeneous conditions of desert ecosystems may be responsible for the high diversity of fungal communities in these habitats [28], [29]. Considering the increase evidence showing that fungal endophytes improve tolerance to drought conditions [30], [31], [32], association of quinoa plants with an array of fungi might be of crucial importance for its survival under the extreme environmental conditions of the Atacama Desert.
Sequencing of the amplified endophyte ITS regions allowed the identification of all the isolates to the genus level, and in several cases enabled us to assign the species classification based on nucleotides conservation. Examples are the sequence Q1 that had a 99% of identity to NCBI sequence ID KX355190, classified as Alternaria alternata, and sequence Q3104 that had 100% match to the NCBI Fusarium acuminatum ID KR051403.
The dominant fungal genera detected in quinoa roots (Penicillium, Phoma and Fusarium) have previously been found as root inhabitants in several plant species [33], [26], [34], and seem to play positive roles in plant tolerance to abiotic stress and plant growth. For example, plant association with Penicillium endophytes can help plants to resist salinity stress and to improve plant growth [33]. Similarly, Phoma spp. are common root endophytes which are able to confer fitness benefits to plants [26], [34]. Previous investigations in Andean C. quinoa have demonstrated the presence of bacterial endophytes associated with this pseudo-cereal [35], [36]. A recent study of Pitzschke [36] showed that 100% of quinoa seeds are inhabited by diverse members of the bacterial genus Bacillus, which are likely to induce a naturally primed state in quinoa plants, enabling them to overcome extreme environmental situations. In contrast to this apparently high bacterial colonization inside of quinoa tissues, another study on fungal root symbionts in roots of Bolivian Andean quinoa plants revealed that fungal endophyte presence was negligible [37]. The present study is the first report on the fungal endophyte community present in Chilean quinoa variety. It highlights the importance of considering root-endophytic fungi as a new additional player, together with endophytic bacteria [36], which promote quinoa's resistance to high-stress environmental conditions.
In Chile, there is a lack of information about endophytic fungi associated with plant roots [37], and about the potential benefits that endophytic fungi can provide to their host plants. Quinoa is well adapted to severe environmental conditions, including extreme aridity [38], low temperatures [39], [40] and high salinity [41]. Further experimental studies are required to elucidate the effects of root endophytic fungi on plant tolerance of C. quinoa to high-stress environmental conditions.
5. Conclusion
Data here showed a relatively high diversity of endophytic fungi associated with roots of C. quinoa. The fungal community was dominated by only three fungal genera: Penicillium, Phoma and Fusarium, which are common endophytes reported in plant roots. This study provided information that roots of C. quinoa harbor a diverse group of endophytic fungi, which might play potential roles on plant host tolerance to stressful conditions.
Funding
This work was partially supported by Fondecyt grants No. 11130039 (MG-T) and No. 11130480 (LB-G).
Competing interests
The authors declare that they have no competing interests.
Acknowledgements
We are very grateful to Mónica Cisternas, Andrea Morales and Carolina Murciano for their valuable help in the lab.
References
- 1.Dighton J., White J.J., Oudemans P. Taylor & Francis Group, CRC Press; Boca Raton, FL: 2005. The Fungal Community: Its Organization and Role in the Ecosystem. [Google Scholar]
- 2.Ananda K., Sridhar K.R. Diversity of endophytic fungi in the roots of mangrove species on west coast of India. Can. J. Microbiol. 2002;48:871–878. doi: 10.1139/w02-080. [DOI] [PubMed] [Google Scholar]
- 3.Khidir H.H., Eudy D.M., Porras-Alfaro A., Herrera J., Natvig D.O., Sinsabaugh R.L. A general suite of fungal endophytes dominate the roots of two dominant grasses in a semiarid grassland. J. Arid Environ. 2010;74:35–42. [Google Scholar]
- 4.Newsham K.K., Upson R., Read D.J. Mycorrhizas and dark septate endophytes in polar regions. Fungal Ecol. 2009;2:10–20. doi: 10.1007/s00572-009-0260-3. [DOI] [PubMed] [Google Scholar]
- 5.Porras-Alfaro A., Herrera J., Sinsabaugh R.L., Odenbach K.J., Lowrey T., Natvig D.O. Novel root fungal consortium associated with a dominant desert grass. Appl. Exp. Microbiol. 2008;74:2805–2813. doi: 10.1128/AEM.02769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toju H., Yamamoto S., Sato H., Tanabe A., Gilbert G., Kadowaki K. Community composition of root-associated fungi in a Quercus dominated temperate forest: “codominance” of mycorrhizal and root-endophytic fungi. Ecol. Evol. 2013;3:1281–1293. doi: 10.1002/ece3.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodríguez R.J., White J.F., Jr., Arnold A.E. Fungal endophytes: Diversity and functional roles. New Phytol. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 8.Crozier J., Thomas S.E., Aime M.C., Evans H.C., Holmes K.A. Molecular characterization of fungal endophytic morphospecies isolated from stems and pods of Theobroma cacao. Plant Pathol. 2006;55:783–791. [Google Scholar]
- 9.Newsham K.K. A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 2011;190:783–793. doi: 10.1111/j.1469-8137.2010.03611.x. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez R.J., Redman R.S., Henson J.M. The role of fungal symbioses in the adaptation of plants to high stress environments. Mitig. Adapt. Strateg. Glob. Chang. 2004;9:261–272. [Google Scholar]
- 11.Khan A.L., Hamayun M., Ahmad N., Hussain J., Kang S.M., Kim Y.H., Adnan M., Tang D.C., Waqas M., Radhakrishnan R., Hwang Y.H., Lee I.J. Salinity stress resistance offered by endophytic fungal interaction between Penicillium minioluteum LHL09 and Glycine max. L. J. Microbiol. Biotechnol. 2011;21:893–902. doi: 10.4014/jmb.1103.03012. [DOI] [PubMed] [Google Scholar]
- 12.Zakaria L., Jamil M.I., Anuar I.S. Molecular characterisation of endophytic fungi from roots of wild banana (Musa acuminata) Trop. Life Sci. Res. 2016;27:153–162. [PMC free article] [PubMed] [Google Scholar]
- 13.Noy-Meyr I. Desert ecosystems: Environment and producers. Annu. Rev. Ecol. Syst. 1973;4:25–51. [Google Scholar]
- 14.Clarke J.D.A. Antiquity of aridity in the Chilean Atacama Desert. Geomorphology. 2006;73:101–114. [Google Scholar]
- 15.Tapia M. IICA; 1982. The Environment, Crops and Agricultural Systems in the Andes of Southern Peru. [Google Scholar]
- 16.FAO . Food and Agricultural Organization of the United; 2011. Quinoa: An Ancient Crop to Contribute to World Food Security. [Google Scholar]
- 17.Bhargava A., Shukla S., Ohri D. Chenopodium quinoa - an Indian perspective. Ind. Crop. Prod. 2006;23:73–87. [Google Scholar]
- 18.INIA Boletín nacional de análisis de riesgos agroclimáticos para las principales especies frutales y cultivos. 2016. http://www2.congreso.gob.pe/sicr/cendocbib/con4_uibd.nsf/32A96E54C255557405257CAD00771034/$FILE/BOLETIN_AGROMETEOROLOGICO_RESUMEN_EJECUTIVOENERO_2014.pdf
- 19.Nicholson T.P., Rudd B.A.M., Dawson M.J., Lazarus C.M., Simpson T.J., Cox R.J. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 2001;8:157–178. doi: 10.1016/s1074-5521(00)90064-4. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 23.Porras-Alfaro A., Bayman P. Hidden fungi, emergent properties: endophytes and microbiomes. Annu. Rev. Phytopathol. 2011;49:291–315. doi: 10.1146/annurev-phyto-080508-081831. [DOI] [PubMed] [Google Scholar]
- 24.Wehner J., Powell J.R., Muller L.A.H., Caruso T., Veresoglou S.D., Hempel S., Rillig M.C. Determinants of root-associated fungal communities within Asteraceae in a semi-arid grassland. J. Ecol. 2014;102:425–436. [Google Scholar]
- 25.Jumpponen A., Trappe J.M. Dark septate endophytes: a review of facultative biotrophic root colonizing fungi. New Phytol. 1998;140:295–310. doi: 10.1046/j.1469-8137.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- 26.Maciá-Vicente J.G., Jansson H.B., Abdullah S.K., Descals E., Salinas J., López-Llorca L.V. Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp. FEMS Microbiol. Ecol. 2008;64:90–105. doi: 10.1111/j.1574-6941.2007.00443.x. [DOI] [PubMed] [Google Scholar]
- 27.Neubert K., Mendgen K., Brinkmann H., Wirsel S.G.R. Only a few fungal species dominate highly diverse mycofloras associated with the common reed. Appl. Environ. Microbiol. 2006;72:1118–1128. doi: 10.1128/AEM.72.2.1118-1128.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chesson P., Gebauer R.L.E., Schwinning S., Huntly N., Wiegand K., Ernest M.S.K., Sher A., Novoplansky A., Weltzin J.F. Resource pulses, species interactions, and diversity maintenance in arid and semi-arid environments. Oecologia. 2004;141:236–253. doi: 10.1007/s00442-004-1551-1. [DOI] [PubMed] [Google Scholar]
- 29.Zak J.C., Sinsabaugh R., MacKay W.P. Windows of opportunity in desert ecosystems: their implications to fungal community development. Can. J. Bot. 1995;73:S1407–S1414. [Google Scholar]
- 30.Redman R.S., Dunigan D.D., Rodríguez R.J. Fungal symbiosis: from mutualism to parasitism, who controls the outcome, host or invader? New Phytol. 2001;151:705–716. doi: 10.1046/j.0028-646x.2001.00210.x. [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez R.J., Redman R.S., Henson J.M. The role of fungal symbioses in the adaptation of plants to high stress environments. Mitig. Adapt. Strateg. Glob. Chang. 2004;9:261–272. [Google Scholar]
- 32.Rodríguez R., Redman R. More than 400 million years of evolution and some plants still can't make it on their own: plant stress tolerance via fungal symbiosis. J. Exp. Bot. 2008;59:1109–1114. doi: 10.1093/jxb/erm342. [DOI] [PubMed] [Google Scholar]
- 33.Khan A.L., Hamayun M., Ahmad N., Hussain J., Kang S.M., Kim Y.H., Adnan M., Tang D.C., Waqas M., Radhakrishnan R., Hwang Y.H., Lee I.J. Salinity stress resistance offered by endophytic fungal interaction between Penicillium minioluteum LHL09 and Glycine max. L. J. Microbiol. Biotechnol. 2011;21:893–902. doi: 10.4014/jmb.1103.03012. [DOI] [PubMed] [Google Scholar]
- 34.Maciá-Vicente J.G., Jansson H.B., Mendgen K., López-Llorca L.V. Colonization of barley roots by endophytic fungi and their reduction of take-all caused by Gaeumannomyces graminis var. tritici. Can. J. Microbiol. 2008;54:600–609. doi: 10.1139/w08-047. [DOI] [PubMed] [Google Scholar]
- 35.Ortuño N., Claros M., Gutiérrez C., Angulo M., Castillo J. Bacteria associated with the cultivation of quinoa in the Bolivian Altiplano and their biotechnological potential. Rev. Agric. 2014;54 [Google Scholar]
- 36.Pitzschke A. Developmental peculiarities and seed-borne endophytes in quinoa: omnipresent, robust bacilli contribute to plant fitness. Front. Microbiol. 2016;7:2. doi: 10.3389/fmicb.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urcelay C., Acho J., Joffre R. Fungal root symbionts and their relationship with fine root proportion in native plants from the Bolivian Andean highlands above 3,700 m elevation. Mycorrhiza. 2011;21:323–330. doi: 10.1007/s00572-010-0339-x. [DOI] [PubMed] [Google Scholar]
- 38.Martínez E.A., Veas E., Jorquera C., San Martín R., Jara P. Reintroduction of quinoa into arid Chile: cultivation of two lowland races under extremely low irrigation. J. Agron. Crop Sci. 2009;195:1–10. [Google Scholar]
- 39.Jacobsen S.E., Monteros C., Corcuera L.J., Bravo L.A., Christiansen J.L., Mujica A. Frost resistance mechanisms in quinoa (Chenopodium quinoa Willd.) Eur. J. Agron. 2007;26:471–475. [Google Scholar]
- 40.Rosa M., Hilal M., González J.A., Prado F.E. Low-temperature effect on enzyme activities involved in sucrose–starch partitioning in salt-stressed and salt-acclimated cotyledons of quinoa (Chenopodium quinoa Willd.) seedlings. Plant Physiol. Biochem. 2009;47:300–307. doi: 10.1016/j.plaphy.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Razzaghi F., Jacobsen S.-E., Jensen C.R., Andersen M.N. Ionic and photosynthetic homeostasis in quinoa challenged by salinity and drought-mechanisms of tolerance. Funct. Plant Biol. 2015;42:136–148. doi: 10.1071/FP14132. [DOI] [PubMed] [Google Scholar]