Abstract
Efficient diagnosis of tuberculosis (TB) is met with multiple challenges, calling for a shift of focus from pathogen-centric diagnostics towards identification of host-based multi-marker signatures. Transcriptomics offer a list of differentially expressed genes, but cannot by itself identify the most influential contributors to the disease phenotype. Here, we describe a computational pipeline that adopts an unbiased approach to identify a biomarker signature. Data from RNA sequencing from whole blood samples of TB patients were integrated with a curated genome-wide molecular interaction network, from which we obtain a comprehensive perspective of variations that occur in the host due to TB. We then implement a sensitive network mining method to shortlist gene candidates that are most central to the disease alterations. We then apply a series of filters that include applicability to multiple publicly available datasets as well as additional validation on independent patient samples, and identify a signature comprising 10 genes — FCGR1A, HK3, RAB13, RBBP8, IFI44L, TIMM10, BCL6, SMARCD3, CYP4F3 and SLPI, that can discriminate between TB and healthy controls as well as distinguish TB from latent tuberculosis and HIV in most cases. The signature has the potential to serve as a diagnostic marker of TB.
Keywords: Tuberculosis, Biomarkers, Network biology, Computational medicine, Diagnostics
Highlights
-
•
An integrated systems biology approach has been adopted to study the host response to tuberculosis.
-
•
A multi-gene host biomarker signature is identified for detecting pulmonary tuberculosis from patient blood samples
-
•
The signature discriminates TB from HIV and latent-TB and can serve as an adjuvant tool in confirming TB diagnosis
Host factors that are altered significantly due to tuberculosis are investigated, with an aim to identify a biomarker panel. A network approach provides a genome-wide view of the molecular interactions, analogous to a road network of a city. By comparing networks between healthy and TB samples, we identify the set of variations in a systematic fashion, analogous to identifying all major variations in the traffic flow in a city between two time points. We then apply a series of filters to identify the most discriminating genes among them. The 10-gene signature is seen to be characteristic of TB.
1. Introduction
Tuberculosis (TB) now ranks along with HIV as the leading cause of death due to an infectious agent worldwide, with approximately 10.4 million people estimated to have acquired TB in 2015, resulting in 1.4 million deaths (World Health Organization, 2016). These deaths are largely preventable by early and efficient diagnosis of the disease. Unfortunately, diagnosis is often delayed due to insensitive and time-consuming methods. Present diagnostic measures rely largely on the detection of Mtb in patient samples together with radiological assessments, and they have several shortcomings. Sputum cultures are the current standard for detecting Mtb, but while sensitive, they take 3–6 weeks to provide conclusive results, thereby delaying the initiation of treatment. Host-based diagnostic methods provide an alternative for early detection of TB onset and enable the monitoring of symptomatic changes. IFN-γ release assays (IGRAs) such as the T-SPOT.TB (Richeldi, 2006, Pai et al., 2014) or the QuantiFERON test (Sultan et al., 2010) measure IFN-γ + production in response to stimulation with Mtb-specific antigens ESAT6 and CFP10 (Mazurek and Villarino, 2003, Ravn et al., 2005). However, IGRAs cannot discriminate between active and latent Mtb infection, and are thus inadequate for marking the disease status. In the clinic, IGRAs are used more often to detect latent tuberculosis than for diagnosis of active disease (Herrera et al., 2011). Existing assays that rely on single-marker readouts, such as that of serum deaminase levels (Gui and Xiao, 2014), also suffer from inadequate sensitivity and/or specificity, calling for more effective host-related multi-marker signatures that hold promise for applications in prognostic research and vaccine trials as well as in monitoring treatment responses. There is thus a current need for a shift from investigations on single markers to high-coverage studies that will reveal signatures consisting of multiple integrated markers (Maertzdorf et al., 2014). Recent years have witnessed an increase in host omics data to identify specific gene variations upon infection with Mtb, including genetic polymorphisms identified by GWAS and linkage and association studies that ascribe host susceptibility to infection (Azad et al., 2012), genome-wide expression variations in patient cohorts as compared to healthy controls, as well as variations over the course of treatment in the same patient.
Transcriptomics provide global coverage into host responses, and are widely used in TB biomarker research (Maertzdorf et al., 2011a, Joosten et al., 2013). One drawback of microarray technologies is the lack of absolute and detailed evaluation of gene expression. Modern deep sequencing technologies provide quantitative and qualitative information on gene expression and genomic composition down to the single-nucleotide level (Normand and Yanai, 2013). RNA sequencing (RNA-Seq) is fast gaining foothold, and provides more accurate measurements of transcript levels and their isoforms with greater sensitivity than microarrays, as it overcomes probe-dependency (Wang et al., 2009). RNA-seq has been applied to study host variations due to mycobacterial infections and has led to rich insights, an example being dual RNA sequencing of host and pathogen in Mtb infected Thp-1 cells that indicated a simultaneous induction of Mycobacterium bovis BCG cholesterol degradation genes and a compensatory upregulation in the host de novo cholesterol biosynthesis genes (Rienksma et al., 2015). Recently, a whole blood signature that could predict the risk of developing active tuberculosis in patients with latent infection was identified by RNA-seq data (Zak et al., 2016).
Although the immunological response against Mtb will be primarily focused in the lung, its pathologic status is reflected in the peripheral blood by circulating immune cells (Weiner et al., 2013). Whole blood transcriptomic profiles provide global insights into host immune responses in tuberculosis and serve as essential tools in determining underlying molecular players of infection.
A multi-marker set of gene classifiers determined from blood transcriptomic data with sufficient discriminatory prowess would thus support current diagnostic measures to enhance early detection of TB in the clinic (Cliff et al., 2015). Gene expression values highlight differentially expressed genes (DEGs), which by themselves are indicative of the variations in disease, but further selection is required to identify a small biomarker set that is characteristic of the disease. Such a selection has been achieved using machine learning methods for a number of diseases including tuberculosis (Blankley et al., 2014). Use of networks, however, provides a different perspective to identify DEGs that may be functionally linked to other differentially regulated genes, either directly or indirectly through other bridging nodes. A systems approach integrating transcriptomic data and genome-wide molecular interaction networks is necessary to provide mechanistic insights into the nature of dynamic responses to infection and help identify the most significant contributors to the disease phenotype. Biological network analysis involves the construction of a pair-wise assembly of molecular interactions among cellular components that will yield a connected network of interactions. The network can be compared to a street-map of a city and provides an overview of the interconnected routes or in other words the topological architecture of the molecular interactions in a cell. Mapping genome-wide expression profiles into molecular networks to construct condition-specific response networks provides an unbiased systematic approach to enable the identification of combinations of host components that can serve as markers for tuberculosis and aid early diagnosis.
India currently leads the world's burden of tuberculosis, accounting for about 2.8 million cases out of the global incidence of 10 million (World Health Organization, 2016). Omics studies on the Indian population have been few and far between. In this study, with an aim to differentiate pulmonary tuberculosis from other conditions, we use a new network-based pipeline for biomarker discovery. We obtain RNAseq data from an Indian cohort and map them onto interaction networks, from which we identify the most influential genes in the host whole blood response network to tuberculosis. We then apply a series of filters to finally discover a 10-gene validated signature that can discriminate TB samples and healthy controls in multiple cohorts from different geographical locations and also discriminate between latent and active tuberculosis. In addition, the signature distinguishes between TB and HIV and has a potential to be used for diagnosis in the clinical setting.
2. Materials and Methods
2.1. Study Participants
Clinical samples were obtained from participants enrolled at the National Institute for Research in Tuberculosis (NIRT), Chennai: active TB (BL), IGRA − ve/healthy control (HC), and IGRA + ve/latent TB (LTB); St. John's Research Institute, Bengaluru (IGRA − ve/healthy controls and HIV +) and Arogyavaram Medical Centre, Madanapalle (IGRA + ve/latent TB). Patients attending the outpatient clinics of NIRT and community controls were enrolled for this study. This was a prospective case control study and we enrolled consecutive patients and controls. The diagnosis of pulmonary tuberculosis (TB) was based on smear and culture positivity. Chest X-rays were used to define cavitary disease as well as unilateral vs bilateral involvement. Smear grades were used to determine bacterial burdens and classified as 1 +, 2 + and 3 +. At the time of enrolment, all active TB cases had no record of prior TB disease or anti-tuberculosis treatment (ATT). Latent tuberculosis (LTB) diagnosis was based on tuberculin skin test (TST) and QuantiFERON TB-Gold in Tube ELISA positivity, absence of chest radiograph abnormalities or pulmonary symptoms. A positive TST result was defined as an induration at the site of tuberculin inoculation of at least 12 mm in diameter to minimize false positivity due to exposure to environmental mycobacteria. NTB individuals were asymptomatic with normal chest X-rays, negative TST (indurations < 5 mm in diameter) and QuantiFERON ELISA results. All participants were BCG vaccinated, HIV negative, non-diabetic and had normal body mass index. All participants did not exhibit signs or symptoms of any associated lung or systemic disease. Standard anti-TB treatment (ATT) was administered to TB individuals using the directly observed treatment, short course (DOTS) strategy. At 6 months following ATT initiation, fresh plasma samples were obtained. All TB individuals were culture negative at the end of ATT. All individuals were examined as part of a study protocol approved by the ‘Internal Ethics Committee’ of NIRT and written informed consent was obtained from all participants (approval number NIRTIEC2010002). Table 1 describes the breakdown of different patient classes. Clinical details of all enrolled participants are provided in Additional File 2. Samples for RNA sequencing were exclusively from NIRT, Chennai. A total of three samples from the IGRA − ve and IGRA + ve categories and 4 from the active TB category were used for RNA sequencing. An additional 13 active TB, 10 IGRA − ve, 9 IGRA + ve and 7 HIV + samples were used for validation of gene expression by qRT-PCR. Classification of IGRA − ve and IGRA + ve individuals was done on the basis of a QuantiFERON assay.
Table 1.
Breakdown of patient classes recruited for this study.
| Condition | No. of samples | Age range (yrs) | Gender male (M), female (F) | Clinical summary |
|---|---|---|---|---|
| Active TB | 19 | 18–52 | 11M, 8F | All 19 cases are sputum smear positive with grade 2 to 3. Chest X-ray is abnormal in all, 13 with bilateral and 6 with unilateral abnormality, with 1 to 6 zones, 6 show cavities and 13 show no cavities. Almost all patients show normal hematology |
| HC | 15 | 30–60 | 8M, 7F | All are sputum smear negative, chest X-rays appear normal, no clinical symptoms of TB and no known prior history of TB |
| LTB IGRA + ve | 13 | 19–34 | 11M, 2F | All are sputum smear negative, chest X-rays appear normal, no clinical symptoms of TB and no known prior history of TB |
| Condition | No. of samples | Age range (yrs) | Gender | CD4 cells/mm | Viral load copies/ml | Clinical summary |
|---|---|---|---|---|---|---|
| HIV | 7 | 25–47 | 4M, 3F | 16–28%; mean: 20.5% (not investigated in 2) | 3.4 × 101 to 8.2 × 104 | All patients are treatment-naive and show normal chest X-rays with no evidence of TB |
2.2. RNA Isolation
Blood (3 ml) from each participant was collected in a Tempus tube, vigorously shaken and transported to IISc, Bangalore, where it was stored at − 80 °C until use. For RNA isolation, frozen Tempus tubes were thawed and RNA was extracted using a Tempus Spin RNA isolation kit (Applied Biosystems) following the manufacturer's instructions. Briefly, blood from the Tempus tube was centrifuged at 3000 g for 30 min at 4 °C to pellet down the RNA which was then re-suspended and loaded onto a spin column for purification. RNA bound to the column was eluted and aliquoted RNA was quantified and either subjected to RNA sequencing or converted to cDNA for gene expression studies by qRT-PCR.
RNA sequencing RNA isolated from Tempus tubes was quantified and subjected to quality control analysis. RNA samples with a RIN > 5 were taken further for RNA sequencing. Library preparation was performed at Genotypic Technology's Genomics facility at Bangalore. Five μg of qubit quantified total RNA was taken and enriched for PolyA using NextFlexpolyA Beads. Transcriptome library for sequencing was constructed as per the NEXTflexRapid Directional RNA-Seqlibrary protocol outlined in “NEXTflex Rapid Directional RNA-Seq sample preparation guide” (Cat # 5138-08). Briefly, the PolyA RNA was fragmented for 10 min at elevated temperature (95 °C) in the presence of divalent cations and reverse transcribed using first strand mix. The RNA–DNA hybrid was cleaned up using HighPrep PCR cleanup beads. Second strand cDNA was synthesized and end repaired using second strand synthesis mix. Directionality is retained by the addition of dUTP at this step. The cDNA was cleaned up using HighPrep PCR cleanup beads. NEXTflex™ RNA-Seq Barcode Adapters were ligated to the cDNA molecules after end repair and addition of “A”-base. SPRI clean-up was performed post-ligation. The library was amplified using 12 cycles of PCR for enrichment of adapter ligated fragments. The prepared library was quantified using qubit and validated for quality by running an aliquot on High Sensitivity Bioanalyzer Chip (Agilent).
2.3. Read Alignment and Transcript Assembly
The Tuxedo protocol was followed for the alignment of reads, transcript assembly and analysis. The paired-end reads were aligned using TopHat2 (version 2.0.9), using the GRCh38 human genome assembly as reference. The discovery of novel junctions was turned off. The transcript assembly using the aligned reads was performed using Cufflinks (version v2.2.1). Cuffmerge followed by Cuffdiff was used to examine the differential expression of transcripts between the different sample groups. The average expression of an individual gene from the fragment per kilobase of transcript per million mapped reads (FPKM) was obtained using Cufflinks. Fold changes were computed for genes in active TB with respect to IGRA − ve healthy controls by taking a ratio of their corresponding FPKM values. Differentially expressed genes in TB were identified by considering fold change values > 2 for upregulated genes and values < 0.5 for downregulated genes.
2.4. cDNA Conversion and qRT-PCR
Approximately 2 μg of RNA from each sample was converted to cDNA using High capacity cDNA conversion kit (Applied Biosystems). Selected genes were validated by qPCR using a SYBR Green master mix (Applied Biosystems) and specific primers (Table S3) except for SLPI which was amplified using a TaqMan master mix and probe. The total reaction volume for SYBR green reactions was 25 μl and for TaqMan it was 10 μl. GAPDH was used as the internal housekeeping control gene in both cases. All reactions were carried out in duplicates along with a no cDNA negative control using the Step One Plus (Applied Biosystems) instrument. Mean CT values were used for calculating relative copy number (RCN) of each gene.
2.5. Construction of the Human Protein–Protein Interaction Network
A network of human protein–protein interactions was constructed based on curating high confidence, experimentally verified interactions from multiple protein–protein interaction sources and pathway databases, as well as from primary literature. This network is termed as human protein–protein interaction network (hPPiN) (Sambarey et al., in press). Briefly, the Search Tool for The Retrieval of Interacting Genes/Proteins (STRING) version 10 (Szklarczyk et al., 2014) was mined to extract all human interactions with a combined score > 900, and the functional nature of these interactions was identified from the protein actions file. Based on these annotations, the edges were assigned directions, however, interactions describing a ‘binding’ event were represented as bidirectional edges. SignaLink v 2.0 (Fazekas et al., 2013) was mined to identify regulatory interactions of transcriptional, post-transcriptional and pathway regulators. Additional interactions present in non-disease conditions were identified from the Cancer Cell Map (Krogan et al., 2015), and the BioGRID database (Chatr-Aryamontri et al., 2015) was mined to identify unique interactions not reported by the other resources used. In addition to PPI databases and resources, primary literature was explored to identify experimentally verified interacting proteins in the human proteome. Interactions were extracted from Multinet (Khurana et al., 2013), and from the macrophage interaction network (Sambarey et al., 2013). From this constructed hPPiN, interactions were extracted for all protein-coding genes present in the RNA-Seq dataset.
2.6. Condition-specific Weighted Networks
The FPKM values for all genes were mapped onto the constructed PPI network in the form of node and edge weights to generate condition-specific response networks. Node and edge weights were modified from Sambarey et al. (2013). The node weight of gene i in condition A is given as
| (1) |
The edge weight We(A) of edge e comprising genes i and j in condition A is computed as:
| (2) |
A lower edge weight is indicative of an active edge, wherein the interacting nodes have high expression values in that condition.
2.7. Shortest Path Analysis
All-vs-all shortest paths were computed for all genes in the network using Dijkstra's algorithm. The algorithm computes minimum weight shortest paths, in which each path begins from a source node and ends with a sink node, through interacting proteins, choosing the least-cost edge in every step. For a path of length n in condition, the PathCost was computed as a summation of the edge weights constituting the path. We have previously performed sensitivity analysis for the response network construction, by modifying expression values of genes in a random manner to reflect the noise that can be introduced in microarray data. Repeating the pipeline over several independent runs incorporating such modifications revealed that the top networks are largely robust to minor variations in differential expression (Sambaturu et al., 2015). Cytoscape v3.1 (Su et al., 2014) was used for network analysis and visualization as it is a well-established software used routinely by researchers working on biological networks. The implementation of Dijkstra's algorithm was done in Python, a well-established method for computing shortest paths in a network.
2.8. Pathway Enrichment
The EnrichR server (Chen et al., 2013) was used to identify significantly enriched KEGG Pathways and WikiPathways, which were pooled and ranked based on the combined score, which is computed as c = log(p) × z, where p is the p-value computed using the Fisher exact test, and z is the z-score computed by assessing the deviation from the expected rank.
3. Results
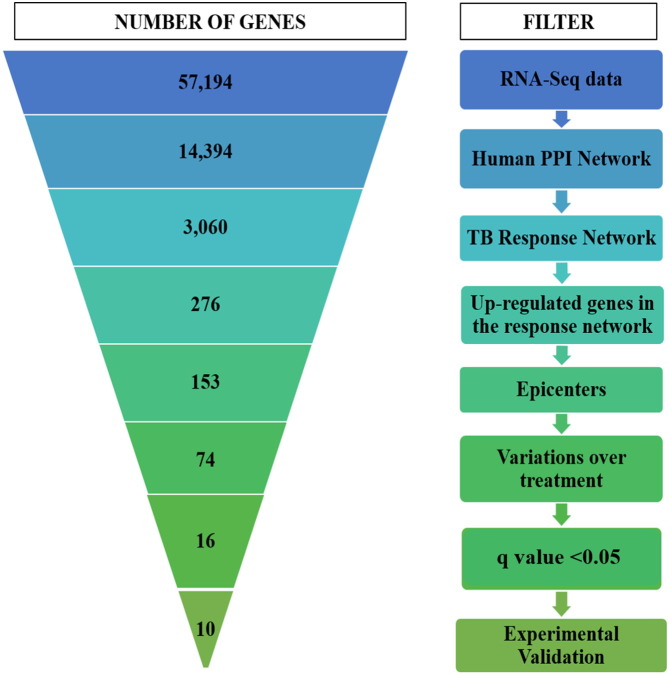
An unbiased computational pipeline was developed to integrate RNA-seq data obtained from TB patients and corresponding healthy controls into a global protein–protein interaction network, resulting in the generation of condition-specific networks. The highest-activity network unique to active TB was identified in which the upregulated genes were shortlisted, and their outward reachability was measured to determine the extent to which these DEGs exert their effects in the network. The expression profiles of these significant and central genes were then monitored over treatment, and those that responded to therapy were further filtered, and among these, those with significant upregulation and a q-value < 0.05 were selected. Additionally, the prediction potential of these genes was tested on existing transcriptomic datasets to assess their ability in discriminating TB samples from corresponding healthy controls (HCs) and other diseases. These predicted markers were then subjected to experimental verification by RT-PCR on additional TB samples, and were also compared with latent infection and HIV to finally shortlist a list of 10 genes that can serve as potential markers for diagnosis in tuberculosis. Fig. 1 illustrates the developed pipeline and the number of genes that were filtered at every step.
Fig. 1.
The computational pipeline used for biomarker discovery in tuberculosis. The figure illustrates the pipeline used in this study, describing multiple filters used to shortlist a minimal set of 16 putative biomarkers, which were then subjected to experimental verification by qRT-PCR, eventually deriving a candidate biosignature of 10 genes in tuberculosis.
3.1. Differentially Abundant Transcripts Highlight an Active Signaling Response in Tuberculosis
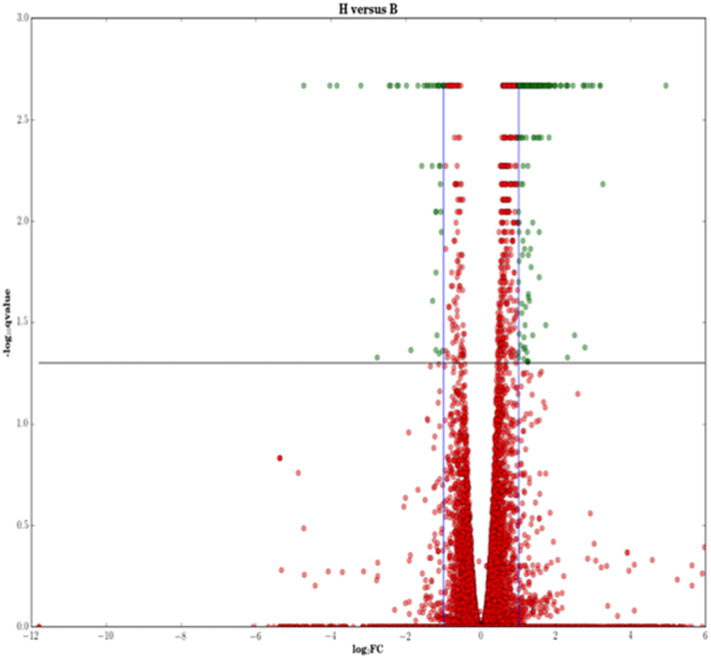
Whole blood samples were taken from three healthy controls (HCs), three patients with baseline tuberculosis (BL) which were subsequently followed up after 6 (FU1) and 12 (FU2) months of treatment. Details about the patient groups, time of sample extraction and number of samples are provided in Table 1. The RNA sequencing was performed as described in the Materials and Methods section. Median FPKM values for the genes across samples were considered as representative expression values for each condition. Differentially expressed genes were identified by computing fold change values relative to the control for baseline tuberculosis (BL/HC), whereas their dynamic changes over treatment were captured by taking fold changes relative to their values in disease (FU1/BL and FU2/BL respectively). The list of fold change values for all genes and their q-value is provided in Additional File 1. There were 525 genes upregulated and 125 genes downregulated in BL, based on a two-fold difference and a q-value < 0.05 (Supplementary Fig. S1). Supplementary Fig. S2 depicts the Gene Ontology (GO) processes that are enriched by these DEGs.
Supplementary Fig. S1.
Identification of differentially expressed genes in baseline tuberculosis (BL) relative to healthy control (HC). An absolute log2 fold change of 2 was considered as a threshold for differential expression. Both up and downregulated genes with a q-value (FDR corrected p-value) < 0.05 are highlighted in green.
Supplementary Fig. S2.
An overview of biological processes enriched in the set of DEGs. The most enriched GO biological processes with a p-value < 0.05 have been represented.
The processes captured by the identified upregulated genes involve the Type I and Type II interferon processes, cytokine signaling mediated by IL-4, IL-6, IL-2, IL-7, and TGF-beta, TNF pathway, chemokine signaling, TLR pathway, and the Age/Rage pathway, among others. These processes broadly capture the innate immune response at play during active infection. To determine the genes that contribute most significantly to these biological processes, we adopted a network approach.
3.2. Response Network Facilitates the Identification of Highest Activities in the Host and Determination of Most Influential Nodes in Disease
The reconstructed hPPiN consisted of 17,063 proteins (nodes) and 208,760 interactions (edges) among them, of which 168,238 were uni-directed and 40,522 were bi-directional interactions. The normalized RNA-seq data had expression information measured in terms of FPKM values for 14,394 protein-coding genes, and the subsequent interaction network for these genes comprised 192,389 interactions. This network is highly dense and largely interconnected, and is seen to follow a scale-free distribution attributed to most biological networks. The hPPiN was constructed based on experimentally validated physical interactions, with directions assigned based on functional annotations. It is thus of higher confidence than similar gene co-expression networks derived based on expression patterns alone.
The computed median FPKM values for conditions HC, BL, FU1 and FU2 were mapped onto hPPiN in the form of node and edge weights, and four corresponding condition-specific networks were generated. Shortest path computation for these networks comprising 14,342 genes and 192,389 interactions resulted in the generation of nearly 1.9 billion paths per network, with only paths of lengths two and higher considered for further analysis. All paths were sorted and ranked based on their cumulative path score, with lower scores implying higher activity. These paths represent the possible routes of signaling from a source gene to a target gene across the topology of the network, and are optimized based on the least costs for each edge traversed to reach a target node. The weights for each edge were formulated to include the gene abundance information of the two connecting nodes, with a lower edge weight correlating with higher node weights of the nodes constituting the edge. The cumulative PathCost, which is a summation of edge weights in a path, is thus reflective of the activity of the nodes involved, with the least-scoring paths taking routes through highly upregulated nodes. The frequency distribution of the path costs followed a long-tail distribution, where the ‘highest-activity’ paths comprising maximally upregulated genes were localized in the beginning, followed by paths of lower activity, and the frequency tailed off asymptotically. The paths at the lower end of the distribution thus have a smaller probability of occurrence in the condition studied. The processes and genes characteristic of any condition are more likely to be represented in the ‘highest-activity’ end of the distribution, hence the paths occurring in the top 99th percentile for each condition-specific network were considered for further analysis.
The highest-activity paths in the BL response network were compared with the corresponding highest-activity paths computed for the HC network, and those that were common to both top networks were eliminated in the process, as they likely represent constitutive activity in the host that is responsible for the regular functioning of the system. Among the 364,965 paths present in the 99th percentile of the BL network, 97,909 paths were seen to be uniquely active in BL relative to the representative HC top network, and the nodes and edges present in these paths resulted in a ‘highest-activity network’ (HAN) for active disease, which we refer to as a TB-specific response network. This response network comprised 3060 genes, and represented about 20% of the total network. Gene enrichment analysis of the nodes in this response network identified a more focused set of pathways that are enriched with greater significance as compared to mere DEG based enrichment, since the network formulation eliminates noise by considering only those genes that are involved in active flows. Cytokine signaling, complement signaling pathway, MAPK signaling, platelet activation, apoptosis, phagocytosis, TNF and TLR signaling as well as the adaptive immune processes such as the T cell receptor signaling pathway are seen to be significantly enriched by the HAN nodes. These processes are reflective of an enhanced inflammatory immune response, as well as the early onset of adaptive immunity in the host.
3.3. The Most Influential Nodes Form a Highly Interconnected Sub-network
The HAN, while reflective of the overall host response in tuberculosis, is still a broad representation of the processes triggered in the host. Interestingly, this network showed high interconnectivity, implying increased cross-talk and concerted action among genes and processes constituting the host's response to infection. An important feature of a good biomarker is its abundance, in that it is measurable in the blood in addition to being differentially regulated in disease. We thus focused on those genes in the HAN that had a fold change of 2 and higher in infection in addition to having a high abundance, both of which are reflected by the computed node weights. There were 276 upregulated nodes present in the top network which contribute to the paths of the highest activity.
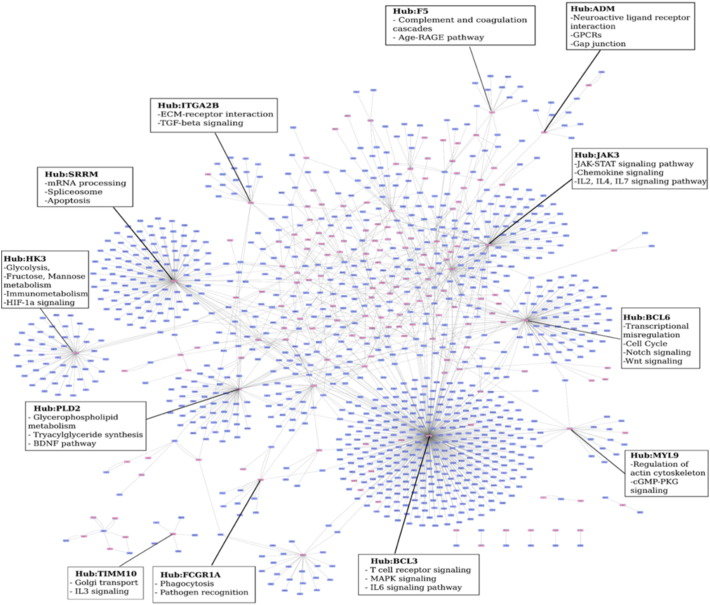
In addition to their node weights, their position in the network as well as the nature and abundance of their connecting partners will dictate the extent to which each of these 276 nodes influences the host response, with some nodes playing a more important role than the others. A node centrality measure termed ripple centrality was computed for each of these nodes. The ripple score determines how the perturbation of a single node impacts its neighbors, and how effectively a change at that node can transmit across the network, leading to the identification of the most influential nodes, also known as epicenters. The ripple centrality measure was computed using the EpiTracer algorithm (Sambaturu et al., 2015), which measures the Outward Reachability of a given node, a property of its connections as well as its condition-specific node weight. Of the 276 upregulated nodes, 153 nodes had a non-zero ripple score, and a sub-network of these nodes and their interacting partners was subsequently analyzed. Fig. 2 illustrates the interconnections and distribution of these 153 upregulated epicenters (highlighted in red) across the network, and the corresponding enrichments of their sub-networks. While the hub nodes which have the highest number of partners in the network are highlighted, it is interesting to note that while most of the partners of these hubs are not upregulated, collectively these individual sub-networks contribute to significantly enrich biological processes, thus emphasizing their importance as downstream effectors of the upregulated gene. Most upregulated genes are seen to occupy the central positions in the network, as demonstrated in Fig. 3. The topological representation of the network is a function of the degree or connectivity of the nodes, with nodes of highest connectivity placed in the center. Observation of DEGs in the network center implies that the infection-induced upregulation is closely connected, with a few molecular players playing a more central role in driving the response to disease.
Fig. 2.
The top response network of the most influential genes (epicenters). Genes that are upregulated (FC > 2) have been marked in red. The hub nodes are highlighted and the most enriched biological processes for their corresponding sub-networks have been illustrated.
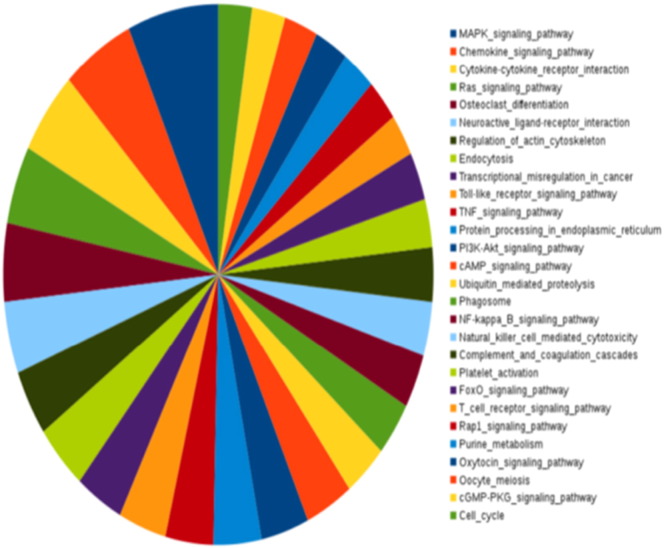
Fig. 3.
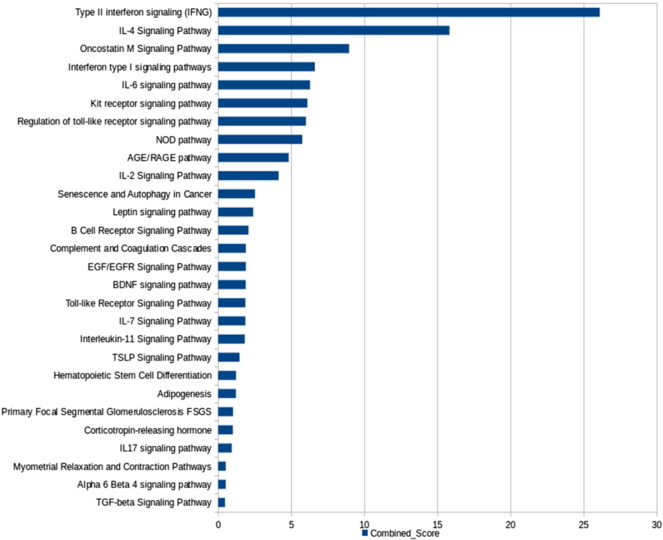
Collective enrichment of the highest-activity network in TB. The enriched KEGG pathways are ranked based on the EnrichR combined score.
The enrichment for the complete network provides deeper insights into the specific processes that are upregulated. In addition to the processes the broader inflammatory processes observed in Supplementary Fig. S2, specific metabolic processes such as glucose catabolism, fructose metabolism, phosphatidylserine, phosphatidylinositol and phosphatidylglycerol metabolism, biosynthesis of glycerolipids and their regulation, NADH regeneration, and purine nucleoside diphosphate metabolism are seen to be over-represented in this network comprising epicenters and their interconnected neighbors of high influence, as described in Fig. 3. Fc-gamma mediated phagocytosis is seen to be upregulated, in addition to signaling mediated by pattern recognition receptors such as Toll-like receptors and stimulatory c-type lectin receptor pathway. Differential uptake of Mtb by its receptors has been reported to play a role in governing the outcome of infection. Cellular responses to stress are notably activated, including regulation of nitric oxide biosynthesis, generation of reactive oxygen species, intrinsic apoptotic signaling in response to DNA damage, response to endoplasmic reticulum stress, necrosis, aging and senescence. Cytokine signaling mediated by pro-inflammatory cytokines IL2 and IL12 as well as the response to interferon gamma, TNF-alpha and Type I interferons are observed. The importance of cytokines in tuberculosis has been well documented, and recent years have placed particular emphasis on the Type I responses, which also show significant enrichment here. The adaptive immune responses are strongly characterized with the activation, differentiation, aggregation and regulation of T and B lymphocytes, that are all observed in the high activity network.
3.4. Reversals in Gene Expression and Pathway Activity Over Treatment
Markers for active infection would ideally get downregulated upon completion of successful anti-TB therapy, indicating that their active expression was reflective of the disease state alone. Nodes belonging to the sub-network of 153 most influential genes were monitored over the corresponding top networks constructed for conditions FU1 and FU2, generated for patients monitored at 6 and 12 months of treatment respectively. As determinants of successful anti-tubercular therapy, genes that are upregulated in active tuberculosis should show a gradual decrease in expression values over treatment, eventually reaching levels closer to that of healthy controls 12 months post-treatment. Markers for active disease alone should therefore rank lower in significance in the corresponding treatment top networks. Of the 153 genes shortlisted in the BL top network, 74 genes showed a linear decrease in expression over 6 and 12 months of therapy, and were completely absent in the FU2 highest-activity network. These 74 genes formed a largely interconnected network where they occupy central positions having a high degree, thereby enabling them to significantly transmit variations in their expression downstream.
3.5. Shortlisting a Minimal Set of Markers for Tuberculosis
Clinical measurements of the 74 genes from patient samples, while feasible, would still pose several constraints in developing countries where tuberculosis is most prevalent. Ranking these genes to shortlist those which are most significant in terms of abundance, and which can discriminate between active tuberculosis patients and healthy controls will lead to the identification of a minimal discriminatory signature. We then applied a filter to these 74 genes to only select a subset of those genes that (a) were most upregulated with a significant FDR-corrected p-value (q-value) < 0.05 and (b) showed at least a two-fold reversal in expression after completion of treatment (FU2). These genes were ranked based on their node weights, which are representative of their relative abundance in disease, making them measurable clinically. A total of 16 candidate markers were selected, which were RAB13, RBBP8, ADM, CECR6, TNNT1, TIMM10, SMARCD3, SLPI, IFI44L, IRF7, BCL6, CYP4F3, HK3, FCGR1A, OSM and MYL9.
The expression of these shortlisted genes was monitored across other reported transcriptomic datasets. There exist several publicly available datasets describing whole blood expression profiles for pulmonary tuberculosis, largely generated by microarray analysis. Table 2 describes the five GEO datasets that were used for comparison in this study. A class prediction step from the linear-discriminant analysis (LDA) method was carried out using these 16 genes to determine if they could sufficiently separate the TB patient samples from those of healthy controls in all five datasets, enabling the assessment of their expression in other measured patient samples across different population cohorts. LDA typically includes a training component to select the features, followed by a classification component using the identified features. The features in this type of study are the expression values of the individual genes. We have bypassed the selection step and instead use the genes selected through the network approach as illustrated in Fig. 1 and have used LDA with a 4-fold cross-validation to show the predictive potential of these genes and their ability to classify TB from HC samples. In other words, we have used LDA to estimate the classification accuracy of the genes that we have already selected through the network-based pipeline.
Table 2.
Whole blood transcriptional profiles used for computational validation.
| Dataset | Number of samples | Population cohort | Reference |
|---|---|---|---|
| GSE19491 | Whole blood samples: 54 TB and 24 HC | UK, SA | Berry et al. (2010) |
| GSE28623 | Whole blood samples: 46 TB and 37 HC | The Gambia | Maertzdorf et al. (2011b) |
| GSE34608 | Whole blood samples: 8 TB and 18 HC | Germany | Maertzdorf et al. (2012) |
| GSE42834 | Whole blood samples: 40 TB and 118 HC | London | Bloom et al. (2013) |
| GSE56153 | Whole blood samples: 18 TB and 18 HC | London | Ottenhoff et al. (2012) |
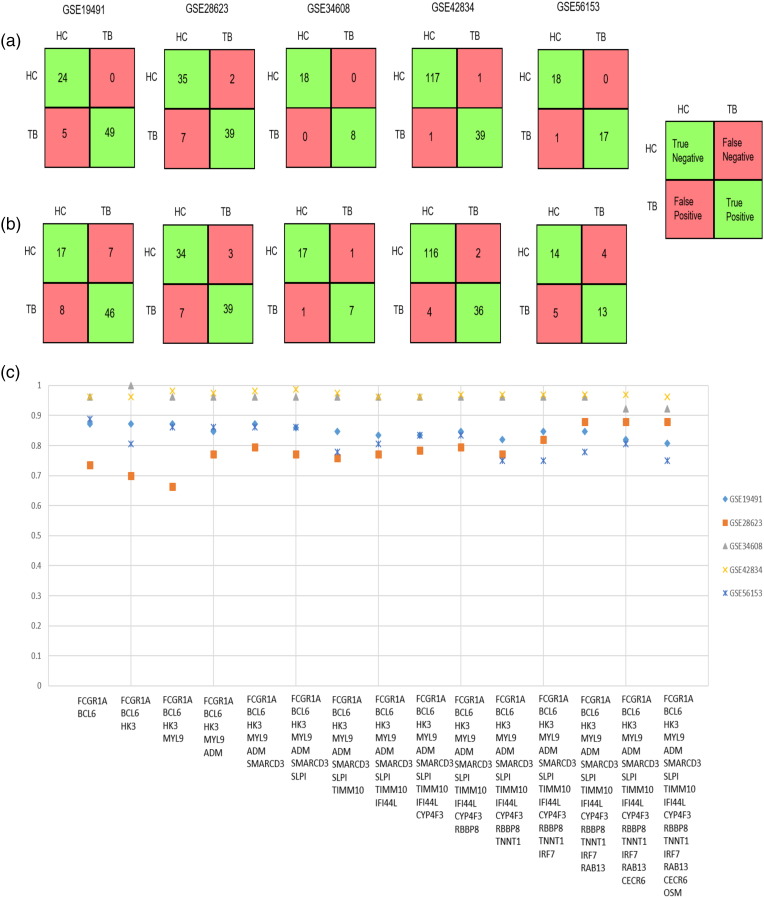
Fig. 4(a) and (b) describe the confusion matrices generated by the 16 genes for five datasets depicting the classification of patient samples into healthy controls and tuberculosis patients, with and without 4-fold cross validation respectively. Fig. 4(c) describes the prediction accuracies for increasing combinations of these 16 genes across the datasets described in Table 2. These genes were ranked based on their node weights in the RNA-Seq data. Increasing linear combinations of these 16 genes (based on their node weights) were used to determine the maximal separability obtained for classification of available TB vs HC samples across different microarray datasets.
Fig. 4.
(a) Confusion matrices describing predictions for TB and HC samples across transcriptomic datasets for 16 genes incorporating a 4-fold cross validation and (b) without cross-validation. True Positives (TP) indicate the TB samples that are correctly identified as TB, whereas True Negatives (TN) refer to the samples that are not TB (which mean that they are HC), that are correctly identified as HC. False Positives (FP) indicate the HC samples that are identified as TB, whereas False Negatives (FN) indicate the TB samples that are identified as HC. (c) Prediction accuracies obtained for increasing linear combinations of 16 genes across 5 whole blood transcriptomic datasets. Datasets GSE34608 and GSE28623 did not have the probe for the gene FCGR1A, and the LDA results for these datasets exclude that gene.
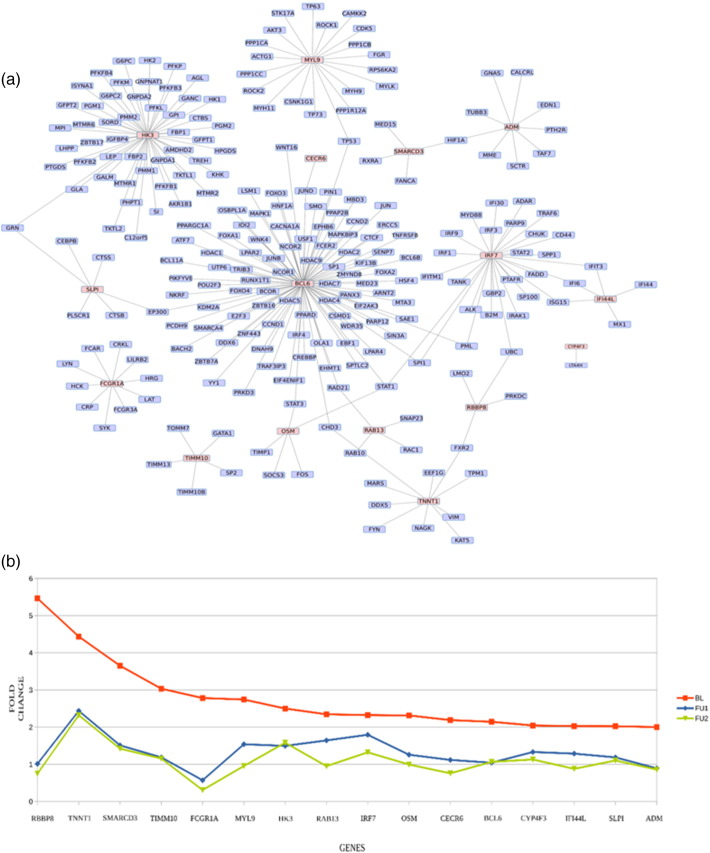
The 16 genes also form a largely interconnected sub-network, as depicted in Fig. 5(a). The variation of fold changes in their expression over the course of treatment is shown in Fig. 5(b).
Fig. 5.
(a) Interaction network of the 16 shortlisted markers. The 16 genes have been highlighted. (b) Reversals in expression over treatment. The fold change values of the 16 markers show downregulation upon 6 (FU1) and 12 (FU2) months of treatment. Fold changes are computed w.r.t a median healthy control.
3.6. Comparison With Published Transcriptome-based Biomarker Studies for Tuberculosis
Several studies have focused on identification of biomarkers via whole blood transcriptomics utilizing machine learning approaches to determine combinations of markers that can accurately distinguish between active TB. Recently, a 3-gene signature was derived by Sweeney et al. (2016). Comprising genes DUSP3, GBP5 and KLF2, a study by Maertzdorf et al. (2016) also determined a four-gene signature by RT-PCR quantitation in the Indian population constituting ID3, GBP1, IFITM3 and P2RY14. These genes were absent in the shortlisted marker list predicted by our study. We assessed the presence of these gene signatures in each step of our computational pipeline to determine where they were filtered out and why they were absent in the final signature predicted in this study. While most of them were reported to be upregulated in our generated dataset, they were not all present in the top network, with none of them forming epicenters. Further, only 2 of those genes show a variation over treatment in our study, as depicted in Table 3.
Table 3.
Assessment of published signatures in this computational pipeline.
| Filter | Khatri signature (2016) |
Maertzdorf signature (2016) |
|||||
|---|---|---|---|---|---|---|---|
| Biomarkers | GBP5 | DUSP3 | KLF2 | ID3 | GBP1 | IFITM3 | P2RY14 |
| DEG-RNAseq | D | D | ✗ | D | D | D | ✗ |
| Top network | ✗ | ✓ | ✗ | ✓ | ✓ | ✗ | ✓ |
| Variation over treatment | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✗ |
| Epicenters | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ |
3.7. Experimental Validation by qRT-PCR
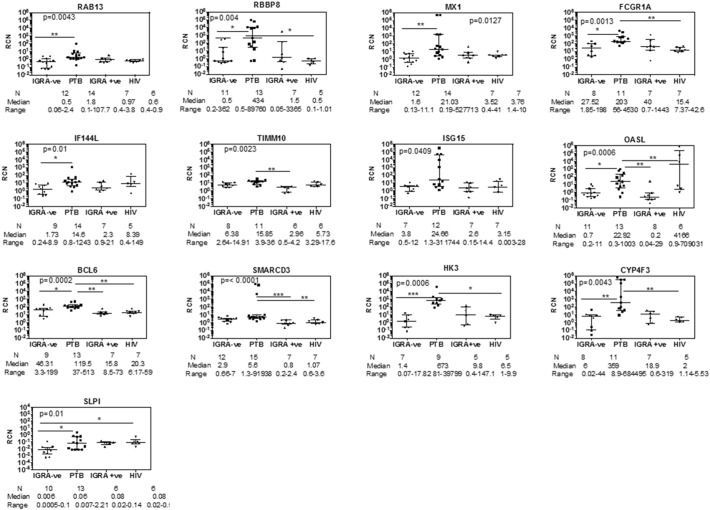
We tested 15 (RAB13, RBBP8, ADM, CECR6, TNNT1, TIMM10, SMARCD3, SLPI, IFI44L, IRF7, BCL6, CYP4F3, HK3, FCGR1A, OSM) genes from a total of 16 shortlisted candidates from the computational pipeline, using qRT-PCR in additional whole blood samples taken from patients freshly diagnosed with TB (Table 1). The probe for gene MYL9 did not work, and hence it was not considered for validation. In addition, we also tested 6 genes (OASL, MX1, ISG15, SOCS3, STAT1, STAT2) belonging to the Type I IFN induced response which was identified to be highly upregulated in active TB as seen in the differentially expressed gene (DEG) list derived from RNA sequencing, as the Type I interferon response has emerged as an important host response in tuberculosis. From the list of potentially most significant genes obtained from the computational pipeline, expression of RAB13, RBBP8, FCGR1A, IFI44L, TIMM10, BCL6, SMARCD3, HK3, CYP4F3 and SLPI was significantly higher in active TB compared to IGRA − ve/healthy controls as determined by the Mann–Whitney U test. OASL, MX1 and ISG15 from the Type I induced gene list could be validated for high expression in TB compared to IGRA − ve/healthy controls. Additionally, expression of genes TIMM10, BCL6, SMARCD3 and OASL was higher in active TB compared to latent TB and expression of FCGR1A, BCL6, SMARCD3, HK3 and SLPI was higher in active TB compared to HIV infection (Fig. 6). Using Dunn's multiple comparisons test, BCL6 and SMARCD3 were most significant in distinguishing active TB from several other groups: healthy control, latent TB and HIV infection. Interestingly, none of the Type I IFN induced genes differentiated between active TB and HIV infection at the N per group studied.
Fig. 6.
qRT-PCR verification of the 16 genes shortlisted by the biomarker identification pipeline. Comparative analysis of IGRA ve/healthy controls, active pulmonary TB, latent TB and HIV infected subjects: All genes listed were verified by qRT-PCR by the SYBR method, with the exception of SLPI, which was verified by TaqMan. GAPDH was used as internal control for calculating relative copy number (RCN) of each gene. Statistical significance for between group RCN differences was first calculated using Mann–Whitney U test between IGRA − ve and active TB subjects. All genes tested were significantly different between these two groups using this test. p-Value shown in each graph was determined by one way ANOVA to correct for multiple comparisons; Dunn's multiple comparisons test was then used to identify significant differences between specific groups as shown by a line. *p = 0.01; **p < 0.01, ***p < 0.0001.
The expression values of the 10 genes were predicted by the pipeline and subsequently verified by qRT-PCR. Expression values for 3 additional genes MX1, OASL and ISG15 were also obtained and subjected to patient-wise analysis in the additional samples obtained for verification. Fold changes per gene in TB patient samples were computed with respect to a median HC. As observed in Fig. 7, in every patient sample, a minimum of 5 genes of the predicted markers were seen to be upregulated, with some patients seeing an upregulation in all the genes measured. The exception was the sample from patient 28, in whom only 5 genes could be measured, of which 2 genes show significant upregulation. Further, every gene showed upregulation in all or most of the patients measured, and is ranked by the proportion of samples in which they are expressed, with respect to healthy controls.
Fig. 7.
Patient-wise fold changes in expression of the candidate markers in TB vs HC. Ranked list of genes validated by qRT-PCR, with upregulation observed in individual PTB samples with respect to a median HC. Genes with a log2FC > 1 have been highlighted.
3.8. Monitoring Expression Changes Upon Treatment
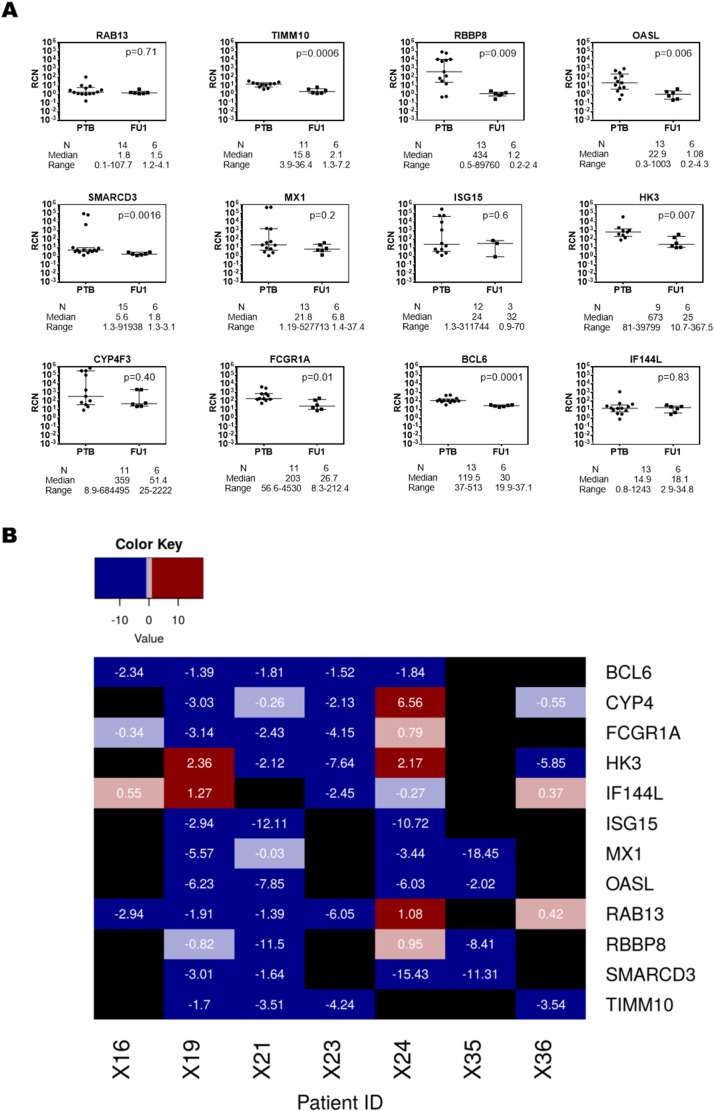
A good biomarker for the disease should show variation over the course of treatment, and the pipeline selected those genes that showed a linear decrease in expression after 6 (FU1) and 12 (FU2) months of treatment. As additional validation, the patients whose blood samples were taken at diagnosis for validation by qRT-PCR were followed up for six months and their samples were tested to determine if there were any reversals in the upregulation of the shortlisted candidate markers. The expression of genes RBBP8, TIMM10, SMARCD3, IFFI44L, BCL6, CYP4F3, HK3 and FCGR1A was shown to have a significant reduction after 6 months of therapy, whereas RAB13 did not show much variation. SLPI was not measured in these patients. Additionally, three other genes upregulated in TB MX1, OASL and ISG15, which were subsequently eliminated at one of the steps of the biomarker identification pipeline, were also measured. Of these, ISG15 showed an increase in expression upon treatment, whereas MX1 and OASL showed significant decrease in expression, indicating changes in the Type I response upon therapy. These results are shown in Fig. 8(a), which indicate that a majority of the genes in the panel showed a decrease in expression values upon treatment, with respect to their expression levels at diagnosis of TB. Patient-wise variation in gene expression over treatment is shown in Fig. 8(b).
Fig. 8.
(a) qRT-PCR verification of the candidate marker genes after six months of treatment (FU1). All genes were verified by qRT-PCR by the SYBR method. GAPDH was used as internal control for calculating relative copy number (RCN) of each gene. Statistical significance for between group RCN differences was calculated using Mann–Whitney U test. p-Value shown in each graph was determined by one way ANOVA to correct for multiple comparisons. (b) Patient-wise fold changes in expression of the candidate markers in FU1 vs TB. Fold changes were computed at FU1 for each gene with respect to their median values in the same patient at diagnosis. Genes with a log2FC < − 1 are highlighted in blue, and those with a log2FC > 1 are highlighted in red. Black indicates those genes that were not measured in a given patient sample.
3.9. Specificity of the Predicted Markers
From the results described above, it is clear that the expression pattern of the 10-gene panel is sufficiently characteristic of the TB condition when compared to HC, latent TB and HIV samples. Beyond this, a frequent requirement in the clinic is to get a differential diagnosis between TB and other diseases whose clinical presentation may resemble pulmonary TB. Transcriptome data for other diseases which elicit a similar inflammatory response in the host as that of TB were publicly available (Berry et al., 2010, Bloom et al., 2013, Kaforou et al., 2013, Haas et al., 2016), and we therefore assessed the predictive power of the 10 genes to distinguish between TB and other diseases. To determine the specificity of our identified signature for these diseases, an LDA was performed on additional samples to determine how many of these samples would be correctly classified into TB and other diseases, using only the 10-gene panel by performing the class prediction step of LDA with 4-fold cross-validation. Table 4 shows the predictive ability of the 10 genes on TB, HIV, LTB and other disease samples reported by Kaforou et al. (2013). Additional File 3 provides the details for the datasets used and the prediction accuracies obtained using both the 16 gene-set and the 10-gene combination to show specificity of TB with respect to other similar diseases.
Table 4.
Predictive potential of the 10-gene combination with and without 4-fold cross-validation on the dataset by Kaforou et al. (2013).
| Condition | #Sample C1 | #Sample C2 | TP | TN | FP | FN | Accuracy | TP noCV | TN noCV | FP noCV | FN noCV | Accuracy noCV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Malawi | ||||||||||||
| TB vs HIVLTB | 51 TB | 35 HIVLTB | 49 | 31 | 4 | 2 | 0.93 | 49 | 31 | 4 | 2 | 0.93 |
| HIVTB vs HIVOD | 50 HIVTB | 35 HIVOD | 41 | 23 | 12 | 9 | 0.75 | 45 | 27 | 8 | 5 | 0.85 |
| TB vs HIVOD | 51 TB | 35 HIVOD | 46 | 23 | 12 | 5 | 0.8 | 48 | 26 | 9 | 3 | 0.86 |
| TB vs HIVTB | 51 TB | 50 HIVTB | 46 | 37 | 13 | 5 | 0.75 | 47 | 41 | 9 | 4 | 0.87 |
| TB vs LTB | 51 TB | 36 LTB | 42 | 33 | 3 | 9 | 0.86 | 43 | 34 | 2 | 8 | 0.89 |
| TB vs OD | 51 TB | 34 OD | 47 | 16 | 18 | 4 | 0.74 | 49 | 22 | 12 | 2 | 0.84 |
| South Africa | ||||||||||||
| TB vs HIVLTB | 47 TB | 49 HIVLTB | 38 | 48 | 1 | 9 | 0.9 | 38 | 49 | 0 | 9 | 0.9 |
| HIVTB vs HIVOD | 47 HIVTB | 57 HIVOD | 29 | 48 | 9 | 18 | 0.74 | 33 | 52 | 5 | 14 | 0.82 |
| TB vs HIVOD | 47 TB | 57 HIVOD | 34 | 49 | 8 | 13 | 0.8 | 38 | 53 | 4 | 9 | 0.88 |
| TB vs HIVTB | 47 TB | 47 HIVTB | 37 | 29 | 18 | 10 | 0.7 | 36 | 35 | 12 | 11 | 0.75 |
| TB vs LTB | 47 TB | 47 LTB | 37 | 43 | 4 | 10 | 0.85 | 40 | 46 | 1 | 7 | 0.91 |
| TB vs OD | 47 TB | 49 OD | 32 | 39 | 10 | 15 | 0.74 | 34 | 42 | 7 | 13 | 0.79 |
C1 and C2 are conditions 1 and 2; TP, TN, FP, and FN are True Positives, True Negatives, False Positives and False Negatives respectively, similar to that in Fig. 4. noCV — no cross-validation. Columns 4 to 7 present the results with a 4-fold cross-validation.
4. Discussion
The host response to Mtb is complex and multifaceted, and involves an elaborate interplay of multiple components of the immune system (Cooper, 2009). The transcriptome offers a global dynamic view of the changes occurring in the host during infection, and provides a list of differentially expressed genes which help characterize specific responses to infection. Whole blood transcriptomes are comprehensively covered genomic data that are reflective of condition specificity. While a global view broadly captures the immune response at play during an active infection, for ease of diagnostics at the clinical level, further filtering of this list will help shortlist the most significant of these genes.
The field of detecting blood biomarkers is rapidly evolving (Haas et al., 2016, O'Bryant et al., 2016, Petruccioli et al., 2016). The key challenge for the success of this approach is the robustness of a minimal biomarker to readily distinguish a disease state. This study was designed to identify a minimal biomarker and to validate the marker to distinguish extremes of disease. To do so we implemented a network approach, by analyzing these DEGs in the context of their connections and the extent to which they can influence their interacting neighbors. Networks provide an overview of the nature of connectedness of each participating gene, enabling the identification of hubs, however, by themselves they only provide a static picture of the system. Integrating condition-specific transcriptomic data into the network results in the generation of weighted response networks which describe how the changes in expression flow across the interconnected routes in a given condition, thereby identifying key regulators of the host response. The hPPiN utilized in this study was constructed using only high scoring experimentally validated physical interactions, with directions assigned based on functional annotations. It is thus of higher confidence than similar gene co-expression networks derived based on expression patterns alone. Each step in the pipeline considers a different aspect, which when put together, provides cumulative insights about multiple aspects. While enrichment of the generated response network provides a better overview of the underlying processes that are activated during infection, further mining of the network is necessary to identify those genes and processes that are primary drivers of the host response, which can serve as putative markers for active tuberculosis.
An ideal biomarker should not only be capable of discriminating between disease and healthy conditions clearly, but should also be functionally relevant. Analysis of network topology has highlighted the central positioning of these predicted markers, which have all shown to exert significant influence on their partners and can thus transmit effects of their expression variation downstream in the host. Additionally, several of these genes have been implicated in the pathobiology of tuberculosis, strengthening their importance as functional biomarkers of disease. Infection with Mtb has been reported to alter immuno-metabolism in the host, with the induction of the Warburg effect primarily by HIF-1 recently observed in mice lungs (Shi et al., 2016). HK3 has been shown to be an important player in the activation of HIF-1 mediated responses, participating in stress-mediated energy metabolism and antimicrobial activity. CYP4F3 encodes leukotriene-B(4) omega-hydrolase 2, and is directly connected to LTA4H in the top response network for TB. The expression profile of LTA4H has been shown to influence the TNF-mediated inflammatory response by regulating the pro-inflammatory lipid leukotriene B4 in tuberculosis as well as in other respiratory diseases such as asthma, and can determine the outcome of infection in macrophages (Tobin et al., 2010, Tobin et al., 2013). Mtb also promotes its survival in host macrophages by dysregulation of lipid mediator balance by enhancing the production of lipoxins, mediated by leukotrienes (Dietzold et al., 2015). Genome-wide associated studies (GWAS) of the African population have reported RBBP8 to be associated with genetic predisposition to tuberculosis (Thye et al., 2010). While its exact mechanism of action in tuberculosis is yet unknown, it has been reported to be upregulated in several TB transcriptomic studies. The expression of genes RBBP8, RAB13, FCGR1A, TIMM10 and SMARCD3 has been demonstrated to significantly distinguish between active TB and other diseases such as sarcoidosis and pneumonia (patent WO2014093872 A1). BCL6 has been shown to mediate a sustained Mtb specific CD4 T cell response in addition to regulating host apoptotic responses (Moguche et al., 2015). Both IFI44L and SLPI participate in the interferon-mediated inflammation in TB (Maji et al., 2015), with IFI44L also implicated in lymph node tuberculosis; SLPI has been shown to exhibit antimicrobial activity and also plays an important role in regulating apoptosis by interacting with membrane phospholipid scramblase (Py et al., 2009).
Experimental validations in a fresh set of patient samples strengthen our predictions, underlining the sensitivity of this methodology which utilized a minimum number of samples to derive a signature, but which still should be substantially validated in an additional pool of samples from existing datasets in literature with significant prediction accuracies, as well as on additional fresh patient cases. The decrease in expression observed for these genes over six months of treatment for the patients followed up further depicts that the candidate genes responded to therapy, and that their high expression was representative of a TB-specific response. Based on these initial results, it seems likely that the signature will hold true for additional patient samples as well.
Difficulties in diagnosis of TB are at times, further compounded by the presence of comorbidities such as HIV infection, as well as by a similar inflammatory response observed in other diseases such as sarcoidosis, pneumonia, SLE and Still's disease. The predicted markers are shown to sufficiently distinguish TB from other diseases, by monitoring their differences in expression in these diseases, as reported in literature. Further, the discriminatory prowess of the signature against HIV was validated experimentally by qRT-PCR. HIV was chosen for comparison as it is well known that a common pathway dysregulated in both HIV and TB is the Type I interferon pathway (Mayer-Barber and Yan, 2016, Pawlowski et al., 2012). We show in this manuscript by validating the minimal marker in additional samples to those used for sequencing that the marker can distinguish TB subjects from healthy controls and HIV infected subjects, with reasonable accuracy. We wish to highlight that the robustness of a marker lies in being successfully validated in samples above and beyond those for its initial identification. We demonstrate that despite the marker being identified using RNA sequencing data from a mere three to four subjects in each group, it was validated in several TB samples collected from an additional clinical site, as well as by comparing it with expression profiles in other diseases from existing literature. Furthermore, we show that the genes in the identified panel respond to treatment and have significantly lower levels of expression.
Mapping omics data into genome-scale interaction networks and analysis in terms of the pathways and processes associated with these genes have served to provide vital clues about the critical differences at a systems level that occur during infection, providing precise suggestions for the development of biomarkers that can not only predict disease risk but also monitor outcome of therapy. Computational system level models serve as platforms for rationalizing available data in both a molecular bottom-up approach and a top-down approach, so as to derive variations in system properties that may reflect disease sub-types and predict response to a particular treatment plan. Such models when supplemented with experimental data can accelerate the progress of system medicine.
The approach implemented in this study is unbiased, in that no other prior knowledge was directly used in the selection of genes to include in the signature. Initially all genes and their known or predicted interactions are considered to reconstruct the network. The transcriptome data has been taken for all genes in a systematic manner. Our pipeline selects genes based on the criteria defined at each stage and hence is unbiased or hypothesis-free. Such an approach thus offers global insights into the multiple changes occurring in the host upon infection, making it feasible to pick among those genes that are topologically significant and functionally relevant, and which demonstrate sufficient discriminatory prowess. Signatures and network patterns specific to different stages of treatment have the potential to drive clinical decisions about the duration of treatment and specific drug combinations, carving a clear roadmap towards precise and personalized medicine. Identification of a marker profile is a step towards aiding early and efficient diagnosis of TB, aimed at enabling more effective management of the disease.
Taken together, this is a comprehensive study showing the identification of a robust minimal marker gene-set for TB that deserves further validation in larger cohort based studies of both TB and other chronic disease states.
The following are the supplementary data related to this article.
The list of fold change values for all genes and their q-value as determined by RNA sequencing.
Clinical details of enrolled participants in this study.
Prediction accuracies of the shortlisted gene signatures in classifying TB and other diseases.
Competing interests
The authors declare that they have no competing interests.
Author's contributions
The project was conceived by NC and the network analysis and identification of the biomarkers was performed by AS and AD. AM performed the RNA-seq data analysis and cross-validation. AA and SN performed the experiments to validate the identified biomarkers and AV supervised the experiments. SS, GS, AJ, CD and SB were involved in the collection and processing of the clinical samples. The manuscript was written and edited by AS, AD, AM, AA, SR, AV and NC.
Acknowledgements
This work was funded in part by a Department of Science and Technology, Govt. of India, grant to the Centre of Excellence in Mathematical Biology (NC) and partly by a Department of Biotechnology, Government of India, Centre of Excellence Award (AV) for research in HIV and TB (DBT/01/CEIB/12/III/09).
The funders had no role in the study design, data collection, data analysis, interpretation, or writing of this report. No one was paid to write this article by a pharmaceutical company or any other agency.
References
- Azad A.K., Sadee W., Schlesinger L.S. Innate immune gene polymorphisms in tuberculosis. Infect. Immun. 2012;80(10):3343–3359. doi: 10.1128/IAI.00443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M.P., Graham C.M., McNab F.W., Xu Z., Bloch S.A., Oni T., Wilkinson K.A., Banchereau R., Skinner J., Wilkinson R.J. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankley S., Berry M.P.R., Graham C.M., Bloom C.I., Lipman M., O'Garra A. The application of transcriptional blood signatures to enhance our understanding of the host response to infection: the example of tuberculosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369(1645):20130427. doi: 10.1098/rstb.2013.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom C.I., Graham C.M., Berry M.P., Rozakeas F., Redford P.S., Wang Y., Xu Z., Wilkinson K.A., Wilkinson R.J., Kendrick Y. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A., Breitkreutz B.-J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43(D1):D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff J.M., Kaufmann S.H., McShane H., van Helden P., O'Garra A. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol. Rev. 2015;264(1):88–102. doi: 10.1111/imr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.M. Cell mediated immune responses in tuberculosis. Annu. Rev. Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzold J., Gopalakrishnan A., Salgame P. F1000prime Reports. 2015. Duality of lipid mediators in host response against Mycobacterium tuberculosis: good cop, bad cop; p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas D., Koltai M., Türei D., Módos D., Pálfy M., Dúl Z., Zsákai L., Szalay-Bekő M., Lenti K., Farkas I.J. SignaLink 2-a signaling pathway resource with multi-layered regulatory networks. BMC Syst. Biol. 2013;7(1):1. doi: 10.1186/1752-0509-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X., Xiao H. Diagnosis of tuberculosis pleurisy with adenosine deaminase (ADA): a systematic review and meta-analysis. Int. J. Clin. Exp. Med. 2014;7(10):3126–3135. [PMC free article] [PubMed] [Google Scholar]
- Haas C.T., Roe J.K., Pollara G., Mehta M., Noursadeghi M. Diagnostic ‘omics’ for active tuberculosis. BMC Med. 2016;14(1):1–19. doi: 10.1186/s12916-016-0583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera V., Perry S., Parsonnet J., Banaei N. Clinical application and limitations of interferon-gamma release assays for the diagnosis of latent tuberculosis infection. Clin. Infect. Dis. 2011;52(8):1031–1037. doi: 10.1093/cid/cir068. [DOI] [PubMed] [Google Scholar]
- Joosten S.A., Fletcher H.A., Ottenhoff T.H.M. A helicopter perspective on TB biomarkers: pathway and process based analysis of gene expression data provides new insight into TB pathogenesis. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0073230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaforou M., Wright V.J., Oni T., French N., Anderson S.T., Bangani N., Banwell C.M., Brent A.J., Crampin A.C., Dockrell H.M. Detection of tuberculosis in HIV-infected and-uninfected African adults using whole blood RNA expression signatures: a case–control study. PLoS Med. 2013;10(10) doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E., Fu Y., Chen J., Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput. Biol. 2013;9(3) doi: 10.1371/journal.pcbi.1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Lippman S., Agard D.A., Ashworth A., Ideker T. The cancer cell map initiative: defining the hallmark networks of cancer. Mol. Cell. 2015;58(4):690–698. doi: 10.1016/j.molcel.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Ota M., Repsilber D., Mollenkopf H.J., Weiner J., Hill P.C., Kaufmann S.H. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0026938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Repsilber D., Parida S.K., Stanley K., Roberts T., Black G., Walzl G., Kaufmann S.H. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12(1):15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- Maertzdorf J., Kaufmann S.H.E., Weiner J 3rd. Toward a unified biosignature for tuberculosis. Cold Spring Harb. Perspect. Med. 2014:a018531. doi: 10.1101/cshperspect.a018531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., McEwen G., Weiner J., 3rd, Tian S., Lader E., Schriek U., Mayanja-Kizza H., Ota M., Kenneth J., Kaufmann S.H. Concise gene signature for point-of-care classification of tuberculosis. EMBO Mol. Med. 2016;8(2):86–95. doi: 10.15252/emmm.201505790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J., Weiner J, 3rd, Mollenkopf H.J., Bauer T., Prasse A., Muller-Quernheim J., Kaufmann S.H. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl. Acad. Sci. U. S. A. 2012;109(20):7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji A., Misra R., Mondal A.K., Kumar D., Bajaj D., Singhal A., Arora G., Bhaduri A., Sajid A., Bhatia S. Expression profiling of lymph nodes in tuberculosis patients reveal inflammatory milieu at site of infection. Sci. Rep. 2015;5 doi: 10.1038/srep15214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber K.D., Yan B. Clash of the Cytokine Titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell. Mol. Immunol. 2016 doi: 10.1038/cmi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek G.H., Villarino M.E. Guidelines for using the QuantiFERON®-TB test for diagnosing latent Mycobacterium tuberculosis infection. Morb. Mortal. Wkly Rep. 2003;52:RR02. [PubMed] [Google Scholar]
- Moguche A.O., Shafiani S., Clemons C., Larson R.P., Dinh C., Higdon L.E., Cambier C.J., Sissons J.R., Gallegos A.M., Fink P.J. ICOS and Bcl6-dependent pathways maintain a CD4 T cell population with memory-like properties during tuberculosis. J. Exp. Med. 2015;212(5):715–728. doi: 10.1084/jem.20141518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand R., Yanai I. Deep Sequencing Data Analysis. 2013. An introduction to high-throughput sequencing experiments: design and bioinformatics analysis; pp. 1–26. [DOI] [PubMed] [Google Scholar]
- O'Bryant S.E., Mielke M.M., Rissman R.A., Lista S., Vanderstichele H., Zetterberg H., Lewczuk P., Posner H., Hall J., Johnson L. Blood-based biomarkers in Alzheimer disease: current state of the science and a novel collaborative paradigm for advancing from discovery to clinic. Alzheimers Dement. 2016 doi: 10.1016/j.jalz.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenhoff T.H., Dass R.H., Yang N., Zhang M.M., Wong H.E., Sahiratmadja E., Khor C.C., Alisjahbana B., van Crevel R., Marzuki S. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0045839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai M., Denkinger C.M., Kik S.V., Rangaka M.X., Zwerling A., Oxlade O., Metcalfe J.Z., Cattamanchi A., Dowdy D.W., Dheda K. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin. Microbiol. Rev. 2014;27(1):3–20. doi: 10.1128/CMR.00034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski A., Jansson M., Skold M., Rottenberg M.E., Kallenius G. Tuberculosis and HIV co-infection. PLoS Pathog. 2012;8(2) doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruccioli E., Scriba T.J., Petrone L., Hatherill M., Cirillo D.M., Joosten S.A., Ottenhoff T.H., Denkinger C.M., Goletti D. Correlates of tuberculosis risk: predictive biomarkers for progression to active tuberculosis. Eur. Respir. J. 2016 doi: 10.1183/13993003.01012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B., Basmaciogullari S., Bouchet J., Zarka M., Moura I.C., Benhamou M., Monteiro R.C., Hocini H., Madrid R., Benichou S. The phospholipid scramblases 1 and 4 are cellular receptors for the secretory leukocyte protease inhibitor and interact with CD4 at the plasma membrane. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0005006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravn P., Munk M.E., Andersen A.B., Lundgren B., Lundgren J.D., Nielsen L.N., Kok-Jensen A., Andersen P., Weldingh K. Prospective evaluation of a whole-blood test using Mycobacterium tuberculosis-specific antigens ESAT-6 and CFP-10 for diagnosis of active tuberculosis. Clin. Diagn. Lab. Immunol. 2005;12(4):491–496. doi: 10.1128/CDLI.12.4.491-496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richeldi L. An update on the diagnosis of tuberculosis infection. Am. J. Respir. Crit. Care Med. 2006;174(7):736–742. doi: 10.1164/rccm.200509-1516PP. [DOI] [PubMed] [Google Scholar]
- Rienksma R.A., Suarez-Diez M., Mollenkopf H.-J., Dolganov G.M., Dorhoi A., Schoolnik G.K., dos Santos V.A.P.M., Kaufmann S.H.E., Schaap P.J., Gengenbacher M. Comprehensive insights into transcriptional adaptation of intracellular mycobacteria by microbe-enriched dual RNA sequencing. BMC Genomics. 2015;16(1):1. doi: 10.1186/s12864-014-1197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambarey A., Devaprasad A., Baloni P., Mishra M., Mohan A., Tyagi P., Singh A., Akshata J.S., Sultana R., Buggi S., Chandra N. NPJ Systems Biology and Applications. 2017. Meta-analysis of host response networks identifies a common core in tuberculosis. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambarey A., Prashanthi K., Chandra N. Mining large-scale response networks reveals ‘topmost activities’ in Mycobacterium tuberculosis infection. Sci. Rep. 2013;3 doi: 10.1038/srep02302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambaturu N., Mishra M., Chandra N. EpiTracer — an algorithm for identifying epicenters in condition-specific biological networks. BMC Genomics. 2015;17(Suppl. 4):543. doi: 10.1186/s12864-016-2792-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Salamon H., Eugenin E.A., Pine R., Cooper A., Gennaro M.L. Infection with Mycobacterium tuberculosis induces the Warburg effect in mouse lungs. Sci. Rep. 2016;5:18176. doi: 10.1038/srep18176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G., Morris J.H., Demchak B., Bader G.D. Biological network exploration with cytoscape 3. Curr. Protoc. Bioinformatics. 2014;47(8.13):8.13.1–8.13.24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan B., Benn P., Mahungu T., Young M., Mercey D., Morris-Jones S., Miller R.F. Comparison of two interferon-gamma release assays (QuantiFERON-TB Gold In-Tube and T-SPOT. TB) in testing for latent tuberculosis infection among HIV-infected adults. Int. J. STD AIDS. 2010;24(10):775–779. doi: 10.1177/0956462413486459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney T.E., Braviak L., Tato C.M., Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir. Med. 2016;4(3):213–224. doi: 10.1016/S2213-2600(16)00048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014:gku1003. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thye T., Vannberg F.O., Wong S.H., Owusu-Dabo E., Osei I., Gyapong J., Sirugo G., Sisay-Joof F., Enimil A., Chinbuah M.A. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat. Genet. 2010;42(9):739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin D.M., Roca F.J., Ray J.P., Ko D.C., Ramakrishnan L. An enzyme that inactivates the inflammatory mediator leukotriene B4 restricts mycobacterial infection. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0067828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin D.M., Vary J.C., Jr., Ray J.P., Walsh G.S., Dunstan S.J., Bang N.D., Hagge D.A., Khadge S., King M.-C., Hawn T.R. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140(5):717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J., Maertzdorf J., Kaufmann S.H.E. The dual role of biomarkers for understanding basic principles and devising novel intervention strategies in tuberculosis. Ann. N. Y. Acad. Sci. 2013;1283(1):22–29. doi: 10.1111/j.1749-6632.2012.06802.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization Global tuberculosis report. 2016. http://www.who.int/tb/publications/globalreport/en/ (Accessed 14 November 2016)
- Zak D.E., Penn-Nicholson A., Scriba T.J., Thompson E., Suliman S., Amon L.M., Mahomed H., Erasmus M., Whatney W., Hussey G.D. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016 doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The list of fold change values for all genes and their q-value as determined by RNA sequencing.
Clinical details of enrolled participants in this study.
Prediction accuracies of the shortlisted gene signatures in classifying TB and other diseases.