Abstract
The efficacy of using Serial Analysis of Gene Expression (SAGE) to analyze the transcriptome of the model dicotyledonous plant Arabidopsis was assessed. We describe an iterative tag-to-gene matching process that exploits the availability of the whole genome sequence of Arabidopsis. The expression patterns of 98% of the annotated Arabidopsis genes could theoretically be evaluated through SAGE and using an iterative matching process 79% could be identified by a tag found at a unique site in the genome. A total of 145,170 reliable experimental tags from two Arabidopsis leaf tissue SAGE libraries were analyzed, of which 29,632 were distinct. The majority (93%) of the 12,988 experimental tags observed greater than once could be matched within the Arabidopsis genome. However, only 78% were matched to a single locus within the genome, reflecting the complexities associated with working in a highly duplicated genome. In addition to a comprehensive assessment of gene expression in Arabidopsis leaf tissue, we describe evidence of transcription from pseudo-genes as well as evidence of alternative mRNA processing and anti-sense transcription. This collection of experimental SAGE tags could be exploited to assist in the on-going annotation of the Arabidopsis genome.
Global gene expression analysis has been widely adopted as a tool to uncover candidate genes and elucidate regulatory pathways controlling important traits in a number of species. Two popular strategies being employed are microarray analysis and Serial Analysis of Gene Expression (SAGE). DNA microarrays (Schena et al., 1995) consisting of amplified cDNAs or synthesized oligonucleotides can be hybridized with different mRNA populations to direct a search toward specific subsets of genes, but the technique is reliant upon either sufficient depth of cDNA coverage or accurate gene annotation. The SAGE procedure isolates a small sequence tag from each transcript within an mRNA population, and the digital count of each tag facilitates a quantitative analysis of the expression of thousands of genes simultaneously (Velculescu et al., 1995). This technique is most effectively used to study gene expression in organisms where a complete genome sequence is available. A comparable technique, massively parallel signature sequencing (MPSS), has also been developed but requires specialized equipment that is presently not commercially available (Brenner et al., 2000).
SAGE analysis has been successfully applied to transcript profiling in a number of eukaryotic species including Saccharomyces cerevisiae (Velculescu et al., 1997), Homo sapiens (Zhang et al., 1997), and more recently in Caenorhabditis elegans (Jones et al., 2001) and Drosophila melanogaster (Gorski et al., 2003). The technique has yet to be widely adopted in plants, although preliminary work has been carried out in rice (Oryza sativa; Matsumura et al., 1999; Gibbings et al., 2003), loblolly pine (Pinus taeda; Lorenz and Dean, 2002), and more recently in Arabidopsis (Lee and Lee, 2003; Fizames et al., 2004).
The most common form of the SAGE procedure isolates a 14-bp tag from the 3′ most NlaIII restriction site of every mRNA found within a sample. In theory, a random tag sequence of this length provides sufficient complexity to uniquely identify its gene of origin since the tag is extracted from a defined position within the transcript. However, the organization of most genomes results in nonrandom DNA sequence that limits the ability of SAGE to unambiguously match the isolated tag to the gene of origin (Lash et al., 2000; Pleasance et al., 2003). In rice, between 23% (Matsumura et al., 1999) and 46% (Gibbings et al., 2003) of tags derived from leaf tissue could be matched to an expressed sequence tag (EST), while in loblolly pine only 32% of the 500 most abundant tags could be matched to the available gene database and none of the differentially expressed SAGE tags could be assigned a match (Lorenz and Dean, 2002). Additionally, in each of these studies the proportion of tag assignments that uniquely matched a single location was not or could not be determined. These studies, particularly the latter, emphasize the importance of performing SAGE analysis in well-characterized genomes. Perhaps more surprising, in recently published SAGE analyses performed on Arabidopsis tissue, between 43% and 73% of the distinct tags remained unmatched to gene sequences (Jung et al., 2003; Lee and Lee, 2003; Fizames et al., 2004), despite the extent of the genomic sequence and annotation available.
Here we describe an assessment of the efficacy of the SAGE technique as a tool for gene expression profiling in Arabidopsis. The assembled annotated whole genome sequence was utilized to generate theoretical SAGE tag data sets for the Arabidopsis nuclear and organelle genomes. An analysis of these data sets established the level of ambiguity associated with theoretical tag-to-gene matching in this species. A comprehensive tag-to-gene mapping analysis was carried out for experimental SAGE tags generated from two Arabidopsis SAGE libraries. A total of 145,170 reliable tags were sequenced, of which 29,632 tags were distinct and 12,988 of these distinct tags were observed more than once. Using a novel iterative matching process, 10,080 (78%) of these tags were unambiguously matched to their representative gene and 6,800 (52%) were matched to the canonical position. A number of the remaining tags that matched noncanonical positions indicate the presence of novel transcripts both in the sense and the anti-sense orientation.
RESULTS
Efficacy of Tag-to-Gene Mapping within the Arabidopsis Genome
To fully exploit the available genome sequence for Arabidopsis we chose to utilize the annotated Arabidopsis sequence available from The Institute for Genomic Research (TIGR). The 30,799 nuclear genes from the annotated Arabidopsis genome sequence (TIGR annotation release v4.0) were partitioned into three categories based on supporting experimental evidence: (1) annotated genes that possess defined untranslated region (UTR) sequences, (2) predicted genes based on numerous gene prediction algorithms, and (3) pseudo-genes.
The SAGE protocol isolates the 10 bp adjacent to the 3′ most anchoring enzyme site in a transcript, and this 14-bp tag, including the enzyme recognition site, is the SAGE tag used to identify the transcript of origin. To facilitate our tag-to-gene matching, the canonical SAGE tag was defined as the 3′ most SAGE tag within each annotated gene. Although the short recognition sequence of the anchoring enzyme will result in a proportion of the canonical tags being derived from 3′ UTR sequence, the random distribution of the restriction site may result in the isolation of canonical tags from exonic or 5′ UTR sequence. A default 5′ and 3′ UTR was determined to effectively analyze the predicted Arabidopsis genes that do not possess defined UTR sequence.
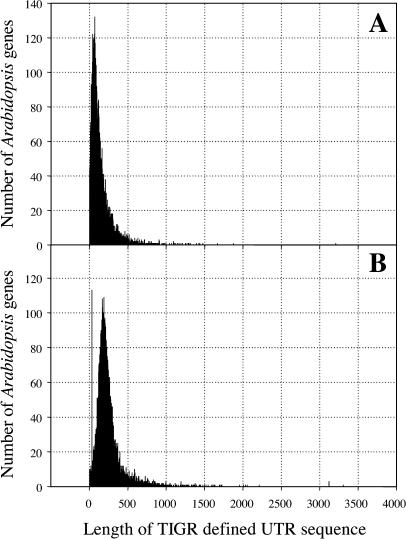
The frequency distributions for the lengths of both the 5′ and 3′ UTR sequences were determined using the 17,754 annotated genes with defined UTRs (Fig. 1). The UTR length that included 95% of each distribution was determined to be 350 bp and 500 bp for the 5′ UTR and 3′ UTR, respectively. Artificial UTRs of these lengths were used to extend those predicted genes lacking sufficient annotation. The distribution for the 3′ UTR length revealed 113 (0.6%) annotated genes with an UTR of 31 bp, which suggested errors in the annotation (Fig. 1B). To limit potential errors, 3′ UTR sequences of insufficient length to be encompassed by 95% of the distribution (less than 90 bp in length) were excluded and treated as having no defined UTR sequence and thus were assigned a UTR of default length. Based on these analyses, the annotated Arabidopsis nuclear genes were divided into 16,550 genes with experimentally defined UTR sequences and 12,031 conceptual transcripts with UTR sequences set to the default lengths. The defined and default nuclear transcripts can be found at http://www.brassica.ca/SAGE/.
Figure 1.
Distributions for the length of TIGR defined Arabidopsis UTR sequences. A, 5′ UTRs; B, 3′ UTRs.
Ideally, each individual SAGE tag should unambiguously identify a unique transcript. However, it is inevitable that a fraction of tags will match multiple locations within the genome and this fraction will increase as the genome size increases. To assess the theoretical efficiency of NlaIII derived SAGE tag-to-gene matching in Arabidopsis, the canonical SAGE tag for each gene or pseudo-gene was extracted from the nuclear and organelle data sets (Table I). Ninety-eight percent of the annotated genes contained an NlaIII site, while the remaining 2% would only be detected by changing the anchoring enzyme. SAGE analysis in Arabidopsis also has the potential to unambiguously differentiate between 79% of all canonical tags, despite the fact that 17% of Arabidopsis genes are duplicated in tandem between 2 and 23 times within the genome (Arabidopsis Genome Initiative, 2000).
Table I.
Efficacy of theoretical SAGE analysis in Arabidopsis based on canonical sense NlaIII derived SAGE tags
| Data Seta | Defined | Default | Pseudo-Gene | Nuclear | Organelle | Total |
|---|---|---|---|---|---|---|
| No. of transcriptsb | 16,550 | 12,031 | 2,218 | 30,799 | 204 | 31,003 |
| No. of transcripts with tag | 16,200 | 11,962 | 2,135 | 30,297 | 129 | 30,426 (98%) |
| No. distinct tagsc | 14,646 | 11,256 | 1,776 | 26,932 | 119 | 27,024 (87%) |
| No. of tags with unique locationd | 13,293 (80%) | 10,745 (89%) | 1,590 (72%) | 24,354 (79%) | 109 (53%) | 24,414 (79%) |
The data sets are separated into nuclear and organelle sets. The nuclear data set is divided into defined, genes with TIGR annotated UTR sequence; default, genes with conceptual UTR sequence added; and pseudo-gene, TIGR annotated pseudo-genes.
The number of transcripts includes all annotated gene iso-forms.
The nonredundant set of SAGE tags.
Number of distinct tags that match unambiguously to a single location within each data set.
In certain instances SAGE may not capture the tags equivalent to the annotated canonical site, for example, due to alternative mRNA processing. Therefore, the current annotation may restrict the matching of legitimate tags. For these reasons, theoretical SAGE tags were also considered from every NlaIII site from exonic sequences and from immature transcript sequences. As expected, this analysis increased the level of ambiguity such that 55% of the 216,669 theoretical SAGE tags could be uniquely assigned to their annotated gene of origin (Supplemental Table I, available at www.plantphysiol.org). This ambiguity was further compounded after including intergenic sequence that resulted in 42% of the 431,518 theoretical tags matching a unique position within the Arabidopsis genomic sequence (Supplemental Table I).
The SAGE protocol also allows the directionality of each tag to be determined, and anti-sense tags have been reported in C. elegans and more recently in plants (Jones et al., 2001; Gibbings et al., 2003). A total of 41% of the 433,578 theoretical tags had the ability to uniquely identify a site within the annotated Arabidopsis genes when all potential Arabidopsis SAGE tags from both strands were assessed (Supplemental Table II). Only 27% of the 863,235 theoretical SAGE tags can uniquely identify the sequence from which they originated when the analysis was extended to make use of the entire genome sequence (Supplemental Table II).
All of the theoretical NlaIII SAGE tags described above form an invaluable resource for tag-to-gene matching in Arabidopsis and these data can be accessed from http://www.brassica.ca/SAGE/.
Generation of Arabidopsis Experimental SAGE Tags
The data from two SAGE libraries generated from Arabidopsis leaf tissue were combined resulting in the extraction of 184,580 tags prior to quality assessment. The Phred sequence trace quality scores were used to remove all tags of low sequence quality (Phred score of <20) due to the 1% error rate associated with single pass sequencing (Hillier et al., 1996). Further quality assessment involved removal of incorrectly sized ditags, duplicate ditags, tags generated from linker sequence, and poly(A) tags. The combined data resulted in the extraction of 145,170 valid experimental tags that describe the transcriptome of Arabidopsis leaf tissue. This analysis identified 29,632 (20%) distinct tags of which 16,644 (11%) tags were observed only once.
Tag-to-Gene Matching of Experimental Tags
We have focused our analysis on tag-to-gene matching for the 12,988 distinct tags that were encountered greater than once and provide limited analysis of singletons. An iterative process was employed to match these tags to their representative Arabidopsis genes that involved assigning the highest level of confidence to tags that match the canonical site within a mature transcript sequence. This analysis assigned 6,800 (52%) tags to a unique canonical position within the Arabidopsis nuclear and organelle data sets (Table II, Canonical Tag Matches). An additional 345 (3%) tags matched canonical positions but could not be unambiguously assigned to a single position. These included instances where a distinct tag identified multiple members of a gene family and where distinct tags were matched to multiple unrelated genes (Table II, Canonical Tag Matches).
Table II.
Tag-to-gene matching for distinct experimental SAGE tags occurring greater than once to canonical tag positions or all possible tag sites
| Canonical Tag Matches | Data Sets
|
|||||
|---|---|---|---|---|---|---|
| Defined | Default | Pseudo-Gene | Nuclear | Organelle | Total | |
| Single matchesa | 6,224 (48%) | 525 (4%) | 29 (0.2%) | 6,778 (52%) | 22 (0.2%) | 6,800 (52%) |
| Multiple matches within data setb | 310 | 30 | 2 | 342 | 3 | 345 (3%) |
| Multiple matches between datasetsc | 301 (2%) | |||||
| Total tags matched | 6,534 | 555 | 31 | 7,120 | 25 | 7,446 (57%) |
| Unmatched tags | 5,542 (43%) | |||||
| Cumulative Tag Matches | Data Sets
|
Data Sets
|
||||||
|---|---|---|---|---|---|---|---|---|
| Defined | Default | Pseudo-Gene | Organelle | Total | Introns | Pseudo-Genomef | Total | |
| Total singlea matches | 7,823 (60%) | 1,260 (10%) | 67 (0.5%) | 36 (0.3%) | 9,186 (71%) | 306 (2%) | 588 (5%) | 10,080 (78%) |
| Total tagsd matched | 8,369 (64%) | 1,414 (11%) | 74 (0.6%) | 41 (0.3%) | 10,765e (83%) | 375 (3%) | 998 (8%) | 12,138e (93%) |
The number of tags that were assigned a match to the different datasets is shown. The percent values are calculated as a proportion of the 12,988 experimental tags that occurred greater than once.
SAGE tags that match a unique location within each of the defined data sets.
SAGE tags that match multiple sites within each of the defined data sets.
SAGE tags that match multiple sites when considering matches to the combined data sets.
These figures include single and multiple matches.
These figures include all matches both within and between data sets.
The pseudo-genome data set consists of the contiguous sequence for each of the five Arabidopsis pseudo-chromosomes.
The unmatched experimental tags were compared to the theoretical tags extracted from all possible NlaIII restriction sites within the nuclear and organelle data sets (Table II, Cumulative Tag Matches). This resulted in a cumulative total of 9,186 (71%) of the unique tags being matched to a single site within either an annotated gene or pseudo-gene (Table II, Cumulative Tag Matches). The remaining unmatched tags were compared to theoretical tags flanking every NlaIII restriction site in data sets generated from all available genomic sequences to allow for incompletely processed hnRNA, misannotation of splice sites, and unannotated transcripts. A further 894 (7%) of the unmatched tags were identical to a unique site within either intronic or intergenic sequence.
These combined analyses matched 12,138 (93%) distinct tags to the Arabidopsis genome, 2,058 matched multiple sites, and 850 tags remained unmatched. In total, 10,080 (78%) of the distinct SAGE tags observed more than once could be unambiguously assigned to a single location within the Arabidopsis genomic sequence. This was in contrast to previous publications where the highest level of tag-to-gene matching in Arabidopsis was 57% (Fizames et al., 2004).
Complexity of Tag-to-Gene Matching
The 30 most abundant SAGE tags identified within this experiment and their genes of origin are presented in Table III. As anticipated, the majority of the identified genes encode proteins involved in photosynthesis, with the remainder classified within oxidative stress, energy production, and cell division categories. The most abundant tag, which comprised 1.7% of the experimental tags, matched At2g34420, a member of a large gene family encoding a Photosystem II chlorophyll a/b binding protein. An additional tag (CATGCTCGGAGCCC) was present at a frequency of 0.4% and matched members of the same gene family including At2g34420. Using the iterative process, 21 of the 30 abundant tags (70%) were unambiguously matched to a single genomic location, although it was noted that two of these tags were unable to distinguish between their possible alternate gene transcripts. The remaining tags matched multiple locations. Three of these could not distinguish duplicate members of gene families that arose presumably via concerted evolution or relatively recent gene duplication events. Four of the SAGE tags matched multiple genes each annotated with a different predicted biological function. It is possible that the use of the conceptual transcripts could generate erroneous matches. However, in only one case was an additional match due to a tag being identified within the UTR of a conceptual transcript.
Table III.
The 30 most abundant SAGE tags in Arabidopsis leaf tissue, their relative frequency, and their genes of origin
| Taga | No. | Frequency % | Tag Position (c/nc)b | Matching Gene Codec | Putative Gene Functionc |
|---|---|---|---|---|---|
| GGAGCTGTTG | 2460 | 1.7 | c | At2g34420.1 | PS II type I chlorophyll a/b binding protein |
| nc | At2g34430.1 | PS II type I chlorophyll a/b binding protein | |||
| GGCCTTCGCC | 2259 | 1.6 | c | At1g29930.1 | PS I light-harvesting chlorophyll a/b binding protein |
| AAGGTGTGGC | 1324 | 0.9 | c | At5g38410.1 | RuBisCO small subunit 3b |
| c | At5g38420.1 | RuBisCO small subunit 2b | |||
| c | At5g38430.1 | RuBisCO small subunit 1b | |||
| CAGGTGTGGC | 1243 | 0.8 | c | At1g67090.1 | RuBisCO small unit, putative |
| TCCGAATCTT | 1162 | 0.8 | nc | At4g21960.1 | Peroxidase – putative |
| AGGAGAAAGA | 1081 | 0.7 | c | At1g29910.1 | PS II type I chlorophyll a/b binding protein |
| AAAGTTCTCG | 910 | 0.6 | c | At5g64040.1 | PS I reaction center subunit psaN precursor |
| TTTGCGATGC | 840 | 0.6 | nc | At4g10340.1 | Light-harvesting chlorophyll a/b binding protein |
| ATAGAACCTT | 800 | 0.6 | c | At2g45180.1 | Unknown protein |
| AACAAATTTG | 777 | 0.5 | nc | At3g47470.1 | Light-harvesting chlorophyll a/b-binding protein |
| GGCAGGCAAG | 772 | 0.5 | nc | rrn23S | 23S Ribosomal subunit - chloroplast genome |
| TGTTTTTATG | 764 | 0.5 | c | At1g61520.1 | Light-harvesting chlorophyll a/b binding protein |
| GAAATGAAAG | 732 | 0.5 | c | AtCg00480 | atpB ATP synthase CF1 β-chain |
| GCACAACAAC | 667 | 0.5 | c | At3g54890.1 | Light-harvesting chlorophyll a/b binding protein |
| c | At3g54890.2 | Light-harvesting chlorophyll a/b binding protein | |||
| c | At3g54890.3 | Light-harvesting chlorophyll a/b binding protein | |||
| GTTTGAAGGA | 656 | 0.5 | nc | At3g16640.1 | Translationally controlled tumor protein |
| TTTCCTTCCT | 649 | 0.5 | c | At1g20620.1 | Catalase 3 |
| CTCGGAGCCC | 616 | 0.4 | c | At1g29920.1 | PS II type I chlorophyll a/b binding protein |
| nc | At1g29930.1 | PS II type I chlorophyll a/b binding protein | |||
| nc | At2g34420.1 | PS II type I chlorophyll a/b binding protein | |||
| c | At2g34420.2 | PS II type I chlorophyll a/b binding protein | |||
| nc | At2g34430.1 | PS II type I chlorophyll a/b binding protein | |||
| GAGGTGGTGA | 575 | 0.4 | c | At3g12690.1 | Protein kinase, putative |
| c | AtCg01310 | rpl2.2 ribosomal protein L2 | |||
| c | AtCg00830 | rpl2.1 ribosomal protein L2 | |||
| CGCCCGCCGC | 554 | 0.4 | Chromosome 2 | 8,286 bp | |
| Chromosome 3 | 14,212,253 bp | ||||
| GTCACTCCTA | 537 | 0.4 | c | At1g06680.1 | 23 kD polypeptide of oxygen-evolving complex |
| GGCGAACGAC | 525 | 0.4 | Unmatched | ||
| ATATTTCTTT | 524 | 0.4 | nc | At3g50450.1* | Hypothetical protein |
| CTCTTTTCTG | 490 | 0.3 | c | At4g25100.3 | Putative Fe superoxide dismutase |
| nc | At4g25100.1 | Putative Fe superoxide dismutase | |||
| nc | At4g25100.2 | Putative Fe superoxide dismutase | |||
| TTTCTATAAA | 477 | 0.3 | c | At1g79040.1 | PS II polypeptide, putative |
| c | At1g80320.1* | Oxidoreductase, 2OG-Fe(II) oxygenase family | |||
| GCGAAAAGGA | 457 | 0.3 | c | At1g32060.1 | Phosphoribulokinase precursor |
| c | AtCg00440 | ndhC NADH dehydrogenase subunit 3 | |||
| GCCGTTCTTA | 447 | 0.3 | c | At2g01010.1 | 18S rRNA |
| c | At3g41768.1 | 18S rRNA | |||
| GGTTTGGTTG | 391 | 0.3 | c | At5g54270.1 | Light-harvesting chlorophyll a/b binding protein |
| c | At1g31335.1 | Expressed protein | |||
| CTTGTGATGG | 388 | 0.3 | c | At2g39730.1 | Auxin-regulated protein |
| c | At2g39730.2 | Auxin-regulated protein | |||
| c | At2g39730.3 | Auxin-regulated protein | |||
| AACACTGCTG | 382 | 0.3 | c | At5g54770.1 | Thiazole biosynthetic enzyme precursor |
| AAAATGAAAA | 382 | 0.3 | c | At3g15590.1 | DNA-binding protein, putative |
The anchoring enzyme sequence CATG has been omitted from the beginning of each tag.
c, Tag is found at the canonical position; nc, tag is found at a noncanonical position.
The gene identifiers and the putative gene functions are those provided by The Arabidopsis Information Resource, http://www.arabidopsis.org.
Indicates that the match to the gene lies within a default UTR sequence.
Alternative Transcript Processing
The precision of gene identification through SAGE allows tags to be unambiguously matched to noncanonical sites within transcripts. This has the potential to identify uncharacterized differential processing events. For example, six of the most highly abundant SAGE tags were unambiguously matched to a noncanonical position (Table III) and in each case the corresponding annotated canonical tag occurred at a low frequency providing evidence for alternative transcription.
The annotation provided by TIGR details differential processing events for only 1,267 Arabidopsis genes resulting in 2,678 alternatively spliced transcripts (Haas et al., 2003). Our experimental SAGE tags identified 966 of these alternative transcripts and multiple alternative mRNA molecules were detected for 66 (5%) of the genes. In total, 9,186 distinct tags were unambiguously matched to 8,293 different annotated Arabidopsis genes. This difference results from 799 instances where multiple distinct tags matched the same Arabidopsis gene, suggesting 10% of the observed genes are subject to alternative processing. It has been proposed that the identification of alternative transcripts could be compromised by the presence of truncated cDNA species due to priming from internal poly(A) tracts within an mRNA molecule (Nam et al., 2002). Analysis of the 8,293 genes identified through SAGE determined that only 190 of these genes contained an internal poly(A) tract (≥8) 5′ of the canonical tag site. For 23 of these genes, we have isolated an alternative tag that could have resulted through priming from the internal poly(A) sequence. Noncanonical tags could also result from incomplete digestion of the cDNA with the anchoring enzyme. If this were the case, the combination of digestion at random NlaIII sites and 3′-end capture employed in SAGE should result in the tag frequency being inversely proportional to the relative position of the tag site to the 3′ end of the molecule. We assessed the SAGE tag count at each NlaIII site for the 799 genes with potential alternative transcripts. The noncanonical tag abundance exceeded that of the canonical tag for 316 of these genes and no internal poly(A) sequence was present. This indicates with greater confidence that these represent genuine examples of alternative mRNA processing.
Current annotation for the gene At3g47470 details a single iso-form, although two distinct SAGE tags were unambiguously matched to this gene. A noncanonical tag (CATGAACAAATTTG) was observed 777 times (Table III) while the canonical tag (CATGTGGCAACAGT) was found 149 times, suggesting this is not an artifact of incomplete digestion. The presence of alternate iso-forms for this gene was corroborated by the presence of full-length cDNA and 3′ EST sequences deposited in GenBank representing these forms (for example AY093080 and AF325012). SAGE also has the ability to detect the presence of alternative transcriptional termination in addition to the identification of alternatively processed transcripts. This was observed for At3g16770 where the canonical tag (CATGTGTAAATAAG) was identified 21 times compared to a noncanonical tag (CATGGCTTATGATG) that was found 340 times. These SAGE tags matched different full length cDNA sequences submitted to GenBank (AY087488 and AY035100) that had a conserved translational stop codon but differed in the length of the 3′ UTR, presumably as the result of differential transcriptional termination. The discovery of this phenomenon through SAGE analysis is particularly interesting in light of recent evidence demonstrating the role of alternative transcriptional termination in the regulation of FCA gene expression (Simpson et al., 2003).
The analysis was extended to include singleton SAGE tags matching a unique position in the genome to establish an estimate for the frequency of alternative mRNA processing in Arabidopsis. This increased the number of genes with multiple tag matches to 3,038. Of these, 2,248, representing 17% of the 12,934 genes unambiguously assigned SAGE tag matches when including singletons, had more abundant noncanonical tags suggesting they were unlikely to be the result of incomplete digestion and could be due to alternate mRNA processing.
Anti-Sense Transcripts
The orientation of each SAGE tag extracted from an mRNA transcript is known, enabling potential anti-sense transcripts to be detected. It is unlikely that all of the remaining 850 unmatched distinct tags result from experimental artifacts since singletons were excluded from the tag-to-gene analysis. The unmatched tags were compared to all possible theoretical tags from the anti-sense strand in all data sets (Table IV). This resulted in the assignment of 259 (2%) tags to a unique anti-sense site within the Arabidopsis nuclear and organelle mRNA data sets and a further 147 (1%) matched a single location in the intronic or intergenic sequences. The remaining 387 (3%) tags that failed to match the data sets could have been derived from sequences spanning an unpredicted intron/exon boundary or from unsequenced regions of the genome or they may represent experimental artifacts.
Table IV.
Tag matching to the anti-sense strand for tags occurring greater than once
| All Tag Sites | Defined | Default | Pseudo-Gene | Nuclear | Organelle | Introns | Pseudo-Genome | Total |
|---|---|---|---|---|---|---|---|---|
| Single matchesa | 201 | 42 | 8 | 251 | 8 | 25 | 122 | 406 (3%) |
| Multiple matchesb | 14 | 1 | 0 | 15 | 0 | 3 | 19 | 37 (0.3%) |
| Between data set matchesc | 20 (0.2%) | |||||||
| Total tags matched | 215 (2%) | 43 (0.1%) | 8 | 266 (2%) | 8 | 28 (0.2%) | 141 (1%) | 463 (4%) |
| Unmatched tags | 387 (3%) |
The percent values are calculated as a proportion of the 12,988 experimental tags that occurred greater than once.
SAGE tags that match a single unique location within each of the defined data sets.
SAGE tags that match multiple sites within each of the defined data sets.
SAGE tags that match multiple sites when considering matches to the combined data sets.
The majority of these anti-sense SAGE tags were detected at low levels that may be expected if anti-sense molecules perform a regulatory function. However, some anti-sense tags were found among the most abundant tags observed. For example, the tag CATGGTCTCTCCAG was present 93 times and was unambiguously matched to At2g37220 in the anti-sense orientation.
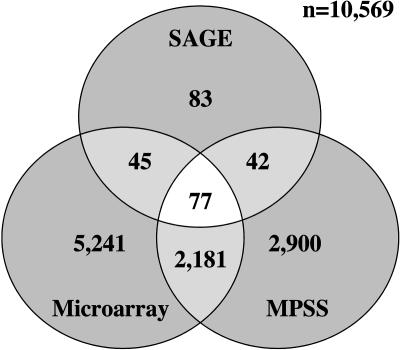
We analyzed leaf tissue expression profiles detected using the alternate platforms of MPSS (Meyers et al., 2004; http://mpss.udel.edu/at/java.html) and microarray (Yamada et al., 2003) to provide further evidence for anti-sense transcription from the 247 Arabidopsis annotated genes identified by the 259 anti-sense SAGE tags. This provided additional evidence for anti-sense transcription for 77 genes where anti-sense transcripts were detected by all three technologies (Fig. 2; Supplemental Table III). A further 87 genes with anti-sense SAGE tags were also corroborated using data from either MPSS or microarray analyses (Supplemental Table III).
Figure 2.
Comparison of the number of genes with anti-sense transcripts detected by three expression profiling technologies. Total number of annotated Arabidopsis genes with anti-sense transcripts expressed in leaf tissue detected using SAGE (247), MPSS (5,200), and microarray (7,544) technologies.
The analysis was extended to include all experimental SAGE tags to provide an estimate for the level of anti-sense transcription in Arabidopsis as some anti-sense molecules may be physiologically active at low concentrations. A total of 1,165 of the remaining unmatched distinct SAGE tags were assigned a match to an annotated Arabidopsis gene in an anti-sense direction after iterative matching to the sense strand. This identified 966 Arabidopsis genes with anti-sense transcription of which 518 were also detected by either MPSS or microarray, with 191 genes common across all three technologies.
DISCUSSION
Efficacy of SAGE Tag-to-Gene Matching within Arabidopsis
Arabidopsis is a widely used model for the study of plant biology due to its small genome size (approximately 120 Mb), low amount of repetitive DNA and fast generation time. Over the years, a large collection of associated genetic and genomics resources has been amassed and these have been augmented by the completion of the whole genome sequence (Arabidopsis Genome Initiative, 2000). cSAGE software and custom Perl scripts were written to extract theoretical SAGE tags from the available assembled and annotated genomic sequence to maximize the amount of information that can be derived from Arabidopsis SAGE libraries.
Conceptual transcripts were constructed for 12,031 nuclear genes by the addition of a 5′ and 3′ UTR of defined length to obtain the correct canonical tag from predicted Arabidopsis genes that have no annotated UTR sequences. This approach has recently been verified by work in D. melanogaster where it was demonstrated that 52% of canonical SAGE tags were located within the UTR sequence (Pleasance et al., 2003). A limitation of this approach is that although the lengths of the artificial UTRs are based on the size distribution of experimentally determined UTRs they will not always accurately reflect the true transcript. In the simplest cases the artificial UTRs will either be too short or too long, which may lead to canonical tags being missed or incorrectly identified. However, tags that originate from these genes would still be matched either as noncanonical tags or as inter-genic tags in the pseudo-genome data set. In more complex situations, the UTR may contain intronic sequence or even an adjacent gene and such atypical UTR structures could impact on the derived SAGE tag.
Almost all Arabidopsis nuclear and organelle genes (98%) can be distinguished using NlaIII based SAGE analysis and 79% of theoretical canonical tags, which are considered the most biologically relevant, can be unambiguously assigned to their gene of origin (Table I). It is necessary to consider noncanonical theoretical tags to identify the origin of unmatched tags in any SAGE experiment. In Arabidopsis, the specificity of tag matching (27%) was marginally lower than that observed in other nonplant model species (35% in D. melanogaster and C. elegans; Pleasance et al., 2003) when comparing all possible canonical and noncanonical tags. The fact that the ambiguity of matching was so similar between these species is perhaps surprising in light of the high level of segmental and tandem duplication within the Arabidopsis genome, with an estimated 60% of the genome occurring as large duplicated segments (Arabidopsis Genome Initiative, 2000).
Previous SAGE analyses in Arabidopsis has utilized the available UniGene data set for tag-to-gene matching (Ekman et al., 2003; Jung et al., 2003; Lee and Lee, 2003). The UniGene sets are constructed not only from well-characterized cDNA sequences but also from sets of error-prone EST sequences that have been estimated to introduce 10% incorrect assignments for tag-to-gene matching (Lash et al., 2000). Additionally, the UniGene sets do not provide a complete representation of the gene repertoire for most species. A further uncharacterized problem is the level of single nucleotide polymorphisms that are present within the Arabidopsis UniGene set due to sequences contributed from different Arabidopsis ecotypes. Canonical tags were extracted from the Arabidopsis UniGene data set (Build no. 41) that contained 25,447 entries, of which 6,318 were singletons and 94% of these singleton sequences terminated with a stop codon. Furthermore, when considering the entire UniGene data set, 33% of the entries had no associated 3′ UTR that may lead to the specious assignment of SAGE tags or the inability to match SAGE tags. This was highlighted through a comparison of canonical tags extracted from annotated Arabidopsis genes common to both the Arabidopsis UniGene data set and the TIGR data set. Although the same tag was extracted from the two databases for 16,381 genes, different tags were extracted for 7,098 genes. A total of 90% of canonical tags extracted from the Arabidopsis UniGene data set could uniquely identify the UniGene contig (or transcript) of origin. In contrast, the same analysis performed using the TIGR annotated Arabidopsis genes, detailing close to the full complement of genes, resulted in the unambiguous assignment of only 79% of the canonical tags. This drop in specificity is in part due to the additional genes present in the TIGR data set, but also indicates that reliance upon the UniGene sequences will underestimate the level of ambiguity for tag-to-gene matching in Arabidopsis as seen in previous analyses (Jung et al., 2003; Lee and Lee, 2003). These data support the use of the TIGR annotated genome sequence for effective matching of SAGE tags in Arabidopsis.
Tag-to-Gene Matching of Experimental SAGE Tags
An iterative process was employed to allow comprehensive tag-to-gene matching for Arabidopsis leaf tissue. The high level of SAGE tag quantitation makes this one of the most comprehensive SAGE analysis of global gene expression in a plant species to date. Conservative sequence quality analysis was also employed to alleviate problems associated with the single pass sequencing of SAGE data, this allowed 145,170 (79%) SAGE tags to be analyzed with confidence, of which 29,632 were unique.
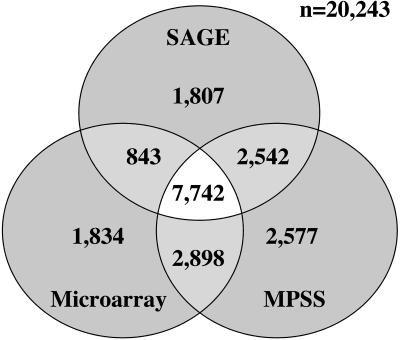
Almost 80% of the 12,988 tags that were observed two or more times could be unambiguously matched to a single genic location in the sense orientation and in the process identified 8,293 different annotated Arabidopsis genes. Of these matches, 84% were to nuclear transcripts comprising annotated UTR sequence. A total of 12,934 genes were uniquely identified by extending the tag-to-gene matching to incorporate the singleton data. Additionally, a greater proportion of these matches were assigned to the conceptual transcripts and pseudogenes perhaps reflecting their relative abundance within the transcriptome. Utilizing a comprehensive set of global arrays for Arabidopsis, Yamada et al. (2003) found 13,317 of the annotated genes were expressed in leaf tissue. Similarly, expression was detected from 15,759 genes in the publicly available MPSS data (Fig. 3). Combining all three analyses it was possible to detect transcription from 20,243 different annotated Arabidopsis genes, which represents approximately 70% of the predicted gene content. Of these, 14,025 were identified by at least two of the three platforms, providing perhaps a more reliable assessment of the leaf tissue transcriptome. A direct comparison between the three data sets is hampered by differences in the growth conditions and developmental stage of material used in each study. We would also expect differences specific to the profiling technologies due to the isolation of MPSS signatures from an alternate restriction site compared to SAGE and the possibility of cross-hybridization on the microarray due to gene and genomic duplication. However, SAGE was able to detect expression in leaf tissue from 64% of the annotated genes identified across the three platforms, which is comparable to 66% for microarray analysis but is lower than the 78% detected using MPSS. To generate a complete representation of the whole transcriptome, SAGE analysis will be limited by the depth of sequencing required since diminishing marginal returns would necessitate sequencing efforts beyond the scope of most projects. The deployment of a technology such as MPSS might be more appropriate to gain such knowledge, although access to this technology is limited to specialized laboratories.
Figure 3.
Comparison of the number of genes detected by three expression profiling technologies. Total number of annotated Arabidopsis genes with sense transcripts expressed in leaf tissue detected using SAGE (12,934), MPSS (15,759), and microarray (13,317) technologies.
In Arabidopsis, 93% of the SAGE tags could be assigned a match. However, 16% of these tags were assigned to more than one location within the genome (Table II, Cumulative Tag Matches). Matching of tags to multiple members of gene families or multiple alternative transcripts of the same gene still allows biological inferences to be made from the SAGE data since the predicted function in each case is identical. However, both the qualitative and quantitative value of the tag data is reduced since further investigation needs to be made on an individual basis. This can be compounded when a tag matches multiple genes with apparently unrelated functions. The majority of tags were matched to a unique genomic location in previously published SAGE analyses (Jones et al., 2001; Lorenz and Dean, 2002; Gibbings et al., 2003; Gorski et al., 2003; Lee and Lee, 2003). This could reflect deficiencies in the available sequence data for the analyses utilizing organisms with much larger genomes than Arabidopsis. However, in Arabidopsis only 5 of the 50 most abundant SAGE tags isolated from pollen were matched to multiple UniGene IDs (Lee and Lee, 2003). Comparing these 50 tags to canonical sites within the TIGR derived nuclear and organelle data sets demonstrated that a further nine tags matched multiple canonical sites, and four previously unmatched tags were also assigned a match, two of which were ambiguous.
A minority of the tags observed greater than once (3%) did not match any sequence within the Arabidopsis genome data sets. These tags may originate from unsequenced heterochromatic regions of the Arabidopsis genome, unpredicted intron splice junctions, or they may be artifacts of the SAGE protocol. These questions should be resolved once the genome is completely sequenced and the depth of EST sequencing increases. Nevertheless, the use of the full genome sequence has achieved a more comprehensive tag assignment than other SAGE studies in Arabidopsis where tags with no matches were as high as 70% (Jung et al., 2003).
It has been suggested that SAGE tags can assist with the annotation of genomic sequence by facilitating the discovery of novel transcripts and providing confirmatory evidence for hypothetical transcripts (Saha et al., 2002). For the experimental SAGE tags either including or excluding singletons, 3,736 or 894 distinct tags were unambiguously assigned to a single location in either intronic or intergenic sequence that may provide evidence for either misannotation or alternative processing of a particular transcript. For the matches to intronic sequence, it is also possible that these tags have been derived from incompletely processed hnRNA. The tags matching intergenic regions could provide evidence for the presence of noncoding RNAs which have been suggested as a new species of regulatory molecule (MacIntosh et al., 2001).
SAGE Tags Identifying Genes Encoded by Extra-Nuclear Genomes
The nuclear genome is almost 250 times larger than the combined organelle genomes. However, 0.3% of the experimental tags were assigned matches to canonical tags from transcripts present on either the mitochondrial (366.9 kb) or the chloroplast genome (154.4 kb). Together these comprised 3% of the total number of experimental tags extracted in this analysis. The highest level of mRNA synthesis for the organelles was detected in the chloroplast, which reflects the fact that they are present at a high copy number in leaf cells and that the leaf is predominantly a photosynthetic organ. However, there is growing evidence that complex signaling and gene regulation between the organelle and nuclear genomes plays a role in controlling a plant's physiological responses, particularly in interactions with the environment (Pfannschmidt et al., 2001; Pfannschmidt, 2003). Since the SAGE protocol captures tags from polyadenylated transcripts, interpreting the significance of any apparent changes in organelle transcript expression is complicated by recent evidence indicating extra-nuclear transcripts are polyadenylated as part of the mRNA degradation pathway (Carpousis et al., 1999; Hayes et al., 1999; Schuster et al., 1999). Therefore, an increase in frequency of an organelle tag could result from an increased rate of transcript degradation and not an increase in gene expression.
Detection of Novel Transcripts
The dichotomy of SAGE analysis lies in the requirement for comprehensive sequence data. Although SAGE tags can be generated from any organism, the ability to analyze and exploit the data is dependant upon the tags being unambiguously matched to their gene of origin. In the present analysis, the annotated Arabidopsis genome sequence provides an excellent resource with which to exploit SAGE data. Although deference was given to canonical tag matches, all possible NlaIII derived tags were considered which allowed a number of novel transcripts to be uncovered.
Alternatively processed transcripts were discovered within the SAGE data at a level of between 4% and 17%, the lower level being consistent with previous observations in plants (Haas et al., 2003; Zhu et al., 2003). However, the previous estimates were based upon available cDNA sequence that may underestimate the presence of alternative transcripts. Although the identification and characterization of such alternative transcripts within the Arabidopsis transcriptome is on-going, the experimental SAGE data provides compelling evidence for a number of as yet unannotated alternative transcripts. This included a number of examples found within the 30 most abundant SAGE tags. It is possible that SAGE data could be used as a tool to further quantify and characterize this phenomenon.
An intriguing observation was the presence of anti-sense transcripts within the SAGE data. The anti-sense tags could be explained by experimental errors such as mispriming from internal poly(A) tracts during second strand synthesis of the cDNA, from spurious promoter regions present on the noncoding strand or from illegitimate transcriptional read through from an adjacent gene or pseudo-gene found on the opposite strand (Elrouby and Bureau, 2001). However, comparing our data with those identified from two other global gene profiling platforms provided corroborating evidence supporting the assignment of the majority of the anti-sense tags. The presence of anti-sense transcripts detected using SAGE has been documented for a number of eukaryotic organisms (Jones et al., 2001; Patankar et al., 2001). In plants, only a small number of anti-sense RNAs have been reported through classical single gene analysis (Dolfini et al., 1993; Cock et al., 1997) and only latterly some evidence for anti-sense transcription was detected in rice through SAGE analysis (Gibbings et al., 2003). Current theory suggests that anti-sense RNAs could perform a regulatory role in gene expression (Vanhee-Brossollet and Vaquero, 1998) in a similar fashion to the more recently detected microRNAs (Reinhart et al., 2002). A conservative estimate for the level of anti-sense transcription in Arabidopsis leaf tissue was 3% of the genes detected by SAGE based on the distinct tags observed greater than once. This estimate increased to 8% when all distinct tags were considered.
SAGE tags were also detected for a number of annotated pseudo-genes. These presumed nonfunctional paralogs have arisen either through gene duplication, segmental genome duplication, or retrotransposition. The occurrence of pseudo-genes has been estimated to be 19% in the human genome (Dunham et al., 1999) and 7% in Arabidopsis (TIGR release v4.0). Although most pseudo-genes are unlikely to be transcribed due to the absence of correct promoter sequences, mRNA sequences have been associated with some of these elements (Olsen and Schechter, 1999). The assignment of experimental SAGE tags to a small percentage of pseudo-genes supports the possibility that they are transcribed but not necessarily that such genes encode a functional protein.
CONCLUSION
SAGE has proved a valuable tool in Arabidopsis to gain insight into the metabolic processes functioning within leaf tissue. Utilizing the available Arabidopsis genome sequence has allowed a comprehensive assignment of SAGE tags to their particular gene of origin. Although qualified by the evident ambiguity of some of the matches, SAGE has uncovered a number of novel transcripts, the biological significance of which has yet to be established. The theoretical SAGE tags made available through this analysis (http://www.brassica.ca/SAGE/) should assist in the effective matching of experimental SAGE tags in future analyses.
The efficacy of SAGE in Arabidopsis has proved to be similar to that of other model systems. However, for the larger crop genomes, the underlying duplication so ubiquitous in recent plant genome evolution could limit the information captured from such data and may warrant the application of techniques that isolate longer sequence tags such as LongSAGE (Saha et al., 2002) or MPSS (Brenner et al., 2000).
MATERIALS AND METHODS
Plant Materials
Leaf tissue was collected from plants grown under axenic conditions to eliminate contaminating sequence from potential pathogens. Arabidopsis (Col-4) seeds were treated with 10 mL of a 30% NaOCl solution for 10 min followed by treatment with an equal volume of 70% ethanol for 5 min before being washed three times for 5 min in an equal volume of dH2O. The axenic seed were germinated on 0.5× Murashige and Skoog media. Control seedlings were grown at 22°C and 125 μE light. The tissue above the media was harvested after 14 d, including leaf and shoot material, and used for RNA extraction. For cold treated tissue, the Arabidopsis plants were grown as described for the control plants but were exposed to low temperature, 4°C and 125 μE light, for a period of 30 min prior to the tissue being harvested and used for RNA extraction.
RNA Isolation and cDNA Synthesis
Total RNA was extracted from 10 g of Arabidopsis plant tissue using the Total RNA extraction kit and mRNA was purified from 600 mg total RNA using an mRNA purification kit according to the manufacturer's protocol (Amersham Biosciences, Baie d'Urfe, Canada). Five micrograms of poly(A) RNA was used to generate double-stranded cDNA using a cDNA synthesis kit (Gibco-BRL, Gaithersburg, MD) according to the manufacturer's protocol with the exception that a biotinylated oligo(dT) primer was used to prime synthesis of the first strand cDNA.
SAGE Library Generation
SAGE procedures were performed according to the original protocol (Velculescu et al., 1995) with the following modifications. To increase the efficiency of subsequent steps, tag concatamers obtained by ligation of the purified ditags were digested for 1 min using the anchoring enzyme NlaIII, before being size fractionated (>500 bp) using PAGE; this process linearized circularized concatamers. The purified concatamers were cloned into the pZero-1 vector (Invitrogen, Carlsbad, CA) and transformed into Escherichia coli strain DH10B. The colonies containing recombinant vectors formed on low salt LB-Zeocin (50 μg/mL) plates were picked, cultured in liquid SOB-Zeocin (50 μg/mL) and plasmid minipreps were performed using an optimized high-throughput 96-well diatomaceous earth protocol (Carter and Milton, 1993; Hansen et al., 1995). The plasmid minipreps were sequenced using BigDye v2 chemistry and the reactions resolved using an ABI3700 DNA analyzer (Applied Biosystems, Foster City, CA).
Extraction of Experimental SAGE Tags
The experimental SAGE tags were extracted from approximately 4,600 primary sequence reads using cSAGE software developed at Agriculture and Agri-Food Canada (AAFC) Saskatoon. cSAGE is a UNIX tool written in C that allows automated analysis of SAGE sequence data from FASTA files and is freely available upon request. The raw sequence data was processed by cSAGE such that ditags were rejected from further analysis if they were less than 24 bp or greater than 28 bp in length or if the mean sequence quality across each ditag had a Phred (Ewing et al., 1998) value of less than 20. Ditags were also removed from the analysis if they were observed more than once within a single library in either orientation in order to limit any PCR bias. Individual tags were extracted, counted, and their frequency within each library was determined once the SAGE ditags were defined. The first SAGE tag was obtained from each ditag by capturing the first 10-bp 3′ of the first CATG site defining a ditag. The second tag was obtained by determining the reverse complement sequence of the 10-bp 5′ to the second CATG site. In addition to the above exclusions, tags that might be derived from linker vector sequence and polyA tags were also removed from further analysis.
Sequence Data Set Construction
A suite of tailored Perl modules was developed to create Arabidopsis sequence data sets for tag matching using the available TIGR XML data (release v4, http://www.tigr.org/tdb/e2k1/ath1/) and GenBank data (http://www.ncbi.nlm.nih.gov) for the chloroplast (accession no. NC_000932) and mitochondrial (accession nos. Y08501 and Y08502) genomes. Listed in order of matching priority, three data sets were generated: (1) the exonic data set, derived from annotated exonic sequences from the nuclear, chloroplast, and mitochondria genomes; (2) the intron data set composed of the annotated immature transcript sequences; and (3) the pseudo-genome data set encompassing the entire Arabidopsis sequence from the nuclear, chloroplast, and mitochondria genomes.
It was necessary to generate conceptual transcripts for a subset of the annotated genes in order to utilize the TIGR data. The 17,754 (58%) Arabidopsis genes with experimentally verified UTR sequence were used to calculate the mean length and distribution of the UTRs after removal of all intronic regions. From these distributions, the length sufficient to encompass 95% of each distribution was determined to specify a default 5′ and 3′ UTR for those genes with limited annotation. These default UTRs were extracted from the Arabidopsis XML files of the pseudo-chromosomes utilizing the coordinates defining each predicted gene to identify the adjacent sequence. The extension of the UTR sequence was terminated at the beginning of the adjacent gene where extension of the predicted gene sequence created overlaps between adjacent genes. However, where data from full-length cDNAs was used to support the gene annotation and overlapping genes were documented these genes remained unaltered.
For comparative purposes, data sets were also generated from the Arabidopsis UniGene Build number 41 (as extracted from http://www.ncbi.nlm.nih.gov/UniGene in November 2003).
Comparison of Expression Profiling Platforms
For MPSS we extracted signature sequences from the LEF and LES libraries, both were derived from 21-d-old leaf material (http://mpss.udel.edu/at/java.html). We only considered MPSS tags of reliable sequence quality (T) that were present at significant levels (>3 tpm) and had only a single hit to the genome. The sense tags matched to annotated Arabidopsis genes were identified by the classes 1 (inside annotated gene), 2 (within 500-bp 3′ of annotated gene), or 7 (within 17 bp of an exon boundary; spliced). The anti-sense tags matched to annotated genes were identified by classes 3 (anti-sense to annotated gene) or 6 (within intron, anti-sense strand).
For global microarray analysis we obtained the expression data described in Yamada et al. (2003) from the NCBI Gene Expression Omnibus site (http://www.ncbi.nlm.nih.gov/geo/). We extracted the expression data generated using 7-d-old leaf tissue denoted by the codes GSM9208-GSM9219 for the whole genome array analysis from both the sense (GSE636) and anti-sense (GSE637) strands.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number GSM30396.
Supplementary Material
Acknowledgments
We thank Diana Bekkaoui and Lian Hao for technical assistance, Dr. Larry Pelcher at NRC-Plant Biotechnology Institute, Saskatoon for use of ELVIS, and Drs. Hossein Borhan, Dwayne Hegedus, and Andrew Sharpe, all at Saskatoon Research Centre, for critical reading of the manuscript.
This work was supported in part by the Genome Prairie project Functional Genomics of Abiotic Stress in Crop Plants and in part by the Agriculture and Agri-Food Canada Canadian Crop Genomics Initiative.
The online version of this article contains Web-only data.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630–634 [DOI] [PubMed] [Google Scholar]
- Carter MJ, Milton ID (1993) An inexpensive and simple method for DNA purifications on silica particles. Nucleic Acids Res 21: 1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ, Vanzo NF, Raynal LC (1999) mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet 15: 24–28 [DOI] [PubMed] [Google Scholar]
- Cock JM, Swarup R, Dumas C (1997) Natural antisense transcripts of the S locus receptor kinase gene and related sequences in Brassica oleracea. Mol Gen Genet 255: 514–524 [DOI] [PubMed] [Google Scholar]
- Dolfini S, Consonni G, Mereghetti M, Tonelli C (1993) Antiparallel expression of the sense and antisense transcripts of maize alpha-tubulin genes. Mol Gen Genet 241: 161–169 [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al (1999) The DNA sequence of human chromosome 22. Nature 402: 489–495 [DOI] [PubMed] [Google Scholar]
- Ekman DR, Lorenz WW, Przybyla AE, Wolfe NL, Dean JF (2003) SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol 133: 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Bureau TE (2001) A novel hybrid open reading frame formed by multiple cellular gene transductions by a plant long terminal repeat retroelement. J Biol Chem 276: 41963–41968 [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Fizames C, Munos S, Cazettes C, Nacry P, Boucherez J, Gaymard F, Piquemal D, Delorme V, Commes T, Doumas P, et al (2004) The Arabidopsis root transcriptome by serial analysis of gene expression. Gene identification using the genome sequence. Plant Physiol 134: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings JG, Cook BP, Dufault MR, Madden SL, Khuri S, Turnbull CJ, Dunwell JM (2003) Global transcript analysis of rice leaf and seed using SAGE technology. Plant Biotechnol J 1: 271–285 [DOI] [PubMed] [Google Scholar]
- Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, Coughlin SM, Zuyderduyn SD, Jones SJ, Marra MA (2003) A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol 13: 358–363 [DOI] [PubMed] [Google Scholar]
- Hansen NJ, Kristensen P, Lykke J, Mortensen KK, Clark BF (1995) A fast, economical and efficient method for DNA purification by use of a homemade bead column. Biochem Mol Biol Int 3: 461–465 [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al (2003) Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes R, Kudla J, Gruissem W (1999) Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem Sci 24: 199–202 [DOI] [PubMed] [Google Scholar]
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al (1996) Generation and analysis of 280,000 human expressed sequence tags. Genome Res 6: 807–828 [DOI] [PubMed] [Google Scholar]
- Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA (2001) Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res 8: 1346–1352 [DOI] [PubMed] [Google Scholar]
- Jung SH, Lee JY, Lee DH (2003) Use of SAGE technology to reveal changes in gene expression in Arabidopsis leaves undergoing cold stress. Plant Mol Biol 52: 553–567 [DOI] [PubMed] [Google Scholar]
- Lash AE, Tolstoshev CM, Wagner L, Schuler GD, Strausberg RL, Riggins GJ, Altschul SF (2000) SAGEmap: a public gene expression resource. Genome Res 10: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Lee DH (2003) Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiol 132: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz WW, Dean JF (2002) SAGE profiling and demonstration of differential gene expression along the axial developmental gradient of lignifying xylem in loblolly pine (Pinus taeda). Tree Physiol 5: 301–310 [DOI] [PubMed] [Google Scholar]
- MacIntosh GC, Wilkerson C, Green PJ (2001) Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol 127: 765–776 [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Terauchi R (1999) Transcript profiling in rice (Oryza sativa L.) seedlings using serial analysis of gene expression (SAGE). Plant J 20: 719–726 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD (2004) Arabidopsis MPSS. An online resource for quantitative expression analysis. Plant Physiol 135: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen T, Rowley JD, Wang SM (2002) Oligo(dT) primer generates a high frequency of truncated cDNAs through internal Poly(A) priming during reverse transcription. Proc Natl Acad Sci USA 99: 6152–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen MA, Schechter LE (1999) Cloning, mRNA localization and evolutionary conservation of a human 5-HT7 receptor pseudogene. Gene 227: 63–69 [DOI] [PubMed] [Google Scholar]
- Patankar S, Munasinghe A, Shoaibi A, Cummings LM, Wirth DF (2001) Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol Biol Cell 12: 3114–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Allen JF, Oelmuller R (2001) Principles of redox control in photosynthesis gene expression. Physiol Plant 112: 1–9 [Google Scholar]
- Pfannschmidt T (2003) Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci 8: 33–41 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Marra MA, Jones SJ (2003) Assessment of SAGE in transcript identification. Genome Res 13: 1203–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE (2002) Using the transcriptome to annotate the genome. Nat Biotechnol 20: 508–512 [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [DOI] [PubMed] [Google Scholar]
- Schuster G, Lisitsky I, Klaff P (1999) Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol 120: 937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787 [DOI] [PubMed] [Google Scholar]
- Vanhee-Brossollet C, Vaquero C (1998) Do natural antisense transcripts make sense in eukaryotes? Gene 211: 1–9 [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Voglestein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270: 484–487 [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Voglestein J, Basrai MA, Bassett DE, Hieter P, Voglestein J, Kinzler KW (1997) Characterisation of the yeast transcriptome. Cell 88: 243–251 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Voglestein B, Kinzler KW (1997) Gene expression profiles in normal and cancer cells. Science 276: 1268–1272 [DOI] [PubMed] [Google Scholar]
- Zhu W, Schlueter SD, Brendel V (2003) Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol 132: 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.