Abstract
Red clover (Trifolium pratense) leaves contain high levels of polyphenol oxidase (PPO) activity and o-diphenol substrates. Wounding of leaves during harvest and ensiling results in browning of leaf tissues from activity of PPO on the o-diphenols. In association with browning, leaf proteins remain undegraded during ensiling, presumably due to PPO-generated o-quinone inhibition of leaf proteases. We cloned three red clover PPO cDNAs, PPO1, PPO2, and PPO3, from a leaf cDNA library. Sequence comparisons among the three red clover PPO clones indicated they are 87% to 90% identical at the nucleotide level (80%–83% amino acid identity). All three encode proteins predicted to localize to the chloroplast thylakoid lumen. RNA-blotting and immunoblotting experiments indicated PPO1 is expressed primarily in young leaves, PPO2 in flowers and petioles, and PPO3 in leaves and possibly flowers. We expressed mature PPO1 in Escherichia coli. A portion of the expressed protein was soluble and functional in an assay for PPO activity. We also expressed the red clover PPO cDNAs under the control of a constitutive promoter in alfalfa (Medicago sativa). The expressed red clover PPO proteins were active in alfalfa extracts as evidenced by o-diphenol-dependant extract browning and quantitative assays of PPO activity. Proteolysis in leaf extracts of alfalfa expressing red clover PPO1 was dramatically reduced in the presence of an o-diphenol compared to controls. Transgenic alfalfa expressing red clover PPO should prove an excellent model system to further characterize the red clover PPO enzymes and PPO-mediated inhibition of postharvest proteolysis in forage plants.
Ensiling crops is a popular method of preserving forage for animal feed, especially in the humid northern regions of the United States. During harvesting and early stages of ensiling, plant membranes are ruptured, releasing proteolytic enzymes that rapidly degrade available substrates. Once fermentation by lactobacteria has progressed sufficiently to lower silage pH to less than about 5, proteolytic activity slows significantly. The wider the window between harvest and reaching a low pH, the more extensive is the proteolytic degradation. Unfortunately, excessive proteolysis in ensiled forages results in economic losses to farmers (Rotz et al., 1993) amounting to approximately $100 million annually. This economic loss is due to purchasing additional protein to supplement diets because of poor utilization of the nonprotein nitrogen products of proteolysis (ammonia, amino acids, and small peptides) by ruminant animals. Ruminants excrete much of this nonprotein nitrogen as urea, resulting in increasing nitrogen burdens upon the environment.
For some ensiled forages, such as alfalfa (Medicago sativa), proteolytic losses are especially high with degradation of 44% to 87% of the forage protein (Papadopoulos and McKersie, 1983; Muck, 1987). By contrast, red clover (Trifolium pratense), a forage with protein content similar to alfalfa, has up to 90% less proteolysis than alfalfa during ensiling (Papadopoulos and McKersie, 1983). Experimental evidence indicates red clover and alfalfa do not differ significantly in inherent proteolytic activity (Jones et al., 1995a). Several observations suggest that the lower extent of postharvest proteolysis in red clover is related to the presence of a soluble polyphenol oxidase (PPO; EC 1.14.18.1 or EC 1.10.3.1) and o-diphenol PPO substrates in its leaves (Jones et al., 1995b, 1995c). Alfalfa leaves and stems, on the other hand, have relatively little if any PPO activity or o-diphenol substrates (Jones et al., 1995b; M. Sullivan and R. Hatfield, unpublished data). Because PPO acts to catalyze the oxidation of o-diphenols to o-quinones, a possible mechanism whereby PPO acts to inhibit postharvest proteolysis could be via PPO-generated o-quinones binding directly to and inactivating endogenous proteases.
PPOs appear to be nearly ubiquitous among plants (Sherman et al., 1995), but the many possible roles they play have not been totally resolved. Oxygen-dependent formation of o-quinones that covalently couple to a number of cellular nucleophiles and consequent secondary quinone reactions lead to the formation of brown and black pigmented polymers (the browning reaction) that are well known. Such reactions are responsible for considerable postharvest losses in fruits and vegetables (Vamos-Vigyazo, 1981). PPO activity is often thought of as a defense mechanism based on the appearance of PPO reaction products upon wounding or pathogenesis and the inducible nature of PPO upon wounding (Mayer and Harel, 1979; Constabel et al., 1995; Thipyapong et al., 1995). Thipyapong and Steffens (1997) showed that antisense down-regulation of constitutive and inducible PPO activity results in a hypersusceptibility to pathogens in tomato (Lycopersicon esculentum). Overexpression of PPO in tomato results in enhanced resistance to bacterial disease (Li and Steffens, 2002). These results strengthen the assertion of a defense role for the enzyme. Although there is speculation as to the mode of action involved in such defensive roles, the actual mechanisms have not been elucidated. Recently, two specific PPO enzymes have been shown to participate in biosynthetic pathways: Nakayama et al. (2001) demonstrated that in snapdragon (Antirrhinum majus), a PPO enzyme is involved in the biosynthesis of yellow floral pigments (aurones), and Cho et al. (2003) have shown a role for a PPO in biosyntheses of 8 to 8′ linked lignans in creosote bush (Larrea tridentate). Because nearly all characterized PPOs appear to be localized to the chloroplast thylakoid lumen, it has been proposed that PPO may also play an indispensable role in chloroplast function with a possible involvement in a Mehler-like reaction detoxifying oxygen species (Sherman et al., 1995). However, our BLAST searches of the Arabidopsis (ecotype Columbia) genome revealed no apparent genes encoding PPO, challenging an indispensable role in chloroplast function.
In an effort to understand the role of PPO in inhibition of postharvest proteolysis, we have been using both biochemical and molecular approaches. To develop a clear picture of all the components and interactions in the observed proteolytic inhibition in red clover, we have sought to develop a model system that allows component roles to be defined. The ideal system would be a plant similar to red clover but without endogenous PPO activity or o-diphenol substrates. Components from red clover, such as PPO activity or o-diphenol substrates, could then be added back to the system. Isolation of PPO genes from red clover would facilitate development of such a model system. Here, we report the isolation of three red clover PPO cDNAs, characterization of their expression patterns, and preliminary characterization of the activities of the encoded enzymes in a plant model system, alfalfa.
RESULTS
Cloning of Red Clover PPO Gene Sequences
In preliminary Southern-blotting experiments using a PPO cDNA from potato (Solanum tuberosum) as a probe (Hunt et al., 1993), we failed to detect hybridization to red clover genomic DNA (data not shown), suggesting red clover PPO genes might not have a sufficiently high degree of sequence similarity to allow use of this heterologous probe for library screening. Consequently, we designed degenerate oligonucleotides corresponding to conserved sequences from several cloned plant PPO genes (see Supplemental Fig. 1, available at www.plantphysiol.org). Oligonucleotide pairs were used in reverse transcription (RT)-PCR reactions with red clover leaf mRNA, and the resulting DNA fragments were cloned and sequenced. Reactions using primers PPO-1 and PPO-3 yielded an approximately 190-bp DNA fragment with a high degree of sequence similarity to previously characterized PPO genes of many plant species. When used alone, the PPO-1 primer amplified a 300-bp DNA fragment, apparently the result of mispriming of the PPO-1 primer on the noncoding DNA strand, that also had a high degree of sequence similarity to several previously cloned PPO genes (see Supplemental Fig. 1). This cloned 300-bp fragment (plasmid designation pPPO-ST2) was ultimately used to screen a red clover leaf cDNA library.
The 300-bp insert of pPPO-ST2 hybridized to three to five bands in a Southern blot of red clover genomic DNA, suggesting the presence of three to five different PPO genes or alleles in the red clover clone used in these experiments (data not shown). Because of the presence of multiple PPO genes or alleles, we decided to construct and screen a red clover leaf cDNA library to obtain full-length PPO clones instead of using a PCR-based approach. Approximately 106 phage from an unamplified library were screened by hybridization under moderate stringency with the 300-bp pPPO-ST2 insert. Following this primary screen, 108 putative PPO clones were picked. Of these, 50 were rescreened by hybridization to the pPPO-ST2 probe, resulting in identification of 23 clones containing hybridizing inserts. Restriction mapping with several enzymes and limited sequence analysis allowed the 23 clones to be divided into three families. The longest member of each family was sequenced on both strands (see Supplemental Fig. 1). We refer to these three cDNA clones as PPO1, PPO2, and PPO3 (GenBank Accession nos. AY017302, AY017303, and AY017304, respectively). PPO1 clones were recovered most frequently (nine full-length clones), whereas PPO2 and PPO3 clones were rarer (two full-length clones each).
Sequence comparison among the three red clover PPO genes indicated their coding regions are 87% to 90% identical at the nucleotide level. Besides base substitutions, there are several small insertions/deletions in the genes relative to each other ranging in size from 3 to 27 bp (see Supplemental Fig. 1). Nucleotide-nucleotide BLAST (BLASTn) searches indicated the clover PPO sequences are unique, although they exhibit sequence similarity to previously cloned plant PPO genes, including those of Vicia faba (Cary et al., 1992), tomato (Newman et al., 1993a), and potato (Hunt et al., 1993). The cloned PPO2 cDNA included 15 bp of 5′ untranslated region (UTR) that included an in-frame stop codon indicating that the ATG at position 16 is used to initiate translation. Although the PPO1 and PPO3 cDNAs have no 5′ UTR and 2 bp of 5′ UTR, respectively, the first ATG of each likely represents the site of translational initiation based on comparison to red clover PPO2 and other cloned plant PPO genes. All three cDNAs are polyadenylated. Sequence analysis of the 3′ ends of the nine full-length PPO1 cDNA clones indicates multiple polyadenylation sites for this gene (data not shown).
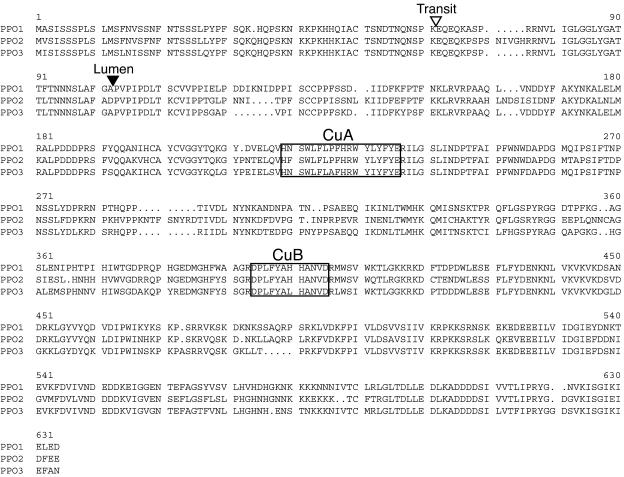
The red clover PPO1, PPO2, and PPO3 cDNAs are predicted to encode protein products of 605, 623, and 599 amino acids residues, respectively (68–71 kD; Fig. 1). The red clover PPO proteins are 76% to 80% identical (80%–83% similar) to each other. BLAST (tBLASTn) searches of the National Center for Biotechnology Information database with the peptide sequences revealed that the most closely related PPOs sequenced to date are those of the legumes V. faba (Cary et al., 1992) and alfalfa (M. Sullivan and R. Hatfield, unpublished data) with 60% to 65% amino acid identity (68%–72% similarity) to the red clover enzymes. Since most other plant PPO enzymes are targeted to the chloroplast thylakoid lumen, we analyzed the red clover proteins for targeting information. The ChloroP algorithm (Emanuelsson et al., 1999) predicts an approximately 60-amino acid chloroplast transit signal for all three red clover PPO proteins (Fig. 1). Red clover PPO protein sequences with the predicted transit peptide removed were further analyzed with the SignalP algorithm (Nielsen et al., 1997) to determine the likelihood of targeting to the thylakoid lumen and identify a potential signal cleavage site (Peltier et al., 2000). The program identified a signal sequence and likely cleavage site for all three red clover PPO gene products (Fig. 1). If the red clover proteins were targeted to the chloroplast thylakoid lumen as predicted, the mature forms of the proteins would be 511, 522, and 504 amino acid residues (or 58.7 kD, 60.3 kD, and 57.3 kD) for the mature products of PPO1, PPO2, and PPO3, respectively. The CuA and CuB copper-binding sites characteristic of tyrosinases and polyphenol oxidases (Steffens et al., 1994) are present in the red clover PPOs (Fig. 1).
Figure 1.
Alignment of predicted amino acid sequences of red clover PPO1, PPO2, and PPO3. Gaps are represented by periods; regions corresponding to conserved CuA and CuB copper-binding sites are boxed; and cleavage sites for the chloroplast transit and thylakoid lumen targeting signals are shown with open and solid arrowheads, respectively. Alignments were created using programs of the Wisconsin Package (Accelrys), and targeting sequences were predicted using ChloroP and SignalP algorithms (Nielsen et al., 1997; Emanuelsson et al., 1999; Peltier et al., 2000).
Red Clover PPO1, PPO2, and PPO3 Are Differentially Expressed
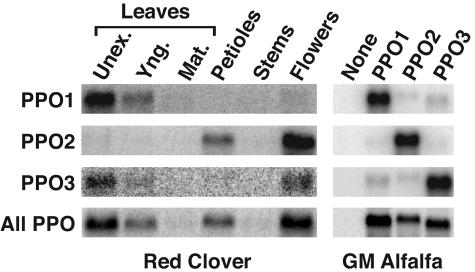
The expression patterns of red clover PPO1, PPO2, and PPO3 were examined by RNA blotting and hybridization using gene-specific probes. Hybridization to blots containing RNA from unexpanded, young, and mature leaves; petioles; stems; and flowers was carried out using probes corresponding to the entire coding regions of the three PPO cDNAs, and blots were washed under relatively high stringency (0.2× SSC, 62°C). Blots containing RNA from transgenic (genetically modified [GM]) alfalfa expressing the individual red clover PPO genes (described in detail below) served as a control of probe specificity. Under the hybridization and wash conditions used, the PPO1, PPO2, and PPO3 probes were quite specific as assessed by the transgenic alfalfa controls (Fig. 2). Given the similar base composition of the PPO coding regions, that the probes were made at the same time using the same amount of template DNA, and that hybridizations were carried out simultaneously, it is reasonable to assume that the probes had similar specific activities and hybridized to their corresponding mRNAs with similar efficiencies. By quantifying the radioactive signals of the transgenic alfalfa control blots, we determined that each transcript was recognized by its corresponding gene-specific probe 7- to 15-fold more efficiently than by the other PPO probes. None of the probes recognized any transcripts in leaves of alfalfa transformed with the vector only. In red clover, all three PPO genes appear to be expressed to some extent in unexpanded leaves (Fig. 2). With the likelihood that the gene-specific probes had similar specific activities and hybridization efficiencies to their corresponding mRNAs, quantitation of radioactive signals on the blots indicate PPO1 is the predominant PPO expressed in unexpanded leaves, with message levels approximately 6-fold higher than those of PPO2 and PPO3. Additionally, PPO1 is expressed in fully expanded young leaves and to a much lesser extent in mature leaves. Besides the low level expression in unexpanded leaves, PPO2 is expressed to a higher degree in petioles and shows the highest level of expression in flowers. The level of expression of PPO2 in flowers is approximately 20-fold higher than that of PPO1 or PPO3. Although PPO1 and PPO3 probes hybridized to mRNA from red clover flowers, this signal could represent cross-hybridization with the relatively abundant PPO2 mRNA. Red clover and control alfalfa RNA blots identical to those described above were also probed with the PPO1 coding region, but hybridization and washes were under lower stringency conditions to allow detection of related red clover PPO transcripts. Under the hybridization and wash conditions used, red clover PPO transcripts were readily detected in RNA from control alfalfa plants expressing PPO1, PPO2, or PPO3. No PPO transcript was detected in alfalfa lacking a PPO transgene. For red clover tissues, PPO transcripts were apparent in unexpanded and young leaves, petioles, and flowers in a pattern consistent with the results of the gene-specific hybridizations. Additionally, very faint signals were detected for RNA from mature leaves and stems.
Figure 2.
Red Clover PPO1, PPO2, and PPO3 are differentially expressed. Total RNA isolated from unexpanded (Unex.), young (Yng.), and mature (Mat.) leaves; mature petioles; stems; and flowers of red clover was used to make multiple RNA blots. Total RNA from GM alfalfa expressing the various red clover PPO genes as indicated or transformed with pILTAB357 only (None) was used to make control blots for probe specificity. The blots were hybridized to 32P-labeled probes corresponding to the entire red clover PPO1-, PPO2-, or PPO3-coding regions under high-stringency conditions as indicated or to the entire red clover PPO1-coding region under low-stringency conditions (All PPO). The resulting blots were imaged using a Phosphoimager. Grayscale ranges for blots were selected to optimize signal-to-noise ratio for each blot.
Expression of Red Clover PPO1 in Escherichia coli
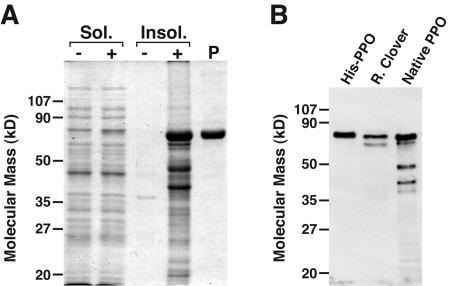
We expressed the red clover PPO1 gene in E. coli, both for a potential source of active PPO enzyme for biochemical studies and to serve as antigen for the production of anti-PPO antiserum. Since our analysis of the PPO protein sequences indicated they are chloroplast thylakoid lumen proteins, we designed pET-based expression constructs that would produce the predicted mature form of PPO1 (i.e. lacking the predicted chloroplast and thylakoid lumen targeting signals; Fig. 1), in both 6×His-tagged and native versions. When expressed in BL21(DE3) RIL cells, 85% to 90% of the expressed protein from either the His-tagged or native construct was insoluble and accumulated in inclusion bodies (Fig. 3; data not shown). For the insoluble protein, both the tagged and native versions migrate with apparent molecular masses of approximately 65 kD on SDS-PAGE. As expected, the tagged version is slightly larger than the native version. The estimated molecular masses are larger than the predicted mature molecular mass of the PPO1 gene product (59 kD) but within the limits of the technique. Attempts to purify the insoluble 6×His-tagged protein by immobilized metal affinity chromatography were ineffective because the protein could not be eluted from the Ni-nitrilotriacetic acid resin. Consequently, milligram quantities of the 6×His-tagged protein were purified by preparative SDS-PAGE (Fig. 3) with an approximate yield of 5 mg protein per liter of culture, and the resulting protein was used for antibody production. In immunoblotting experiments, the resulting anti-PPO1 antiserum recognized two bands in red clover leaf extracts (Fig. 3). The larger of the two bands is, as expected, slightly smaller than the 6×His tagged PPO but comigrates with the native (i.e. nontagged), mature version of PPO expressed in E. coli. The slightly smaller band recognized by the anti-PPO1 antiserum in the red clover leaf extract is likely the PPO3 gene product (discussed in more detail below).
Figure 3.
Expression of red clover PPO1 in E. coli and generation of anti-red clover PPO1 antiserum. A, Stained SDS-PAGE gel of portions of the soluble (Sol., 5 μg [equivalent to approximately 50 μL induced culture]) and insoluble (Insol., equivalent to approximately 100 μL of induced culture) fractions from E. coli expressing His-tagged red clover PPO1 (+). Comparable fractions from E. coli transformed with the empty expression vector served as a negative control (−). A sample of the insoluble His-tagged red clover PPO1 protein purified by preparative SDS-PAGE and used to immunize a rabbit (P, approximately 2 μg) was also run on the gel. B, Immunoblot of E. coli and red clover extracts. Purified protein used to prepare the anti-PPO1 antiserum (His-PPO, 50 ng), red clover leaf extract (R. Clover, 10 μg), or insoluble fraction from E. coli expressing a PPO corresponding to the predicted native (i.e. nontagged), mature form (Native PPO, approximately 50 ng) were used to make an immunoblot, and PPO was detected using anti-red clover PPO1 antiserum as described in “Materials and Methods.”
Immunoblotting of soluble fractions from E. coli expressing the native PPO1 protein indicate 10% to 15% of the PPO is soluble with migration indistinguishable from that present in the insoluble fraction (data not shown). Further, compared to extracts of E. coli containing the pET28a vector only, extracts of E. coli expressing PPO1 contain low but significant and reproducible levels of PPO activity as detected by a 5,5′-dithio-bis-(2-nitrobenzoic acid) quinone trap assay (Esterbauer et al., 1977) using a caffeic acid substrate (up to 6.7 × 10−1 versus 4.0 × 10−3 nkat mg−1 protein for PPO1-expressing and control E. coli extracts, respectively). This result indicates that at least some fraction of PPO produced in E. coli is correctly folded to produce soluble active protein.
Expression of Red Clover PPO Genes in Alfalfa
Because the red clover PPOs are predicted to be targeted to the chloroplast thylakoid lumen, expression in a plant and targeting to a plastid may be required for optimal folding and activity. The entire coding regions, including chloroplast-targeting signals from PPO1, PPO2, and PPO3, were inserted behind the cassava vein mosaic virus promoter (Verdaguer et al., 1996) in the pILTAB357 transformation vector, and the resulting constructs were transformed into alfalfa. For each red clover PPO cDNA, independent alfalfa transformants containing the nptII selectable marker gene were identified (Table I). Protein extracts were made from leaves of the transgenic alfalfa plants and used immediately in quantitative assays of PPO activity using caffeic acid as a substrate. Extracts of leaves of red clover or alfalfa transformed with the pILTAB357 vector only served as positive and negative controls, respectively. Most of the tested plants had PPO activity detectible above the range of background values seen for the control alfalfa plant (Table I). In the case of PPO1-alfalfa plants, the median level of activity was similar to the lower range of PPO activity measured for red clover. Pairwise comparison of measured activities between the three groups of transgenic plants (i.e. those expressing PPO1, PPO2, or PPO3) by the Mann-Whitney test revealed they were significantly different (P < 0.01). Two to three plants having the highest activity from each group were used in the remaining analyses.
Table I.
Enzyme activity in transgenic alfalfa expressing red clover PPO cDNAs
| Tissue Source | No. of Plants PPO Positivea/Total | PPO Activity
|
|
|---|---|---|---|
| Range | Median ± MADb | ||
| nkat mg−1 | |||
| PPO1-alfalfa | 4/4 | 1.45–4.90 | 3.57 ± 1.18 |
| PPO2-alfalfa | 21/27 | 0–0.38 | 0.10 ± 0.07 |
| PPO3-alfalfa | 12/13 | 0.02–4.18 | 0.67 ± 0.53 |
| Red cloverc | – | 6.28–72.81 | 41.41 ± 27.47 |
| Control alfalfac | – | 0–0.02 | 0.02 ± 0.00 |
PPO-positive plants are defined as those having PPO activities above the measured range for control alfalfa.
Median absolute deviation.
Values represent multiple measurements made on a single genotype (red clover) or transformant (control alfalfa). Average PPO activity ±se measured was 34.54 ± 11.96 and 0.02 ± 0.00 nkat mg−1 for red clover and control alfalfa, respectively.
Interestingly, red clover PPO activity was not only stable in alfalfa extracts; the measured activity increased upon a 2- to 7-d incubation at 30°C, with 5- to 10-fold increases for PPO1 and PPO2 and 40- to 50-fold increases for PPO3. No activity increases were detected in the vector-only control alfalfa, and red clover exhibited a total loss in activity upon extended 30°C incubation.
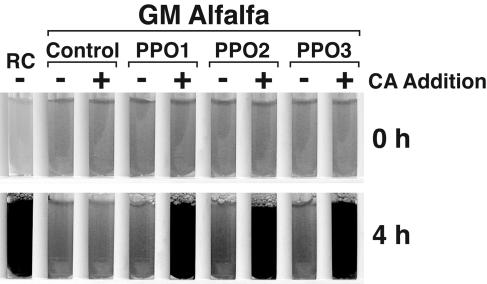
We also assessed whether the red clover PPO gene products expressed in alfalfa could cause extract browning, a hallmark of PPO activity (Fig. 4). When an o-diphenol substrate, caffeic acid, was added to extracts of alfalfa expressing the red clover PPO transgenes, browning was apparent within 2 to 15 min. Not surprisingly, extracts of alfalfa expressing PPO1 and PPO3 showed the most rapid browning, presumably because of the higher levels of PPO activity toward caffeic acid present in these plants. No browning was seen without the exogenously added substrate, and no browning occurred in extracts of alfalfa transformed with the pILTAB357 vector alone, even after a 24-h incubation in the presence of caffeic acid.
Figure 4.
Expression of active PPO in transgenic alfalfa. Leaf extracts of alfalfa transformed with red clover PPO1, PPO2, or PPO3 were incubated at room temperature with (+) or without (−) exogenously added PPO substrate (caffeic acid [CA] 3 mm final concentration). Leaf extracts of red clover (RC) and alfalfa transformed with the pILTAB357 vector only (Control) served as positive and negative controls, respectively. Browning was apparent within 2 to 5 min for GM alfalfa expressing red clover PPO genes. The control alfalfa extract showed no apparent browning, even after 24 h.
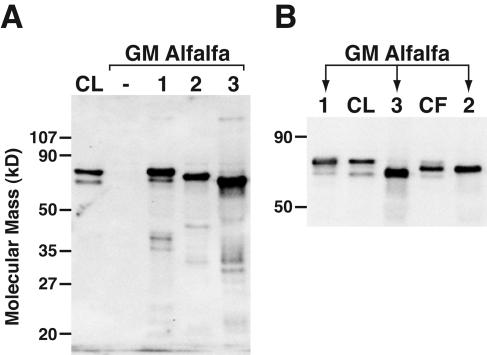
Leaf extracts of red clover or transgenic PPO alfalfa were also used in immunoblotting experiments with anti-PPO1 antiserum (Fig. 5). The antiserum failed to detect any protein in leaves of alfalfa transformed with pILTAB357 only. Extracts of alfalfa expressing red clover PPO1, PPO2, or PPO3, however, each showed a major band that was recognized with anti-PPO1 antiserum. The proteins had slightly different and distinguishable migrations on SDS-PAGE that were not entirely consistent with the molecular masses of the predicted mature form of each of the PPO proteins (e.g. although PPO2 protein is predicted to be the largest of the three red clover PPO proteins, it has an intermediate migration on SDS-PAGE). Immunoblotting of leaf extracts of additional independent transgenic lines expressing each of the red clover PPOs showed the same pattern of differential migration. Since the protein extractions were carried out on ice in the presence of a protease inhibitor cocktail, it seemed unlikely that the unexpected migrations were due to extensive in vitro proteolysis. Additionally, given the slightly anomalous migration of the PPO1 protein derived from E. coli, the anomalous migration of alfalfa-derived proteins may not be surprising. The PPO1 and PPO3 proteins derived from GM alfalfa comigrated with the two major cross-reacting protein bands detected in red clover leaves. This was especially evident when the samples were run side by side and the SDS-PAGE gel was run to optimize resolution in this size range (Fig. 5). It is not clear if a faint cross-reacting band detected in red clover leaves corresponded to the PPO2 gene product. A prominent protein band was detected in clover flowers, however, that comigrated with the PPO2 gene product expressed in alfalfa. The protein expression patterns revealed by immunoblotting are consistent with the results of the RNA-blotting experiments.
Figure 5.
Red clover PPO1, PPO2, and PPO3 gene products are distinguishable by immunoblotting and show tissue-specific expression. A, An immunoblot of 10 μg leaf extract from red clover (CL) and GM alfalfa transformed with vector only (−), or red clover PPO1, PPO2, or PPO3 as indicated was prepared and PPO was detected with anti-red clover PPO1 antiserum as described in “Materials and Methods.” B, A higher resolution immunoblot including an extract of red clover flowers (CF, approximately 10 μg).
Postharvest Proteolysis Is Inhibited in Alfalfa Expressing Red Clover PPO1
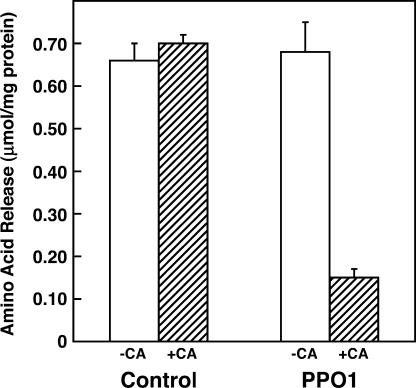
To determine if red clover PPO and an o-diphenol are capable of inhibiting postharvest proteolysis in alfalfa, we assessed the extent of proteolysis in extracts of leaves of control alfalfa (transformed with the pILTAB357 vector only) or alfalfa expressing red clover PPO1 (PPO1-alfalfa) in the presence or absence of the o-diphenol caffeic acid. Proteolysis was measured as the release of free amino acids following a 4-h incubation at 37°C. For extracts of control alfalfa, caffeic acid had no effect on amino acid release. For extracts of PPO1-alfalfa, however, an approximately 80% caffeic acid-dependent decrease in amino acid release was observed (Fig. 6). Without caffeic acid, amino acid release in the PPO1-alfalfa extract was indistinguishable from that of the control extract.
Figure 6.
PPO inhibits postharvest proteolysis in an o-diphenol-dependent manner. Amino acid release during a 4-h incubation at 37°C was used to measure proteolysis in extracts of control or PPO1-expressing alfalfa as indicated in the presence (+CA) or absence (−CA) of 3 mm caffeic acid. Amino acid release is expressed as μmol mg−1 extract protein, and values are the average of three experiments ±se.
DISCUSSION
Understanding the basic biochemistry underlying postharvest proteolytic inhibition in red clover is critical to developing strategies that would improve ensiling methods to minimize true protein loss. Such improvements would lead to better utilization of silage nitrogen by ruminant animals and consequently decrease nitrogen excretion into the environment. As a first step in advancing our understanding of postharvest proteolytic inhibition, we have isolated PPO genes from red clover.
Screening a leaf cDNA library made from a single red clover genotype with high levels of PPO, we isolated cDNAs corresponding to three unique PPO genes. Given that the clover clone used is diploid, the three genes identified must represent at least two genetic loci. Although there are small insertions and deletions relative to each other, the coding regions of the three genes show a relatively high degree of sequence identity (87%–90%). This level of sequence identity falls within the upper range of sequence identity seen for the seven-member PPO gene family of tomato (70%–90%; Newman et al., 1993a). The 3′ UTRs of PPO1 and PPO3 share 83% sequence identity, while that of PPO2 shares only limited sequence identity with PPO1 and PPO3 (approximately 50%), suggesting that PPO1 and PPO3 are more closely related to each other than to PPO2. It is not clear whether PPO1 and PPO3 represent different alleles of the same genetic locus, however.
The red clover PPO genes encode proteins with characteristics similar to many other PPO proteins, including conserved copper-binding motifs and predicted amino-terminal signals to target the mature enzyme to the chloroplast thylakoid lumen. We used ChloroP and SignalP algorithms to predict the cleavage sites of the chloroplast transit and thylakoid lumen targeting signals (Fig. 1; Nielsen et al., 1997; Emanuelsson et al., 1999; Peltier et al., 2000). Two lines of evidence suggest that the predicted cleavage is accurate. First, the predicted cleavage corresponds well to the experimentally determined amino termini of multiple PPO proteins purified from other plant species (Newman et al., 1993a; Gerdemann et al., 2001; Cho et al., 2003). Second, expression of the SignalP-predicted mature, thylakoid lumen-targeted form of PPO1 in E. coli produces a protein that comigrates on SDS-PAGE with PPO protein present in the leaves of red clover.
The red clover PPO genes show differential expression based on both RNA-blotting experiments with gene-specific probes (Fig. 2) and on immunoblot experiments, which take advantage of the differential migration on SDS-PAGE of the red clover PPO gene products (Fig. 5). Although all three PPO genes must be expressed to some degree in young leaves (since the cDNAs were isolated from a young leaf cDNA library), PPO1 is the predominant PPO in this tissue based on its more frequent recovery from the screen of an unamplified library and the results of the RNA-blotting experiments (Fig. 2). Additionally, preliminary experiments indicate the red clover PPO gene products have differing substrate specificities and that the substrate specificity of PPO1 most closely parallels that seen for extracts of young red clover leaves (Sullivan et al., 2004). The differences in expression patterns and substrate specificity suggest that the PPO1, PPO2, and PPO3 gene products have different functional roles in vivo. Given its high level of expression and activity in leaves, it is attractive to speculate that the PPO1 gene product plays a defensive role. Despite a decline in PPO1 transcript levels with leaf maturity, PPO activity in mature red clover leaves remains relatively high and is adequate to promote rapid browning upon tissue disruption (M. Sullivan and R. Hatfield, unpublished data), indicating the PPO1 protein might be quite stable. The high level of floral expression of PPO2 might indicate a biosynthetic role analogous to that of the PPO involved in aurone synthesis in snapdragon (Nakayama et al., 2001). PPO3 and PPO1 have similar expression patterns, although PPO3 is expressed at a lower level and seems to differ from PPO1 with respect to substrate specificity (Sullivan et al., 2004). PPO3 may be allelic with PPO1 and/or share overlapping functions with PPO1.
We detected low but reproducible amounts of PPO activity in extracts of E. coli expressing PPO1, indicating that at least some fraction of the protein is soluble and enzymatically active. We have not found other reports of active PPO being expressed in E. coli. Our result suggests that modifications of this bacterial expression system could result in greater yields of soluble, active PPO protein. The crystal structure of a PPO enzyme from sweet potato (Ipomoea batatas) indicates the presence of two disulfide bridges in the protein (Klabunde et al., 1998). The Cys residues forming these bridges are highly conserved among several plant PPOs, suggesting they may be important in proper folding of the enzyme. If so, bacterial expression under conditions that favor disulfide bond formation may substantially increase the yield of active PPO produced, which would provide a convenient means for carrying out structure/function analyses on PPO proteins.
We also expressed the red clover PPO cDNAs in alfalfa, where targeting of the expressed protein to the chloroplast thylakoid lumen should facilitate proper folding and production of soluble active protein. Despite some reports of endogenous PPO expression in alfalfa (Jiang and Miles, 1993; Bellucci et al., 1999), we have detected little, if any, PPO activity in the leaves of the regenerable clone Regen-SY27 and little, if any, o-diphenol PPO substrates. This finding is consistent with a previous study that found little, if any, PPO activity and relatively low levels of soluble phenols in field-grown alfalfa (Pioneer 5432; Jones et al., 1995b). This lack of endogenous PPO and o-diphenol substrates continues to facilitate our analyses of the red clover PPOs expressed from transgenes in alfalfa. Expression of the red clover PPOs in alfalfa was easily detected at the RNA, protein, and enzyme activity levels. Pairwise comparison of measured PPO activity between groups of transgenic plants expressing PPO1, PPO2, or PPO3 by the Mann-Whitney test revealed significant differences between all three groups (P < 0.01). This difference could be due to differences in expression, either at the level of transcription or due to posttranscriptional events, or due to inherent differences in enzyme activity. Because of the way the expression constructs were made, any differences in expression would have to be due to differences in the coding regions of the PPO genes. Although mRNA and PPO protein levels were not measured for all the transgenic plants, for plants with the highest levels of PPO activity for each gene, these appear similar (Figs. 3 and 5), though their PPO activities differed substantially, at least between PPO2 and PPO1 or PPO3 (Table I). These findings are consistent with recent preliminary experiments indicating differences in PPO substrate specificity among the red clover enzymes (Sullivan et al., 2004).
When expressed in alfalfa, the red clover PPOs were highly stable, with PPO activity detectible for more than 2 weeks following incubation at 30°C. Crude extracts of red clover show rapid inactivation of PPO upon incubation, presumably due to inactivation of PPO by the o-quinones it forms, since removal of low-Mr compounds by gel filtration stabilizes the PPO activity (K. Frost and R. Hatfield, unpublished data). This rapid inactivation of PPO activity in red clover extracts may account for the variability seen in measured activity (Table I), although other factors, such as tissue maturity, environment, etc., could contribute to the variability as well. Surprisingly, PPO activity in alfalfa extracts actually increased 5- to 50-fold during extended (2- to 7-d) incubations at 30°C in the absence of added o-diphenol substrate. We have seen postincubation enzyme activities as high as 58.2, 4.33, and 55.5 nkat mg−1 protein for PPO1, PPO2, and PPO3 transgenic alfalfa extracts, respectively. For PPO1 and PPO3 transgenic alfalfa, this level of enzyme activity approximates that measured in red clover leaf extracts. The observed increase in activity for alfalfa-expressed PPO may be the release of latency that has been reported for several other PPOs when exposed to small amounts of denaturants or proteases (Steffens et al., 1994). The PPO activity in red clover extracts subjected to gel filtration (to prevent inactivation by o-quinones) shows only a modest (less than 2-fold) increase upon incubation (M. Sullivan, unpublished data), suggesting the enzyme is either already fully activated in red clover or that red clover lacks a component required for full activation in vitro.
Given the lack of endogenous PPO and PPO substrates and the similar levels of endogenous proteases compared to red clover, transgenic alfalfa expressing red clover PPO1 provided an excellent opportunity to examine the role PPO plays in inhibition of proteolysis during ensiling of red clover. Assessment of proteolysis in extracts of control and PPO1-expressing alfalfa demonstrated that PPO is capable of substantially inhibiting postharvest proteolysis in an o-diphenol-dependent manner. This finding indicates that PPO and o-diphenols likely are responsible for the low extent of proteolysis seen for ensiled red clover. Transgenic alfalfa should prove an excellent model to evaluate additional aspects of PPO-mediated proteolytic inhibition, including factors affecting the efficiency of inhibition and elucidation of the mechanism of inhibition.
MATERIALS AND METHODS
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Plant Materials and Transformation
A single red clover (Trifolium pratense) genotype was used in all experiments involving red clover. The clone (lab designation PPO) was selected from a population of WI-2 (lot C136) germplasm (Smith and Maxwell, 1980) as having relatively high levels of PPO in its leaves. For alfalfa (Medicago sativa) transformation, leaf explants from a highly regenerable clone of Regen-SY (Bingham, 1991) were cocultured with Agrobacterium tumefaciens containing each binary vector, and somatic embryos were produced essentially as described by Austin et al. (1995). After establishing plants in soil, putative transformants were initially screened for the presence of nptII by PCR as previously described (Saruul et al., 2002). Because of the out-crossing nature of alfalfa, all experiments were performed with primary transformants (T0) propagated from stem cuttings in vermiculite in a greenhouse. Both red clover and transformed alfalfa were maintained in a greenhouse year-round and fertilized weekly (Peter's soluble 20-20-20; Scotts, Marysville, OH). Supplemental lighting (13 h/d) was used during all but summer months.
RNA Methodologies
Total RNA was prepared from plant tissues using an RNeasy kit (Qiagen, Valencia, CA). Alternatively, total RNA was prepared by the method of Chang et al. (1993). mRNA was prepared from total RNA using the PolyATtract Kit (Promega, Madison, WI). Formaldehyde agarose gels were run and RNA blots prepared as previously described (Newman et al., 1993b) using 10 μg of each total RNA sample.
Generation of Red Clover PPO Gene Fragments by PCR and cDNA Library Construction and Screening
Degenerate oligonucleotide primers were designed corresponding to well-conserved regions of several plant PPOs, including spinach (Spinacia oleracea; GenBank accession nos. Z66559, U19270, and X90869), potato (Solanum tuberosum; M95196, M95197, U22922, U22921, and U22923), tomato (Lycopersicon esculentum; Z12837, Z12838, Z12834, Z12836, Z12833, and Z12835), Vicia faba (Z11702), grape (Vitis vinifera; Z27411), and pokeweed (Phytolacca americana; D45386). These primers included PPO-1 (5′-CAACAAGCTARKRTHCATTGTGCTT-3′) and PPO-3 (5′-ATCCCAATTCCARWAHGG-3′; see Supplemental Fig. 1). Primers and red clover leaf mRNA were used in RT-PCR reactions using the Access RT-PCR system (Promega) according to the manufacturer's protocol with 40 cycles of 30 s at 94°C, 1 min at 51°C, and 2 min at 68°C. Putative amplified PPO gene fragments were cloned into the EcoRV site of pBluescript II SK(−) (Stratagene, La Jolla, CA) using a T-tailing cloning strategy (Ausubel et al., 1998).
A cDNA library was constructed in the Lambda ZAP II vector using the ZAP-cDNA Synthesis kit (Stratagene). cDNA was prepared using mRNA from newly expanded red clover leaves, cloned as EcoRI-XhoI fragments into the Lambda ZAP II vector and packaged according to the manufacturer's protocol. The resulting cDNA library had >99% recombinants as assessed by blue/white screening using X-gal (5-bromo-4-chloro-3-indollyl β-d-galactopyranoside) and isoproplyl-β-d-thiogalactoside (IPTG). Approximately 106 recombinant phage from the unamplified library were plated and lifted to nylon or nitrocellulose filters using standard protocols (Sambrook et al., 1989) then screened by hybridization with a cloned 300-bp PPO fragment derived by PCR using the PPO-1 primer.
DNA and RNA Hybridization
32P-labeled DNA probes were made by the random primed method (Feinberg and Vogelstein, 1983) using DNA fragments derived from restriction digestion of plasmids or PCR reactions as indicated. RNA blots and library filters were preincubated for at least 1 h with hybridization buffer (50 mm NaPO4, pH 7.0, 0.8 m NaCl, 1 mm EDTA, 10× Denhardt's, 0.25 mg/mL sheared herring sperm DNA, 0.5% SDS). For RNA blots, hybridization buffer included 50% formamide. Blots and filters were incubated overnight with approximately 106 cpm/mL of 32P-labeled probe as indicated. For library filters, hybridization was at 50°C, and posthybridization washes were carried out in 2× SSC (1× SSC is 0.15 m NaCl, 0.015 m sodium citrate), 0.1% SDS at 55°C. For RNA blots, hybridization was at 60°C and posthybridization washes were carried out in 0.2× SSC, 0.1% SDS at 62°C or hybridization was at 45°C and posthybridization washes were carried out in 2× SSC at 60°C for high- and low-stringency conditions, respectively. Washed filters were exposed to autoradiography film (library screening) or imaged and radioactive signals quantified using a Phosphoimager and ImageQuant software (Amersham Biosciences, Piscataway, NJ).
DNA Sequencing and Sequence Analysis
DNA sequence was determined by cycle sequencing using Big Dye chemistry (Applied Biosystems, Foster City, CA). Sequencing reactions were outsourced to the University of Wisconsin Biotechnology Center to be run on automated DNA sequencers (ABI models 377XL and 377-96). Sequence analyses were carried out using the Wisconsin Package, Version 10 (Accelrys, San Diego). Chloroplast transit and thylakoid lumen targeting signals were analyzed using the ChloroP (Emanuelsson et al., 1999) and SignalP (Nielsen et al., 1997) algorithms available online at http://www.cbs.dtu.dk/services. Unless otherwise indicated, BLAST searches were carried out using the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov).
Construction of Plasmids for Expression of Red Clover PPO in Escherichia coli and Alfalfa
For all gene constructions, standard molecular biology techniques were used (Sambrook et al., 1989; Ausubel et al., 1998). When fragments for cloning were generated via PCR, the cloned insert was sequenced to ensure that no mutations were introduced that would alter the sequence of translated protein.
For expression in E. coli, two plasmids were made to express the predicted mature form of red clover PPO1 either with or without a 6×His amino-terminal tag. The red clover PPO1 cDNA as an EcoRI-XhoI fragment in pBluescript SKII(−) served as a template for PCR using either the primer 5′-GCGCGCGCCATATGGCTCCAGTGCCAATTCCAG-3′ (for His-tagged PPO1) or 5′-GCGCGCGCCATGGCTCCAGTGCCAATTCCAG-3′ (for native PPO1) and the T7 primer (5′-GTAATACGACTCACTATAGGGC-3′). The resulting PCR products were digested with either NdeI and XhoI (for His-tagged PPO1) or NcoI and XhoI (for native, nontagged PPO1) and inserted into pET28a (Novagen, Madison, WI) digested with either NdeI and XhoI or NcoI and XhoI. The resulting plasmids, pMLS46 and pMLS56, were designed to produce 6×His-tagged and nontagged versions of the predicted mature form of red clover PPO1, respectively.
For expression in transgenic alfalfa, PCR primer pairs were designed to introduce XbaI restriction endonuclease sites on either side of the coding regions of red clover PPO1, PPO2, and PPO3. Additionally, the forward primers provided the proposed dicot consensus sequence AAACA (Joshi et al., 1997) immediately upstream of the initiating Met codon. The primer pairs were: PPO1, 5′-GGGTCTAGAAACAATGGCATCTATCTCATCC-3′ and 5′-GGGTCTAGATCAATCTTCAAGCTCTATC-3′; PPO2, 5′-GGGTCTAGAAACAATGGTATCTATCTCATCC-3′ and 5′-GGGTCTAGATCATTCTTCAAAGTCTATC-3′; and PPO3, 5′-GGGTCTAGAAACAATGATATCTATTTCATCC-3′ and 5′-GGGTCTAGATCAATTTGCAAATTCTATC-3′. The red clover PPO1, PPO2, and PPO3 cDNAs as EcoRI-XhoI fragments in pBluescript SKII(−) served as templates for PCR. The resulting PCR products were digested with XbaI and cloned into pBluescript SKII(−) as an intermediate step. The PPO coding regions were finally cloned behind the cassava vein mosaic virus promoter in the plant transformation vector pILTAB357 (Verdaguer et al., 1996). The red clover PPO1 coding region was cloned as an XbaI-PstI (blunted with T4 DNA polymerase) fragment between XbaI and EcoRI (filled in with T4 DNA polymerase) sites of pILTAB357. The red clover PPO2 and PPO3 coding regions were cloned as XbaI-EcoRI fragments between XbaI and EcoRI sites of pILTAB357.
Expression of Red Clover PPO1 in E. coli and Preparation of Antigen for Antibody Production and Soluble Extracts for Activity Assays
Plasmids pMLS46 and pMLS56 were transformed into the codon-enhanced E. coli strain BL21(DE3) RIL (Stratagene). Cultures were grown with shaking at 37°C to midlogrithmic phase (optical density at 600 nm approximately 0.5) in Luria-Bertani medium supplemented with 50 μg/mL kanamycin, induced by adding IPTG to 1 mm, then grown an additional 2 to 3 h. The cells were harvested by centrifugation at 4,000g for 10 min. Inclusion bodies containing the expressed PPO protein were prepared by resuspending the cells in one-tenth the original culture volume of inclusion body isolation buffer (IBIB; 20 mm Tris-HCl, pH 7.5, 10 mm EDTA, 1% Triton X-100). Lysozyme was added to 100 μg/mL and the suspension incubated for 15 min at room temperature. All subsequent steps were carried out at 4°C or on ice. The suspension was sonicated briefly to shear DNA, and the inclusion bodies were separated from the soluble extract by centrifuging at 10,000g for 10 min. The pellet was washed once by resuspending in one-tenth the original culture volume of IBIB and centrifugation at 10,000g for 10 min. The pellet was resuspended in one-one hundredth the original culture volume of 50 mm Tris, pH 8.0. Aliquots of inclusion bodies corresponding to 100 mL of culture were collected by centrifugation (14,000g for 5 min) and stored at −80°C until ready for further purification. Soluble extracts for activity assays were prepared essentially as above, except CuCl2 (20 μm final concentration) was added to the culture medium upon IPTG induction, EDTA was omitted from IBIB, and the soluble fraction was aliquotted and stored at −80°C following the post-lysis centrifugation. For samples to be run on SDS-PAGE, a protease inhibitor cocktail (P-8849; Sigma-Aldrich, St. Louis) was added to IBIB at a rate of 1% (v/v).
To purify protein for antibody production, inclusion bodies from 50 mL of culture were solubilized in SDS-PAGE sample buffer at 70°C and run on 1-mm-thick SDS-PAGE (10% acrylamide; 37.5:1 acrylamide:bis-acrylamide) minigels (8.5 × 6 cm). The prominent PPO1 gene product band was visualized by staining with CuCl2 (Lee et al., 1987) and excised. Excess Cu2+ was removed by washing the fragment twice with 10 mL of washing solution (250 mm EDTA, 200 mm Tris, pH 8.0, 0.1% SDS) 15 to 20 min per wash. Gel fragments from multiple gels were combined, crushed by passing through a syringe without a needle, and the protein was eluted by adding two volumes of elution buffer (50 mm Tris, pH 8.0, 10 mm EDTA, 0.1% SDS) and incubating at room temperature with gentle shaking for 24 to 48 h. The eluate was concentrated using a microconcentrator (Centricon 10; Millipore, Billerica, MA), and the buffer was exchanged for 50 mm Tris, pH 8.0, 0.1% SDS. The resulting protein was used for antibody production in rabbits at a commercial facility (Harlan Bioproducts for Science, Indianapolis) using standard protocols.
Plant Protein Extraction and PPO Activity Assays
Fully expanded young alfalfa or clover leaves were powdered in liquid nitrogen using a mortar and pestle and extracted into 100 mm NH4OAc, 20 mm Tris, pH 7.5, using 3 mL of buffer per g fresh weight. For red clover tissue, 50 mm ascorbic acid was included in the extraction buffer to prevent extensive browning by endogenous PPO and o-diphenol substrates. For samples to be run on SDS-PAGE, a protease inhibitor cocktail (Sigma P-8849) was added to 1% (v/v). Extracts were kept on ice or at 4°C throughout. Extracts were centrifuged at 10,000g for 5 min, filtered through Miracloth (Calbiochem, San Diego), and centrifuged again at 10,000g for 10 min. Aliquots of the clarified extracts were frozen in liquid nitrogen and stored at −80°C. Protein concentration of extracts was determined using the BCA Protein Assay Reagent (Pierce Biotechnology, Rockford, IL) and bovine serum albumin as a standard.
Simple browning assays were carried out by addition of 100 mm caffeic acid in ethanol to 3 mm final concentration or ethanol to 3% (v/v) as a negative control and incubation at room temperature. Reactions to quantify PPO activity were carried out essentially as described by Esterbauer et al. (1977) and consisted of 950 μL of 20 mm Tris-acetate, pH 7.0, 20 μL of 2-nitro-5-thio-benzoic acid solution [prepared by suspending 19 mg 5,5′-dithio-bis(2-nitrobenzoic acid) (D-8130, Sigma-Aldrich) in 13.3 mL water and adding 30 mg NaBH4], and 20 μL of 100 mm caffeic acid in ethanol. Reactions were initiated by adding 10 μL of tissue extract (diluted when appropriate), incubated at 25°C, and A412 was measured over time using a Beckman DU50 spectrophotometer (Beckman Instruments, Fullerton, CA) and data capture software. Reaction rates were determined by plotting A412 versus time, determining the slope of the linear portion of the reaction by linear regression and converting to nmol substrate using the conversion 91.0 nmol/A412 nm (Esterbauer et al., 1977). Specific activity of a given extract was calculated by dividing reaction rate by the amount of extract protein present in the reaction and expressed as nkat mg−1. Statistical analysis of transgenic plant PPO activity was carried out using the Mann-Whitney test (Nap et al., 1993). PPO activity measurements for E. coli extracts were essentially the same, except Tris-acetate buffer was reduced to 860 μL and 100 μL of E. coli extract was used.
SDS-PAGE and Immunoblotting
SDS-PAGE (10% acrylamide; 37.5:1 acrylamide:bis-acrylamide) was carried out using standard methodologies (Harlow and Lane, 1988). Gels were stained using Gel Code Blue Stain Reagent (Pierce Biotechnology). E. coli and leaf extracts for SDS-PAGE were prepared as detailed elsewhere in “Materials and Methods” and denatured for SDS-PAGE by boiling with an equal volume of SDS-PAGE sample buffer for 5 min. Due to the large amount of interfering carbohydrate present, red clover flower protein samples were prepared by extraction with phenol essentially as described at http://plantpath.unl.edu/llane/text/proteinphenol.html (L. Lane, personal communication). Precipitated protein from 2 g of mature red clover flowers was resuspended in 0.5 mL of SDS-PAGE sample buffer, boiled 10 min with intermittent vortexing, and insoluble material was removed by centrifugation (5 min at 14,000g). Protein concentration was estimated by comparison to a sample of E. coli extract of known protein concentration on SDS-PAGE. For immunoblotting, following electrophoresis proteins were transferred to polyvinylidene fluoride (Bio-Rad Laboratories, Hercules, CA), and the blots were processed and developed according to the manufacturer's instructions. Detection was accomplished using anti-PPO1 antiserum diluted 1:1,000, alkaline phosphotase conjugated goat anti-rabbit alkaline phosphotase diluted 1:3,000 as the secondary antibody, and Immun-Star Chemiluminescent Substrate (Bio-Rad Laboratories). Blots were imaged using a Lumi-Imager (Roche Molecular Biochemicals, Indianapolis) or a Chemidoc System (Bio-Rad Laboratories).
In Vitro Proteolysis Assay
Extracts of control or PPO1-expressing alfalfa leaves were prepared as described elsewhere in “Materials and Methods,” except the extraction buffer was 50 mm MES, pH 6.5. Proteolysis assay reactions in 50 mm MES, pH 6.5, contained 2 mg mL−1 leaf extract protein and either caffeic acid (3% [v/v] of a 100 mm stock in ethanol, 3 mm final) or ethanol (3% [v/v]) as a control. Duplicate samples of each reaction were removed before and after a 4-h incubation at 37°C. Once removed, the samples were immediately mixed with one-half volume 15% trichloroacetic acid (TCA, 5% [w/v] final) and placed on ice for at least 30 min. TCA-insoluble material was removed by centrifugation at 16,000g for 5 min. Amino acid concentration of the 5% TCA supernatants was determined using Nin-Sol AF ninhydrin reagent (Pierce Biotechnology) with Gly as the standard, and values for duplicate samples were averaged. Amino acid release following the 37° incubation was determined by subtracting the amino acid concentration of the initial samples from the amino acid concentration of the final samples and expressing the result as μmol amino acid released per mg of extract protein. The results of three experiments using independently prepared extracts were averaged.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY017302 to AY017304.
Supplementary Material
Acknowledgments
We thank Sara Rierson and Mindy Dornbusch for excellent technical assistance and Dr. Jane Marita for comments on the manuscript.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.047449.
References
- Austin S, Bingham ET, Matthews D, Shahan M, Will J, Burgess RR (1995) Production and field performance of transgenic alfalfa expressing alpha-amylase and manganese-dependent lignin peroxidase. Euphytica 85: 381–393 [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Deidman JG, Smith JA, Struhl K, editors (1998) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Bellucci M, Pupilli F, Arcioni S (1999) Variation for polyphenol oxidase activity in stems of Medicago species. Agronomie 19: 73–77 [Google Scholar]
- Bingham ET (1991) Registration of alfalfa hybrid Regen-SY germplasm for tissue culture and transformation research. Crop Sci 31: 1098 [Google Scholar]
- Cary JW, Lax AR, Flurkey WH (1992) Cloning and characterization of cDNAs coding for Vicia faba polyphenol oxidase. Plant Mol Biol 20: 245–253 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11: 113–116 [Google Scholar]
- Cho MH, Moinuddin SGA, Helms GL, Hishiyama S, Eichinger D, Davin LB, Lewis NG (2003) (+)-Larreatricin hydroxylase, an enantio-specific polyphenol oxidase from the creosote bush (Larrea tridentata). Proc Natl Acad Sci USA 100: 10641–10646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel CP, Bergey DR, Ryan CA (1995) Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octdecanoid defense signaling pathway. Proc Natl Acad Sci USA 92: 407–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterbauer H, Schwarzl E, Hayn M (1977) A rapid assay for catechol oxidase and laccase using 2-nitro-5-thio benzoic acid. Anal Biochem 77: 486–494 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Gerdemann C, Eicken C, Magrini A, Meyer HE, Rompel A, Spener F, Krebs B (2001) Isozymes of Ipomoea batatas catchol oxidase differ in catalase-like activity. Biochim Biophys Acta 1548: 94–105 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Hunt MD, Eannetta NT, Yu H, Newman SM, Steffens JC (1993) cDNA cloning and expression of potato polyphenol oxidase. Plant Mol Biol 21: 59–68 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Miles PW (1993) Responses of a compatible lucerne variety to attack by spotted alfalfa aphid: changes in the redox balance in affected tissues. Entomol Exp Appl 67: 263–274 [Google Scholar]
- Jones BA, Hatfield RD, Muck RE (1995. a) Characterization of proteolysis in alfalfa and red clover. Crop Sci 35: 537–541 [Google Scholar]
- Jones BA, Hatfield RD, Muck RE (1995. b) Screening legume forages for soluble phenols, polyphenol oxidase and extract browning. J Sci Food Agric 67: 109–112 [Google Scholar]
- Jones BA, Muck RE, Hatfield RD (1995. c) Red clover extracts inhibit legume proteolysis. J Sci Food Agric 67: 329–333 [Google Scholar]
- Joshi CP, Zhou H, Huang XQ, Chiang VL (1997) Context sequences of translation initiation codon in plants. Plant Mol Biol 35: 993–1001 [DOI] [PubMed] [Google Scholar]
- Klabunde T, Eicken C, Sacchettini JC, Krebs B (1998) Crystal structure of a plant catechol oxidase containing a dicopper center. Nat Struct Biol 5: 1084–1090 [DOI] [PubMed] [Google Scholar]
- Lee C, Levin A, Branton D (1987) Copper staining: a five-minute protein stain for sodium dodecyl-sulfate polyacrylamide gels. Anal Biochem 166: 308–312 [DOI] [PubMed] [Google Scholar]
- Li L, Steffens JC (2002) Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta 215: 239–247 [DOI] [PubMed] [Google Scholar]
- Mayer AM, Harel E (1979) Polyphenol oxidase in plants. Phytochemistry 18: 193–215 [Google Scholar]
- Muck RE (1987) Dry matter level effects on alfalfa silage quality. I. Nitrogen transformation. Trans ASAE 30: 7–14 [Google Scholar]
- Nakayama T, Soto T, Fukui Y, Yonekura-Sakakibara K, Hayashi H, Tanaka Y, Kusumi T, Nishino T (2001) Specificity analysis and mechanism of aurone synthesis catalyzed by aurenusidin synthase, a polyphenol oxidase homolog responsible for flower coloration. FEBS Lett 499: 107–111 [DOI] [PubMed] [Google Scholar]
- Nap JP, Keizer P, Jansen R (1993) First-generation transgenic plants and statistics. Plant Mol Biol Report 11: 156–164 [Google Scholar]
- Newman SM, Eannetta NT, Yu HF, Prince JP, Devicente MC, Tanksley SD, Steffens JC (1993. a) Organization of the tomato polyphenol oxidase gene family. Plant Mol Biol 21: 1035–1051 [DOI] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ (1993. b) DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell 5: 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Papadopoulos YA, McKersie BD (1983) A comparison of protein degradation during wilting and ensiling of six forage species. Can J Plant Sci 63: 903–912 [Google Scholar]
- Peltier JB, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ (2000) Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12: 319–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotz CA, Pitt RE, Muck RE, Allen MS, Buckmaster DR (1993) Direct-cut harvest and storage of alfalfa on the dairy farm. Trans ASAE 36: 621–628 [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Saruul P, Srienc F, Somers DA, Samac DA (2002) Production of a biodegradable plastic polymer, poly-beta-hydroxybutyrate, in transgenic alfalfa. Crop Sci 42: 919–927 [Google Scholar]
- Sherman TD, Gardeur TL, Lax AR (1995) Implications of the phyogenetic distribution of polyphenol oxidase in plants. In CY Lee, JR Whitaker, eds, Enzymatic Browning and Its Prevention. American Chemical Society, Washington, DC, pp 103–119
- Smith RR, Maxwell DP (1980) Registration of WI-1 and WI-2 red clover. Crop Sci 20: 831 [Google Scholar]
- Steffens JC, Harel E, Hunt MD (1994) Polyphenol oxidase. In BE Ellis, GW Kuroki, HA Stafford, eds, Genetic Engineering of Plant Secondary Metabolism, Vol 28. Plenum Press, New York, pp 275–312
- Sullivan M, Thoma S, Samac D, Hatfield R (2004) Cloning of red clover and alfalfa polyphenol oxidase genes and expression of active enzymes in transgenic alfalfa. In A Hopkins, ZY Wang, R Mian, M Sledge, R Barker, eds, Molecular Breeding of Forage and Turf. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 189–195
- Thipyapong P, Hunt MD, Steffens JC (1995) Systemic wound induction of potato (Solanum tuberosum) polyphenol oxidase. Phytochemistry 40: 673–676 [Google Scholar]
- Thipyapong P, Steffens JC (1997) Tomato polyphenol oxidase. Differential response of the polyphenol oxidase F promoter to injuries and wound signals. Plant Physiol 115: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamos-Vigyazo L (1981) Polyphenol oxidase and peroxidase in fruits and vegetables. CRC Crit Rev Food Sci Nutr 15: 49–127 [DOI] [PubMed] [Google Scholar]
- Verdaguer B, de Kochko A, Beachy RN, Fauquet C (1996) Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol 31: 1129–1139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.