Abstract
In tobacco (Nicotiana tabacum), hyperosmotic stress induces rapid activation of a 42-kD protein kinase, referred to as Nicotiana tabacum osmotic stress-activated protein kinase (NtOSAK). cDNA encoding the kinase was cloned and, based on the predicted amino acid sequence, the enzyme was assigned to the SNF1-related protein kinase type 2 (SnRK2) family. The identity of the enzyme was confirmed by immunoprecipitation of the active kinase from tobacco cells subjected to osmotic stress using antibodies raised against a peptide corresponding to the C-terminal sequence of the kinase predicted from the cloned cDNA. A detailed biochemical characterization of NtOSAK purified from stressed tobacco cells was performed. Our results show that NtOSAK is a calcium-independent Ser/Thr protein kinase. The sequence of putative phosphorylation sites recognized by NtOSAK, predicted by the computer program PREDIKIN, resembled the substrate consensus sequence defined for animal and yeast (Saccharomyces cerevisiae) AMPK/SNF1 kinases. Our experimental data confirmed these results, as various targets for AMPK/SNF1 kinases were also efficiently phosphorylated by NtOSAK. A range of protein kinase inhibitors was tested as potential modulators of NtOSAK, but only staurosporine, a rather nonspecific protein kinase inhibitor, was found to abolish the enzyme activity. In phosphorylation reactions, NtOSAK exhibited a preference for Mg2+ over Mn2+ ions and an inability to use GTP instead of ATP as a phosphate donor. The enzyme activity was not modulated by 5′-AMP. To our knowledge, these results represent the first detailed biochemical characterization of a kinase of the SnRK2 family.
The yeast (Saccharomyces cerevisiae) SNF1 complex and its homologue in mammals, the AMP-activated protein kinase (AMPK), represent signaling kinases that are highly conserved in eukaryotic cells. They are involved in protection of cells against nutritional or environmental stress, especially those causing depletion of cellular ATP (for review, see Hardie et al., 1998; Hardie, 1999; Hardie and Hawley, 2001). AMPK is activated by heat shock, hypoxia, inhibitors of oxidative phosphorylation, and exercise in muscle, whereas SNF1 is activated by glucose starvation of yeast. In plants, three subfamilies of SNF1-related kinases (SnRKs), i.e. SnRK1, SnRK2a/2b, and SnRK3, have been identified (Halford and Hardie, 1998; Hardie, 2000). The SnRK1 subfamily represents direct structural and functional homologues of the SNF1/AMPK family (Halford and Hardie, 1998). Remarkably, disruption of both genes encoding SnRK1s in the moss Physcomitrella patens yields plants that require continuous illumination and are unable to survive in a normal day-night light cycle (Thelander et al., 2004). The SnRK2 and SnRK3 groups are unique to plants and may be involved in the response to other environmental stresses. Several kinases of the SnRK3 subfamily have been described. One is the wheat (Triticum aestivum) protein kinase WPK4, whose expression is up-regulated by light, cytokinins, and low temperature, and down-regulated by sucrose (Sano and Youssefian, 1994; Ikeda et al., 1999). Transcription of the rice (Oryza sativa) WPK4 homologue (OsPK4) is induced by illumination, nutrient deprivation, and treatment with cytokinins, whereas the maize (Zea mays) homologue (ZmPK4) is constitutively expressed regardless of light, nutrient, and cytokinin status but is increased upon exposure to low temperature (Ohba et al., 2000). It has been suggested that the physiological roles of WPK4 and Zmpk4 are to maintain carbon assimilation rates at appropriate levels during periods of low temperature. There are several reports indicating that a group of kinases interacting with calcineurin B-like proteins, which belong to the SnRK3 subfamily, take part in protection of plant cells against abiotic stresses (for review, see Luan et al., 2002; Chinnusamy et al., 2004; Gong et al., 2004). The best known member of this group is the SOS2 kinase, which is required for sodium and potassium ion homeostasis and salt tolerance (Liu et al., 2000). There are also results suggesting that kinases of the SnRK2 subfamily are involved in abiotic stress signaling. Expression of protein kinase induced by abscisic acid (PKABA1) from wheat, the first kinase of the SnRK2a subfamily to be identified, is induced by dehydration, cold, salinity, and osmotic stress (Anderberg and Walker-Simmons, 1992; Holappa and Walker-Simmons, 1995). One of the functions of PKABA1 is to suppress the GA induction of genes encoding hydrolytic enzymes, by repressing the GA induction of GAMyb (Gomez-Cadenas et al., 2001). The activity of the ABA-activated and Ca2+-independent protein kinase (AAPK), a kinase related to PKABA1 in fava bean (Vicia faba), is induced by ABA in guard cells in response to drought (Li and Assmann, 1996; Li et al., 2000). This enzyme regulates stomatal closure and anion channels (Li et al., 2000). One of the substrates of AAPK is AAPK-interacting protein (AKIP), which is implicated in the ABA regulation of posttranscriptional RNA metabolism (Li et al., 2002). OST1 (Open Stomata 1) protein kinase in Arabidopsis, most probably a homologue of AAPK, is activated by ABA, and mediates not only the regulation of stomatal aperture by ABA but also acts upstream of production of reactive oxygen species (Mustilli et al., 2002). In parallel, the same kinase was discovered by Shinozaki and co-workers as a kinase activated by ABA and required for dehydration stress signaling in Arabidopsis (Yoshida et al., 2002). Expression of wound- and ABA-induced protein kinase (WAPK) from tobacco (Nicotiana tabacum), closely related to PKABA1, is regulated by wounding, ABA, and methyl jasmonate (Lee et al., 1998).
Several lines of evidence suggest that kinases of the SnRK2b subfamily are also involved in stress signaling in plants. Expression of the genes encoding soybean (Glycine max) protein kinases SPK-3 and SPK-4, both members of this subfamily, are induced by dehydration and high salinity (Yoon et al., 1997). Two other soybean protein kinases, i.e. SPK-1 and SPK-2 from the same subfamily, are able to phosphorylate soybean unusual phosphatidyl inositol transfer-like protein (Ssh1p). Ssh1p is phosphorylated upon exposure of plant tissues to hyperosmotic stress (Kearns et al., 1998; Monks et al., 2001).
We identified in tobacco cells a 42-kD protein kinase that is rapidly activated by osmotic stress (Mikołajczyk et al., 2000). The enzyme was assigned to the SnRK2b subfamily based on sequence analysis of internal tryptic peptides obtained by microsequencing of the purified kinase. A similar kinase described by Hoyos and Zhang was named HOSAK because its activation was observed in tobacco cells exposed to hyperosmotic stress (Hoyos and Zhang, 2000).
Recently, Kobayashi et al. demonstrated that members of the SnRK2 family from rice are activated by hyperosmotic stress and only some of them are regulated also by ABA (Kobayashi et al., 2004). Their results suggest that the SnRK2 protein kinase family has evolved specifically for hyperosmotic stress signaling.
For studies of enzyme function, knowledge of the biochemical features of the enzyme is very important. Systematic biochemical analysis has been performed for animal AMPK and yeast SNF1 kinases (for review, see Hardie et al., 1998; Hardie, 1999). In the case of plant SnRKs, the best characterized are the enzymes of the SnRK1 subfamily, whose structure and biochemical features (e.g. substrate specificity) are similar to those described for SNF1/AMPK (for review, see Halford and Hardie, 1998). Recently, the biochemical characterization of several Arabidopsis protein kinases of the SnRK3 subfamily was described: the SOS2 kinase that functions in salt tolerance (Gong et al., 2002a); four SOS2-like protein kinases (PKSs); PKS11, probably involved in sugar sensing (Gong et al., 2002b); PKS6 and PKS18, involved in ABA signaling (Gong et al., 2002c, 2002d); and AtCIPK1/PKS13 (Shi et al., 1999). In contrast to SnRK1 and SnRK3, the biochemical properties of enzymes of the SnRK2 subfamily are still largely unknown (for review, see Harmon, 2003; Halford et al., 2004). The elucidation of the substrate specificity of members of the SnRK2 subfamily is of major importance for establishing simple, sensitive, and specific assays for monitoring their activity. A knowledge of the substrate specificity of these kinases may also be helpful in the search for endogenous SnRK2 target proteins, while investigation of their biochemical properties and mechanisms of regulation may give insight into their cellular function. Therefore, the biochemical characterization of the tobacco 42-kD osmotic stress-activated protein kinase (NtOSAK), a member of the SnRK2 subfamily, was undertaken.
RESULTS
Cloning of cDNA Encoding NtOSAK
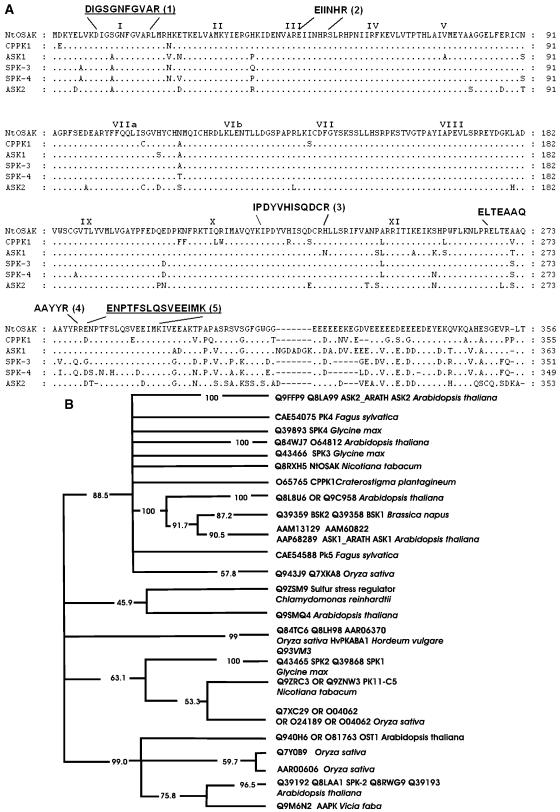
NtOSAK, purified to near-homogeneity, was subjected to microsequencing. Partial amino acid sequences of five internal tryptic peptides were obtained (Mikołajczyk et al., 2000). The sequences of two peptides (peptide 1 and 5, see Fig. 1A) were used to design primers for reverse transcription (RT)-PCR, with total RNA isolated from tobacco BY-2 cells (treated for 15 h with 250 mm NaCl) as a template. To minimize primer degeneracy, the same codon usage as in cDNAs encoding N. tabacum protein kinases of the SnRK2 subfamily, i.e. WAPK and PK11-C1 (GenBank accession nos. AF032465 and AAD00239, respectively) was chosen. In the RT-PCR reaction, a 960-bp DNA product encoding a fragment of a kinase belonging to the SnRK2 family was generated. The fragment encoded the sequences of all previously determined NtOSAK peptides. A full-length cDNA encoding NtOSAK was obtained by screening a BY-2 cells cDNA library using the RT-PCR product as a probe. The cDNA sequence obtained was submitted to GenBank (accession no. AY081175). The predicted kinase contains all 11 conserved kinase subdomains characteristic of Ser/Thr kinases (Hanks et al., 1988) plus a stretch of acidic amino acids at the C terminus, present in most of the known kinases of the SnRK2 subfamily sequenced to date. Analysis of the deduced amino acid sequence by a standard National Center for Biotechnology Information (NCBI)-BLAST homology search shows the greatest similarity to protein kinases assigned to the SnRK2b subfamily according to Halford and Hardie (1998), i.e. CPPK1 from Craterostigma plantagineum, SPK3 and SPK4 from soybean, and ASK1 and ASK2 from Arabidopsis (Fig. 1A), suggesting that NtOSAK is a member of this subfamily. Generation of a phylogenetic tree confirmed that the NtOSAK kinase belongs to the branch containing the ASK1 and ASK2 kinases from Arabidopsis (Fig. 1B).
Figure 1.
NtOSAK belongs to the SnRK2 subfamily. A, Comparison of the predicted amino acid sequence of NtOSAK with sequences of other protein kinases. The sequences of five protein kinases exhibiting sequence similarity are aligned with that of NtOSAK. References for the sequences of the listed kinases are as follows: CPPK1-protein kinase 1 from Craterostigma plant (P. Heino, M. Nylander, T. Palva, and D. Bartels, unpublished data; Swiss-Prot accession no. O65765); soybean protein kinases SPK-3 (Swiss-Prot accession no. Q43466) and SPK-4 (Swiss-Prot accession no. Q39893; Yoon et al., 1997); and Arabidopsis protein kinases ASK1 (Swiss-Prot accession no. P43291) and ASK2 (Swiss-Prot accession no. P43292; Park et al., 1993). The conserved Ser/Thr protein kinase subdomains (Hanks et al., 1988) are indicated by roman numerals. Gaps are marked with dashes and were introduced to maximize alignment. The peptide sequences obtained by microsequencing are shown above the NtOSAK sequence. The peptide sequences used for designing PCR primers are underlined. B, Phylogenetic tree of plant SNF1-related protein kinases that are closely related to NtOSAK. The tree is a consensus of 100 maximum parsimony “bootstrap” trees. Percentage values on each branch represent the corresponding bootstrap probabilities. The multiple alignment was created using the CLUSTALW computer program (Thompson et al., 1994). Columns with gaps and places where alignment seemed doubtful were excluded. The multiple alignment after edition contains 282 columns. The consensus maximum parsimony tree was calculated using the PHYLIP package (Felsenstein, 1989).
Immunodetection of NtOSAK in BY-2 Cells Extracts
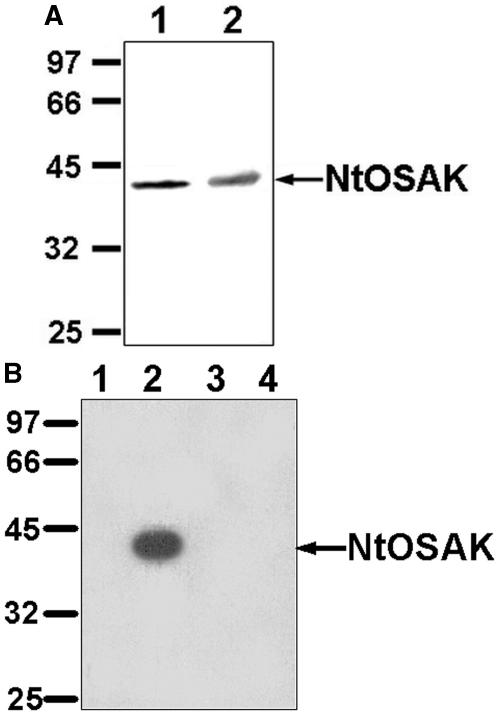
To verify that the p42-kD kinase rapidly activated by osmotic stress is encoded by the cloned cDNA, specific anti-NtOSAK antibodies were generated. The antibodies were directed against the C-terminal peptide of the protein kinase sequence deduced from the cloned cDNA. Protein extracts prepared from BY-2 cells (untreated and treated for 5 min with 250 mm NaCl) were subjected to western-blot analysis. In both extracts, the anti-NtOSAK antibodies recognized a single protein band with a molecular mass of 42 kD (Fig. 2A). Subsequently, cell extracts from untreated and 250 mm NaCl-treated cells were immunoprecipitated with anti-NtOSAK antibodies. The resulting immunocomplexes were analyzed by in-gel kinase assay using myelin basic protein (MBP) as substrate. The antibodies immunoprecipitated the 42-kD protein kinase activated by osmotic stress, thus confirming that the cloned cDNA encodes the studied enzyme (Fig. 2B).
Figure 2.
Antibodies specific to the C terminus of the protein kinase encoded by the cloned cDNA (anti-NtOSAK) immunoprecipitate the 42-kD OSAK. A, Western-blot analysis. Protein extracts from BY-2 cells untreated (lane 1) and treated for 5 min with 250 mm NaCl (lane 2) were probed with anti-NtOSAK antibodies. B, Immunoprecipitation followed by immunocomplex kinase assay. Protein extracts (50 μg) from untreated (lane 1) and from cells subjected for 5 min to 250 mm NaCl (lane 2) were immunoprecipitated with anti-NtOSAK antibodies, and the resulting immunocomplexes were analyzed by in-gel kinase assay with MBP as substrate. Lanes 3 and 4 represent controls where the preimmune serum was used instead of the antibodies for immunoprecipitation; lane 3, protein extracts (50 μg) from untreated, and lane 4, from cells subjected for 5 min to 250 mm NaCl. Molecular mass markers are given in kilodaltons at left.
Recombinant GST-NtOSAK Does Not Exhibit Any Enzymatic Activity
NtOSAK was expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli and purified on Glutathione-Sepharose resin (Amersham Pharmacia Biotech AB, Uppsala). The recombinant protein migrated at the expected molecular mass upon gel electrophoresis (data not shown). However, the protein displayed no detectable kinase activity when analyzed with MBP or casein as substrate. Results from other laboratories show that several protein kinases belonging to the SnRK2 subfamily (e.g. SPK-3, SPK-4, PKABA1, AAPK, and OST1) are also inactive when they are expressed as recombinant proteins in bacteria (Yoon et al., 1997; Li et al., 2000; Mustilli et al., 2002; Hrabak et al., 2003). In the case of AAPK and OST1, the authors claimed that both kinases require the ABA signaling cascade(s) in plants for their activation. Our results suggest that GST-NtOSAK expressed in bacteria is not active because phosphorylation of NtOSAK is required for its activation (Mikołajczyk et al., 2000; A. Kelner, I. Pękala, J. Sikora, M. Dadlez, M. Bucholc, P. Siedlecki, P. Zielenkiewicz, and G. Dobrowolska, unpublished data).
Purification of Active NtOSAK from BY-2 Cells
NtOSAK was isolated from BY-2 cells treated for 5 min with 250 mm NaCl. The enzyme was purified to near-homogeneity (about 12,000-fold) by four-step chromatography on Source 15Q, phenyl-Sepharose, heparin-Sepharose, and, finally, MonoQ according to a previously described procedure (Mikołajczyk et al., 2000). The purified enzyme exhibited a specific activity of about 200 nmol min−1 mg−1.
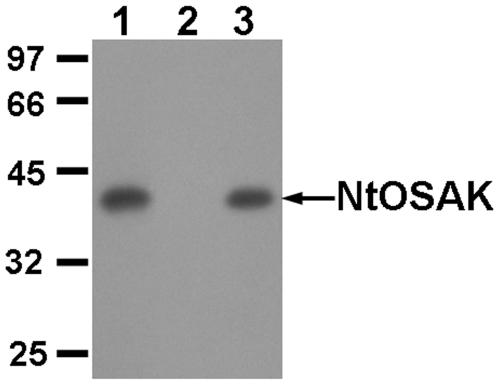
To demonstrate rigorously that the enzymatic preparation used for the biochemical studies consisted of only one kinase, i.e. NtOSAK, proteins eluted from the MonoQ column were immunoprecipitated using anti-NtOSAK antibodies. The kinase activity of the immunocomplexes and of the postimmunoprecipitation supernatant was analyzed by in-gel kinase assays using MBP as substrate (Fig. 3). Our results show that the kinase activity was completely immunoprecipitated, with no activity in the supernatant, confirming that the purified protein used for biochemical analysis is NtOSAK and not another protein kinase. Additionally, the activity of the resulting immunocomplexes was analysed by in-solution kinase assays with all of the substrates used in the studies described below. The immunocomplexes phosphorylated all of these substrates (data not shown).
Figure 3.
Immunological analysis of the 42-kD NtOSAK purified from tobacco cells. Proteins eluted from the MonoQ column were subjected to immunoprecipitation with anti-NtOSAK activity. Activity of the preparation before immunoprecipitation (lane 1) and after immunoprecipitation; immunocomplex (lane 3) and remaining supernatant (lane 2) were analyzed using in-gel kinase assay with MBP. Molecular mass markers are given in kilodaltons at left.
Determination of Km for ATP as a Phosphate Donor
Kinetic constants for ATP as a phosphate donor in the phosphorylation reaction catalyzed by NtOSAK were determined by analysis of casein phosphorylation in the presence of 1 to 500 μm ATP. The Km value was estimated to be 14 ± 2 μm (Table I). To determine if NtOSAK can use another nucleotide triphosphate (e.g., GTP) as a phosphate donor, unlabeled GTP in concentrations from 0 to 100 μm was added to 2 and 5 μm [γ32P]ATP. Addition of GTP did not change the rate of casein phosphorylation (data not shown), showing that GTP did not compete with ATP. The results indicate that GTP is not utilized by NtOSAK as a phosphate donor.
Table I.
Kinetic parameters for NtOSAK
| Substrate | Km | Vmax |
|---|---|---|
| μm | nmol/min/mg | |
| ATP | 14.4 ± 2.0 | 573 ± 34 |
| MBP | 23.2 ± 4.8 | 220 ± 18 |
| Casein | 26 ± 3 | 130 ± 6 |
| GST-ACC | 17 ± 6 | 18.1 ± 3.4 |
| SAMS | 125 ± 19 | 399 ± 27 |
| AMARA | 63 ± 10 | 300 ± 19 |
Km and maximal velocity (Vmax) for protein and peptide substrates, as well as for ATP, were determined for three independent experiments. Parameters were estimated by fitting of data directly to the Michaelis-Menten equation using GraphPad Prism 3.0, and are quoted ±se.
NtOSAK Prefers Mg2+ over Mn2+ Ions in the Phosphorylation Reaction
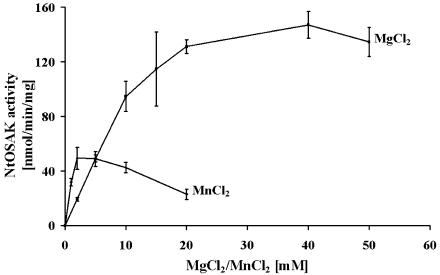
To define the divalent cation preference in phosphorylation reactions catalyzed by NtOSAK and to establish the optimal reaction conditions, the effect of various concentrations of Mg2+ and Mn2+ ions on the rate of phosphorylation was investigated. In these experiments, the substrate concentrations were constant: 40 μm for ATP and 1.2 mg/mL for casein. Our results showed that NtOSAK prefers MgCl2 over MnCl2. Casein phosphorylation was about 3 times more efficient in the presence of MgCl2 than MnCl2 (Fig. 4). The optimal concentration of MgCl2 was in the range of 20 to 40 mm, and of MnCl2 at 2.5 mm, whereas at higher concentration MnCl2 was inhibitory. Similar results were obtained when MBP instead of casein was used as the phosphate acceptor (data not shown).
Figure 4.
Dependence of Mg2+ and Mn2+ concentration on casein phosphorylation by NtOSAK. Phosphorylation of casein was performed at various Mg2+ (as MgCl2) and Mn2+ (as MnCl2) concentrations as indicated.
Substrate Specificity of NtOSAK
Our previous results (Mikołajczyk et al., 2000) showed that NtOSAK was able to phosphorylate several proteins, e.g. MBP, casein, histone H1, GST-ATF-2 (96), and GST-c-Jun (76), routinely used as substrates for activity assay of different protein kinases. Among them, the best substrates for NtOSAK were MBP and casein. Analysis of the phosphorylation kinetics of these two proteins showed that both were phosphorylated almost equally well by NtOSAK (Table I).
Recently, the computer program PREDIKIN was developed for the prediction of the substrate specificity of protein Ser/Thr kinases (Brinkworth et al., 2003; see http://smms.uq.edu.au/kinsub). The program is based on available data on crystal structures, molecular modeling, and sequence analyses of kinases and substrates. Knowing the amino acid sequence of NtOSAK, we were able to apply the program for prediction of NtOSAK substrate specificity. The predicted sequence motif recognized by NtOSAK on substrates was [KR]-[QMTAS]-X-[ST]-[VILMF]-[SQN]-[FLIRK]. This motif resembles to some extent the consensus sequence for substrates of SNF1/AMPK protein kinases (Dale et al., 1995). However, the computer analysis prediction is limited to heptapeptide sequences containing Ser/Thr, and, hence, at least two important determinants of AMPK/SNF1 substrates were missing, i.e. the hydrophobic residues at positions −5 and +4. These positions were not considered in the PREDIKIN analysis. Recently, it has also been shown that there are important determinants for recognition of substrates by AMPK N-terminal to the hydrophobic residue at −5 (Scott et al., 2002). Therefore, the phosphorylation of protein and peptide substrates for the AMPK/SNF1 kinase was also examined. We determined the phosphorylation kinetics for the following substrates: a fusion protein between GST and part of the sequence of rat acetyl-CoA-carboxylase containing the primary site for AMPK (GST-ACC; Scott et al., 2002); a synthetic peptide based on the primary site phosphorylated on rat ACC by AMPK, i.e. SAMS (HMRSAMSGLHLVKRR); and a peptide named AMARA (AMARAASAAALARRR)—an artificially designed peptide—which is an excellent substrate for AMPK and plant SnRK1 (Davies et al., 1989; Dale et al., 1995).
The kinetic parameters obtained using these substrates showed that NtOSAK phosphorylated casein, MBP, and AMPK/SNF1 substrates with nearly equal efficiency (Table I). The Km values for GST-ACC, SAMS, and AMARA in the reaction catalyzed by NtOSAK, i.e. 17 ± 6 μm, 125 ± 19 μm, and 63 ± 10 μm, respectively, are comparable to those obtained with mammalian AMPK or plant SnRK1 (Davies et al., 1989; Dale et al., 1995; Scott et al., 2002).
NtOSAK Is a Ca2+-, CI−-, and AMP-Independent Protein Kinase
It has been suggested that the stretch of acidic amino acids in the C-terminal domain of SnRK2 might be responsible for the binding of calcium ions and may provide a mechanism of enzymatic regulation (Harmon, 2003). To establish whether NtOSAK activity is modulated by calcium ions, phosphorylation of MBP and casein was measured in the presence or absence of Ca2+ (5 mm Ca2+ acetate or 1 mm EGTA). NtOSAK activity was the same in both cases (data not shown), indicating that NtOSAK is a calcium-independent protein kinase.
Since mammalian AMPK is activated by 5′-AMP (Carling et al., 1987) whereas the yeast SNF1, plant SnRK1, and SOS2 and the SOS2-related enzymes are not, we tested whether NtOSAK was AMP sensitive. NtOSAK activity was measured in the presence of 5′-AMP in concentration from 1 to 200 μm. No AMP dependence was seen even at the highest AMP concentration used (data not shown), indicating that NtOSAK is not affected by AMP.
Because NtOSAK is activated in cells exposed to 250 mm NaCl and several protein kinases in vitro are either activated or inhibited by salts, NtOSAK activity was analyzed in the presence of 0 to 250 mm chloride salts, i.e. NaCl, KCl, and NH4Cl, using casein as substrate. None of these salts, even at the highest concentration used, had any direct effect on NtOSAK activity (data not shown).
Effect of Temperature on NtOSAK Activity
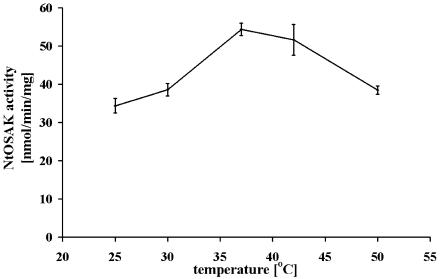
To determine the optimal temperature for measuring NtOSAK activity, the effect of temperature from 25°C to 50°C on casein phosphorylation was analyzed. The temperature optimum was found to be 37°C (Fig. 5). The enzyme was relatively insensitive to temperature changes within the range studied.
Figure 5.
Effect of temperature on the phosphorylation rate catalyzed by NtOSAK. Phosphorylation of casein was performed at each temperature indicated. Results shown are the average of three independent assays plotting the initial rates against incubation temperature.
Effect of Protein Kinases Inhibitors on NtOSAK Activity
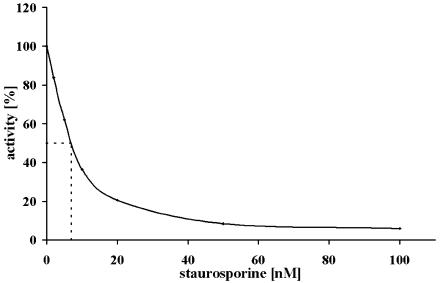
Specific protein kinase inhibitors are useful in the functional analysis and monitoring of protein kinases activity. Therefore, several compounds that are inhibitors of different protein kinases were analyzed as potential effectors of NtOSAK. These included heparin, 1-(β-d-ribofuranosyl)-5,6-dichlorobenzimidazole, and tetrabromoimidazole (potent inhibitors of CK2, kindly provided by Professor D. Shugar, Institute of Biochemistry and Biophysics PAS, Warsaw), bisindolylmaleimide, Ro 318220, and W-7 (inhibitors of Ca2+-dependent protein kinases), SB 203580 (an inhibitor of mammalian p38 mitogen-activated protein kinase [MAPK]), PD98059 and LY294002 (inhibitors of MAPK kinases), H-89 (developed as a selective PKA inhibitor, although inhibiting other kinases including AMPK), quercetin (an inhibitor of several protein kinases including CK2, S6K1, GSK3, and AMPK), and staurosporine (a nonspecific inhibitor of many Ser/Thr protein kinases). These compounds were tested on NtOSAK activity at concentrations used to inhibit other protein kinases as described by Davies et al. (2000). Apart from staurosporine, none of the compounds tested was able to abolish NtOSAK activity. The IC50 for staurosporine was estimated as 7 nm (Fig. 6).
Figure 6.
Inhibition of NtOSAK activity by staurosporine. The effect of staurosporine was estimated using casein phosphorylation in the presence of various concentrations of the inhibitor, as indicated. The activity measured in the absence of staurosporine was taken as 100%.
DISCUSSION
The 42-kD protein kinase, named NtOSAK, which is rapidly activated in tobacco cells exposed to osmotic stress, has now been analyzed at both the molecular and biochemical levels. The sequences of five internal peptides of the purified kinase suggested that the enzyme is closely related to kinases that belong to the SnRK2b subfamily of SNF1-related kinases (Mikołajczyk et al., 2000). The cDNA cloned on the basis of the sequences of those peptides encodes a protein kinase with an amino acid sequence showing 89%, 84%, and 81% identity with the following members of the SnRK2 group: CPPK1 from the resurrection plant C. plantagineum (P. Heino, M. Nylander, T. Palva, and D. Bartelss, unpublished data; GenBank accession no. CAA06503), and ASK1 and ASK2 from Arabidopsis (Park et al., 1993) kinases, respectively. A phylogenetic tree of plant SNF1-related protein kinases showed that NtOSAK belongs to the SnRK2 branch containing ASK1 and ASK2. To prove that the cloned cDNA encoded NtOSAK, and not a different member of the SnRK2 subfamily, immunoprecipitation of cell extracts of untreated and 250 mm NaCl-treated tobacco cells with specific antibodies raised against the C-terminal peptide deduced from the cloned cDNA sequence was performed. In tobacco cells, the antibodies recognized the 42-kD protein kinase activated by osmotic stress, confirming that the cloned cDNA encodes NtOSAK.
There are several lines of evidence indicating that kinases of the SnRK2 subfamily play an important role in abiotic stress signal transduction. However, so far their biochemical features, and biochemical assays for monitoring their activity, have not been described. Difficulties with obtaining active enzymes of the SnRK2 subfamily suitable for such characterization are the main reason for the lack of such data. Expression of several SnRK2 family members, including NtOSAK, in E. coli yields inactive enzymes. Therefore, the biochemical characterization of NtOSAK purified from tobacco cells subject to osmotic stress has been undertaken. The data presented here describe the substrate specificity, divalent cation preferences, phosphate donor specificity, and effect of potential modulators on NtOSAK activity.
Our previously published results (Mikołajczyk et al., 2000) showed that NtOSAK is a monomeric 42-kD enzyme, phosphorylating a broad spectrum of proteins (casein, histone H1, MBP, and transcription factors c-Jun and ATF-2). In this paper a more precise substrate specificity study has been performed. Computer analysis using the program PREDIKIN (Brinkworth et al., 2003) predicted a heptapeptide sequence that may be recognized by NtOSAK. Because this sequence resembled to some extent the recognition motif for SNF1/AMPK substrates, the phosphorylation of specific substrates of these kinases by NtOSAK was analyzed. The results showed that the kinetic parameters for GST-ACC (a fusion protein containing a phosphorylation site sequence from a natural AMPK substrate, acetyl-CoA carboxylase), as well as the peptides SAMS and AMARA (both designed as specific substrates for SNF1/AMPK), were comparable with those previously described for phosphorylation catalyzed by AMPK. This indicates that the substrate recognition motif for NtOSAK is similar to that for AMPK (hydrophobic-X-basic-X-X-Ser/Thr-X-X-X-hydrophobic) or plant SnRK1 (hydrophobic-X-basic-X-X-Ser-X-X-X-hydrophobic), where the hydrophobic residue can be M, V, L, I, or F, and the basic residue can be R, K, or H (Dale et al., 1995). The same consensus recognition motif as for SnRK1 was recently described also for SOS2 protein kinase and SOS2-like protein kinases (Gong et al., 2002a, 2002b, 2002c, 2002d), members of the SnRK3 subfamily. The GST-ACC substrate also contains additional determinants, i.e. an amphipathic helix, N-terminal to the hydrophobic residue at P-5 (Scott et al., 2002). These results indicate that all known plant SNF1-related enzymes might have substrate recognition motifs similar to each other, and to animal and yeast AMPK/SNF1 kinases. However, our studies on casein and MBP phosphorylation by NtOSAK have shown very similar kinetic parameters for these substrates. All the substrates have one common feature: in casein, MBP, as well as in histone, and in other NtOSAK substrates, there are sequences similar to those predicted as NtOSAK phosphorylation sites, although they are not perfect matches. Therefore, we do not exclude some differences in the sequence recognized by NtOSAK; perhaps a basic residue at position −3 with respect to phosphorylated Ser/Thr is the main substrate determinant.
All kinases require divalent cations (Mg2+ or Mn2+) to coordinate the phosphate groups of the phosphate donor. NtOSAK, like most Ser/Thr kinases, exhibits a strong preference for Mg2+ over Mn2+ in the phosphorylation reaction. Phosphorylation of casein, as well as of other substrates, performed at optimal concentration of the cations was about 3 times more efficient using Mg2+ than for Mn2+. The cation preference of NtOSAK is similar to that of WPK4 kinase (Ikeda et al., 1999), but is different than that of calcineurin B-like interacting kinases, such as SOS2 protein kinase (Gong et al., 2002a) and PKSs (Shi et al., 1999; Gong et al., 2002b, 2002c, 2002d), which prefer Mn2+ over Mg2+. The various cation preferences indicate differences among the above kinases in binding of the metal ion-ATP substrate. In fact, SOS2 and PKSs kinases exhibited about 10 times lower Km values for ATP than NtOSAK (Gong et al., 2002a, 2002b, 2002c, 2002d). The differences in affinity for ATP could be exploited in designing specific NtOSAK (SnRK2) inhibitor(s), which could be helpful in distinguishing between phosphorylation catalyzed by NtOSAK (SnRK2) and by other kinases exhibiting similar substrate specificity (for example, SOS2, SOS2-like kinases, and SnRK1). It should be mentioned that most of the specific protein kinase inhibitors that have been developed to date are competitive with respect of ATP (Cohen, 1999).
In our studies, we have not yet found a specific NtOSAK inhibitor. Even inhibitors like H89 and quercetin, which inhibit mammalian AMPK (Davies et al., 2000), did not inhibit NtOSAK. Only staurosporine, a relatively nonspecific protein kinase inhibitor, was able to abolish NtOSAK activity. Our results do provide information concerning protein kinase inhibitors that do not have an effect on NtOSAK. These might be helpful in future studies, e.g. for elimination of protein kinases which copurify with NtOSAK (or other kinases of the SnRK2 subfamily) and can mask its activity. For example, during NtOSAK purification we monitored activity using casein as substrate (the cheapest one available) in the presence of 100 μm heparin to abolish protein kinase CK2 activity, which partially copurifies with NtOSAK during the first steps of purification.
NtOSAK is a calcium-independent protein kinase whose activity is not regulated by AMP. In contrast to mammalian AMPK, none of the plant SnRK kinases tested to date have been found to be modulated by AMP.
Results previously presented showed that NtOSAK is regulated by phosphorylation because its activity can be fully abolished by treatment with the Ser/Thr protein phosphatase, PP2A (Mikołajczyk et al., 2000). This is the most likely explanation as to why recombinant NtOSAK expressed in E. coli is not active. This feature is common to many protein kinases involved in signal transduction (Johnson et al., 1996). It is known that the activity of kinases from the SNF1/AMPK family is regulated by phosphorylation on specific Thr residues within the “T-loop” by upstream kinases (Hawley et al., 1996; McCartney and Schmidt, 2001). Recently, several kinases phosphorylating and activating SNF1/AMPK were identified (Hawley et al., 2003; Hong et al., 2003; Nath et al., 2003; Sutherland et al., 2003). Taking into account that NtOSAK is a rather small protein with no evident regulatory domain apart from a stretch of acidic amino acids at the C terminus, and that the enzyme is activated very rapidly (it is fully active after 1 min of osmotic stress), phosphorylation/dephosphorylation seems to be the main regulatory mechanism of its activity. The phosphorylation-mediated activation mechanism of SnRK2 members was recently suggested (Kobayashi et al., 2004).
To our knowledge, the results presented here are the first data describing the biochemical features of protein kinases from the SnRK2 subfamily involved in stress signaling pathways in plants. The results indicate that SnRK2 kinases exhibit similar substrate specificity to the yeast and animal SNF1/AMPK kinases, and to plant kinases belonging to the other SnRK subfamilies (SnRK1 and SnRK3), even though they have different structures outside of the kinase domain. Our results show that they differ in their requirement for divalent cations and affinity for ATP. The results obtained should be useful for designing specific kinase inhibitors and for monitoring SnRK2 activity in physiological studies.
MATERIALS AND METHODS
Cloning of cDNA Encoding NtOSAK
Two primers, 5′-GGGAATTTTGGGGTGGCTAGG-3′ (forward primer) and 5′-CATGATTTCCTCCACACTCTG-3′ (reverse primer; synthesized at IBB PAS, Warsaw), which correspond to peptide 1 and peptide 5 (Fig. 1A), respectively, were used to amplify DNA encoding NtOSAK by PCR. The first strand cDNA was obtained by RT using SUPERSCRIPT II (Invitrogen, Carlsbad, CA) on RNA prepared from BY-2 cells treated for 15 h with 250 mm NaCl. RNA was isolated using TRI REAGENT (MRC, Cincinnati) according to the procedure recommended by the manufacturer. The RT-PCR product (about 960 bp) was cloned into pGEM-T Easy vector (Promega, Madison, WI) and sequenced. A clone whose deduced amino acid sequence matched the internal peptides was labeled with [α-32P]dCTP (Amersham Pharmacia AB) and was used to screen a BY-2 cells cDNA library (obtained from Professor Chaubet, University of Strasbourg, France) under high stringency. Briefly, phage plaques were transferred onto nylon membranes (Hybond N; Amersham Pharmacia Biotech AB) and the bound DNA hybridized with the labeled probe at 42°C overnight in hybridizing solution containing 50% formamide, 5× Denhardt's solution, 5× sodium chloride/sodium phosphate/EDTA, 0.1% SDS, and 100 μm denatured herring sperm DNA. After hybridization, the filters were washed twice (30 min each) with 2× SSC, 0.1% SDS at room temperature, once (30 min) with 0.1× SSC, 0.1% SDS at room temperature, once (30 min) with 0.1× SSC, 0.1% SDS at 55°C. Two positive clones were obtained by screening about 5 × 105 plaque-forming units. The DNA insert of one of them was sequenced on both strands.
Expression of GST-NtOSAK in Escherichia coli
For expression of GST-NtOSAK in bacteria, full-length NtOSAK cDNA was cloned as XhoI/NotI fragment into pGEX-4T-3 (Amersham Pharmacia Biotech AB) using the following primers: 5′-CCCCTCGAGTGGTGGTGCGGATAAATACGAGCTTGTGAAAGATATAGGGTCA-3′ (forward primer) and 5′-ATAGTTATGCGGCCGCTACCACCTGCTGTGAGACGAACTTCTCCGCTTTCATG-3′ (reverse primer).
The sequence of the plasmid was confirmed by DNA sequencing.
The GST-NtOSAK construct was transformed into E. coli BL21-DE3, and the GST fusion protein was expressed and purified using gluthathione-agarose beads according to the manufacturer's instruction (Amersham Pharmacia Biotech AB).
Cell Culture and Treatment
BY-2 tobacco (Nicotiana tabacum) cells, kindly provided by Professor Witold Filipowicz (Friedrich-Miescher Institute, Basel), were cultured under conditions described elsewhere (Nagata et al., 1992) in Murashige and Skoog medium supplemented with 100 mg/L myoinositol, 1 mg/L thiamine HCl, 255 mg/L KH2PO4, 0.2 mg/L 2,4-D, and 3% Suc. They were subcultured every 7 d.
The cells were treated with 250 mm NaCl for 5 min, harvested by centrifugation, quickly frozen in liquid nitrogen, and stored at −80°C until analyzed.
Preparation of Protein Extracts from Tobacco Cells
The cells were sonicated three times for 20 s in the extraction buffer (20 mm Tris, pH 7.5, 2 mm EDTA, 2 mm EGTA 50 mm β-glycerophosphate, 100 μm Na3VO4, 2 mm dithiothreitol [DTT], 500 μm phenylmethylsulfonyl fluoride, 1 μm pepstatin, 1 μm leupeptin, 1 μm aprotinin, and 250 mm Suc) with a Sonifier 250 Sonicator (Branson Ultrasonics, Danburg, CT) using approximately 1 mL of the extraction buffer for each 2 mL of cells. After sonication, the extracts were centrifuged at 100,000g for 60 min at 4°C, and the supernatants were used for further studies.
Immunoblot Analysis
Anti-NtOSAK specific antibodies were raised against the C-terminal peptide (KQVQQAHESGEVRLT) of the kinase, the sequence of which was predicted from the cloned cDNA (see “Cloning of cDNA Encoding NtOSAK”). Polyclonal antisera were raised in rabbits and purified by affinity chromatography (BioGenes, Berlin).
Western-blot analysis was performed as described previously (Mikołajczyk et al., 2000). Proteins from crude extracts were separated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose by electroblotting. The membrane was blocked for 2 h at room temperature in Tris-buffered saline plus Tween 20 buffer (10 mm Tris, pH 7.5, 100 mm NaCl, 0.1% Tween 20) containing 5% dry milk and then incubated for 2 h in Tris-buffered saline plus Tween 20 buffer with primary anti-NtOSAK antibodies at a dilution of 1:1,000. After removing unbound antibodies by extensive washing, the blots were incubated with an alkaline phosphatase-conjugated secondary antibody and visualized using nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate.
Immunoprecipitation and Immunocomplex Kinase Activity Assay
Immunoprecipitation was performed as described previously (Mikołajczyk et al., 2000). Proteins from crude extracts (50 μg) or from fractions obtained after MonoQ chromatography (0.1 μg) were incubated with anti-NtOSAK antibody (24 μg) in immunoprecipitation buffer (20 mm Tris, pH 7.5, 2 mm EDTA, 2 mm EGTA, 50 mm β-glycerophosphate, 100 μm Na3VO4, 2 mm DTT, 500 μm phenylmethylsulfonyl fluoride, 1 μm pepstatin, 1 μm leupeptin, 1 μm aprotinin, 1% Triton X-100, 150 mm NaCl) at 4°C for 4 h on a rocker. Approximately 25 μL of packed volume of protein A-agarose (Santa Cruz Biotechnology, Santa Cruz, CA) was added, and the incubation was continued for another 2 h. Agarose bead-protein complexes were pelleted by brief centrifugation and washed three times with the immunoprecipitation buffer.
Ten microliters 3× sample buffer was added to the pelleted agarose bead-protein complex, and the sample was heated at 95°C for 3 min. After brief centrifugation, the supernatant was analyzed by the in-gel kinase activity assay using MBP or casein as substrates. In gel kinase activity assays were performed as described below.
In-Gel Kinase Activity Assay
In-gel kinase activity assays were performed as described previously (Mikołajczyk et al., 2000). Aliquots of protein were electrophoresed in 10% SDS-polyacrylamide gels containing 0.5 mg/mL of MBP or casein. After denaturation with 6 m guanidine HCl followed by renaturation of the separated proteins, the kinase activity was assayed in the reaction buffer (10 mm Tris, pH 7.5, 2 mm DTT, 0.1 mm EGTA, 15 mm MgCl2, 20 μm ATP) supplemented with 50 μCi of [γ32P]ATP (Amersham Pharmacia Biotech AB). The unincorporated [γ32P]ATP was removed by extensive washing of the gels in 5% TCA with 1% sodium phosphate. Finally, the gels were stained with Coomassie Brilliant Blue R 250, dried, and exposed to Amersham Hyperfilm MP (Amersham Pharmacia Biotech AB).
Protein Kinase Assays in Solution
Phosphorylation of protein and peptide substrates for NtOSAK kinetic studies: 10-μL aliquots of purified enzyme (about 0.08 μg) were incubated with one of the protein (MBP, casein, GST-ACC) or peptide (SAMS, AMARA) substrates in concentrations as indicated in “Results” and with 50 μm ATP supplemented with 1 μCi [γ32P]ATP (Amersham Pharmacia Biotech AB) in kinase buffer (25 mm β-glycerophosphate, pH 7.5, 1.25 mm EGTA, 1 mm DTT, 30 mm MgCl2, and 150 μm Na3VO4). The final volume of an incubation sample was 25 μL. After 10 min of incubation at 37°C, 20 μL of the mixture was spotted onto a P81 phosphocellulose (in case of MBP and the peptides) or onto a 3 MM filter strip (in case of casein), and the reaction was terminated by washing the filter with 0.85% H3PO4 or with 5% TCA, respectively. The radioactivity incorporated into protein/peptide was measured by scintillation counting. In the case of GST-ACC, the phosphorylation reaction was stopped by addition of Laemmli sample buffer and incubation at 100°C for 5 min. Proteins were separated by SDS-PAGE, and phosphorylated GST-ACC was visualized by autoradiography. The radioactive band of GST-ACC was excised from the dried gel, and the radioactivity incorporated was measured by scintillation counting. For evaluation of Km for ATP as a phosphate donor substrate, concentration (casein) was held constant (1.2 mg/mL, approximately 60 μm), whereas ATP concentration was varied from 1 to 500 μm.
For evaluation of the effect of temperature, protein kinase inhibitors, Cl− or Ca2+ ions, the reaction mixture contained the following: 1.2 mg/mL of casein, 30 mm MgCl2, and 50 μm ATP in kinase buffer.
For determination of the optimal temperature of substrate phosphorylation, reaction mixtures were incubated at different temperatures in the range of 25°C to 50°C.
NtOSAK Purification
NtOSAK was purified to near-homogenity from about 500 g of BY-2 cells treated for 5 min with 250 mm NaCl, according to the procedure described previously (Mikołajczyk et al., 2000). Briefly, protein extract from about 500 g of BY-2 cells treated for 5 min with 250 mm NaCl was loaded onto a 15-mL SOURCE 15Q (Amersham Pharmacia Biotech AB) column previously equilibrated with buffer A (the extraction buffer without Suc). After washing with 150 mL of buffer A, the column was developed with a 300-mL linear gradient (0–0.4 m NaCl) in the same buffer. The 42-kD kinase was eluted at about 300 mm NaCl. The fractions containing the highest kinase activity were pooled and, after adjusting the conductivity to 500 mm NaCl, loaded onto a 10-mL phenyl-Sepharose CL-6B (Amersham Pharmacia Biotech AB) column equilibrated with buffer A supplemented with 500 mm NaCl. The column was washed with 50 mL of the above buffer, followed by 20 mL of buffer A plus 20% ethylene glycol. The enzyme was eluted with buffer A containing 60% ethylene glycol, and 2-mL fractions were collected. Active fractions were pooled and directly applied onto a 2-mL heparin-Sepharose CL-6B (Amersham Pharmacia Biotech AB) column equilibrated with buffer A. The column was washed with 20 mL of buffer A, and for enzyme elution an NaCl step gradient was applied to −5 mL of buffer A plus 200 mm NaCl followed by 5 mL of buffer A plus 500 mm NaCl. The kinase was eluted with 200 mm NaCl. The peak fractions were pooled, desalted using PD10 column (Amersham Pharmacia Biotech AB), and applied onto a Mono Q H5/5 column (Amersham Pharmacia Biotech AB) equilibrated with buffer A. After washing with 30 mL of buffer A, the column was developed with a 100-mL linear gradient (0–0.4 m NaCl) in the same buffer. The flow rate was 1 mL/min, and 1-mL fractions were collected. The active fractions were pooled, concentrated to about 8 μg/mL by using a Centricon 30 (Amicon, Beverly, MA), and stored at −80°C until analyzed.
Calculation of Kinetic Parameters
Km and maximal velocity Vmax for protein and peptide substrates, as well as for ATP, were determined for three independent experiments. Parameters were estimated by fitting of data directly to the Michaelis-Menten equation using GraphPad Prism 3.0 and are quoted ±se. Since the NtOSAK preparation was not homogeneous we could not readily calculate values for kcat.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all parts of the material. Obtaining any permission will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers O65765, Q43466, Q39893, P43291, P43292, AF032465, AAD00239, AY081175, and CAA06503.
Supplementary Material
Acknowledgments
We thank Professor David Shugar for providing DRB and tetrabromoimidazole. We are grateful to Professor Chaubet for the BY-2 cells cDNA library, and Professor Witold Filipowicz for the BY-2 cells. We are also very grateful to all members of our laboratory for stimulating discussions and especially to Arkadiusz Ciesielski for help in the preparation of the manuscript.
This work was supported by the Polish State Committee for Scientific Research (grant no. 6PO4A2719 to G.D.) and by a Programme Grant from the Wellcome Trust (to D.G.H.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046151.
References
- Anderberg RJ, Walker-Simmons MK (1992) Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc Natl Acad Sci USA 89: 10183–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkworth RI, Breinl RA, Kobe B (2003) Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc Natl Acad Sci USA 100: 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling D, Zammit VA, Hardie DG (1987) A common bicyclic protein kinase cascade inactivates the regulatory enzymes of fatty acid and cholesterol biosynthesis. FEBS Lett 223: 217–222 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu J-K (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Cohen P (1999) The development and therapeutic potential of protein kinase inhibitors. Curr Opin Chem Biol 3: 459–465 [DOI] [PubMed] [Google Scholar]
- Dale S, Wilson WA, Edelman AM, Hardie DG (1995) Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase. I. FEBS Lett 361: 191–195 [DOI] [PubMed] [Google Scholar]
- Davies SP, Carling D, Hardie DG (1989) Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur J Biochem 186: 123–128 [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P (2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J 351: 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Gomez-Cadenas A, Zentella R, Walker-Simmons MK, Ho T-HD (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13: 667–679 [PMC free article] [PubMed] [Google Scholar]
- Gong D, Gong Z, Gou Y, Chen X, Zhu J-K (2002. b) Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J Biol Chem 277: 28340–28350 [DOI] [PubMed] [Google Scholar]
- Gong D, Gong Z, Gou Y, Zhu J-K (2002. c) Expression, activation, and biochemical properties of a novel Arabidopsis protein kinase. Plant Physiol 129: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Gou Y, Jagendorf AT, Zhu J-K (2002. a) Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiol 130: 256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Zhang C, Chen X, Gong Z, Zhu J-K (2002. d) Constitutive activation and transgenic evaluation of the function of an Arabidopsis PKS protein kinase. J Biol Chem 277: 42088–42096 [DOI] [PubMed] [Google Scholar]
- Gong D, Guo Y, Schumaker KS, Zhu J-K (2004) The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol 134: 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hardie DG (1998) SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol 37: 735–748 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Zhang Y, Paul MJ (2004) Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. J Exp Bot 55: 35–42 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- Hardie DG (1999) Roles of the AMP-activated/SNF1 protein kinase family in the response to cellular stress. Biochem Soc Symp 64: 13–27 [PubMed] [Google Scholar]
- Hardie DG (2000) Plant protein-serine/threonine kinases: classification into subfamilies and overview of function. In M Kreis, JC Walker, eds, Plant Protein Kinases. Academic Press, San Diego, pp 1–44
- Hardie DG, Carling D, Carlson M (1998) The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem 67: 821–855 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Hawley SA (2001) AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays 23: 1112–1119 [DOI] [PubMed] [Google Scholar]
- Harmon AC (2003) Calcium-regulated protein kinases of plants. Gravit Space Biol Bull 16: 1–8 [PubMed] [Google Scholar]
- Hawley SA, Bordeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG (2003) Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J Biol 2: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG (1996) Characterisation of the AMP-activated protein kinase kinase from rat liver, and identification of threonine-172 as the major site at which it phosphorylates and activates AMP-activated protein kinase. J Biol Chem 271: 27879–27887 [DOI] [PubMed] [Google Scholar]
- Holappa LD, Walker-Simmons MK (1995) The wheat abscisic acid-responsive protein kinase mRNA, PKABA1, is up-regulated by dehydration, cold temperature, and osmotic stress. Plant Physiol 108: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-P, Leiper FC, Woods A, Carling D, Carlson M (2003) Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci USA 100: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos ME, Zhang S (2000) Calcium-independent activation of salicylic acid-induced protein kinase and a 40-kilodalton protein kinase by hyperosmotic stress. Plant Physiol 122: 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman M R, et al (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Koizumi N, Kusano T, Sano H (1999) Sucrose and cytokinin modulation of WPK4, a gene encoding a SNF1-related protein kinase from wheat. Plant Physiol 121: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LN, Noble MEM, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Kearns MA, Monks DE, Fang M, Rivas MP, Courtney PD, Chen J, Prestwich GD, Theibert AB, Dewey RE, Bankaitis VA (1998) Novel developmentally regulated phosphoinositide binding proteins from soybean whose expression bypassed the requirement for an essential phosphatidyl inositol transfer protein in yeast. EMBO J 17: 4004–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of rice sucrose nonfermenting 1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16: 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lee MH, Chung WI, Liu JR (1998) WAPK, a Ser/Thr protein kinase gene of Nicotiana tabacum, is uniquely regulated by wounding, abscisic acid and methyl jasmonate. Mol Gen Genet 259: 516–522 [DOI] [PubMed] [Google Scholar]
- Li J, Assmann SM (1996) An abscisic acid-activated and calcium-independent protein kinase from guard cells of fava bean. Plant Cell 8: 2359–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kinoshita T, Pandey S, Ng CK, Gygi SP, Shimazaki K, Assmann SM (2002) Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418: 793–797 [DOI] [PubMed] [Google Scholar]
- Li J, Wang X-Q, Watson MB, Assmann SM (2000) Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287: 300–303 [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfer U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14 (Suppl): S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney RR, Schmidt MC (2001) Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem 276: 36460–36466 [DOI] [PubMed] [Google Scholar]
- Mikołajczyk M, Awotunde OS, Muszyńska G, Klessig DF, Dobrowolska G (2000) Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell 12: 165–178 [PMC free article] [PubMed] [Google Scholar]
- Monks DE, Aghoram K, Courtney PD, DeWald DB, Dewey RE (2001) Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog trough activation of the protein kinases SPK1 and 2. Plant Cell 13: 1205–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A-C, Merlot S, Vavasseur A, Frenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cells line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Nath N, McCartney RR, Schmidt MC (2003) Yeast Pak1 kinase associates and activates Snf1. Mol Cell Biol 23: 3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba H, Steward N, Kawasaki S, Berberich T, Ikeda Y, Koizumi N, Sano H (2000) Diverse response of rice and maize genes encoding homologs of WPK4, an SNF1-related protein kinase from wheat, to light, nutrients, low temperature and cytokinins. Mol Gen Genet 263: 359–366 [DOI] [PubMed] [Google Scholar]
- Park YS, Hong SW, Oh SA, Kwak JM, Lee HH, Nam HG (1993) Two putative protein kinase genes from Arabidopsis thaliana contain highly acidic domains. Plant Mol Biol 22: 615–624 [DOI] [PubMed] [Google Scholar]
- Sano H, Youssefian S (1994) Light and nutritional regulation of transcripts encoding a wheat protein kinase homolog is mediated by cytokinins. Proc Natl Acad Sci USA 91: 2582–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Norman DG, Hawley SA, Kontogiannis L, Hardie DG (2002) Protein kinase substrate recognition studies using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J Mol Biol 317: 309–323 [DOI] [PubMed] [Google Scholar]
- Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11: 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJR, Schmidt MC, Hardie DG (2003) Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol 13: 1299–1305 [DOI] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H (2004) Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J 23: 1900–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HW, Kim MC, Shin PG, Kim JS, Kim CY, Lee SY, Hwang I, Bahk JD, Hong JC, Han C, et al (1997) Differential expression of two functional serine/threonine protein kinases from soybean that have an unusual acidic domain at the carboxy terminus. Mol Gen Genet 255: 359–371 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.