Abstract
The kinetics of genome-wide responses of gene expression during the acclimation of cells of Synechocystis sp. PCC 6803 to salt stress were followed by DNA-microarray technique and compared to changes in main physiological parameters. During the first 30 min of salt stress, about 240 genes became induced higher than 3-fold, while about 140 genes were repressed. However, most changes in gene expression were only transient and observed among genes for hypothetical proteins. At 24 h after onset of salt stress conditions, the expression of only 39 genes remained significantly enhanced. Among them, many genes that encode proteins essential for salt acclimation were detected, while only a small number of genes for hypothetical proteins remained activated. Following the expression of genes for main functions of the cyanobacterial cell, i.e. PSI, PSII, phycobilisomes, and synthesis of compatible solutes, such as ion homeostasis, distinct kinetic patterns were found. While most of the genes for basal physiological functions were transiently repressed during the 1st h after the onset of salt stress, genes for proteins specifically related to salt acclimation were activated. This gene expression pattern reflects well the changes in main physiological processes in salt-stressed cells, i.e. transient inhibition of photosynthesis and pigment synthesis as well as immediate activation of synthesis of compatible solutes. The results clearly document that following the kinetics of genome-wide expression, profiling can be used to envisage physiological changes in the cyanobacterial cell after certain changes in growth conditions.
Microorganisms have often been used as model organisms to study basic physiological processes in living cells. Acclimation to high-salt concentrations is of high importance for basic as well as applied research, since a high percentage of irrigated land suffers from increasing levels of salts and can no longer be used to grow crop plants, which have generally a rather low halotolerance. The best-characterized photoautotrophic microorganism in relation to salt and osmotic stress response is the moderately halotolerant cyanobacterium Synechocystis sp. strain PCC 6803 (henceforth referred to as Synechocystis). Studies using this cyanobacterium are strongly promoted by the knowledge of its complete genome sequence (Kaneko et al., 1996). On this basis, DNA microarrays have been developed (CyanoCHIP from TaKaRa in Japan; Postier et al., 2003), which allow us to characterize transcriptional changes during environmental stress on the whole-genome level.
The basic response of Synechocystis cells to salt stress is well understood (Hagemann et al., 1999). As in most other living cells, it can be distinguished into three phases. The first reaction after a sudden rise in concentration of NaCl includes a rapid influx of Na+ and Cl− within seconds into the cytoplasm after turgor collapse. The second phase, which occurs in up to 60 min, is characterized by an exchange of Na+ by K+, leading to the elimination of toxic effects of high concentrations of Na+ on the cell metabolism. In the third phase, which lasts for several hours, the synthesis or uptake of compatible solutes takes place to further stabilize the osmotic potential of the cytoplasm and to maintain the highly ordered structure of complex proteins and biological membranes at enhanced concentrations of toxic ions (Reed et al., 1985). Synechocystis is able to acclimate to high concentrations of NaCl up to 1.2 m by synthesis de novo of the compatible solutes glucosylglycerol (GG) and Suc (Reed and Stewart, 1985). Additionally, salt stress activates an ATP-binding cassette (ABC)-type transporter for compatible solutes, such as GG, trehalose, and Suc (Mikkat et al., 1996) and ion-exchangers, such as Na+/H+-antiporters (Inaba et al., 2001; Elanskaya et al., 2002). Furthermore, this organism acclimates to salt stress by tuning the main bioenergetic processes, namely photosynthesis and respiration, in enhancement of cyclic electron-transport activity via PSI and the cytochrome-oxidase activity (Jeanjean et al., 1993), respectively.
Recently, the complete set of salt- and osmo-regulated genes after exposure of Synechocystis cells to salt stress for a short term (30 min) has been identified by DNA-microarray technique. In this study, 28 genes were found to be more than 3-fold induced specifically by salt, 11 genes by osmotic, and 34 genes by both stress treatments, respectively (Kanesaki et al., 2002). Among them, about 50% of the up-regulated genes encode so-called hypothetical proteins, which are not functionally characterized. Interestingly, homologs to the salt-induced genes found in Synechocystis are regulated by salt stress also in higher plants, suggesting that this cyanobacterium serves as a good model system in investigation of the response of organisms to salt stress (Bohnert et al., 2001). DNA microarrays were also successfully applied to study high light, UV, cold, and iron-starvation stress-induced genes in this model strain (Suzuki et al., 2000; Hihara et al., 2001; Huang et al., 2002; Singh et al., 2003) or for identification of regulatory proteins, which are involved in the stress-induced activation of gene expression. Main focus has been directed to His kinases as sensory compounds of two-component signal transduction systems, which coordinate responses to environmental changes in prokaryotes as well as some plants, fungi, protozoa, and archaea. The Synechocystis genome encodes 42 His kinases (Mizuno et al., 1996), and some of them have been characterized as sensors involved in the acclimation to cold, phosphate starvation, and light stress as well as osmotic and salt stress (Hirani et al., 2001; Suzuki et al., 2001, 2004; Mikami et al., 2002; Marin et al., 2003).
In this study, we intended to evaluate the kinetics of acclimation to salt stress by following changes in salt-regulated expression of genes by DNA-microarray analysis. Since this technique measures relative changes in mRNA levels and biological functions are fulfilled by proteins, it is a matter of discussion whether or not microarray data can shed light on physiological processes. Therefore, in parallel changes in main physiological parameters during salt acclimation were analyzed to evaluate the relationship between transcriptional and physiological changes. In general, surprisingly good correlations between kinetics of gene expression and of activities of corresponding processes have been found. However, even after comparisons of gene expression and physiological processes, it remains difficult to assign functions to the increasingly expressed genes, which encode proteins of unknown function during salt acclimation of Synechocystis cells.
RESULTS
Salt-Induced Gene Expression Is a Dynamic Process
During the experiments, cells were harvested at various time points (0 h or control before NaCl addition, 0.25, 0.5, 2, 6, and 24 h, respectively) after addition of 684 mm NaCl that corresponds to 4% (w/v). Since gene expression and physiological data of cyanobacterial cells depend critically on growth parameters, all experiments were performed at exactly the same light, CO2, and nutrient conditions. From a subset of samples, total RNA was isolated and used for genome-wide transcriptional analyses using a DNA-microarray technique. In a control experiment, total RNA samples from two independent control cultures were used for cDNA synthesis with Cy5 and Cy3, respectively. In the subsequent DNA-microarray experiment with these two cDNAs from control cells, for all genes induction values around 1 were found, which were always inside the range of 0.5 to 2.0 (data not shown). Since for the time course one representative experiment was chosen, a gene was regarded as induced when the induction factor was higher than 3.0 and regarded as repressed when the induction factor was lower than 0.33. In long-term salt-acclimated cells, genes were regarded as induced or repressed when exceeding the factors 2.0 and 0.5 which were found to represent significant changes (Kanesaki et al., 2002).
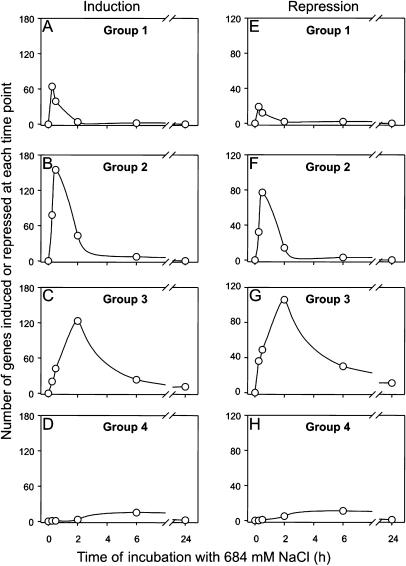
The salt stress conditions used here induced the expression of approximately 360 genes and repressed the expression of approximately 200 genes, which revealed different kinetics. According to the time of maximum induction or repression, the salt-stress-regulated genes were categorized into four groups: Group 1 included all genes that revealed a maximum of induction or repression at 0.25 h (64/19 genes), group 2 included all genes with a maximum of induction or repression at 0.5 h (155/77 genes), group 3 included genes with a maximum of induction or repression at 2 h (123/106 genes), and group 4 included genes with a maximum of induction or repression at 6 h (15/11 genes) after salt shock, respectively. In Figure 1, the induction or repression of genes from the different groups was followed for each time point during salt stress acclimation. The top 10 genes, which showed the highest induction factor in each group, are listed in Table I.
Figure 1.
Global changes in gene expression during the acclimation of cells of Synechocystis to salt stress due to 684 mm NaCl. In a genome-wide DNA-microarray experiment, the number of genes found to be maximum induced (A–D) or repressed (E–H) at the time points 0.2, 0.5, 2, and 6 h after the onset of salt stress, respectively, was calculated, and these genes were assigned to different groups (1–4). For all genes of the different groups, the induction was followed in time, and the results are presented for each group of genes regarded as induced or repressed. A gene was regarded as induced or repressed if the induction factor was higher than 3 or lower than 0.33, respectively. The data set represented here was obtained from one representative time course experiment. For each time point, one array was hybridized.
Table I.
Top 10 genes that showed the highest induction factors in each of the groups of maximally induced genes at 0.2, 0.5, 2, and 6 h during acclimation of Synechocystis to 684 mm NaCl (4%, w/v) from DNA-microarray analysis
| ORFa | Gene | Function | Adb | Ind-Factorc | Exp Devd |
|---|---|---|---|---|---|
| Group1 | |||||
| sll1192 | CRCB-like protein | 6.9 | 0.8 | ||
| sll0871 | Unknowne | 6.7 | 0.3 | ||
| sll1082 | ATP-binding protein of an ABC transport system | 6.6 | 0.2 | ||
| slr0941 | Unknown | 6.2 | 0.3 | ||
| sll1917 | hemN | Oxygen-independent coprophorphyrinogen III oxidase | 6.1 | 0.2 | |
| slr1332 | fabF | β-Ketoacyl-acyl carrier protein synthase | 5.6 | 0.1 | |
| slr0041 | cmpB | Permease of ABC-type bicarbonate transport system | 5.3 | 1.0 | |
| sll1634 | Glucosyltransferase, family 3 | 5.1 | 0.6 | ||
| sll0474 | Sensory transduction His kinase (Hik28) | 5.0 | 0.2 | ||
| slr1647 | Unknown | 4.8 | 0.5 | ||
| Group2 | |||||
| sll0939 | Unknown | a | 86.1 | 9.9 | |
| sll0938 | Asp transaminase | a | 71.7 | 16.3 | |
| slr0895 | Transcriptional regulator | 44.8 | 9.6 | ||
| sll1653 | menG | 2-Phytyl-1,4-benzoquinone methyltransferase | b | 40.0 | 3.5 |
| slr0959 | Probable metal dependent protease | 39.8 | 6.6 | ||
| slr1932 | Probable α/β-hydrolase | 28.6 | 3.4 | ||
| sll1652 | Unknown | b | 26.9 | 1.7 | |
| slr0967 | Unknown | a | 25.3 | 0.8 | |
| sll1389 | Unknown | 21.2 | 0.2 | ||
| sll1541 | Lignostilbene-α,β-dioxygenase | 21.1 | 0.5 | ||
| Group3 | |||||
| sll1862 | Unknown | c | 265.2 | 51.1 | |
| sll1863 | Unknown | c | 231.5 | 14.3 | |
| sll1566 | ggpS | Glucosylglycerol phosphate synthase | d | 84.5 | 2.4 |
| ssr2595 | hliB | High light-inducible polypeptide | 50.5 | 9.3 | |
| sll0528 | Probable metallo-protease | 50.3 | 1.9 | ||
| sll1085 | glpD | Glycerol-3-phosphate dehydrogenase | d | 30.4 | 2.0 |
| sll1514 | hsp17 | 16.6-kD Small heat shock protein | 23.9 | 5.8 | |
| slr1670 | Unknown | d | 23.2 | 3.5 | |
| slr1687 | nblB | Phycocyanin α-subunit phycocyanobilin lyase | 21.8 | 2.5 | |
| slr1675 | hypA | Hydrogenase expression/formation protein | 21.3 | 0.2 | |
| Group4 | |||||
| slr0765 | mscS | Small-conductance mechano-sensitive channel | 11.3 | 0.7 | |
| slr0423 | rlpA | Rare lipoprotein A | 9.7 | 0.8 | |
| sll1450 | nrtA | Substrate-binding protein of ABC-type NO3-transporter | 6.3 | 0.1 | |
| slr2048 | Unknown | 5.0 | 0.5 | ||
| sll1898 | ctaA | Probable cytochrome oxidase assembly protein | e | 3.8 | 0.7 |
| sll0471 | Unknown | 3.7 | 0.4 | ||
| sll1899 | ctaB | Cytochrome c oxidase folding protein | e | 3.4 | 0.2 |
| slr0678 | exbD | Probable biopolymer transport protein | 3.4 | 0.1 | |
| slr0757 | kaiB1 | Circadian clock protein KaiB homolog | 3.3 | 0.3 | |
| slr1031 | tyrS | Tyrosyl tRNA synthetase | 3.3 | 0.1 |
See also Figure 3.
Open reading frame (ORF) designations according to Kaneko et al. (1996).
Adjacent genes (Ad) on the chromosome are indicated by identical letters.
Induction factor for each gene in a typical experiment
Experimental deviation in a typical experiment (see “Materials and Methods”).
ORFs for proteins of unknown function, so-called hypothetical proteins.
The genes of group 1 were only shortly induced, and after 2 h most of them returned to the original level of expression, while at 24 h the expression of none of these genes was still regarded to be enhanced (Fig. 1A). Similar kinetics were found for the 155 genes of group 2, which showed their maximal induction at 30 min. At the first time point (15 min), 78 genes of group 2 were already regarded as induced, whereas the other 77 genes were newly induced at 30 min. Two hours after salt addition, only 43 genes of group 2 were still regarded as induced and the number decreased further, so that at 24 h the expression of no gene from this group was still enhanced (Fig. 1B). In the same manner, the induction of gene expression was followed for groups 3 and 4. In contrast to groups 1 and 2, the expression of 12 genes of group 3 and 1 gene of group 4 remained higher than 3-fold induced after salt treatment for 24 h (Fig. 1, C and D).
Comparable kinetics of the salt-regulated expression of genes regarded as repressed during salt stress was observed among the groups 1 to 4 (Fig. 1, E–H). However, the number of genes observed as repressed was lower than the number of induced genes, in particular in group 1. In the case of induction and repression, a large number of genes were transiently induced and only a small number of genes were still differently expressed 24 h after salt addition (Fig. 1). As was found previously (Kanesaki et al., 2002; Marin et al., 2003), among the transiently induced genes of the groups 1 to 4, many encode so-called hypothetical proteins of unknown function (for examples of the highest induced genes, see Table I).
After Long-Term Salt Acclimation, Only a Small Number of Genes Remained Induced
The expression of 39 genes was enhanced more than 2-fold after salt treatment for 24 h. Most of them remained enhanced in fully salt-acclimated cells (Table II). The increased expression of these genes was confirmed in three independent experiments encompassing culture of cells, RNA isolation, cDNA synthesis, and DNA-microarray hybridization and evaluation. Most of the stably up-regulated genes belonged to group 3 with the maximum induction around 2 h during salt stress. Many of these genes encode proteins for salt acclimation, namely the ggpS, stpA, glpK, and glpD genes for enzymes involved in GG biosynthesis; the ggtABC genes for the ABC-type translocator for compatible solutes; the gene nhaS6 (sll0556) for one of the six Na+/H+ antiporters in Synechocystis; and the gene slr0765 for one of the nine mechano-sensitive channel-like proteins, such as the gene slr1894 for a probable DNA-binding stress protein. Some other stress-related genes also revealed an enhanced expression 24 h after the salt acclimation but returned to lower values during further cultivation in high salt medium. They include ssr2595 and ssl2542 for high light-induced proteins, sll0430 for the heat shock protein HtpG, and slr0193 for the RNA-binding protein Rbp3 (Table II). Three genes for proteins acting in basic carbohydrate metabolism (pfkA for phosphofructokinase, cbbA for Fru-1,6-biphosphate aldolase, and cfxE for pentose-5-phoshate-3-epimerase) were also found to be up-regulated after salt acclimation for 24 h, but they showed an unchanged expression after long-term salt acclimation. In addition, the genes sll1898 and sll1899 for proteins involved in cytochrome oxidase folding or assembly were detected. The enhanced cytochrome oxidase activity is characteristic for salt-stressed Synechocystis cells (Jeanjean et al., 1993). Two genes for proteins involved in polypeptide maturation, which remove the formyl group of the N-formyl-Met at the N terminus (peptide deformylase, slr1549, pdf) and the N-terminal Met (Met aminopeptidase, slr0786, map), respectively, also showed an increased level of mRNA 24 h after addition of NaCl (Table II).
Table II.
Genes whose salt-induced expression was enhanced with the induction factor higher than 2.0 after salt treatment with 684 mM NaCl (4%, w/v) for 24 h or 5 d
| 24 h
|
5 d
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ORFa | Gene | Function | Adb | Ind-Factorc | Stdevd | Groupe | Ind-Factorf | Stdevg |
| sll1862h | Unknowni | a | 49.87 | 4.51 | 3 | 9.55 | 1.65 | |
| sll1863h | Unknown | a | 25.44 | 1.17 | 3 | 11.37 | 1.04 | |
| sll1566h | ggpS | Glucosylglycerol Phosphate synthase | b | 7.56 | 0.36 | 3 | 3.97 | 1.18 |
| slr0765h | mscS | Mechano-sensitive channel | c | 6.82 | 0.07 | 4 | 3.42 | 0.80 |
| slr0747h | ggtA | ATP-binding subunit of the osmolyte transporter | d | 5.19 | 0.30 | 3 | 2.79 | 0.02 |
| sll0528 | Unknown | 5.04 | 0.34 | 3 | 1.59 | 0.30 | ||
| slr1672h | glpK | Glycerol kinase | b | 5.01 | 0.35 | 3 | 2.74 | 0.71 |
| slr0786h | map | Methionine aminopeptidase | 4.01 | 0.83 | 3 | 2.33 | 0.15 | |
| slr0903 | moaE | Molybdopterin (MPT) converting factor, subunit 2 | 3.73 | 0.20 | 3 | 2.58 | 0.61 | |
| sll1085h | glpD | Glycerol-3-phosphate dehydrogenase | b | 3.53 | 0.55 | 3 | 2.66 | 0.28 |
| slr1894h | Probable DNA-binding stress protein | 3.22 | 0.43 | 3 | 2.51 | 0.24 | ||
| slr1687 | Probable phycobilin lyase | e | 3.14 | 0.12 | 3 | 1.05 | 0.21 | |
| ssr2595 | hliB | High light-inducible protein | h | 3.01 | 0.36 | 3 | 0.85 | 0.02 |
| slr1670h | Unknown | b | 2.89 | 0.08 | 3 | 2.00 | 0.62 | |
| sll1898 | Probably involved in cytochrome aa3 oxidase assembly | f | 2.89 | 0.02 | 4 | 2.06 | 0.14 | |
| slr0898 | nirA | Ferredoxin-nitrite reductase | 2.80 | 0.20 | 3 | 0.82 | 0.13 | |
| slr0746h | stpA | Glucosylgycerol phosphate phosphatase | d | 2.75 | 0.54 | 3 | 2.24 | 0.08 |
| slr0529 | ggtB | Substrate-binding protein of the osmolyte transporter | g | 2.63 | 0.13 | 3 | 1.48 | 0.28 |
| sll1071 | Unknown | 2.61 | 0.67 | * | 0.99 | 0.04 | ||
| slr1673 | spoU | rRNA methylase | b | 2.48 | 0.05 | 3 | 1.74 | 0.36 |
| ssl3044 | Probable ferredoxin | e | 2.44 | 0.68 | 3 | 0.95 | 0.01 | |
| slr1674h | Unknown | b | 2.41 | 0.20 | 3 | 2.94 | 0.22 | |
| slr0193 | rbp3 | RNA-binding protein | 2.24 | 0.51 | 3 | 0.83 | 0.03 | |
| sll1330 | rre37 | Response regulator | 2.23 | 0.13 | 3 | 1.89 | 0.21 | |
| sll0745 | pfkA | Phosphofructokinase | c | 2.23 | 0.24 | 3 | 1.41 | 0.00 |
| sll0018 | cbbA | Fructose-1,6-bisphosphate aldolase | 2.18 | 0.58 | * | 0.88 | 0.04 | |
| sll0556 | nhaS6 | Na+/H+ antiporter | 2.16 | 0.32 | 2 | 0.89 | 0.01 | |
| slr1938 | Putative translation initiation factor EIF-2b subnuit 1 | 2.16 | 0.01 | 3 | 1.64 | 0.21 | ||
| sll1980 | trxA | Thiol-disulfide interchange protein (thioredoxin) | 2.16 | 0.09 | 4 | 1.48 | 0.23 | |
| sll0807 | cfxE | Pentose-5-phosphate-3-epimerase | 2.14 | 0.07 | * | 2.15 | 0.12 | |
| slr1544 | Unknown | h | 2.14 | 0.17 | 2 | 0.99 | 0.27 | |
| sll0430 | htpG | Heat shock protein | 2.13 | 0.07 | 3 | 1.36 | 0.11 | |
| slr0530 | ggtC | Permease subunit of the osmolyte transporter | g | 2.13 | 0.21 | 3 | 1.42 | 0.20 |
| sll1899 | ctaB | Cytochrome c oxidase folding protein | f | 2.09 | 0.24 | 4 | 2.02 | 0.59 |
| slr1932 | Unknown | a | 2.09 | 0.05 | 2 | 1.36 | 0.10 | |
| slr1549 | Polypeptide deformylase | 2.09 | 0.04 | 3 | 1.35 | 0.06 | ||
| sll0828 | nylA | 6-Aminohexanoate-cyclic-dimer hydrolase | 2.07 | 0.47 | 1 | 1.40 | 0.01 | |
| ssl2542 | hilA | High light-inducible protein | 2.05 | 0.14 | 3 | 0.92 | 0.16 | |
| sll1653 | menG | 2-Phytyl-1,4-benzoquinone methyltransferase | 2.04 | 0.41 | 2 | 1.01 | 0.16 | |
Experimental conditions for salt treatment and DNA-microarray analysis are described in “Materials and Methods.”
Open reading frame (ORF) designations according to Kaneko et al. (1996).
Adjacent genes (Ad) on the chromosome are indicated by identical letters.
Induction factor for each gene.
Experimental deviation in a typical experiment.
Group number according to Figure 1 (*, genes maximally expressed 24 h after salt addition).
Induction factor for each gene in cells acclimated for 5 days to 684 mM NaCl.
Standard deviation from 3 independent experiments.
ORFs increasingly expressed in cells acclimated to increasing salt concentrations.
ORFs for proteins of unknown function, so-called hypothetical proteins.
In order to evaluate the salt-dependent expression of genes, RNA was isolated from cells completely acclimated to salt concentrations of 1%, 2%, 3%, and 4% NaCl (w/v), respectively, and analyzed by DNA-microarray technique. Thereby the expressions of 13 genes shown in Table II were stepwise enhanced in correlation to external salt concentrations. In contrast to the most transiently induced genes of groups 1 and 2, only 8 genes existed among the up-regulated genes at 24 h, which encode proteins of unknown function (Table II).
Northern-Blotting Analysis Validates DNA-Microarray Results
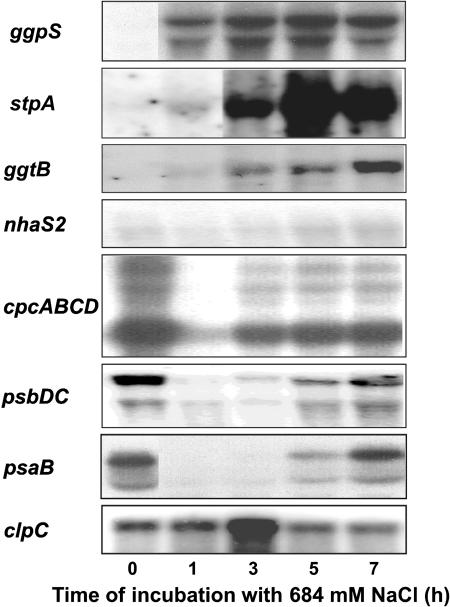
In order to evaluate whether or not DNA-microarray experiments gave a reliable reflection of gene expression pattern in salt-shocked Synechocystis cells, the expression of several genes was followed by northern-blotting experiments (Fig. 2). In all cases, a good correspondence of both methods was found: genes whose expression was induced or repressed by microarray experiments showed the same behavior in northern-blotting assays (compare Fig. 2 and Figs. 3, 4, or 5 for selected genes). For genes encoding enzymes involved in osmoregulation (ggpS, stpA, and ggtB—all group 3 genes) and the stress-related protease subunit ClpC, an induction of the mRNA content was observed, while the mRNA contents of genes for subunits of the PSI, PSII, or phycobiliosmes (psaB, psbCD, and cpcABCD, respectively, all group 3 genes) were drastically repressed. The gene nhaS2 was selected as an example which shows almost no significant changes in the mRNA level in northern-blotting analysis as in DNA-microarray experiments. Furthermore, the comparison of both methods clearly indicates that changes found in DNA-microarray experiments do not only reflect relatively higher or lower levels of mRNA, but also reveal real induction and repression, respectively, of genes.
Figure 2.
Confirmation of the expression profiles obtained through DNA-microarray analysis by northern-blotting analysis of selected salt-regulated genes in Synechocystis cells during acclimation to salt stress due to 684 mm NaCl. With the exception of the nhaS2, all selected genes belong to group 3. RNA was isolated from cells before salt stress and at time points indicated after the addition of NaCl. Five micrograms of RNA were transferred to a nylon membrane and allowed to hybridize with a radioactively labeled probe specific for the genes indicated left. Details of experimental conditions are described in “Materials and Methods.”
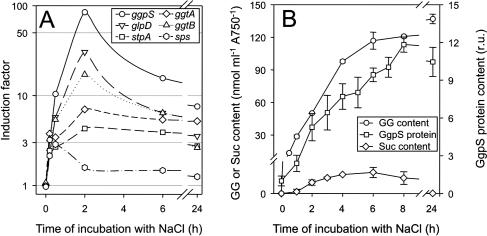
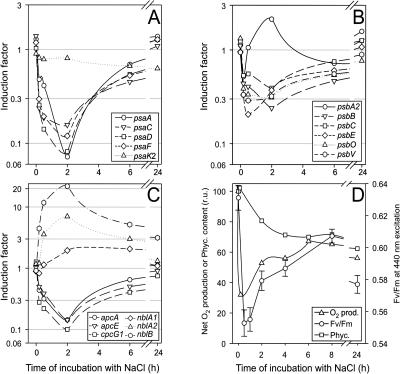
Figure 3.
Comparison of gene expression examined by DNA-microarray analysis with physiological parameters of the synthesis of compatible solutes in cells of Synechocystis during the acclimation to salt stress of 684 mm NaCl. The data set represented here was obtained from one representative time course experiment. For each time point, one array was hybridized. The numbers represent the mean and sds from the two complete sets of genes onto one DNA microarray. Besides the expression of selected genes for proteins of GG synthesis (ggpS, glpD, stpA), Suc synthesis (sps), and uptake of GG (ggtA, ggtB; A), changes of GG-content, GgpS-protein content, and Suc-content (B) were followed during the salt-acclimation process.
Figure 4.
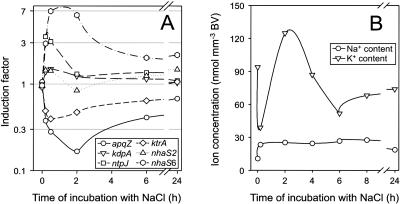
Comparison of gene expression examined by DNA-microarray (see Fig. 3) analysis with physiological parameters related to ion homeostasis in cells of Synechocystis during the acclimation to salt stress of 684 mm NaCl. Besides the expression of selected genes for proteins involved in potassium-uptake systems (kdpA, ntpJ, ktrA), Na+/H+-antiporter (nhaS2, nhaS6), and the water channel (aqpZ; A), changes in the concentration of potassium and sodium ions were followed during the salt-acclimation process.
Synthesis and Uptake of Compatible Solutes
During acclimation to salt stress, very high values of induction were observed for genes, which encode for proteins involved in GG synthesis (ggpS) and in the metabolism of glycerol-3-phosphate (glpD, glpK), a precursor of GG (Fig. 3A). The enhanced level of ggpS-mRNA corresponded to the increased level of GgpS protein as demonstrated by immunoblotting analysis and was also accompanied by an immediate increase in the GG content (Fig. 3B). The transient accumulation of low levels of Suc during salt stress was reflected by the transient induction of the sps gene for the key enzyme in Suc synthesis. Besides the synthesis, the uptake system Ggt of compatible solutes was activated at the level of transcription (Fig. 3A). After 24 h of salt stress, a steady-state level of GG is adjusted; the expression of all genes involved in GG synthesis or GG uptake was still found to be enhanced (Table II), whereby the level of sps transcript returned to the original level and no Suc was further detected.
Ion Transport
Sodium export, potassium uptake, and water flux across the membrane are involved in ion homeostasis during salt stress response in Synechocystis. A transient induction of the genes nhaS1 (slr1727; data not shown), nhaS3 (sll0689; data not shown), and nhaS6 (sll0556) was observed, whereby other genes of this multigene family like nhaS2 (sll0273) were not found to be salt-regulated (Fig. 4A) as also demonstrated by northern-blotting analysis (Fig. 2). By flame photometry, we could not detect the very fast influx of Na+ after the onset of salt treatment as reported for other cyanobacterial strains (Blumwald et al., 1984; Reed et al., 1985). However, the transient efflux and subsequent uptake of K+ is shown and accompanied by a transient induction of expression of the ntpJ (ktrB) gene for a subunit of the Ktr-type potassium-uptake systems, respectively. Interestingly, the ktrA (sll0493) gene for the soluble subunit of the KtrAB system, which is mainly responsible for potassium uptake, in particular after salt shock (Berry et al., 2003), is slightly repressed at the level of transcription. Genes for subunits of the Kdp potassium-uptake system are not induced at the level of transcription as exemplarily shown for kdpA (Fig. 4). For the apqZ gene for the water channel protein, a strong repression of the mRNA content was observed by DNA-microarray analysis (Fig. 4A) and confirmed by northern-blotting analysis (data not shown). In contrast to the genes involved in synthesis or uptake of compatible solutes, all genes involved in ion or water transport were no longer differentially expressed 24 h after salt shock with the exception of the nhaS6 gene (sll0556; Table II).
A Transient Decrease in Photosynthetic Activity
After addition of NaCl, the photosynthetic evolution of oxygen and the maximal efficiency of PSII photochemistry (Fv/Fm) decreased immediately (Fig. 5D). On the level of gene expression, there is also a pronounced decrease in mRNA levels of numerous genes for subunits of PSI (Fig. 5A) and PSII (Fig. 5B). For all main components of PSI, a transient decrease of the RNA levels was observed during salt stress response. Only a few genes for small subunits, such as psaK, showed no significant changes in transcript level. However, 24 h after salt addition, levels of mRNA of all psa genes did not differ from control levels.
Figure 5.
Comparison of gene expression examined by DNA-microarray analysis (see Fig. 3) with physiological parameters of the photosynthetic apparatus in cells of Synechocystis during the acclimation to salt stress of 684 mm NaCl. Besides the expression of selected genes for proteins of PSI (A), PSII (B), and phycobilisomes (C), physiological parameters (D), such as net O2 evolution, yield of PSII fluorescence Fv/Fm, and phycocyanin content were followed during the salt-acclimation process.
Most genes for main components of PSII showed a decreased level of mRNA with the only exception of the psbA genes (Fig. 5B). The psbA2 (slr1311) and psbA3 (sll1867) genes, which were found to be induced after high light stress (Hihara et al., 2001), showed a decreased mRNA content immediately after shock, while a transient increase up to 2.0 was observed at 2 h after salt shock (Fig. 5B). Contrarily, for the psbA1 gene (slr1181), which is not transcribed in Synechocystis (Bouyoub et al., 1993), no change in the signal ratio was found after the salt shock treatment (data not shown). The psbV and psbO genes for proteins associated with the oxygen-evolving complex at the lumenal site of PSII displayed a decrease in levels of mRNA. Compared to the lowered expression of genes for PSI subunits, the decrease in the levels of mRNA was less pronounced among genes for PSII subunits. However, 24 h after salt addition, all genes for PSII subunits showed levels of mRNA comparable to those of control cells (Fig. 5B) as observed for PSI genes.
Levels of light-harvesting pigments, chlorophyll a (data not shown), and phycocyanin (Fig. 5D) changed during salt acclimation of Synechocystis cells. Corresponding to the decrease in the phycocyanin level, a strong decrease in transcript level of genes for phycocyanin and allo-phycocyanin linker proteins (cpc, apc; Fig. 5C) was found. Furthermore, an induction of the nblA genes for polypeptides involved in phycobilisome degradation and an nblB homolog (slr1687) for phycocyanin α-subunit phycocyanobilin lyase was measured. However, 24 h after salt addition, all genes for subunits of phycobilisomes with the exception of nblB displayed unchanged transcript levels in comparison to control cells, whereby the phycocyanin content in salt-acclimated cells remained lower (Fig. 5D).
DISCUSSION
The analyses of salt-induced gene expression at various time points revealed that salt-regulated genes could be categorized into four groups regarding their times of maximal induction. While the early induced genes are mostly induced only transiently, the genes that belong to groups characterized by maximum expression at later time usually showed a longer enhanced expression, and some were also observed as induced in cells acclimated for 24 h or several days to elevated salt concentrations (Table II). Among them are genes that are recognized to be essential for salt acclimation, such as ggpS and stpA involved in GG synthesis (Hagemann et al., 1999) and cfxE (Karandashova et al., 2002).
How far some of the early salt-induced genes are involved in the induction of genes appearing later during salt acclimation should be analyzed in further experiments. Among the early salt-induced genes, two for group 2 sigma factors (SigB, SigD) have been found (Kanesaki et al., 2002; Marin et al., 2003). It has been shown that these sigma factors seem to cooperate with the primary sigma factor by the expression of different subsets of genes (Imamura et al., 2003). However, the expression of the group 3 sigma factor SigF, which was found to be involved in the expression of salt stress proteins (Huckauf et al., 2000), was not significantly changed during salt acclimation. This corresponds to the finding that group 3 sigma factors seem to be constitutively expressed and their activity seems to be regulated on the protein level (Imamura et al., 2003). Nevertheless, the different gene induction kinetics clearly indicate that for studies analyzing putative regulatory proteins, the expression pattern at different time points should be analyzed.
For the investigation of long-term physiological acclimation in relation to gene expression under our conditions, incubation for at least 24 h at higher concentrations of salt was necessary. Thereby a rather small number of genes were recognized that show changes in the gene expression. Among them are only a few genes with unknown function. To investigate their function for salt acclimation, most of the genes (sll1862/63, sll0528, slr1687, slr1670, slr1544, and slr1932), which encode for hypothetical proteins and were found to be increasingly expressed at 24 h after salt shock, were mutated. In each case, completely segregated mutants were obtained. However, none of them showed any significant reduction in salt resistance level compared to cells of the wild type (M. Hagemann and K. Marin, unpublished data). Therefore, even a stable and high up-regulation of genes for unknown proteins such as for the highly induced sll1862/1863 genes does not necessarily indicate that these proteins are essential for salt acclimation. Additional analyses are necessary to elucidate the possible function of these and other hypothetical proteins. In Saccharomyces cerevisiae, only 0.88% of the genes that exhibited a significant increase in mRNA expression after salt shock also showed after mutation a significant decrease in fitness under NaCl stress conditions (Giaever et al., 2002). In Synechocystis, we found that only 15 (3.6%) of the 360 salt-induced genes displayed a stable induction in long-term salt-acclimated cells (Table II), while only 4 of them (1.1%) are essential for salt acclimation according to Karandashova et al. (2002). Contrarily, genes that do not show a stable increase in expression were found to be essential for high salt resistance, such as the ntpJ gene (Berry et al., 2003).
Salt Stress Response Includes Activation of Ion Transport and Synthesis of Compatible Solutes on the Level of Gene Expression
As a reason for the inhibition of photosynthesis and growth after the sudden increase in salt concentration, the influx of high concentrations of sodium ions within seconds into the cytoplasm was discussed (Blumwald et al., 1984; Reed et al., 1985). As a first line of defense, the activation of Na+-export mechanisms such as Na+/H+-antiporters and Na+/ATPases results in the reduction of the sodium concentration to control levels within minutes (Nitschmann and Packer, 1992). By flame photometry, we could not follow the fast influx and efflux of sodium ions. Obviously, such fast transport events could not rely on induction of new channels but are based on the activation of existing ion channels. An immediate activation of transporters by salt has been shown in Corynebacterium glutamicum and also liposomes (Rübenhagen et al., 2001). Nevertheless, on the level of gene expression, we could show an induction of a few ion channels, the Na+/H+ antiporter genes nhaS1, 3, and 6. Thereby, nhaS3 encodes an essential protein homologous to NhaA in Escherichia coli, the induction of which was observed to be essential during salt stress response (Dover et al., 1996; Inaba et al., 2001; Wang et al., 2002). Inactivation of the stably induced genes sll0556 for NhaS6 did not change the salt resistance level of Synechocystis (T. Ogawa and M. Hagemann, unpublished data). The induction of nhaA in E. coli was accompanied by an activation of potassium-uptake systems. Besides the activation of the Kdp, Trk, and Kup systems on the biochemical level, the regulation on the transcriptional level was demonstrated (Csonka and Epstein, 1996; Weber and Jung, 2002). In Synechocystis, Kdp- and Ktr-type potassium transporters are present, whereby the Ktr system was found to be essential for a successful salt acclimation (Berry et al., 2003). In correlation, after a sudden efflux of the accumulated potassium we could follow the reuptake of this ion during salt stress acclimation (Fig. 4B). However, only the expression of the membrane subunit NtpJ (KtrB) became transiently induced, while that of the soluble subunit KtrA did not. Therefore, it is difficult to evaluate the importance of ion transport activities from the expression pattern of the corresponding genes, since regulation of transport activity seems to be the dominating regulatory mechanism.
Two hours after salt addition, the potassium concentration decreased whereby the content of the compatible solute GG increased in parallel. An exchange of high concentrations of potassium and chloride ions by GG was also demonstrated for Synechocystis sp. strain PCC 6714 (Reed et al., 1985). As a fast response, the synthesis of the compatible solutes is activated biochemically in Synechocystis (Hagemann et al., 1999). In addition, all genes involved in GG synthesis were found to be induced at high levels (ggpS, stpA, glpD; Figs. 2 and 3), and the ggpS mRNA was translated to increase the level of GgpS protein (Fig. 3). Furthermore, the expression of all genes for proteins involved in GG synthesis was still being enhanced at 24 h and also several days after salt addition, underlining the meaning of the transcriptional regulation for long-term acclimation (Table II). A comparable observation was made in DNA-microarray experiments for E. coli, whereby the otsAB genes involved in trehalose synthesis were strongly induced (Weber and Jung, 2002). The accumulation of the compatible solutes seems to be the prerequisite for restoration of the efficiency of photochemistry and photosynthetic oxygen evolution as well as the reduced gene expression, since all the transiently repressed processes recover only when a substantial amount of the compatible solute GG was accumulated (compare Figs. 3B and 5D).
Besides genes for proteins of de novo synthesis of GG, also genes for the GG-uptake system were induced and showed an enhanced long-term expression. This observation is in agreement with the results of Mikkat and Hagemann (2000) and observations made for other bacterial-uptake systems for different compatible solutes (Bremer and Krämer, 2000). A general observation in bacteria during acclimation to enhanced salt concentrations is a decrease in growth rate. Besides the inhibition of the metabolism, turgor collapse was discussed as a reason for growth delay. After accumulation of GG, the water potential of the cytoplasm is stabilized and turgor reestablished. Besides turgor regulation, GG accumulation seems to be involved also in regulation of cell division (Ferjani et al., 2003).
Gene Expression Profiles Reflect the Effects of Salt Stress on Photosynthesis
For a functional interpretation of our results, we compared physiological data obtained during salt stress response with the expression of genes for components involved in the acclimation process. During salt stress, a long known observation in cyanobacteria is the breakdown of photosynthetic oxygen evolution and a decrease in the efficiency of photochemistry (Fig. 5; Reed et al., 1985; Allakhverdiev et al., 2000). This process is also evident on the level of gene expression by a strong decrease in mRNA content of almost all genes for main subunits of PSI and PSII. As an exception, the psbA transcript levels were not significantly affected as shown for psbA2 in Figure 5. In agreement, Allakhverdiev et al. (2002) could demonstrate an inhibition of psbA translation at salt concentrations of 0.5 m NaCl in Synechocystis, which is one of the few published examples for a lack of correspondence between transcript and protein levels. Generally, it is assumed that transcription and translation in bacteria are closely linked; therefore, mRNA and protein level should correspond as shown here for ggpS.
During a decrease in the photosynthetic electron-transport rate, an unchanged absorption of excitation energy could lead to a harmful production of reactive oxygen species. The degradation of the light-harvesting antenna complex, the phycobilisomes, can minimize this problem and was first observed in cyanobacteria during starvation for sulfur or nitrogen (Collier and Grossman, 1992). In Synechococcus sp. strain PCC 7942, the decomposition of phycobilisomes was triggered by a strong induction of the nblA gene during nutrient starvation. In addition, the NblB protein, which displays homologies to phycocyanin α-subunit phycocyanobillin lyase, was involved in regulation of this process (Collier and Grossman, 1994; Dolganov and Grossman, 1999). In Synechocystis, a reduction of the phycocyanin content was also demonstrated during salt acclimation (Fig. 5; Schubert et al., 1993), and the expression of two nblA genes were found to be necessary for phycobilisome degradation (Baier et al., 2001). During salt stress response, we observed a decrease of the mRNA content of genes for allo-phycocyanin and phycocyanin linker proteins (apcA and cpcB; see also Figs. 2 and 5) as well as phycobilisome rod linker or core proteins (cpcCG and apcF). The repression of genes for structural components was accompanied by the induction of both genes for the NblA polypeptides as well as a strong induction of slr1687, an NblB homolog in Synechocystis. Therefore, reduced expression of genes for structural components of phycobilisomes and an increased expression of genes for degrading activities seem to be the basis for the down-regulation of phycobilisome content in salt-acclimated cells.
CONCLUSION
The dynamic process of salt acclimation is multiphasic, and the physiological responses correlate well in many cases with changes in the expression pattern of corresponding genes. Thereby, many known and numerous unknown genes were regarded to be differently expressed. In a functional description, main processes of the cyanobacterial metabolism like light harvesting and photosynthetic electron transport are affected, which is at least accompanied by the repression of genes for main components of the PSI and PSII such as the phycobilisomes. Since in bacteria, transcription and translation are closely connected, one may assume that a significant increase or decrease in mRNA levels should induce similar changes in protein amount, as has been shown for GgpS. The stress response comprises the regulation of ion homeostasis by activation of sodium exporters and potassium-uptake systems such as the synthesis of the compatible solute GG. Most of the processes are activated on the gene expression level only transiently, whereas during long-term acclimation the changed expression of only a few mostly known genes is necessary. It cannot be excluded that among them exist some genes for proteins whose up-regulation does not necessarily mean that they exhibit a function during salt acclimation. Overall, the genome-wide gene expression data obtained by DNA-microarray technique seem to be a valuable tool to predict concerted regulatory and physiological alterations at least during acclimation of a cyanobacterium to an environmental stress.
MATERIALS AND METHODS
Strains and Culture Conditions
A strain of Synechocystis sp. PCC 6803, which was selected for higher transformation efficiency, kindly provided by Dr. S. Shestakov (Moscow State University, Department of Genetics), served as the wild type. Axenic cells were cultivated in batch cultures at 30°C bubbling with CO2-enriched air (5% [v/v]) and permanent illumination (170 μmol m−2 s−1; Osram L58W/32 Lumilux deluxe; Munich) using BG11 buffered with [tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid to pH 8.0 that contained 20 mm Na+ as a standard medium (Rippka et al., 1979). Glass tubes with a diameter of 3.5 cm were used for the cultivation. Cells were harvested daily by centrifugation (4,000g, 20°C, and 10 min) and inoculated into fresh medium at a defined OD750 of 0.7.
Mutation of Selected Genes
Interposon mutagenesis was used to inactivate genes that showed an enhanced expression in salt-acclimated cells (for a detailed description, see Huckauf et al., 2000). The coding sequences of the ORFs were amplified by PCR and cloned into the plasmid pGEMT (Promega, Madison, WI). Into unique restriction sites of the ORFs, antibiotic resistances cartridges were inserted. Such inactivated gene constructs were transformed into cells of Synechocystis, and mutants were selected onto antibiotica-containing selective media. Changes in their genotypes were evaluated by PCR.
DNA-Microarray Analysis
Exponentially growing cells (OD750 approximately 1.0) were harvested from 10 mL of the culture by centrifugation at 4,000g for 5 min at 2°C and then immediately frozen in liquid nitrogen and stored at −80°C. RNA was extracted with hot phenol and chloroform and purified using the High Pure RNA Isolation kit (Roche Diagnostics, Mannheim, Germany).
A Synechocystis DNA microarray (CyanoCHIP) was obtained from TaKaRa (Kyoto). This microarray covered 3,079 of the 3,168 ORFs of Synechocystis, excluding genes for transposases. As hybridization probes, Cy3 dye-labeled or Cy5 dye-labeled cDNAs were used, which were synthesized by reverse transcription of 20 μg of total RNA using an RNA Fluorescence Labeling kit (M-MLV) provided by TaKaRa. After incubation for 16 h, the microarrays were rinsed with 2× SSC (1× SSC is 150 mm NaCl and 15 mm Na-citrate) at room temperature. They were washed with 2× SSC at 60°C for 10 min and 0.2× SSC, 0.1% SDS at 60°C for 5 min. Finally, they were rinsed with distilled water at room temperature. Moisture was removed with an air sprayer before analysis with an array scanner (GMS418; Affymetrix, Santa Clara, CA).
For quantification with the ImaGene version 4.0 program (BioDiscovery, El Segundo, CA), the local background of each spot was subtracted and the signal was normalized by transforming it to the ratio of the spot-specific intensity relative to the total intensity of signals from all genes with the exception of rDNA genes. Therefore, changes in the level of transcript of each gene relative to the total level of mRNAs were calculated. Each gene is spotted twice on the microarray, allowing signal evaluation and error exclusion.
The time-dependent expression of genes such as the analysis of gene expression in cells acclimated to 1%, 2%, or 3% NaCl, respectively, was analyzed by one DNA microarray per sample. The gene expression in cells acclimated for 5 d to 4% NaCl was analyzed by three independent biological replications and three DNA-microarray experiments.
Northern-Blotting Analysis
Cells for these RNA extractions were cultivated in exactly the same conditions that were used for cells in DNA-microarray experiments. However, cells were harvested from cultures independent from that used for RNA isolations in microarray experiments. RNA was extracted and purified as above. Five micrograms of total RNA was transferred onto a nylon membrane (Roti-Nylon plus; Carl Roth GmbH, Karlsruhe, Germany) by semidry blotting. Gene-specific DNA probes for the northern-blotting experiments were obtained by PCR amplification of sequences that encode the genes ggpS (sll1566), psbDC (sll0849/0851), ggtB (slr0529), psaB (slr1834), stpA (slr0746), nhaS2 (sll0273), clpC (sll0020), and cpcBAC2C1 (sll1577/1578/1579/1580). Primers (18-mers) corresponding to the start and stop of the encoding sequences of the corresponding genes were designed according to the known genome sequence of Synechocystis (Kaneko et al., 1996). The DNA was labeled by α-32P-dATP (Amersham Bioscience, Freiburg, Germany) using a random prime labeling kit (MBI Fermentas, St. Leon-Rot, Germany). Hybridization signals were recorded and quantified with a phosphoimager (BAS1000; Fuji, Tokyo).
Physiological Characterization
Cells for physiological characterization were cultivated in exactly the same conditions that were used for cells in DNA-microarray experiments. Salt stress experiments were performed in batch cultures by addition of crystalline NaCl to obtain the final concentration of 684 mm (4% [w/v]). In acclimation experiments, cells were used after precultivation for 5 d at the desired NaCl concentration with daily medium exchange. The content of GG and Suc was analyzed by HPLC (Schoor et al., 1996). The GgpS protein content was analyzed by immunoblotting experiments using a GgpS-specific antibody as described (Marin et al., 2002). Growth and cell density were monitored by the OD750 of cyanobacterial cultures after dilution using a double-beam UV/VIS spectrophotometer (U2000; Hitachi, Ort, Germany). Phycocyanin content was estimated as described by Sigalat and de Kouchkowsky (1975). PSII fluorescence with and without 10 μm 3-(3,4-dichlorophenyl)-1,1-dimethylurea was recorded using a microplate fluorescence reader (Lambda Fluoro 320, Bio-Tek instruments, MWG, Munich) after excitation at 440 nm and emission at 680 nm, respectively. Photosynthetic oxygen evolution was measured with a Clark-type electrode at saturating light intensity of about 150 μmol photons m−2 s−1 inside the cuvette. The quantitative analysis of Na+ and K+ was performed by flame photometry as described (Mikkat et al., 2000).
This work was supported by the Deutsche Forschungsgemeinschaft (grant to M.H.), by the Russian Foundation for Basic Research (grant no. 03–04–48581), by the Russian Science Support Foundation (grant to D.L.), by Grants-in-Aid for Scientific Research (grant no. 13854002 to N.M. and I.S.) and for Exploratory Research (grant no. 14654169 to I.S.), by the Japan Society for the Promotion of Science, by Grants-in-Aid for Scientific Research on Priority Areas (grant nos. 14086207 to N.M. and 15013260 to I.S.), by the Ministry of Education, Science, Sports and Culture of Japan, and by the Salt Science Research Foundation (grant no. 03S1 to I.S.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.045047.
References
- Allakhverdiev SI, Nishiyama Y, Miyairi S, Yamamoto H, Inagaki N, Kanesaki Y, Murata N (2002) Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in Synechocystis. Plant Physiol 130: 1443–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N (2000) Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123: 1047–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier K, Nicklisch S, Grundner C, Reinecke J, Lockau W (2001) Expression of two nblA-homologous genes is required for phycobilisome degradation in nitrogen-starved Synechocystis sp. PCC6803. FEMS Microbiol Lett 195: 35–39 [DOI] [PubMed] [Google Scholar]
- Berry S, Esper B, Karandashova I, Teuber M, Elanskaya I, Rögner M, Hagemann M (2003) Potassium uptake in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 depends on a Ktr-like system encoded by slr1509 (ntpJ). FEBS Lett 548: 53–58 [DOI] [PubMed] [Google Scholar]
- Blumwald E, Wolosin JM, Packer L (1984) Na+/H+ exchange in the cyanobacterium Synechococcus 6311. Biochem Biophys Res Commun 122: 452–459 [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Ayoubi P, Borchert C, Bressan RA, Burnap RL, Cushman JC, Cushman MA, Deyholos M, Fischer R, Galbraith DW, et al (2001) A genomics approach towards salt stress tolerance. Plant Physiol Biochem 39: 295–311 [Google Scholar]
- Bouyoub A, Vernotte C, Astier C (1993) Functional analysis of the two homologous psbA gene copies in Synechocystis PCC 6714 and PCC 6803. Plant Mol Biol 21: 249–258 [DOI] [PubMed] [Google Scholar]
- Bremer E, Krämer R (2000) Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in bacteria. In G Storz, R Hengge-Aronis, eds, Bacterial Stress Responses. ASM Press, Washington, DC, pp 79–97
- Collier JL, Grossman AR (1992) Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: Not all bleaching is the same. J Bacteriol 174: 4718–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier JL, Grossman AR (1994) A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J 13: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka LN, Epstein W (1996) Osmoregulation. In FC Neidhardt, ed, Escherichia coli and Salmonella Cellular and Molecular Biology. ASM Press, Washington, DC, pp 1210–1223
- Dolganov N, Grossman AR (1999) A polypeptide with similarity to phycocyanin alpha-subunit phycocyanobilin lyase involved in degradation of phycobilisomes. J Bacteriol 181: 610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover N, Higgins CF, Carmel O, Rimon A, Pinner E, Padan E (1996) Na+-induced transcription of nhaA, which encodes an Na+/H+ antiporter in Escherichia coli, is positively regulated by nhaR and affected by hns. J Bacteriol 178: 6508–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elanskaya I, Karandashova I, Bogachev A, Hagemann M (2002) Physiological characterization of mutants of the cyanobacterium Synechocystis sp. strain PCC 6803 affected in putative Na+/H+ antiporter encoding genes. Biochemistry 67: 432–440 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Mustardy L, Sulpice R, Marin K, Suzuki I, Hagemann M, Murata N (2003) Glucosylglycerol, a compatible solute, sustains cell division under salt stress. Plant Physiol 131: 1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Hagemann M, Schoor A, Mikkat S, Effmert U, Zuther E, Marin K, Fulda S, Vinnemeier J, Kunert A, Milkowski C, et al (1999) The biochemistry and genetics of the synthesis of osmoprotective compounds in cyanobacteria. In A Oren, ed, Microbiology and Biogeochemistry of Hypersaline Environments. CRC Press, Boca Raton, FL, pp 177–186
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirani TA, Suzuki I, Murata N, Hayashi H, Eaton-Rye JJ (2001) Characterization of a two-component signal transduction system involved in the induction of alkaline phosphatase under phosphate-limiting conditions in Synechocystis sp. PCC 6803. Plant Mol Biol 45: 133–144 [DOI] [PubMed] [Google Scholar]
- Huang L, McCluskey MP, Ni H, LaRossa RA (2002) Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J Bacteriol 184: 6845–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckauf J, Nomura C, Forchhammer K, Hagemann M (2000) Stress responses of Synechocystis sp. strain PCC 6803 mutants impaired in genes encoding putative alternative sigma factors. Microbiology 146: 2877–2889 [DOI] [PubMed] [Google Scholar]
- Imamura S, Yoshihara S, Nakano S, Shiozaki N, Yamada A, Tanaka K, Takahashi H, Asayama M, Shirai M (2003) Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J Mol Biol 325: 857–872 [DOI] [PubMed] [Google Scholar]
- Inaba M, Sakamoto A, Murata N (2001) Functional expression in Escherichia coli of low-affinity and high-affinity Na+(Li+)/H+ antiporters of Synechocystis. J Bacteriol 83: 1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanjean R, Matthijs HCP, Onana B, Havaux M, Joset F (1993) Exposure of the cyanobacterium Synechocystis PCC 6803 to salt stress induces concerted changes in respiration and photosynthesis. Plant Cell Physiol 34: 1073–1079 [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3: 109–136 [DOI] [PubMed] [Google Scholar]
- Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N (2002) Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem Biophys Res Commun 290: 339–348 [DOI] [PubMed] [Google Scholar]
- Karandashova I, Elanskaya I, Marin K, Vinnemeier J, Hagemann M (2002) Identification of genes essential for growth at high salt concentrations using salt-sensitive mutants of the cyanobacterium Synechocystis sp. PCC 6803. Curr Microbiol 44: 184–188 [DOI] [PubMed] [Google Scholar]
- Marin K, Huckauf J, Fulda S, Hagemann M (2002) Salt-dependent expression of glucosylglycerol-phosphate synthase, involved in osmolyte synthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 184: 2870–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin K, Suzuki I, Ribbeck K, Kanesaki Y, Hagemann M, Murata N (2003) Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA 100: 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami K, Kanesaki Y, Suzuki I, Murata N (2002) The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp. PCC 6803. Mol Microbiol 46: 905–915 [DOI] [PubMed] [Google Scholar]
- Mikkat S, Hagemann M (2000) Molecular analysis of the ggtBCD operon of Synechocystis sp. strain PCC 6803 encoding the substrate-binding protein and the transmembrane proteins of an ABC transporter for the osmoprotective compound glucosylglycerol. Arch Microbiol 174: 273–282 [DOI] [PubMed] [Google Scholar]
- Mikkat S, Hagemann M, Schoor A (1996) Active transport of glucosylglycerol is involved in salt adaptation of the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 142: 1725–1732 [DOI] [PubMed] [Google Scholar]
- Mikkat S, Milkowski C, Hagemann M (2000) The gene sll0273 of the cyanobacterium Synechocystis sp. strain PCC 6803 encodes a protein essential for growth at low Na+/K+ ratios. Plant Cell Environ 23: 549–559 [Google Scholar]
- Mizuno T, Kaneko T, Tabata S (1996) Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res 3: 407–414 [DOI] [PubMed] [Google Scholar]
- Nitschmann WH, Packer L (1992) NMR studies on Na+ transport in Synechococcus PCC 6311. Arch Biochem Biophys 294: 347–352 [DOI] [PubMed] [Google Scholar]
- Postier BL, Wang HL, Singh A, Impson L, Andrews HL, Klahn J, Li H, Risinger G, Pesta D, Deyholos M, et al (2003) The construction and use of bacterial DNA microarrays based on an optimized two-stage PCR strategy. BMC Genomics 4: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RH, Stewart WDP (1985) Osmotic adjustment and organic solute accumulation in unicellular cyanobacteria from freshwater and marine habitats. Mar Biol 88: 1–9 [Google Scholar]
- Reed RH, Warr SRC, Richardson DL, Moore DJ, Stewart WDP (1985) Multiphasic osmotic adjustment in a euryhaline cyanobacterium. FEMS Microbiol Lett 28: 225–229 [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–16 [Google Scholar]
- Rübenhagen R, Morbach S, Krämer R (2001) The osmoreactive betaine carrier BetP from Corynebacterium glutamicum is a sensor for cytoplasmic K+. EMBO J 20: 5412–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoor A, Hagemann M, Erdmann N (1996) Nonradiometric assay for glucosylglycerol-synthesizing enzymes involved in the cyanobacterial salt adaptation. J Microbiol Methods 27: 139–145 [Google Scholar]
- Schubert H, Fulda S, Hagemann M (1993) Effects of adaptation to different salt concentrations on photosynthesis and pigmentation of the cyanobacterium Synechocystis sp. PCC 6803. J Plant Physiol 142: 291–295 [Google Scholar]
- Sigalat C, de Kouchkowsky Y (1975) Fractionnement et caracterisation de l'algue bleue unicellulaire Anacystis nidulans. Physiol Veg 13: 243–258 [Google Scholar]
- Singh AK, McIntyre LM, Sherman LA (2003) Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 132: 1825–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I, Kanesaki Y, Mikami K, Kanehisa M, Murata N (2001) Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol 40: 235–244 [DOI] [PubMed] [Google Scholar]
- Suzuki I, Los DA, Kanesaki Y, Mikami K, Murata N (2000) The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J 19: 1327–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Ferjani A, Suzuki I, Murata N (2004) The SphS-SphR two-component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J Biol Chem 279: 13234–13240 [DOI] [PubMed] [Google Scholar]
- Wang HL, Postier BL, Burnap RL (2002) Polymerase chain reaction-based mutageneses identify key transporters belonging to multigene families involved in Na+ and pH homeostasis of Synechocystis sp. PCC 6803. Mol Microbiol 44: 1493–1506 [DOI] [PubMed] [Google Scholar]
- Weber A, Jung K (2002) Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J Bacteriol 184: 5502–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]