Abstract
Osimertinib, third-generation epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI), has been approved in the US and EU for the treatment of EGFR mutant T790M-positive non-small cell lung cancer (NSCLC) patients resistant to first- or second-generation EGFR-TKIs, such as gefitinib, erlotinib and afatinib. Although exciting survival data and response rates have been registered in patients treated with this and other third-generation EGFR-TKIs, unfortunately acquired resistance still occurs after approximately 10 months. Mechanisms determining progression of disease are heterogeneous and not fully understood. EGFR-dependent resistance mechanisms (such as new EGFR mutations), bypass pathway activation [as erb-b2 receptor tyrosine kinase 2 (HER2) or MET amplification] and histological transformation [in small cell lung cancer (SCLC)] have been reported, similarly to previous generation TKIs. Here, we review principle mechanisms of innate and acquired resistance described in literature both in clinical and preclinical settings during NSCLC treatment with third-generation EGFR-TKIs.
Keywords: Epidermal growth factor receptor, non-small cell lung cancer (NSCLC), third-generation tyrosine kinase inhibitor, T790M, resistance
Introduction
EGFR mutated lung cancer represents approximately 10–15% of non-small cell lung cancer (NSCLC) in Caucasian population. Exon 19 deletion (del19) and exon 21 p.L858R mutation account for about 85–90% of all EGFR activating mutations and are the most relevant predictive factors of response to EGFR-TKI (1). To date, gefitinib, erlotinib and afatinib are the best therapeutic choice in first-line treatment of patients with advanced EGFR mutated NSCLC (2). However, acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) is an unavoidable process and usually appears after 10–12 months of therapy. The occurrence of a second EGFR mutation p.T790M in exon 20 represents the most frequent mechanisms of acquired resistance with a prevalence ranging between 49% and 63% (3-5). The secondary T790M point mutation increases receptor affinity for ATP binding with a consequent drastic reduction in drug activity. New EGFR-TKIs with specific capability to bind T790M mutated receptor have been developed and successfully tested in patients with acquired resistance (6-8). Moreover, thanks to the higher ability to spare EGFR wild-type counterpart, third-generation TKIs have demonstrated high tolerability. With these evidences, AZD9291 (osimertinib), CO-1686 (rociletinib), HM61713 (olmutinib) and others (EGF816, ASP8273) are object of several clinical trials and osimertinib has already obtained FDA and EMA approval for the treatment of EGFR mutant T790M-positive NSCLC.
Although exciting survival data and response rates have been registered in patients treated with third-generation EGFR-TKIs, unfortunately acquired resistance still occurs after about 10 months (6,7). Mechanisms determining progression of disease are various and not fully understood. Patients who failed treatment with third-generation EGFR-TKIs showed EGFR modifications, alternative pathway activation or histologic transformation, suggestive of overlapping mechanisms of resistance occurring under the intensive pressure of EGFR inhibition.
The aim of this review is to elucidate resistance mechanisms to third-generation EGFR-TKIs that have been described both in clinical and preclinical settings, giving perspectives on possible future therapeutic options to overcome them.
EGFR-dependent
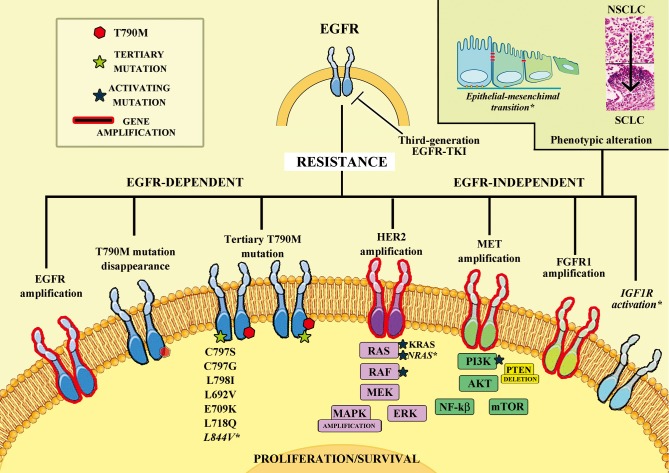
To date, the main mechanisms of resistance to third-generation EGFR-TKIs reported involve EGFR, with new tertiary mutations (C797S and others), similarly to T790M for first- and second-generation TKIs, with EGFR gene amplification and with reduction or disappearance of T790M cell clones (Table 1 and Figure 1).
Table 1. EGFR-dependent mechanisms of resistance to third-generation EGFR-TKIs.
| Mechanism | Author | Sample | N° of patients | T790M | Method | Other mechanisms associated | 3rd TKI |
|---|---|---|---|---|---|---|---|
| C797S | Yu et al. [2015] (9) | Tissue | 1 | Present | NGS | — | Osimertinib |
| Thress et al. [2015] (10) | Plasma/Tissue | 6 | Present | NGS, ddPCR | — | Osimertinib | |
| Chabon et al. [2016] (11) | Plasma | 1 | Present | CAPP-Seq | — | Rociletinib | |
| Song et al. [2016] (12) | Tissue | 1 | Present | NGS | — | Olmutinib | |
| Ortiz-Cuaran et al. [2016] (13) | Tissue | 1 | Present | NGS | Intermediate MET amp [1] | Osimertinib | |
| Other mutations | |||||||
| C797G | Menon et al. [2016] (14) | Tissue | 1 | Present | NGS | EGFR and MYC amp [1] | Osimertinib |
| L798I | Chabon et al. [2016] (11) | Plasma | 1 | Present | CAPP-Seq | EGFR amp [1] | Rociletinib |
| E709K | Plasma | 1 | Present | CAPP-Seq | — | Rociletinib | |
| L692V | Plasma | 1 | Present | CAPP-Seq | — | Rociletinib | |
| L718Q | Bersanelli et al. [2016] (15) | Tissue | 1 | Present | NGS | — | Osimertinib |
| T790M reduction or disappearance; T790M reduction, T790M loss | Chabon et al. [2016] (11) | Plasma | 28 | Reduced | CAPP-Seq | Several mechanisms associated | Rociletinib |
| Piotrowska et al. [2015] (16) | Tissue | 6 | Absent | NGS | SCLC [2] | Osimertinib | |
| Thress et al. [2015] (10) | Plasma | 4 | Absent | ddPCR | — | Osimertinib | |
| Chia et al. [2016] (17) | Tissue | 2 | Absent | ddPCR | MET amp [1] | Osimertinib | |
| EGFR amplification | Menon et al. [2016] (14) | Tissue | 1 | Present | NGS | EGFR C797G and MYC amp [1] | Osimertinib |
| Chabon et al. [2016] (11) | Plasma | 4 | Present | CAPP-Seq | EGFR L798I [1], PIK3CA mut [1], CDKN2A mut [1] | Rociletinib | |
| Piotrowska et al. [2015] (16) | Tissue | 3 | Present | NGS | — | Rociletinib | |
| L844V | Ercan et al. [2015] (18) | Ba/F3 cells | Pre-clinical | — | Site direct mutagenesis | — | WZ4002 |
The number of patients with each specific associated resistance mechanism is indicated in parenthesis. amp, amplification; CAPP-Seq, cancer personal profiling by deep sequencing; ddPCR, droplet digital polymerase chain reaction; mut, mutation; NGS, next generation sequencing; SCLC, small cell lung cancer; 3rd TKI, third-generation tyrosin kinase inhibitor; EGFR-TKI, epidermal growth factor receptor-tyrosine kinase inhibitor; CDKN2A, cyclin dependent kinase inhibitor 2A; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha.
Figure 1.
Mechanisms of resistance to third-generation EGFR TKIs. Schematic representation of innate and acquired resistance described both in clinical and preclinical settings during treatment of non-small cell lung cancer with third-generation epidermal growth factor receptor tyrosine-kinase inhibitors. Mechanisms listed in Italic and with * were observed only in pre-clinical setting. Amp, amplification; del, deletion; EGFR-TKI, epidermal growth factor receptor-tyrosine kinase inhibitor; FGFR1, fibroblast growth factor receptor 1; HER2, erb-b2 receptor tyrosine kinase 2; IGF1R, insulin-like growth factor-1 receptor; EMT, epithelial-mesenchymal transition.
Tertiary EGFR mutations
C797S mutation
The emergence of a new EGFR mutation is one of the first mechanisms described in patients with acquired resistance to third-generation EGFR-TKIs. Similarly to p.T790M, p.C797S occurs in EGFR exon 20 determining the substitution of a cysteine with a serine in the position 797. The aminoacid cysteine located at the position 797 represents the site used by all third-generation EGFR-TKIs for the covalent binding to the receptor, which is necessary to contrast the increased affinity for ATP determined by p.T790M (19). Therefore, the aminoacidic substitution caused by the point mutation translates in the TKI inability to suppress EGFR activity.
Several authors documented the appearance of p.C797S in preclinical setting (18,20). Ercan and colleagues published a study in which mutagenesis was applied to evaluate EGFR mutations conferring resistance to osimertinib, rociletinib or WZ4002 (18). Their results confirm that C797 represents the most common site of acquired mutations conferring resistance to third-generation TKIs. Interestingly, basing on their models, T790M-negative cells with p.C797S could maintain sensitivity to quinazoline-based EGFR inhibitors, such as gefitinib or afatinib. Similarly, Niederst et al. present a study conducted on cell lines treated with increasing doses of WZ4002 and found out that resistant cells expressed C797S point mutation, in cis with p.T790M in 85% of cases (20). They observed that cells with mutations in trans could be sensitive to a combined therapy with first- and third-generation TKI, while those with mutations in cis are resistant to any EGFR-TKI both alone and combined. Finally, they described the emergence of p.C797S in the absence of p.T790M, a possible scenario in case of first-line therapy with third-generation EGFR-TKI; in preclinical models, these cells retained sensitivity to afatinib or gefitinib.
The first evidence of p.C797S isolated in NSCLC patients was documented by Thress et al. (10). The authors analyzed plasmatic samples from 19 patients with acquired resistance to osimertinib and identified the emergence of p.C797S in 6 of them (31%). Considering only patients with p.T790M detectable in pre-treatment samples, the prevalence of p.C797S raises to 40% (6 out of 15). All patients with post-osimertinib p.C797S retained p.T790M after progression and presented EGFR del19 as activating mutation; p.C797S occurred both in cis and in trans with p.T790M. Moreover, in two patients undergone to tumor re-biopsy, they described, by using Next Generation Sequencing (NGS), two different plasmatic DNA alterations encoding for p.C797S (T→A and G→C), while the biopsy only revealed one of them (T→A), highlighting the ability of plasmatic analysis to reflect different tumoral clones.
Similar results were reported in other patients series treated with osimertinib (9,13), while some differences were evidenced after rociletinib treatment (11,16). By using cancer personal profiling by deep sequencing (CAPP-seq), Chabon and colleagues analyzed pre- and post-treatment plasma samples collected from 43 patients treated and progressed to rociletinib (11). The results evidenced a high heterogeneity in acquired resistance mechanisms, stressing the importance of plasmatic monitoring to obtain a wider spectrum of developed alterations. In particular, only one patient out of 43 (2%) presented p.C797S in cis with p.T790M, a lower frequency if compared to osimertinib series (10). These findings were confirmed by Piotrowska et al. who found no p.C797S in a group of 12 patients progressed to rociletinib (16). This raises the hypothesis of different pattern of resistance between rociletinib and osimertinib.
Finally, to our knowledge, only a case report has been published demonstrating the presence of p.C797S, along with p.T790M and EGFR del19, in the lymph node re-biopsy of a patient progressed to olmutinib (12).
Interestingly, recently a variant of C797 mutation has been described in a patient progressed to osimertinib with massive pleural effusion (14). Authors isolated a new p.C797G mutation in cis with T790M and associated with focal MYC and EGFR amplifications.
Other EGFR mutations
In their report, Chabon et al. pointed out the occurrence of rare tertiary mutations in plasma samples of patients progressed to rociletinib (11). Beyond p.C797S mentioned above, they reported subsequent EGFR mutations: p.L798I, p.L692V and p.E709K. Whilst p.E709K and p.L692V have been previously described as activating mutations occurring in EGFR exon 18, this report for the first time describes the point mutation L798I, never isolated before neither in vitro nor in vivo (21,22). L798 residue is located nearby C797 and its modification could theoretically interfere with drug binding. In this patient the mutation was associated to EGFR CNG (Copy Number Gain) and, accordingly with previous observations, coexisted with p.T790M in cis.
Our group published a case report of a patient with activating EGFR L858R initially treated with gefitinib and, after T790M-mediated resistance, with osimertinib (15). When patient progressed to osimertinib, the re-biopsy showed the presence of a new p.L718Q mutation, not detectable in the pre-osimertinib tissue specimen. This mutation has been described before in third-generation TKI-resistant cells and, similarly to p.C797S, cells harboring p.L718Q but p.T790M negative were sensitive to quinazoline-based EGFR-TKIs (18). Another tertiary EGFR mutation was described in preclinical models, p.L844V, responsible of resistance due to interference with drug binding (18). In cell models, when associated to p.T790M, p.L718Q and p.L844V determined resistance to all EGFR-TKIs.
T790M reduction/disappearance
The selective pressure determined by third-generation TKI treatment could result in a reduction or disappearance of T790M mutated neoplastic clones, with consequent acquired resistance, as observed by different authors, including Piotrowska and colleagues (16). Of 64 patients treated with rociletinib in a phase I/II trial, 12 presented sufficient paired pre- and post-therapy biopsy. Six out of 12 patients showed absence of T790M mutation in post-therapy biopsy but 2 of these presented small cell histology transformation. Longitudinal observation, through plasmatic monitoring with BEAMing (beads, emulsion, amplification, and magnetics), allowed to distinguish two different resistance pathways: one with increasing plasmatic levels of p.T790M and activating mutation, reflecting the emergence of a resistant clone still carrying p.T790M and probably with new acquired mechanisms; the other with plasmatic T790M disappearance, suggesting the prevalence of T790M-negative clones no more sensitive to drug inhibition. Plasmatic findings in this study always corresponded to post-progression biopsy results and anticipated evidence of radiological progression, as previously observed with first-generation TKIs (23). An interesting correlation between high baseline plasmatic p.T790M levels and better tumor shrinkage was reported, suggesting that high p.T790M burden, expressed as T790M/activating mutation ratio, could represent a useful tool to predict benefit from rociletinib therapy. Similar results were obtained also by Chabon et al. (11).
In addition, also Thress et al. reported that 4 of 15 T790M-positive patients lost T790M plasmatic expression after progression to osimertinib, remaining positive for EGFR activating mutation, which levels increased after progression (10). T790M disappearance was reported also by Chia et al. in a short communication describing two patients treated with osimertinib (17). At the time of progression to osimertinib, both underwent re-biopsy and p.T790M was not detectable; pre- and post-osimertinib biopsies sites were different for both patients and inter-metastatic heterogeneity may have played a role. In fact, despite T790M-negative biopsy, a patient presented increasing p.T790M plasmatic levels before progression to osimertinib.
EGFR amplification
EGFR amplification was known as a potential mechanism of acquired resistance of first-generation TKI (3,24), but emerging clinical evidences demonstrated that could mediate acquired resistance also after third-generation TKI treatment.
Piotrowska and colleagues observed that three patients developed EGFR amplification in the resistance biopsy, not identified in pre-treatment specimens (16). All three patients maintained activating EGFR and p.T790M mutations along with EGFR amplification. Interestingly one patient presented intrinsic resistance, even if had a significantly lower CNG (6.4) if compared with the other two patients (both reporting CNG >25) progressed after initial response. Moreover, in one of the last two patients, the second post-progression biopsy, in a different anatomic site, showed histological transformation with no EGFR amplification. Also Chabon and colleagues identified somatic copy number alteration (SNCA) involving EGFR gene in plasmatic samples from 4 out of 43 (9%) patients progressed to rociletinib (11). Three of them presented others detectable genetic alterations: EGFR L798I mutation, cyclin dependent kinase inhibitor 2A (CDKN2A) mutation and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) mutation plus ERBB2 SCNA. To determine if EGFR CNG can mediate drug-resistance, they transfected EGFR L858R/T790M double positive cells with lentiviral vectors encoding EGFR and observed a significant decreased of rociletinib inhibitory potency. Moreover, these authors demonstrated that patients with CNGs in pre-rociletinib samples presented higher risk to develop primary resistance. These observations suggest that CNGs could represent negative predictive factor for third-generation TKI therapy. In vitro, the presence of EGFR amplification was reported also by Niederst et al. in cell lines derived from a pleural effusion of an erlotinib resistant patient and exposed to increasing dose of WZ4002 (20).
EGFR-independent
Bypass pathway activation
Similarly to first- and second-generation EGFR-TKIs and ALK-inhibitors, also in case of third-generation TKIs, alternative mechanism of resistance can occur involving bypass pathway. Alterations of several pathways have been evidenced in clinical and/or preclinical studies, such as erb-b2 receptor tyrosine kinase 2 (HER2) and MET amplification, PIK3CA activating mutations, PTEN deletion, RAS mutations and others (Table 2 and Figure 1).
Table 2. EGFR-independent mechanisms of resistance to third-generation EGFR-TKIs.
| Mechanism | Author | Sample | N° of patients | T790M | Method | Other mechanisms associated | 3rd TKI |
|---|---|---|---|---|---|---|---|
| HER2 amplification | Planchard et al. [2015] (25) | Tissue | 1 | Absent | CGH/FISH | — | Osimerinib |
| Oxnard et al. [2015] (26) | Plasma/tissue | 2 | Absent | NGS/CGH | — | Osimertinib | |
| Chabon et al. [2016] (11) | Plasma | 4 | Present [3]; absent [1] | CAPP-Seq | MET amp [1], CDKN2A mut [1], EGFR amp and PIK3CA mut [1] |
Rociletinib | |
| Ortiz-Cuaran et al. [2016] (13) | Tissue | 3 | Present | FISH | MET amp [1] | Rociletinib/Osimertinib | |
| MET amplification | Planchard et al. [2015] (25) | Tissue | 1 | Absent | NGS/CGH/IHC | — | Osimertinib |
| Ou et al. [2016] (27) | Tissue | 1 | 3% | NGS | — | Osimertinib | |
| Chia et al. [2016] (17) | Tissue | 1 | Absent† | ddPCR | — | Osimertinib | |
| Ortiz-Cuaran et al. [2016] (13) | Tissue | 3 | Present | FISH | HER2 amp [1] | Osimertinib | |
| Chabon et al. [2016] (11) | Plasma | 11 | Present [7]; absent [4] | CAPP-Seq | CDKN2A mut [1]; PIK3CA mut [1]; PIK3CA, KRAS and MET mut [1]; HER2 amp [1] |
Rociletinib | |
| PIK3CA mutations | Chabon et al. [2016] (11) | Plasma | 5 | Present [4]; absent [1] | CAPP-Seq | MET amp [1]; MET amp, KRAS and MET mut [1]; EGFR and HER2 amp [1] |
Rociletinib |
| Oxnard et al. [2015] (26) | Biopsy | 1 | Absent | NGS | — | Osimertinib | |
| PTEN loss | Kim et al. [2015] (28) | Tissue | 1 | Present | NGS | — | Osimertinib |
| RAS-MAPK pathway activation | |||||||
| KRAS mut | Ortiz-Cuaran et al. [2016] (13) | Tissue | 1 | Absent | NGS | C797S in plasma | Osimertinib |
| Chabon et al. [2016] (11) | Plasma | 3 | Present | CAPP-Seq | MET amp, PIK3CA mut and MET mut [1]; KIT mut [1] |
Rociletinib | |
| BRAF mut | Oxnard et al. [2015] (26) | Tissue | 1 | Absent | NGS | — | Osimertinib |
| MAPK1/AKT3 overexpression | Kim et al. [2015] (28) | Tissue | 1 | Absent | NGS | — | Osimertinib |
| FGF2-FGFR1 autocrine-loop | Kim et al. [2015] (28) | Tissue | 1 | Absent | NGS | — | Osimertinib |
| SCLC transformation | Piotrowska et al. [2015] (16) | Tissue | 2 | Absent | NGS | — | Rociletinib |
| Kim et al. [2015] (28) | Tissue | 1 | Absent | NGS | — | Osimertinib | |
| Ham et al. [2016] (29) | Tissue | 2 | Absent | NGS | EGFR amp [1] | Osimertinib | |
| EMT | Walter et al. [2013] (30) | NCI-H1975 cells | Pre-clinical | Present | RNA-seq | — | Rociletinib |
| NRAS mutation/CNG | Eberlein et al. [2015] (31) | PC9 cell lines | Pre-clinical | — | NGS | — | Osimertinib |
| IGF1R activation | Park et al. [2016] (32) | PC9 cell lines | Pre-clinical | — | Western blot | — | WZ4002 |
The number of patients with each specific associated resistance mechanism is indicated in parenthesis. †, absent also in plasma sample. amp, amplification; CAPP-Seq, cancer personal profiling by deep sequencing; CGH, comparative genomic hybridization; ddPCR, droplet digital polymerase chain reaction; CNG, copy number gain; mut, mutation; FISH, fluorescent in situ hybridization; IHC, immunohistochemistry; NGS, next generation sequencing; SCLC, small cell lung cancer; EGFR-TKI, epidermal growth factor receptor-tyrosine kinase inhibitor; IGF1R, insulin-like growth factor-1 receptor; EMT, epithelial-mesenchymal transition; FGFR1, fibroblast growth factor receptor 1; HER2, erb-b2 receptor tyrosine kinase 2.
HER2 and MET amplification
HER2 and MET amplification may be considered the second most common findings of acquired resistance under first-generation EGFR-TKIs, seen in 10–20% of patients (3-5).
Planchard et al. reported for the first time HER2 amplification as a potential mechanism of acquired resistance to third-generation TKI (25). One patient treated with osimertinib for more than 12 months developed acquired resistance due to significant HER2 amplification found by comparative genomic hybridization (CGH) analysis in the lung sample and confirmed by fluorescent in situ hybridization (FISH) (HER2/CEP17 ratio: 6.65). NGS analysis showed the absence of EGFR T790M mutation in presence of activating del19 mutation. Absence of HER2 amplification was assessed on pre-treatment samples. The EGFR T790M mutation and HER2 amplification appear to be mutually exclusive as described for first-generation TKIs (33). Similar findings were also presented by Oxnard et al. in 2 of 40 patients treated with osimertinib (26).
The same mechanism of resistance was observed also in cohort of patients treated with rociletinib (11). Four patients presented HER2 amplification in post-treatment specimen: two of these were concurrent with other SCNA and single nucleotide variation (SNV). Despite of the cases treated with osimertinib, the cohort with HER2 amplification treated with rociletinib seems to retain the T790M mutation; only in one patient was not detectable, but he presented a very low level of T790M also at baseline.
Ortiz-Cuaran et al. described in their cohort two cases of HER2 amplification (13). In a patient treated with rociletinib HER2 amplification was detectable already after three weeks of treatment, while for the patient treated with osimertinib was detectable in lung sample biopsy collected before treatment. The authors described another patient treated with osimertinib with concurrent amplification of HER2 and MET, but lacking of pre-treatment sample. These findings lead the authors to hypothesize that HER2 amplification might substitute for EGFR signaling and explain the lack of response to third-generation TKIs occurred in these patients.
Regarding MET amplification, Planchard et al. reported first evidence in a patient treated with osimertinib (25). This case, treated with osimertinib for 10 months until progression of pulmonary disease, showed significant amplification of MET (cMET/CEP7: 5.32) confirmed with CGH analysis and by immunochemistry. NGS analysis showed presence of activating mutation L858R but no EGFR T790M mutation. Due to unavailability of the pre-osimertinib tissue, the authors were not able to demonstrate if MET amplification was absent prior to osimertinib treatment. Instead, Ou et al. compare genomic profile of the pre- and post-osimertinib tumor demonstrating MET amplification as mechanism of acquired resistance to third-generation EGFR-TKI (27). In fact, they reported one osimertinib treated patient that presents high level of MET amplification (30 copies). EGFR T790M mutation was detected at 21% reads immediately prior to starting osimertinib, but only present in about 3% of the sequencing reads in the post-osimertinib progression sample. Clinically, the tumor grew rapidly within two months, indicating MET amplification as a potential potent driver of rapid tumor growth.
Also Ortiz-Cuaran et al. showed high-level amplification of MET either in tumor biopsy collected before treatment in a patient that experienced primary resistance to rociletinib and in the post-treatment biopsy of a patient that developed resistance after stable disease to osimertinib (13). Thanks to in vitro models they could provide functional evidence that HER2 and MET amplification may induce innate and acquired resistance to this new class of EGFR inhibitors, confirming clinical observations (13). Other pre-clinical studies confirmed the role of MET amplification as resistance mechanism to third-generation TKI, suggesting a potential role of MET-inhibitor, alone or in combination, to overcome this resistance (34,35).
In the cohort of patients treated with rociletinib presented by Chabon et al. MET copy number gain was the most frequent mechanism of acquired resistance (11). Among the 43 patients, 11 (26%) had MET amplifications; of these, 7 patients presented only MET amplification, 3 had also SNV in other genes (PIK3CA and CDKN2A) and 1 presented concurrent HER2 amplification, similarly to Ortiz-Cuaran et al. (13). The authors, analyzing an expanded cohort of 16 patients T790M-positive and with MET copy number gain in pre-treatment biopsies or plasma, observed that this group displayed significantly less tumor shrinkage and shorter median progression-free survival (PFS) than patients without MET alterations. These findings underlying that the presence of different mechanisms at the baseline of third-generation TKIs is associated with an inferior therapeutic response to EGFR-TKI.
PIK3CA activating mutations
Activating mutations of the catalytic subunit alpha (PIK3CA) of PI3K lipid kinases family through PI3K/AKT/mTOR pathway characterize 2–4% of adenocarcinoma of the lung in a not-mutually-exclusive manner to other oncogenic driver mechanisms (36,37). Shorter median survival has been described in patients with coexistence of PIK3CA and EGFR mutations, suggesting synergistic effects likely due to stronger activation of the relevant downstream signals (36,37).
Chabon et al. identified two activating mutations, p.E542K and p.E545K, of PIK3CA gene as potential mechanism of acquired resistance in 5 patients treated with rociletinib (11). Only two patients present activating mutations in PIK3CA alone, while the others presented also SCNA in MET, EGFR and HER2 genes. In particular, in a patient that presented concurrence of the p.E542K and MET amplification, the SCNA was presenting also prior to start rociletinib. This patient was classified to have an innate resistance to rociletinib, according to a PFS shorter than 3 months. The subclone with MET copy-number gain increased over the course of therapy while the abundance of two different activating PIK3CA mutations varied over the time. p.E545K was described also in a patient of Oxnard’s cohort (26).
PTEN deletion
PTEN loss was previously described as a mechanism of resistance to EGFR first-generation TKI (38). Recently, Kim et al. reported a case of a patient with EGFR p.T790M mutation and a PTEN deletion before osimertinib therapy and with a following increase of the proportion of tumors with PTEN deletions and EGF mRNA levels in post-treatment tumors (28). This gradual increase of PTEN deletions and EGF overexpression might contribute to focal progression to osimertinib. EGFR mutational analysis confirms the retention of activating and resistance mutations. The limited panel of genes studied and therefore the potential genetic alterations underestimated and the presence of PTEN deletions before osimertinib treatment in a patient with tumor response should be considered in the interpretation of real potential role of PTEN deletion as resistance mechanism.
RAS-MAPK pathway activation
The emergence of KRAS activating mutation in patients treated with first-generation EGFR-TKIs was previously described and postulated as a potential mechanism of escape from EGFR-TKI inhibition (39). Ortiz-Cuaran and colleagues described a patient treated with osimertinib that presented p.C797S in a plasma sample with corresponding re-biopsy C797S and T790M-negative but KRAS G12S-positive (13). EGFR inhibition through osimertinib may functionally deplete oncogenic EGFR signaling to a level that would allow the emergence of cells harboring KRAS mutations. These data are supported by the results of Hata et al. and Unni et al. (40,41). Also Chabon et al. observed the emergence of three KRAS activating mutations (p.G12A, p.Q61H and p.A146T) as a potential mechanism of acquired resistance to rociletinib (11). Only the patient with KRAS p.G12A mutation presented a single mechanism of acquired resistance, while the other two showed heterogeneous mechanisms: concurrent KRAS p.Q61H with PIK3CA p.E81K, MET p.D1304H point mutations and MET amplification and concurrent KRAS p.A146T with KIT p.L576P mutation.
Another gene involved in pathway of RAS-MAPK and associated to acquired resistance was described by Oxnard et al. (26). In a cohort of 40 patients treated with osimertinib NGS analysis performed on tumor biopsy revealed that one patient presented loss of T790M and the presence of p.V600E BRAF mutation.
MAPK1 amplification was described as a resistance mechanism to WZ4002 in pre-clinical study performed by Ercan et al. (42). Kim et al. presented amplification of MAPK1 gene in a patient treated with osimertinib (28).
Eberlein and colleagues conducted a very meaningful pre-clinical study regarding the involvement of RAS-MAPK pathway in acquired resistance to third-generation TKIs (31). With a comparison across 32 populations of cell lines with acquired resistance to different EGFR-TKIs, the authors detected, as frequent mechanisms of resistance to osimertinib, NRAS missense mutations (including a novel E63K mutation) or NRAS copy number gain. All these resistant cell lines were sensitive to inhibition by MEK inhibitor selumetinib in combination with EGFR-TKI. Similar results were registered by Ortiz-Cuaran et al. that observed in vitro that PC9KRAS-G12S treated with osimertinib and trimetinib showed a full inhibition of MAPK signaling (13). Combined therapy was also tested in study published by Tricker et al. where the authors observed a mechanism of WZ4002 acquired resistance mediated by the rapidly reactivation of ERK1/2 (43). Combination of third-generation TKI with trametinib prevents ERK1/2 reactivation, increases WZ4002-induced apoptosis and inhibits the emergence of resistance in WZ4002-sensitive models.
These results support use of MEK inhibitors, such as selumetinib and trametinib, in combination with new EGFR-TKIs to overcome acquired resistance mechanisms or to delay/prevent resistance to EGFR-TKI. A phase I trial (NCT02143466) testing the combination of osimertinib and selumetinib is ongoing (Table 3).
Table 3. Up-coming combination trials with third generation EGFR-TKIs.
| Eudract Number | No. of arms | Trial phase | No. of estimated patients | Inclusion of patients pre-treated with 3rd generation TKI | EGFR-TKI | Combined drug | Target of the combined drug |
|---|---|---|---|---|---|---|---|
| NCT02496663 | 1 | 1 | 30 | No | Osimertinib | Necitumumab | EGFR |
| NCT02503722 | 1 | 1 | 36 | Yes (in dose escalation phase) | Osimertinib | INK128 | TORC1/2 |
| NCT02520778 | 1 | 1 | 50 | Yes (in dose escalation phase) | Osimertinib | Navitoclax | Bcl2 family |
| NCT02335944 | 1 | 1b/2 | 80 | No | EGF816 | INC280 | MET |
| NCT02323126 | 2 | 2 | 100 | No | EGF816† | Nivolumab† | PD-1 |
| NCT02789345 | 2 | 1 | 74 | No | Osimertinib | Ramucirumab | VEGFR2 |
| Osimertinib | Necitumumab | EGFR | |||||
| NCT02143466 | 3 | 1b | 198 | Yes (depending on the specific cohort) | Osimertinib | Selumetinib | MEK |
| Osimertinib‡ | Durvalumab‡ | PD-L1 | |||||
| Osimertinib | AZD6094 | MET |
†, the other arm will test nivolumab plus INC280; ‡, arm closed due to toxicity. EGFR-TKI, epidermal growth factor receptor-tyrosine kinase inhibitor; PD-1, programmed cell death 1; VEGFR2, vascular endothelial growth factor receptor 2; PD-L1, programmed cell death 1 ligand 1.
FGF2-fibroblast growth factor receptor 1 (FGFR1)
FGF2-FGFR1 autocrine loop-mediated resistance mechanism was described by Kim et al. in one patient treated with osimertinib (28). Osimertinib-resistant tumor harbored focal FGFR1 amplification and displayed approximately 20-fold higher FGF2 mRNA compared with baseline tumor. NGS analysis showed the loss of EGFR T790M mutation in post-osimertinib tumor. This mechanism was supported also by in vitro analysis, where a FGF2 supplement conferred resistance to osimertinib in EGFR-mutant NSCLC cells.
Insulin-like growth factor-1 receptor (IGF1R) pathway
Recently, a preclinical study evidenced, in two cell lines resistant to WZ4002, an aberrant activation of IGF1R accompanied by loss of IGF binding protein-3 (IGFBP3) (32). Down-regulation of IGF1R by shRNA, as well as inhibition of IGF1R activity either by a small molecule or a monoclonal antibody restored the sensitivity to WZ4002 both in vitro and xenograft. These results suggest that activation of the IGF1R pathway associated with IGFBP3 loss can induce an acquired resistance to EGFR-TKI, as WZ4002. Therefore, a combined therapy of IGF1R inhibitors and EGFR-TKIs might be a viable treatment strategy for overcoming acquired resistance or delay/prevent resistance.
Phenotypic alterations
SCLC transformation
Piotrowska et al. reported, for the first time, two patients treated with rociletinib that developed acquired resistance via small cell lung cancer (SCLC) transformation (16). Consistent with previous reports referred to acquired resistance to first-generation of TKI (20), the transformed SCLCs continued to harbor their original EGFR-activating mutations, but not T790M; one patient developed a mutation in RB1 and the other lost expression of RB1, evaluated by immunohistochemistry.
Kim et al. and Ham et al. published the same mechanism of acquired resistance for osimertinib separately (28,29). Ham et al. reported two cases of acquired resistance mediated by SCLC transformation after osimertinib therapy. The two patients presented disease progression after 14 and 18 months, respectively, and histological analysis of tissue biopsies of both showed SCLC, positive for CD56. NGS analysis showed persistence of EGFR activating mutation (L858R mutation for first patient and Del19 for the second one) but loss of T790M. The authors reported for first patient also EGFR gene amplification that is not clear if present before osimertinib treatment. Kim et al. described post-osimertinib tumor with neuroendocrine morphology and expression of CD56, chromogranin A and synaptophysin, not present in pre-treatment. Also in this case NGS analysis revealed the depopulation of EGFR T790M-mutant clones in post-osimertinib tumor with a loss of RB1, similarly to patients described by Piotrowska et al.
Epithelial-mesenchymal transition (EMT)
EMT has been previously associated to EGFR-TKIs resistance in NSCLC (44) and it was firstly presented as a potential in vitro mechanism of resistance to third-generation TKIs by Walter and colleagues (30). They treated cell lines harboring L858R and T790M for several months with increasing doses of rociletinib until developed of resistance. Comparison results of RNA-seq from cell lines that developed acquired resistance with the parental ones underlying a significant enrichment of genes involved in EMT. This finding was also confirmed with qPCR and Western Blot analysis showing an up-regulation of vimentin, AXL, ZEB1, CDH5 and FN1 expression and a down-regulation of E-Cadherin, MIR200B, CLDN4, EPCAM and CLDN7 consistent with a mesenchymal signature in the resistant clones. EGFR expression was moderately reduced in the resistant cell clones compared with the parental cell line and no additional EGFR mutations were observed.
Discussion
Basing on results discussed in this review, the pattern of acquired resistance to third-generation EGFR-TKIs seems to be extremely various and heterogeneous, probably more complex than that of first- and second-generation EGFR-TKIs. Higher heterogeneity may be the result of wider sequencing approaches employed, of more sensitive molecular analysis techniques used and also of the assessment of plasmatic samples in several studies.
In particular, liquid biopsy appears to be the more promising source to fully understand mechanisms of acquired resistance, bypassing the limit of inter-metastatic heterogeneity. This concept is clearly evidenced by Chabon and colleagues who found out evidence of multiple resistance mechanisms at a very high frequency (46% of T790M-mutant patients) (11). However, liquid biopsy presents a relevant limitation, related to the impossibility to detect histological transformation, described as resistance mechanism of all generations EGFR-TKIs (16,28). Invasive and non-invasive biopsy methods have areas of overlap as well as distinct advantages or disadvantages in the evaluation of patients with disease progression on targeted therapies, being together able to highlight multiple mechanisms, as reported by Ortiz-Cuaran et al. (13).
Despite the typology of emerged resistance mechanisms, all studies evidenced the original EGFR activating mutation as detectable at the time of resistance, except only one patient in Kim et al. cohort (28), suggesting that EGFR remains the principal driver for neoplastic clones even after drug selective pressure. For this reason, new EGFR inhibitors and combined therapies with other target agents are under evaluation (Table 3). Jia et al. have recently published the results of preclinical tests of a new molecule, EAI045, obtained from the EGFR allosteric inhibitor EAI001 (45). Whilst EAI045 seems to be inactive towards del19 variants, it demonstrated, when combined to cetuximab, to potently inhibit both double mutant L858R/T790M and triple mutant L858R/T790M/C797S cells.
In a preclinical model of acquired resistance to rociletinib via MET amplification, Chabon and colleagues raised the hypothesis that combination of target therapy for both EGFR an MET genes could overcome drug resistance (11). Rociletinib resistant cells were treated with rociletinib and crizotinib, MET inhibitor, with consequent restoration of rociletinib sensitivity. Similar results were obtained also with a new third-generation EGFR-TKI, as EGF816 combined INC280, a cMET inhibitor (46). Moreover, to address resistance via MET amplification recently a bispecific EGFR-cMET antibody was developed with very encouraging results in vitro and in vivo (47). Similarly, as mentioned above, different studies, presenting activation of RAS-MAPK pathway as mechanism of acquired resistance, provide results of a combination of third-generation TKI with a MEK inhibitor (13,31,43). Overall, these data support the use of a combination of EGFR-TKIs with an inhibitor of a different pathway (MET, MEK, IGFR, etc.) to delay or prevent resistance to EGFR-TKI or to treat patients who have progressed with a specific resistance mechanism. Several trials have been developed and are now recruiting patients, offering combined therapies with third-generation EGFR-TKIs (Table 3).
Other ongoing studies were initiated evaluating combination EGFR-TKIs with a programmed cell death 1 (PD-1) axis inhibitors, based on a presumption that a highly active therapy as an EGFR-TKI could induce immune priming and up-regulation of PD-L1 (48).
About C797S point mutation, the most frequent mechanism of acquired resistance to osimertinib, preclinical data suggested that the presence of the mutation in cis or in trans with p.T790M might have important implications in therapeutic decisions (20). In fact, giving that C797S positive cells seem to retain sensitivity to quinazoline-based EGFR-TKIs, the occurrence in trans is the premise for a combined therapy with first and third-generation TKIs, aiming to suppress C797S and T790M positive alleles respectively. Unfortunately, more frequently the two resistance mutations occur in cis, a condition that determines resistance to all available EGFR-TKIs, even if combined. In this situation, new generation of irreversible and reversible mutant EGFR inhibitors with strong noncovalent binding properties and with high inhibitory activities against the cysteine-mutated L858R/T790M/C797S are in development (49).
These findings raise questions regarding the best treatment sequence in clinic practice. Trials currently ongoing comparing first- with third-generation EGFR inhibitors in TKI-naive patients will be critical to determine not only the clinical efficacy but also the resistance mechanisms to these drugs when used in this setting. In fact, the sequential treatment of a third-generation followed by first-generation TKI should be considered for those patients developing C797S mutation without T790M. Combinations with other target agents (see above), combination of multiple generations EGFR-TKIs as well as of EGFR-TKIs plus EGFR antibodies (18,20) could be more effective than single agent therapy, but it has not been tested in clinic yet. Clinical trials evaluating these different approaches are awaited to further improve the treatment of EGFR-mutated NSCLC.
The acquisition of C797S is more frequent in patients progressed to osimertinib, approximately one third of treated patients (10), than in patients progressed to rociletinib, raising the hypothesis that acquired resistance could be drug-specific. These differences may be due to different potencies or pharmacokinetics of the two drugs, as well as potential off-target activities. Therefore, in case of resistance to rociletinib, combined or sequential therapeutic approaches with first-third generation TKIs may be not so relevant. Sequist et al. published interesting results from a group of patients progressed to rociletinib and successfully treated with osimertinib, opening a possible scenario of sequential strategy with third-generation TKIs (50). This scenario may be analogous to observations in NSCLC ALK positive patients, in whom the next-generation ALK inhibitors (ceritinib, alectinib or brigatinib) can induce responses in patients who developed resistance to the less potent crizotinib (51). Thus, rational sequencing of drugs with different patterns of resistance mechanisms may be a generalizable strategy for maximizing therapeutic benefits. However, recently the clinical development of rociletinib and also of olmutinib was interrupted.
Potential predictive factor of EGFR-TKI resistance were also indicated in this review. The ratio of T790M/activating-mutations (11,16) could predict the patients able to obtain a longer benefit from third-generation TKI, just as the pre-existing copy number gains in some genes like MET, HER2 and EGFR (11,13). In particular, amplification of these genes could lead to an innate resistance to third-generation TKIs and justify a combination therapy. Piotrowska et al. also observed that EGFR amplification is very common findings especially if drug concentration is not above the level needed to suppress adequately the target (16). They speculate that higher drug concentrations or a more potent TKI-agent could not be as susceptible to this resistance mechanism.
In conclusion, the availability of third-generation EGFR-TKIs targeting T790M-mutant-specific NSCLC represents a significant development in the treatment of EGFR-mutated patients. As indicated in this review, escape mechanisms EGFR-dependent or -independent are likely to emerge, highlighting the importance of repeat tumor biopsies and/or to collect plasma circulating tumor DNA (ctDNA) at the time of disease progression. An understanding of the mechanisms of resistance is key in the future development of the next-generation of EGFR-TKIs and of new agent combinations.
Acknowledgements
We thank Lorenzo Cainelli for support in creating figure.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Fang S, Wang Z. EGFR mutations as a prognostic and predictive marker in non-small-cell lung cancer. Drug design, development and therapy. 2014;8:1595-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masters GA, Temin S, Azzoli CG, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2015;33:3488-515. 10.1200/JCO.2015.62.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011;17:1169-80. 10.1158/1078-0432.CCR-10-2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 7.Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med 2015;372:1700-9. 10.1056/NEJMoa1413654 [DOI] [PubMed] [Google Scholar]
- 8.Kim ES. Olmutinib: First Global Approval. Drugs 2016;76:1153-7. 10.1007/s40265-016-0606-z [DOI] [PubMed] [Google Scholar]
- 9.Yu HA, Tian SK, Drilon AE, et al. Acquired Resistance of EGFR-Mutant Lung Cancer to a T790M-Specific EGFR Inhibitor: Emergence of a Third Mutation (C797S) in the EGFR Tyrosine Kinase Domain. JAMA Oncol 2015;1:982-4. 10.1001/jamaoncol.2015.1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. 10.1038/ncomms11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song HN, Jung KS, Yoo KH, et al. Acquired C797S Mutation upon Treatment with a T790M-Specific Third-Generation EGFR Inhibitor (HM61713) in Non–Small Cell Lung Cancer. J Thorac Oncol 2016;11:e45-7. 10.1016/j.jtho.2015.12.093 [DOI] [PubMed] [Google Scholar]
- 13.Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous Mechanisms of Primary and Acquired Resistance to Third-Generation EGFR Inhibitors. Clin Cancer Res 2016;22:4837-47. 10.1158/1078-0432.CCR-15-1915 [DOI] [PubMed] [Google Scholar]
- 14.Menon R, Müller J, Schneider P, et al. A Novel EGFR(C797) Variant Detected in a Pleural Biopsy Specimen from an Osimertinib-Treated Patient Using a Comprehensive Hybrid Capture-Based Next-Generation Sequencing Assay. J Thorac Oncol 2016;11:e105-7. 10.1016/j.jtho.2016.04.005 [DOI] [PubMed] [Google Scholar]
- 15.Bersanelli M, Minari R, Bordi P, et al. L718Q Mutation as New Mechanism of Acquired Resistance to AZD9291 in EGFR-Mutated NSCLC. J Thorac Oncol 2016;11:e121-3. 10.1016/j.jtho.2016.05.019 [DOI] [PubMed] [Google Scholar]
- 16.Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. 10.1158/2159-8290.CD-15-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chia PL, Do H, Morey A, et al. Temporal changes of EGFR mutations and T790M levels in tumour and plasma DNA following AZD9291 treatment. Lung Cancer 2016;98:29-32. 10.1016/j.lungcan.2016.05.003 [DOI] [PubMed] [Google Scholar]
- 18.Ercan D, Choi HG, Yun CH, et al. EGFR Mutations and Resistance to Irreversible Pyrimidine-Based EGFR Inhibitors. Clin Cancer Res 2015;21:3913-23. 10.1158/1078-0432.CCR-14-2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070-4. 10.1038/nature08622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niederst MJ, Hu H, Mulvey HE, et al. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin Cancer Res 2015;21:3924-33. 10.1158/1078-0432.CCR-15-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yam I, Lam DC, Chan K, et al. EGFR array: uses in the detection of plasma EGFR mutations in non-small cell lung cancer patients. J Thorac Oncol 2012;7:1131-40. 10.1097/JTO.0b013e3182558198 [DOI] [PubMed] [Google Scholar]
- 22.Cheng C, Wang R, Li Y, et al. EGFR Exon 18 Mutations in East Asian Patients with Lung Adenocarcinomas: A Comprehensive Investigation of Prevalence, Clinicopathologic Characteristics and Prognosis. Sci Rep 2015;5:13959. 10.1038/srep13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bordi P, Del Re M, Danesi R, et al. Circulating DNA in diagnosis and monitoring EGFR gene mutations in advanced non-small cell lung cancer. Transl Lung Cancer Res 2015;4:584-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ercan D, Zejnullahu K, Yonesaka K, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene 2010;29:2346-56. 10.1038/onc.2009.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Planchard D, Loriot Y, André F, et al. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann Oncol 2015;26:2073-8. 10.1093/annonc/mdv319 [DOI] [PubMed] [Google Scholar]
- 26.Oxnard G. Mechanisms of acquired resistance to AZD9291 in EGFRT790 M positive lung cancer. IASLC 16th World Conf Lung Cancer; September 6-9, 2015; Denver, Colorado 2015. Available online: http://library.iaslc.org/
- 27.Ou SH, Agarwal N, Ali SM. High MET amplification level as a resistance mechanism to osimertinib (AZD9291) in a patient that symptomatically responded to crizotinib treatment post-osimertinib progression. Lung Cancer 2016;98:59-61. 10.1016/j.lungcan.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 28.Kim TM, Song A, Kim DW, et al. Mechanisms of Acquired Resistance to AZD9291: A Mutation-Selective, Irreversible EGFR Inhibitor. J Thorac Oncol 2015;10:1736-44. 10.1097/JTO.0000000000000688 [DOI] [PubMed] [Google Scholar]
- 29.Ham JS, Kim S, Kim HK, et al. Two Cases of Small Cell Lung Cancer Transformation from EGFR Mutant Adenocarcinoma During AZD9291 Treatment. J Thorac Oncol 2016;11:e1-4. 10.1016/j.jtho.2015.09.013 [DOI] [PubMed] [Google Scholar]
- 30.Walter AO, Sjin RT, Haringsma HJ, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discov 2013;3:1404-15. 10.1158/2159-8290.CD-13-0314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eberlein CA, Stetson D, Markovets AA, et al. Acquired Resistance to the Mutant-Selective EGFR Inhibitor AZD9291 Is Associated with Increased Dependence on RAS Signaling in Preclinical Models. Cancer Res 2015;75:2489-500. 10.1158/0008-5472.CAN-14-3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JH, Choi YJ, Kim SY, et al. Activation of the IGF1R pathway potentially mediates acquired resistance to mutant-selective 3rd-generation EGF receptor tyrosine kinase inhibitors in advanced non-small cell lung cancer. Oncotarget 2016;7:22005-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012;2:922-33. 10.1158/2159-8290.CD-12-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi P, Oh YT, Zhang G, et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 2016;380:494-504. 10.1016/j.canlet.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 35.Mizuuchi H, Suda K, Murakami I, et al. Oncogene swap as a novel mechanism of acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor in lung cancer. Cancer Sci 2016;107:461-8. 10.1111/cas.12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaft JE, Arcila ME, Paik PK, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther 2012;11:485-91. 10.1158/1535-7163.MCT-11-0692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludovini V, Bianconi F, Pistola L, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol 2011;6:707-15. 10.1097/JTO.0b013e31820a3a6b [DOI] [PubMed] [Google Scholar]
- 38.Sos ML, Koker M, Weir BA, et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res 2009;69:3256-61. 10.1158/0008-5472.CAN-08-4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del Re M, Tiseo M, Bordi P, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget 2016. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hata AN, Niederst MJ, Archibald HL, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 2016;22:262-9. 10.1038/nm.4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unni AM, Lockwood WW, Zejnullahu K, et al. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. Elife 2015;4:e06907. 10.7554/eLife.06907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ercan D, Xu C, Yanagita M, et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov 2012;2:934-47. 10.1158/2159-8290.CD-12-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tricker EM, Xu C, Uddin S, et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer Discov 2015;5:960-71. 10.1158/2159-8290.CD-15-0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Byers LA , Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279-90. 10.1158/1078-0432.CCR-12-1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 2016;534:129-32. 10.1038/nature17960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia Y, Juarez J, Li J, et al. EGF816 Exerts Anticancer Effects in Non-Small Cell Lung Cancer by Irreversibly and Selectively Targeting Primary and Acquired Activating Mutations in the EGF Receptor. Cancer Res 2016;76:1591-602. 10.1158/0008-5472.CAN-15-2581 [DOI] [PubMed] [Google Scholar]
- 47.Moores SL, Chiu ML, Bushey BS, et al. A Novel Bispecific Antibody Targeting EGFR and cMet Is Effective against EGFR Inhibitor-Resistant Lung Tumors. Cancer Res 2016;76:3942-53. 10.1158/0008-5472.CAN-15-2833 [DOI] [PubMed] [Google Scholar]
- 48.Gettinger S, Politi K. PD-1 Axis Inhibitors in EGFR- and ALK-Driven Lung Cancer: Lost Cause? Clin Cancer Res 2016;22:4539-41. 10.1158/1078-0432.CCR-16-1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Günther M, Juchum M, Kelter G, et al. Lung Cancer: EGFR Inhibitors with Low Nanomolar Activity against a Therapy-Resistant L858R/T790M/C797S Mutant. Angew Chem Int Ed Engl 2016;55:10890-4. 10.1002/anie.201603736 [DOI] [PubMed] [Google Scholar]
- 50.Sequist LV, Piotrowska Z, Niederst MJ, et al. Osimertinib Responses After Disease Progression in Patients Who Had Been Receiving Rociletinib. JAMA Oncol 2016;2:541-3. 10.1001/jamaoncol.2015.5009 [DOI] [PubMed] [Google Scholar]
- 51.Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. 10.1158/2159-8290.CD-13-0846 [DOI] [PMC free article] [PubMed] [Google Scholar]