Abstract
Ultraviolet-B (UV-B) photons can cause substantial cellular damage in biomolecules, as is well established for DNA. Because RNA has the same absorption spectrum for UV as DNA, we have investigated damage to this cellular constituent. In maize (Zea mays) leaves, UV-B radiation damages ribosomes by crosslinking cytosolic ribosomal proteins S14, L23a, and L32, and chloroplast ribosomal protein L29 to RNA. Ribosomal damage accumulated during a day of UV-B exposure correlated with a progressive decrease in new protein production; however, de novo synthesis of some ribosomal proteins is increased after 6 h of UV-B exposure. After 16 h without UV-B, damaged ribosomes were eliminated and translation was restored to normal levels. Ribosomal protein S6 and an S6 kinase are phosphorylated during UV-B exposure; these modifications are associated with selective translation of some ribosomal proteins after ribosome damage in mammalian fibroblast cells and may be an adaptation in maize. Neither photosynthesis nor pigment levels were affected significantly by UV-B, demonstrating that the treatment applied is not lethal and that maize leaf physiology readily recovers.
The evolution of terrestrial life was made possible by formation of a stratosphere that screens solar UV radiation, absorbing UV-C (<280 nm) and the most energetic UV-B (280–315 nm). Recent depletion of stratospheric ozone by chlorofluorocarbons and other pollutants has increased terrestrial UV-B levels with potentially deleterious consequences for all living organisms and particularly for plant development and physiology (Ballaré et al., 2001; Searles et al., 2001; Paul and Gwynn-Jones, 2003). In response to the inevitable exposure to damaging terrestrial UV-B, plants have evolved UV-induced mechanisms of protection and repair, such as accumulation of UV-absorbing pigments and use of UV-A photons to repair UV-B induced DNA damage (Stapleton and Walbot, 1994; Britt, 1996). Because of its absorption spectrum, DNA is a major target of UV-B radiation (Britt, 1996). This radiation can also damage proteins and lipids directly (Gerhardt et al., 1999); RNA molecules strongly absorb UV-B, but less is known about UV-B mediated damage to this cellular constituent.
Experimentally, UV has been extensively used to analyze ribosome structure in vitro, because crosslinks can be introduced at points of close contact between proteins, within ribosomal RNA (rRNA), and between proteins and rRNA, tRNA, or mRNA (Brimacombe et al., 1990; Noah et al., 2000). Furthermore, in vivo crosslinks are generated in rRNA by UV treatment of bacterial cells (Stiege et al., 1986). In mammalian cells, UV-C and UV-B cause specific damage to the 3′-end of the 28S rRNA activating a response called ribotoxic stress (Iordanov et al., 1997, 1998). Protein synthesis in plant cells is also sensitive to UV-C. Murphy et al. (1973) demonstrated that 254 nm radiation disrupted amino acid incorporation into protein in a wheat germ in vitro synthesis reaction and that the most sensitive target was mRNA. Subsequent studies in cultured tobacco (Nicotiana tabacum) cells (Murphy et al., 1974) indicated that 254 nm UV-C slowed amino acid incorporation in vivo within 15 min of exposure at a dose of 1 W m−2. Recently, an Arabidopsis mutant with elevated sensitivity to UV-C was identified; this mutant generated by T-DNA insertion caused a chromosomal deletion in the promoter of one of three S27 ribosomal protein genes preventing its expression (Revenkova et al., 1999). In this paper, the authors propose that S27 protein is required for the elimination of possibly damaged mRNA after UV irradiation.
Of greater biological relevance are the responses of plants to UV-B, because UV-C is absent from the terrestrial spectrum. Previously, using microarray hybridization to assess maize (Zea mays) acclimation responses to UV-B, we found that genes encoding protein translation components were the largest functional group up-regulated by UV-B (Casati and Walbot, 2003). On a microarray containing approximately 2,500 distinct maize cDNA types, there was significant up-regulation of 44 transcripts associated with translation or ribosome assembly. This is very likely an underestimate of the contribution of translation-associated transcripts for two reasons: the cDNAs were sequenced from libraries built from poly(A+) selected transcripts, meaning that organellar transcripts are not represented, and the array contains only about 5% of the expected number of maize genes. Moreover, similar experiments done using additional maize organs exposed to 8-h UV-B supplementation and analyzed on microarray slides with 5,600 cDNA types identified transcripts for other ribosomal proteins as significantly increased in abundance after radiation treatment (Casati and Walbot, 2004). Similarly, Valery et al. (2001) reported up-regulation of transcripts encoding several ribosomal proteins after UV-B treatment of cultured human melanocytes; similar results were shown in experiments with primary human keratinocytes (Sesto et al., 2002).
An increased translation of mRNAs encoding ribosomal proteins and translation factors has been suggested as an additional mechanism for recovery from ribosome damage in animal cells (Meyuhas et al., 1997; Brenneisen et al., 2000). Previously, phosphorylation of the 40S ribosomal protein S6 has been implicated in the translational up-regulation of mRNAs coding for the components of protein synthetic apparatus in specific cultured cell types (Thomas, 2002). Selectively translated mRNAs contain an oligopyrimidine tract at their 5′ termini (TOP); this 5′ TOP motif is essential for increased translation (Meyuhas et al., 1997). Phosphorylation of the ribosomal protein S6 and of the corresponding ribosomal protein S6 kinase signaling pathway occurs upon UV-B irradiation (Brenneisen et al., 2000; Nomura et al., 2001). Brenneisen et al. (2000) demonstrated that the activity of p70 ribosomal S6 kinase is increased in cultured human dermal fibroblasts after UV-B irradiation; they hypothesized that the p70 ribosomal S6 kinase is an essential component of a DNA damage-dependent signaling pathway. Furthermore, Nomura et al. (2001) confirmed that UV-B induces activation of ribosomal p70 S6 kinase in cultured mouse epidermal cells. Recently, the accumulation of hyper-phosphorylated isoforms of S6 were described in root tips of maize; four Ser and one Thr residue can be phosphorylated, and the extent of modification is modulated by environmental conditions (Williams et al., 2003). Consequently, a similar pathway as well as other mechanisms could exist in plant cells that mediate selective translation of ribosomal proteins and translation factors after UV-B damage.
To pursue the observation that genes encoding components of the ribosome were up-regulated by UV-B exposure, we have investigated the possible effects of this irradiation on ribosomes and their function in translation. Maize plants were exposed under conditions that simulate terrestrial wavelengths and UV-B fluence in areas experiencing ozone depletion. Here we show that UV-B inhibits protein synthesis in maize leaves and that crosslinks form between RNA and specific ribosomal proteins. Translation competence is restored after a dark period of 16 h and is correlated with the disappearance of RNA-ribosomal protein crosslinked species. We also show that UV-B specifically increases translation of ribosomal proteins and that ribosomal protein S6 and the S6 kinase are phosphorylated during the irradiation treatment. Consequently, plants cells can increase selective translation of transcripts for ribosomal proteins despite ribosomal damage and the deleterious impact of UV-B radiation on overall protein synthesis.
RESULTS
Ribosomal Proteins and RNA Are Crosslinked by UV-B Radiation in a Dosage-Dependent Manner
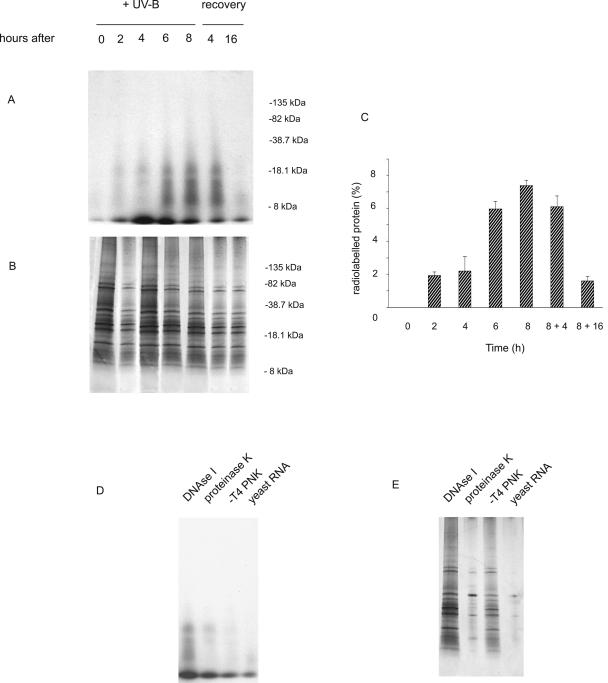
Previously, using microarray hybridization to assess maize acclimation responses to UV-B, we found that transcripts encoding components of the protein synthetic apparatus increase after radiation treatments in leaves (Casati and Walbot, 2003, 2004), paralleling results from cultured human melanocytes (Valery et al., 2001; Sesto et al., 2002). As mentioned above, during in vitro irradiation UV can crosslink proteins and RNA molecules (rRNA, tRNA, or mRNA) within ribosomes. Given these observations, we investigated if similar effects of UV-B radiation can occur in vivo in maize ribosomes. Total ribosomes were purified from adult maize leaves after different periods of UV-B irradiation. Purified ribosomes were treated extensively with single- and double-strand specific RNases and then incubated with [γ32P] dATP in the presence of T4 polynucleotide kinase to end-label RNA as described in “Materials and Methods.” Proteins crosslinked with ribonucleotides should be labeled with [32P] (Kozlov et al., 1998). Proteins were resolved on SDS-PAGE gels and visualized by Coomassie Blue stain; radiolabeled species were detected by autoradiography (Fig. 1). As shown in Figure 1A, UV-B radiation crosslinked proteins with RNA in a dose-dependent manner when normalized to equal amounts of total protein analyzed at each point (Fig. 1C). After 2 h of UV-B exposure, several crosslinked products were detected, and these increased with longer UV-B exposure. Crosslinking is cumulative for at least 8 h. Faint bands were detected when the same experiment was done with ribosomes incubated with proteinase K after the treatment with RNases, probably because some protection against proteinase digestion exists when proteins are associated with RNA (Fig. 1D). No bands were observed when the labeling reaction was completed in the absence of T4 polynucleotide kinase, or when the experiments were done using yeast RNA instead of maize ribosomes (Fig. 1D). The same protein size classes were detected when the reactions were done in the presence of DNAse I (Fig. 1D), demonstrating that the bands observed by autoradiography correspond to protein-RNA species. We also studied the kinetics of elimination of the crosslinked proteins; 4 h after the UV-B treatment, the crosslinked species were still present (Fig. 1A), but after a dark period of 16 h the crosslinked protein-RNA moieties were almost undetectable, similar to control tissues from plants grown without UV-B.
Figure 1.
Crosslinking of ribosomal proteins and RNA in a dosage-dependent manner. A and D, Autoradiography of [32P]-labeled RNA crosslinked to ribosomal proteins; B and E, Coomassie Blue-stained gel. A and B, After UV-B exposure of leaves for 2, 4, 6, and 8 h, purified ribosomes were treated as described in “Materials and Methods.” Recovery: After 8 h of UV-B, leaves were maintained without UV-B for 4 or 16 h in the dark. D and E, Controls: Proteins were incubated with DNAse I or with proteinase K after the treatment with RNases; the labeling reaction was done without T4 polynucleotide kinase (−T4 PNK); and yeast RNA was used instead of purified ribosomes. C, Quantification of radioactive bands by densitometry of the autoradiograph. The percentage of labeling was corrected for loading differences per lane. The molecular mass of marker proteins is indicated in the right of A and B. Figure 1A shows the result of one experiment, the same bands were detected in all eight experiments.
Identification of RNA-Crosslinked Ribosomal Proteins
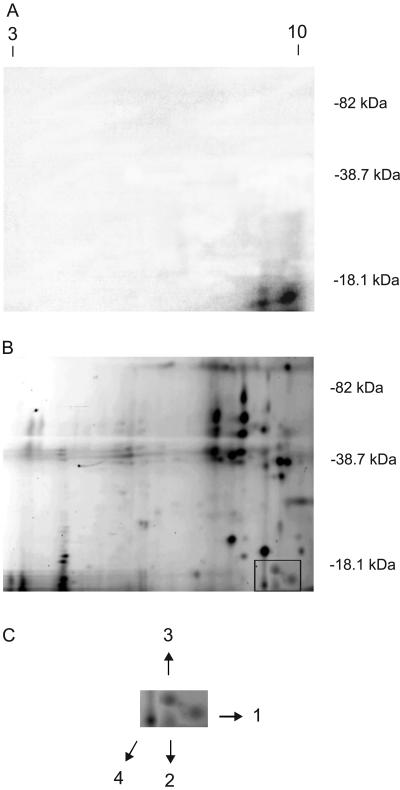
To identify which proteins crosslinked with RNA, proteins from ribosomes after 8 h leaf exposure to UV-B were treated with RNases and then incubated with [γ32P] dATP in the presence of T4 polynucleotide kinase to end-label RNA and then fractionated by two-dimensional (2D) gel electrophoresis. Given the extensive RNase treatment of proteins prior to the labeling reaction, we would expect only a single nucleotide to be present making the labeled proteins very close in molecular mass to the unlabeled form. The first dimension was an isoelectric focusing gel (IEF); the second dimension gel was a 12% SDS-PAGE gel in which fractionation was based on apparent molecular mass. After the second electrophoretic step, the purified ribosomal proteins were visualized by staining with SYPRO ruby fluorescent dye (Bio-Rad, Hercules, CA), from which we estimate that approximately 70 distinguishable staining spots were resolved, which are a mixture of cytoplasmic and plastid ribosomes; radiolabeled species were detected by autoradiography (Fig. 2). Four prominent 32P-labeled proteins were observed (Fig. 2A), corresponding to four fluorescent stained proteins (Fig. 2B). These spots were excised from the 2D gels, and high-resolution tandem mass spectrometry (MS) analysis was used to identify the proteins by sequencing the peptide fragments. Protein 1 has identity to Brassica napus cytosolic 40S ribosomal protein S14 (GenBank accession no. AAO41731; Table I). Protein 2 has identity to Oryza sativa 60S ribosomal protein L32 (GenBank accession no. BAC75414; Table I), and protein 3 to Arabidopsis 60S ribosomal protein L23a (GenBank accession no. AAB87692; Table I). All three of these proteins are components of cytosolic ribosomes. In contrast, protein 4 corresponds to Zea mays chloroplastic 50S ribosomal protein L29 (GenBank accession no. AAD50383; Table I). Therefore, both cytosolic and chloroplastic ribosomes are affected by crosslinking after maize leaves are exposed to UV-B. All four proteins have a very basic pI predicted from the primary amino acid sequences, a feature verified by their migration in the 2D gel (Fig. 2; Table I). Although L29 has a higher molecular mass than L23a, it has a higher mobility in SDS-PAGE in our experiments (Fig. 2; Table I). This could reflect effects of the nucleotides crosslinked onto the proteins or unknown characteristics of these proteins.
Figure 2.
Crosslinking of ribosomal proteins and RNA after 8 h of UV-B by 2D gel electrophoresis. Fifty micrograms of ribosomal protein was separated in the first dimension by IEF electrophoresis (pH 3–10), and in the second dimension by 12% [w/v] SDS-PAGE. A, Autoradiography of [32P]-labeled RNA crosslinked to ribosomal proteins in a 2D gel; B and C, SYPRO ruby fluorescent-stained proteins corresponding to the crosslinked [32P]-labeled RNA species. The molecular mass of marker proteins is indicated in the right of A and B, and pH values on the gel are indicated on the top of A. The proteins corresponding to RNA-radiolabeled species, as detected by fluorescence after staining with SYPRO ruby dye and by autography, are identified on C with numbers 1, 2, 3, and 4. These correspond to the proteins listed in Table I.
Table I.
Crosslinked ribosomal proteins identified by mass spectrometry
| Band | Protein Database Match | Predicted Molecular Mass | pI | GenBank Accession No. | Peptides Matched | Measured Mass | Computed Mass | Peptide Sequence |
|---|---|---|---|---|---|---|---|---|
| Species | kD | D | D | |||||
| 1 | 40S ribosomal protein S14 (Brassica napus) | 16.305 | 10.14 | AAO41731 | 2 | 1,053.53 | 1,053.56 | TPGPGAQSALR |
| 1,584.81 | 1,584.81 | IEDVTPIPSDSTRR | ||||||
| 2 | 60S ribosomal protein L32 (O. sativa) | 15.707 | 10.43 | BAC75414 | 1 | 1,510.88 | 1,510.88 | AAQLDIVVTNKLAR |
| 3 | 60S ribosomal protein L23a (Arabidopsis) | 17.462 | 10.20 | AAB87692 | 2 | 1,433.71 | 1,433.70 | LTPDYDALDVANK |
| 1,829.94 | 1,829.98 | LTPDYDALDVANKIGII | ||||||
| 4 | 50S ribosomal protein L29 (maize) | 18.082 | 10.85 | AAD50383 | 2 | 2,612.21 | 2,612.27 | AMTTEQMEEEVVDL KGELFLLR |
| 1,044.52 | 1,044.51 | EQELEEIR |
The highest statistically significant match for each protein is shown, with the determined molecular mass, calculated pI and GenBank accession numbers of these matches. Proteins were extracted from an SDS-PAGE gel and treated as described in “Materials and Methods” before mass spectrometry. Peptide masses obtained experimentally and calculated from composition are shown, along with the amino acid sequences deduced from patterns of fragmentation.
UV-B Crosslinking of Ribosomal Proteins Is Correlated with a Decrease in Amino Acid Incorporation into Protein
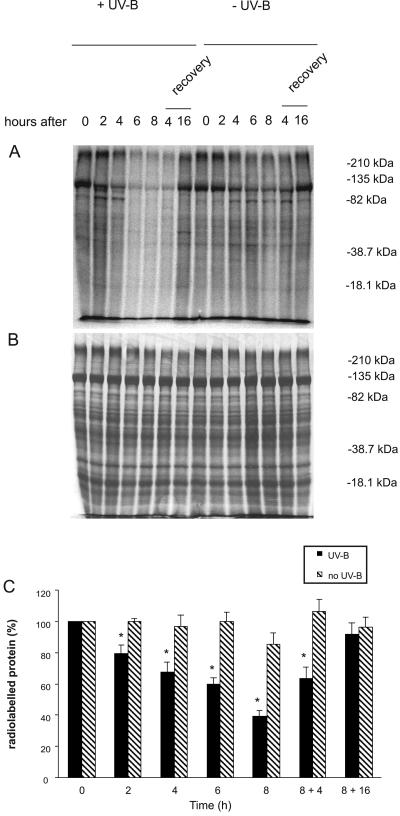
To study the possible impact of ribosome crosslinking on translation, we performed in vivo labeling of leaf proteins with [35S]Met at the same UV-B irradiation periods used to investigate crosslinked ribosomes. As shown in Figure 3A, there is a significant and progressive decrease in the amount of newly synthesized protein after UV-B exposure, in relation to equal amounts of total protein analyzed at each point (Fig. 3B). To accommodate known differences in protein labeling from circadian effects on the synthesis of major leaf proteins in maize (Hartwell et al., 1996; Polidoros and Scandalios, 1998), the levels of new proteins were compared with newly synthesized proteins in control leaves without UV-B at the same times of day. Labeling of all the prominent proteins is reduced after UV-B exposure (Fig. 3A). Quantification of radioactive bands by densitometry (Fig. 3C) showed that translation in UV-B irradiated plants decreased up to 60% after 8 h of UV-B exposure. Normal amino acid incorporation levels were restored after a dark period of 16 h. Thus, both translation inhibition and recovery correlate with the levels of crosslinked products (Fig. 1A), and the observed decrease in translation is probably related to this damage.
Figure 3.
UV-B inhibition of protein synthesis is correlated with ribosomal damage. Thirty micrograms of total cellular proteins were resolved by 10% SDS-PAGE after in vivo [35S]Met labeling, visualized by autoradiography (A) and staining with Coomassie Blue (B) following the UV-B treatments and recovery periods indicated. Treatments started at 8 am. As a control for circadian differences in translation, proteins were monitored from untreated plants at the same time points, as shown on the right half of the gel. C, Quantification of radioactive bands by densitometry of the autoradiograph. The percentage of labeling was corrected for loading differences per lane. UV-B treatments different from the control at P < 0.05 are marked with an asterisk, based on a paired t test.
UV-B Radiation Increases Translation of Ribosomal Proteins
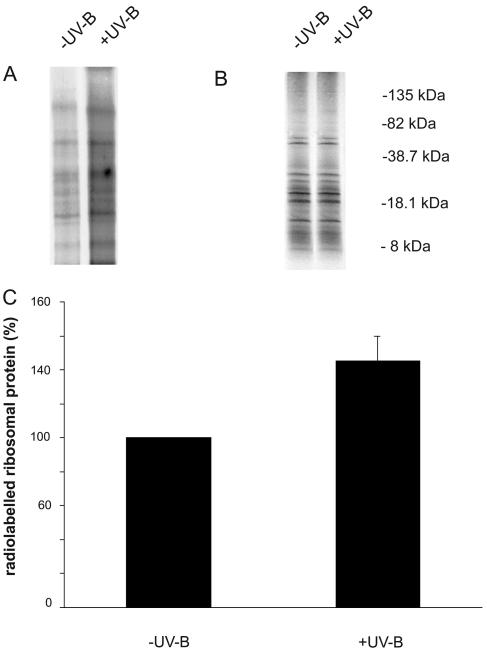
Because transcript abundance of translation-related proteins is increased in maize (Casati and Walbot, 2003, 2004) and in cultured human melanocytes and keratinocytes (Valery et al., 2001; Sesto et al., 2002) and selective translation of such factors has been documented in cultured animal cells, it is logical to ask if translation of ribosomal proteins and translation factors is differentially affected by UV-B irradiation when overall translation in impaired (Fig. 4A). To evaluate the effect of UV-B exposure on ribosomal protein synthesis, we performed in vivo labeling of leaf ribosomal proteins with [35S]Met after 6 h UV-B irradiation and in control plants not exposed to UV-B. For this experiment, leaf samples were incubated with [35S]Met for 2 h after the UV-B irradiation treatment, and ribosomes were purified as described in “Materials and Methods.” Results after SDS-PAGE gels and autoradiography showed a 45% increase in de novo ribosomal proteins synthesis after UV-B exposure compared to synthesis in control plants when equal amounts of proteins were compared (Fig. 4). These data confirm that UV-B induces the de novo synthesis of ribosomal proteins, and this is observed despite the overall decrease in translation (Fig. 3).
Figure 4.
Ribosomal protein synthesis stimulated by UV-B irradiation. Ribosomal proteins were resolved by 10% to 20% SDS-PAGE after in vivo [35S]Met labeling, visualized by autoradiography (A) and staining with Coomassie Blue (B) following the 6-h UV-B treatment (+UV-B) or in control conditions without UV-B (−UV-B). C, Quantification of radioactive labeled proteins by densitometry of the autoradiograph. The experiment was repeated three times and normalized to equal amounts of ribosomal protein. The molecular mass of marker proteins is indicated in the right of B.
Phosphorylation of Ribosomal Protein S6 and Its Protein Kinase after UV-B Exposure
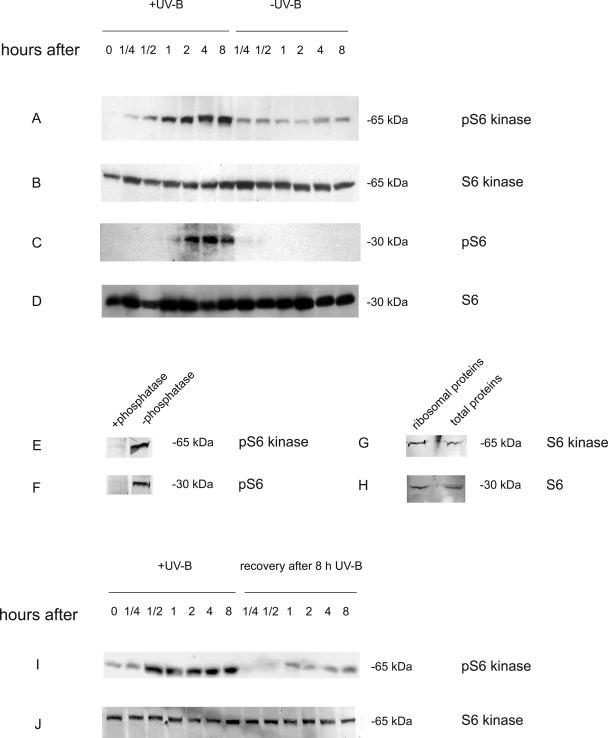
In animal cells ribosomal protein S6 phosphorylation has been implicated as one route to translational up-regulation by UV-B of mRNAs coding for the components of protein synthetic apparatus. Phosphorylation of ribosomal protein S6 and p70 S6 kinase has also been reported in maize (Williams et al., 2003; de la Cruz et al., 2004). For ribosomal protein S6, phosphorylated isoforms in Ser-238 and Ser-241 have been described (Williams et al., 2003), while for p70 S6 kinase, residue Thr-389 was found to be phosphorylated in correlation with its activation (de la Cruz et al., 2004).
To determine the status of these components in UV-B treated maize we conducted western blot analysis using antibodies raised against conserved peptides of ribosomal protein S6, phospho-S6, S6 kinase, and phospho-S6 kinase. Protein extracts were prepared from leaves after different time periods of UV-B radiation. As the control, samples were collected at the same times from plants that were not exposed to UV-B. Because activation of signal cascades is usually very rapid, samples were collected beginning 15 min after the initiation of UV-B exposure. The quantification of proteins shown in Figure 5 indicates that within 1 h of UV-B exposure, high levels of phosphorylated S6 kinase are detected (Fig. 5A) compared to the equal loading control, which demonstrates that both the irradiated and unirradiated samples have similar levels of S6 kinase (Fig. 5B). The phosphorylation state of the kinase persists with longer exposure to UV-B. No phosphorylation was detected if the protein extracts were incubated with Lambda protein phosphatase (Fig. 5E). Low levels of the phosphorylated forms of S6 kinase are also observed in control plants. This could reflect antibody cross-reaction with the nonphosphorylated form of S6 kinase or result from tissue wounding during leaf maceration that could induce some phosphorylation of this protein. Figure 5G shows that a band with the same molecular mass reacts with the antibodies against S6 kinase when purified ribosomal proteins are loaded on a gel, showing that the protein recognized by these antibodies copurify with the ribosomes.
Figure 5.
UV-B induction of phosphorylation of ribosomal protein S6 and S6 kinase and dephosphorylation of S6 kinase during recovery. Total cellular proteins were resolved by 10% or 12% SDS-PAGE after different times of UV-B exposure. Western-blot analysis of protein samples was performed with antibodies against pS6 kinase (A), S6 kinase (B), pS6 (C), or S6 (D). Protein extracts from plants that were exposed under UV-B for 8 h were treated with or without Lambda protein phosphatase for 30 min and western-blot analysis of protein samples was performed with antibodies against pS6 kinase (E) or pS6 (F). Western-blot analysis of purify ribosomal protein and total cellular proteins after UV-B exposure was performed with antibodies against S6 kinase (G) or S6 (H). Western-blot analysis of protein samples after different times of UV-B exposure and during recovery after 8 h UV-B exposure was performed with antibodies against pS6 kinase (I) or S6 kinase (J). Fifty or five micrograms of total soluble or purified ribosomal proteins, respectively, were loaded in each lane.
Using a similar protocol, phosphorylation of ribosomal protein S6 was detected during UV-B irradiation using antibodies directed against phospho-Ser-238 (Fig. 5C). No phosphorylation was detected if the protein extracts were incubated with Lambda protein phosphatase (Fig. 5F) or when antibodies against phospho-S6 with only the Ser-241 modification were used. This shows that antibodies only recognize the antigenic peptide when Ser-238 is phosphorylated. These results demonstrate phosphorylation of maize S6 at residue 238, but the lack of cross-reactivity of the antibodies used to monitor phospho-S6 at Ser-241 could be negative because the antibody fails to recognize the maize protein (see “Materials and Methods”). Collectively, the results show that UV-B radiation can activate phosphorylation of ribosomal protein S6 and the corresponding S6 kinase. To confirm that the protein recognized by the antibodies is in fact a ribosomal protein, purified ribosomal proteins were analyzed by western blot using antibodies against ribosomal protein S6. Figure 5H shows that the antibodies recognize a band with the same molecular mass when either purified ribosomal proteins or total soluble proteins are loading on the gel, demonstrating that the protein detected on the western blot is a ribosomal protein.
Dephosphorylation of S6 kinase is also very fast: 15 min after an 8 h-UV-B treatment the levels of phosphorylation of S6 kinase were similar to the levels in control plants, and this status remained constant during the 8-h recovery period (Fig. 5I). This result was confirmed in an independent experiment (Fig. 5). In this experiment enhanced phosphorylation of pS6 kinase was also detected within 15 min, indicating that modulation of S6 kinase by phosphylation is a rapid response to UV-B exposure and to the removal of this environmental factor.
Decrease of Translation Does Not Result from Widespread Cell Death
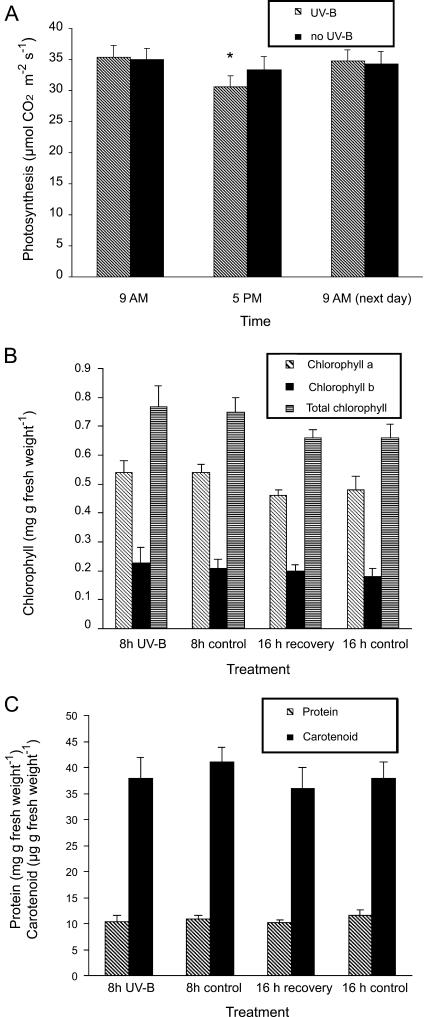
To rule out that the decrease in translation during UV-B irradiation results from cell death, physiological properties were assessed immediately after and 16 h after the 8 h of UV-B treatment. CO2 fixation shows a small but significant decline (with P > 0.95 in a paired-t test) in photosynthesis rate after UV-B irradiation, but recovery is complete by the next day (Fig. 6A). No changes in the levels of chlorophylls a and b (Fig. 6B), carotenoids, or total proteins (Fig. 6C) were detected. Therefore, we find no evidence of UV-B-induced cell death at the wavelengths and fluence rate used in our experiments.
Figure 6.
UV-B treatment is not lethal to maize plants. A, Photosynthetic CO2 fixation before UV-B treatment (9 am), after 8 h of UV-B (5 pm), and 16 h posttreatment (next day 9 am) compared to matched, untreated plants. Total chlorophylls, chlorophyll a and b (B), and total proteins and carotenoids (C) after 8 h UV-B (8 h UV-B) and 16 h posttreatment (16 h recovery) compared to untreated controls. Measurements are the average of six adult leaves from six different plants. Statistical significance was analyzed using a paired-t test with P < 0.05; differences from the control are marked with an asterisk.
DISCUSSION
Using a label transfer method, we demonstrated that UV-B photons cause ribosomal damage by crosslinking RNA to ribosomal proteins in irradiated maize leaves. We have also found that UV-B radiation inhibits protein synthesis in vivo while specifically increasing translation of some ribosomal proteins. Because amino acid incorporation into protein so closely parallels the levels of ribosomal proteins crosslinked to RNA, UV-B inhibition of protein synthesis is probably a consequence of the accumulation of crosslinks between cytosolic ribosomal proteins S14, L32, and L23a, and chloroplastic ribosomal protein L29 and RNA that impairs ribosome function. Given that only proteins from purified ribosomes were analyzed by 2D gel electrophoresis, we consider it unlikely that a suite of minor, nonribosomal proteins of low Mr and high IEF would routinely crosslink to RNA and be purified with ribosomes in this analysis.
S14 is a cytosolic ribosomal protein; it is a member of Group I when eukaryote ribosomal proteins are classified based on homology to proteins of archae- and eubacteria (Wool et al., 1995). Proteins in Group I have orthologs in eubacteria, archaebacteria, and eukaryotes. Eukaryote ribosomal protein S14 is related to the archaebacterial Halobacterium marismortui S11 and to the eubacterial Escherichia coli S11 (Wool et al., 1995). E. coli ribosomal protein S11 can be in vitro crosslinked to nucleotides 693 to 697 of 16S rRNA using bis-(2-chloroethyl)-methylamine (Greuer et al., 1987) and to nucleotides 702 to 705 of 16S rRNA using S11-methyl p-azidophenyl acetimidate (Osswald et al., 1990); therefore, this protein is in close contact with ribosomal RNA in eubacteria. A similar interaction probably occurs with the eukaryote counterpart and 18S rRNA in cytosolic ribosomes. Such an interaction has been demonstrated for yeast (Saccharomyces cerevisiae) ribosomal protein S14, which not only binds to a conserved helix in 18S rRNA, but also directly binds to an RNA stem-loop structure in S14B pre-mRNA that is necessary for S14B regulation (Fewell and Woolford, 1999). These results support the idea that S14 can directly bind different RNA molecules, and at least one of these species can be crosslinked by UV-B. Because of the extensive digestion of crosslinked RNA with RNases, it would not be possible to identify the type of RNA involved in the crosslinking. L32 is a cytosolic ribosomal protein; it is a member of Group II in which proteins have orthologs in archaebacteria and eukaryotes but not in eubacteria (Barakat et al., 2001). This protein is orthologous to L32 in other eukaryotes and to the archaebacterial H. marismortui L5 (Wool et al., 1995). In yeast, the transcript of L32 interacts with L32 protein to form a structure that regulates translation (Li et al., 1996). Thus, as occurs with S14, L32 binds mRNAs in addition to RNA within the ribosomes. L23a is a Group I cytosolic ribosomal protein (Wool et al., 1995). Arabidopsis L23a is related to rat ribosomal protein L23a, to yeast L25, to the archaebacterial Methanococcus vannielii L23, to the eubacterial E. coli L23, and to other members of the L23 family of ribosomal proteins (Suzuki and Wool, 1993; Barakat et al., 2001; McIntosh and Bonham-Smith, 2001). The crystal structure of the large ribosomal subunit from Haloarcula marismortui resolved to 2.4 Å shows that L23 interacts with 23S rRNA in two different domains, I and III; the interaction with domain III is very substantial (Ban et al., 2002). Moreover, E. coli L23 can be crosslinked in vitro to nucleotides 61 to 68 of 23S rRNA using nitrogen mustard (Osswald et al., 1990) and to nucleotides 137 to 141 of 23S rRNA using nitrogen mustard and 2-iminothiolane (Wower et al., 1981; Osswald et al., 1990). A similar contact probably occurs with L23a and 28S rRNA in plants.
L29 is a chloroplastic ribosomal protein with high similarity to other chloroplastic and eubacterial L29 ribosomal proteins. Ribosomal protein L29 has been shown to be covalently attached to nucleotides 99 to 107 of 23S rRNA by 2-iminothiolane (Wower et al., 1981; Osswald et al., 1990) and by nitrogen mustard (Osswald et al., 1990) after in vitro treatment of E. coli ribosomes. Therefore, this protein is in close contact with 23S rRNA. In summary, all four proteins detected after label transfer from RNA are in intimate contact with rRNA in intact ribosomes. In maize leaves these interactions (or other RNA-protein contacts) can be converted into covalent bonds by natural levels of UV-B radiation.
The build-up of RNA-protein crosslinking involving ribosomal proteins is paralleled by an overall decrease in translation. These observations suggest that the crosslinking observed results in severe ribosomal damage. Previous studies have reported inhibition of translation in plant cells by highly energetic UV-C. For example, when Murphy et al. (1973) isolated protein synthesis components from wheat embryos and irradiated them with UV-C, the in vitro translation rate decreased substantially. In addition, irradiation of liquid-cultured tobacco cells strongly and quickly inhibited their ability to incorporate labeled amino acids into protein (Murphy et al., 1974). The translation block also resulted in formation of larger polysomes, likely indicating that the ribosomes were intact but stalled on the mRNA. Murphy et al. (1973, 1974) used highly energetic UV-C, which may result in a damage spectrum different from UV-B. UV-C is absent from the solar spectrum, while our experiments were done using UV-B, a more environmentally relevant perturbation.
Damaged ribosomes may be degraded followed by rapid synthesis of new ribosomes during recovery from UV-B damage. In this study we demonstrate that UV-B radiation induces de novo synthesis of ribosomal proteins. This observation can be explained in part by the large increase in levels of transcripts encoding ribosomal proteins detected in microarray experiments after UV-B exposure of mammalian (Valery et al., 2001; Sesto et al., 2002) and plant (Casati and Walbot, 2003, 2004) tissues. Selective translation mediated by changes in ribosome preferences may also contribute. This increase in synthesis of ribosomal proteins after UV-B irradiation is not the result of a general stress response in plants, as other abiotic stresses such as dehydration cause the coordinate down-regulation of the translation of the ribosomal protein gene mRNAs in Arabidopsis (Kawaguchi et al., 2004). The phosphorylation status of the S6 ribosomal protein has been implicated as one of the ribosome constituents that modulates translational selectivity (Thomas, 2002). We also found that both ribosomal protein S6 and an S6 kinase are phosphorylated after UV-B exposure. Phosphorylation of these proteins is activated by UV-B in some types of cultured animal cells (Brenneisen et al., 2000; Nomura et al., 2001) where S6 phosphorylation has been implicated in the translational up-regulation of mRNAs containing the 5′ TOP motif; exemplar transcripts encode components of the protein synthetic apparatus (Thomas, 2002). One mechanism permitting selective translation of eukaryotic mRNA requires an oligopyrimidine tract at the 5′ terminus. This element usually consists of a cytidine residue at the cap site followed by an uninterrupted sequence of 7 to13 pyrimidine nucleotides (Levy et al., 1991). The 5′ TOP sequence is essential for the translational regulation observed (Meyuhas et al., 1997). The role of the 5′ TOP motif has not been demonstrated in plants cells; however, we found 8 of 12 full-length transcripts for maize in GenBank, nuclear-encoded ribosomal proteins have this type of polypyrimidine track in the 5′ leader (see Supplemental Table I, available at www.plantphysiol.org). The remainder contained shorter oligopyrimidine tracts (between 5–6 bases, not shown); as oligonucleotide tracks can be difficult to sequence accurately, the tracks could be longer. As a control, we searched GenBank for similar sequences in randomly selected full-length sequenced maize transcripts. Among 19 complete gene transcripts encoding proteins with different cell functions, only 3 have 5′oligopyrimidine sequences with characteristics of 5′ TOP sequences (see Supplemental Table II). This demonstrates that the TOP sequences are not common. In evaluating the oligopyrimidine tracks we found no strict consensus motif, although some transcripts have the same 5′ TOP sequence. This is also true for the vertebrate ribosomal protein 5′ TOP sequences (Levy et al., 1991). Combined with the observations that UV-B stimulates phosphorylation of S6 and its kinase and selective translation of some ribosomal proteins, the presence of 5′ TOP sequences in some maize ribosomal protein transcripts suggests that translational control of these proteins by UV-B in plants could utilize mechanisms similar to those in mammalian cells. Proof that this pathway permits selective translation of transcripts for ribosomal proteins during conditions of ribosomal damage and the consequent low translation rates will require a detailed evaluation of the 5′ TOP and of the mechanism(s) by which UV-B treatment results in rapid phosphorylation of S6 kinase and S6. The results presented define the kinetics of response and serve as a platform for further biochemical studies. When catalogues of full-length maize cDNAs are available, a more thorough statistical analysis of 5′ TOP sequence frequency will also be possible.
Phosphorylation of S6 was described previously in plant ribosomes. In response to several stress treatments, hyperphosphorylated isoforms of S6 accumulate in maize root tips (Williams et al., 2003). Of particular interest are the Ser-238 and Ser-241 residues, because these are differentially phosphorylated after specific environmental changes. The phosphorylation status of S6 also changes in response to heat shock in cultured tomato cells and in maize embryos (Scharf and Nover, 1982; Beltrán-Peña et al., 2002) and to oxygen deprivation in maize roots (Bailey-Serres and Freeling, 1990). In addition, in cultured cells of Arabidopsis, S6 accumulates as three partially phosphorylated and one nonphosphorylated isoform under standard growth conditions; after heat shock hypophosphorylated isoforms accumulate (Turck et al., 1998). Moreover, Turck et al. (2004) showed that activation of S6 kinase is regulated by phytohormones in Arabidopsis, and this regulation is, at least in part, via a lipid kinase-dependent pathway. In these experiments, they demonstrated that the activation of S6 kinase correlates with the translational up-regulation of ribosomal protein S6 and S18A mRNAs without affecting global translation. In our experiments, we found that both S6 and S6 kinase are phosphorylated after UV-B exposure. These results confirm the rapidity of response to S6 and S6 kinase to environmental perturbations and extend the observations that UV-B mediates phosphorylation changes of these proteins in animal tissue culture cells (Brenneisen et al., 2000; Nomura et al., 2001).
If phosphorylation of S6 and S6 kinase results in translational preference for mRNAs encoding some ribosomal proteins, this UV-B activated pathway could be one mechanism to mediate the increase in de novo synthesis of ribosomal proteins observed in our experiments (Fig. 4). In other words, in a situation of ribosomal damage, phosphorylation of these proteins mediated by a stress-induced signaling pathway can selectively promote translation of proteins required to restore this crucial cellular function. As shown in Figure 3, overall translation in maize leaves recovers within 16 h after the cessation of UV-B irradiation; therefore, new ribosomes that are synthesized within this period can overcome the results of UV-B mediated crosslinking within organellar and cytosolic ribosomes. Mouse ribosomal protein L32 mRNA, the ortholog of corn ribosomal protein L32 that we identified as crosslinked to RNA, contains a TOP element in its 5′ untranslated region and is translationally regulated by p70 S6 kinase (Kakegawa et al., 2002). Complete transcript and maize gene sequences are not yet available for the three cytosolic ribosomal protein genes crosslinked to RNA by UV-B; however, a complete transcript sequence of the L29 plastid ribosome protein is available. Only 28 nucleotides of 5′ untranslated region have been sequenced; and there is no sequence with similar characteristics as the described 5′ TOP tracts in this region.
In addition to the de novo synthesis of ribosomes, restoration of translation competence by repair of RNA-protein crosslinks could exist. Currently, there is very limited information about RNA repair in plants. UV-C and short wavelength UV-B (<285 nm) induce uracil-uracil pyrimidine and cyclobutane dimers in RNA (Wacker, 1961). Paralleling enzymatic photoreactivation of such lesions in DNA, this type of RNA damage is repaired in light-grown but not in dark-grown plants. RNA repair was established in a series of elegant studies with tobacco mosaic and other RNA viruses. Purified, infectious viral genomes were irradiated and then applied to plant leaves, and the treated plants were maintained under normal greenhouse conditions or in the dark. Only the light-grown plants supported lesion development from irradiated viral RNA, while untreated RNA produced disease lesions in the dark control plants (Bawden and Kleczkowski, 1959). Similar results were obtained by irradiation of infected leaves (Bawden and Sinha, 1961). Although the enzyme(s) responsible for photoreactivation repair of intra-strand RNA damage has not been isolated, it is clear that UV-induced lesions in purified RNA can be repaired in plants. In contrast to the repair of lesions induced in purified RNA, irradiation of intact virions resulted in damage that could not be repaired in plants grown in the light or the dark (Bawden and Kleczkowski, 1959). The nature of this damage was not elucidated, but it seems highly likely that RNA-protein and protein-protein crosslinking occurred and that this damage prevented viral replication.
There are a handful of additional reports on mechanisms of RNA repair in eukaryotic cells. Two human DNA demethylases can also demethylate RNA in vivo, illustrating that RNA repair can be a potentially important cellular defense mechanism after exposure to alkylating agents (Aas et al., 2003). Enzymes similar to other DNA repair systems may participate in ribosome repair, but such functions have not been identified. There are also reports of photorepair of UV-inactivated RNAs in insect cells (Jäckle and Kalthoff, 1978, 1980; Kalthoff et al., 1978).
Crosslinking within ribosomes as reported here is a direct mechanism that could reduce translation, but indirect effects such as UV-induced changes in translation factors cannot be ruled out. Highly energetic UV-C can induce phosphorylation of the α-subunit of eukaryotic translation initiation factor 2 in mammalian cells; this was correlated to a decrease of translation (Wu et al., 2002). A similar regulation of translation by UV-B could exist in maize.
Despite the presence of significant ribosome damage and a decrease in translation, physiological parameters, such as photosynthesis and pigment levels, were not significantly affected by the UV-B treatment employed. The fluence rate simulates the ozone depletion currently experienced in some terrestrial habitats. There is a small but significant decline in photosynthesis rate after 8 h of UV-B irradiation, but recovery is complete by the next day. Therefore, we find no evidence that UV-B induces cell death at the wavelengths and fluence rate used. Collectively, our results together with microarray data for UV-B exposure experiments (Valery et al., 2001; Sesto et al., 2002; Casati and Walbot, 2003, 2004) suggest that new synthesis of ribosomes occurs as a response to UV-B damage. Restoration of protein synthetic capacity, like rapid repair of UV-induced DNA damage, is a plant adaptation to daily solar UV-B exposure.
MATERIALS AND METHODS
Plant Material and Radiation Treatments
The b, pl W23 inbred line was used for all experiments. This line is considered a wild-type maize (Zea mays) inbred similar to most commercial maize lines in lacking anthocyanin and other UV-B absorbing flavonoid pigments in adult leaves. UV-B treatments were done in a greenhouse illuminated for 14 h daily with a combination of sodium vapor, metal halide, and UV-A bulbs to a fluence approximately 20% of noon summer white sunlight. Adult plants were illuminated using UV-B lamps for different periods of time (F40UVB 40 W and TL 20 W/12; Philips, Eindhoven, The Netherlands) using fixtures mounted 30 cm above the plants with a biologically effective UV irradiance of 0.36 W m−2 (9 kJ m−2 d−1) normalized to 300 nm (Caldwell, 1971). The bulbs were covered with cellulose acetate filters (100 μm extra clear cellulose acetate plastic, Tap Plastics, Mountain View, CA); the cellulose acetate sheeting does not remove any UV-B radiation from the spectrum but excludes wavelengths lower than 280 nm. Control, no UV-B plants were exposed for the same period of time under the same lamps covered with polyester filters (100 μm clear polyester plastic; Tap Plastics). This polyester filter absorbs both UV-B and wavelengths lower than 280 nm. The output of the UV-B source in the greenhouse was recorded using an Optronics model 752 spectroradiometer (Optronics Laboratories, Orlando, FL) that was calibrated against a National Bureau of Standards certified radiation source before each use. The spectrum under each treatment was recorded periodically with 1-nm resolution across the entire sunlight spectrum (290–800 nm). The experiments were repeated four times.
Ribosome Purification and Crosslinking Studies
Total ribosomes were purified according to Yusupov and Spirin (1988). A total of 0.5 mg of purified ribosomes, quantified as described in Williams et al. (2003), were incubated overnight at 37°C with RNase T1 and RNase V1 (4,000 units mL−1 each; Ambion, Austin, TX) plus RNase A (4,000 μg mL−1; Qiagen, Valencia, CA) in a buffer containing 10 mm Tris-HCl, pH 8.1, 0.5 mm EDTA, and 8 m UREA, and Complete protease inhibitor cocktail (Roche, Indianapolis). The next day, 8,000 units mL−1 each of RNase T1 and V1 plus 8,000 μg of mL−1 RNase A were added followed by 2 h incubation. This combination of nucleases should result in monoribonucleotide or very short oligoribonucleotides attached to protein. Ribosomal proteins were precipitated with 2 volumes of ethanol to facilitate resuspension of proteins and 0.3 m NaAc overnight at −20°C. The pellet was resuspended in the polynucleotide kinase buffer (New England Biolabs, Beverly, MA), and 5 μg of protein was incubated with 50 μCi of [γ32P] dATP in the presence of 10 units of T4 polynucleotide kinase (New England Biolabs) for 1.5 h at 37°C. The mixture was desalted using a MicroSpin S-300 column (Amersham Biotech, Piscataway, NJ), and protein concentration was determined using the Bradford reagent (Bio-Rad); 5 μg was loaded onto a precast 10% to 20% SDS-PAGE gel (Bio-Rad). After electrophoresis, the gel was stained with Coomassie Blue dye, and radioactive-labeled ribonucleotides crosslinked to proteins were detected by autoradiography. Quantification was achieved by densitometry of the autoradiograms and Coomassie Blue stained gels using a Kodak ds 1D Digital Science, Scientific Imaging System (New Haven, CT), and the amount of radiolabeled protein was normalized to equal amounts of proteins on the gels after quantification; the percentage of labeling was calculated for the same amount of protein loaded on the gel. The experiment was repeated eight times. Figure 1A shows the result of one experiment, the same three bands were detected in all experiments.
2D PAGE
Fifty micrograms of ribosomal protein pellet was resuspended in 50% (v/v) glycerol before electrophoresis. Separation in the first dimension was an IEF polyacrylamide gel (pH 3–10), using 0.1 m H3PO4 in the cathode and 0.1 mm NaOH in the anode; the solutions were prepared immediately before use. Electrophoresis was carried out to equilibrium for 1 h at 100 V, 1 h at 250 V, and 1 h at 300 V with migration toward the cathode. After the run, first-dimension gels were incubated in the equilibration buffer for 15 min (0.05 m Tris-HCl, pH 6.8, 5% [v/v] β-mercaptoethanol, 10% [v/v] glycerol, 2% [w/v] SDS, and 0.1% [w/v] bromphenol blue); the soaked gel was then embedded at the top of the second-dimension gel with agarose (0.05 m Tris-HCl, pH 6.8, 5% [v/v] β-mercaptoethanol, 10% [v/v] glycerol, 2% [w/v] SDS, and 1% [w/v] agarose). Separation in the second dimension was in a 12% [w/v] SDS-PAGE gel using Mini-PROTEAN II cell (Bio-Rad). Gels were stained with SYPRO Ruby protein gel stain (Bio-Rad) for 16 h.
ESI-QUAD-TOF Mass Spectrometry
Proteins corresponding to the bands crosslinked to RNA as detected by autoradiography in the 2D PAGE were digested with trypsin for MS/MS and analyzed by ESI-QUAD-TOF MS in the Stanford mass spectrometry facility. For database searches after Tandem MS analysis, Mascot MS/MS Ions Search from Matrix Science was used (www.matrixscience.com). No restrictions were used for this search, and Cys alkylation (propionamide) and Met oxidation were entered as differential modifications.
In Vivo Labeling and Extraction of Proteins
After UV-B treatments, in vivo labeling of proteins was done on 0.5 g of leaf segments cut into 1-mm strips perpendicular to the veins. Leaf pieces were incubated with 20 μCi mL−1 [35S]Met in 0.02% Tween 40 for 1.5 h at 25°C. After the labeling period, the leaf pieces were rinsed extensively with water, powdered with liquid N2, and extracted with buffer containing 100 mm Tris-HCl, pH 7.3, 1 mm EDTA, 10 mm MgCl2, 50 mm KH2PO4, 15 mm β-mercaptoethanol, 20% (v/v) glycerol, and Complete protease inhibitor cocktail (Roche); or were used for ribosome purification as described above. Thirty micrograms of total protein or 10 μg of ribosomal protein determined using Bradford reagent (Bio-Rad) was loaded on a 10% SDS-PAGE gel or on a 10% to 20% SDS-PAGE gel, respectively. Gels were stained with Coomassie Blue stain and radioactive proteins were detected by autoradiography. Quantification was achieved by densitometry of the autoradiograms and Coomassie Blue stained gels using a Kodak ds 1D Digital Science, Scientific Imaging System (New Haven, CT). Normalization was done as described above.
Western-Blot Analysis
For western-blot analysis, protein extracts were prepared as described for in vivo labeling, except that 20 μL of phosphatase inhibitor cocktails I and II (Sigma-Aldrich, St. Louis) was added to the buffer before extraction. As a control, protein extracts prepared with extraction buffer without the phosphatase inhibitors were incubated with Lambda protein phosphatase for 30 min following the manufacturer's instructions (CalBiochem, San Diego). SDS-PAGE was performed with 10% (w/v) polyacrylamide gels for S6 kinase and phospho-S6 kinase detection; 12% (w/v) polyacrylamide gels were used for S6 and phospho-S6 detection. Proteins on the gels were either stained with Coomassie Blue or electroblotted onto a PDF membrane for immunoblotting according to Burnette (1981). p70 S6 kinase, phospho-p70 S6 kinase (Thr-389), S6 ribosomal protein, and phospho-S6 ribosomal protein (Ser 235/236) and (Ser 240/244) commercial IgG fractions were used for detection (Cell Signaling Technology, Beverly, MA). For p70 S6 kinase detection, polyclonal antibodies against a synthetic peptide derived from human p70 S6 kinase. For phospho-p70 S6 kinase detection, polyclonal antibodies against a synthetic peptide corresponding to the residues around Thr-389 of human p70 S6 kinase was used. For S6 ribosomal protein detection, polyclonal antibodies against a synthetic peptide corresponding to residues 185 to 205 of human S6 protein was used. For phospho-S6 ribosomal protein detection, polyclonal antibodies against a synthetic peptide corresponding to residues around Ser-235 and Ser-236, or Ser-240 and Ser-244 of human S6 protein was used. For phospho-p70 S6 kinase and phospho-S6 ribosomal protein, antibodies detect endogenous levels only when the proteins are phosphorylated. The antibodies were raised in rabbit, and all cross react with the proteins from different species (for more information, see www.cellsignal.com). Bound antibody was visualized by linking to alkaline phosphatase-conjugated goat anti-rabbit IgG according to the manufacturer's instructions (Promega, Madison, WI). The molecular masses of the polypeptides were estimated from a plot of the log of molecular mass of marker standards (Bio-Rad) versus migration distance.
CO2 Exchange and Photosynthetic Pigment Measurements
CO2 exchange measurements were done using a LI-COR 6400-02B portable photosynthesis system (LI-COR, Lincoln, NE). The CO2 was maintained constantly at 400 μL L−1. Leaf temperature was kept at 27°C, and the photosynthetic photon flux density was 800 μmol m−2 s−1. Total chlorophylls and carotenoids were determined by standard procedures (Wintermans and de Mots, 1965).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AAO41731, BAC75414, AAB87692, and AAD50383.
Supplementary Material
Acknowledgments
We thank Cathy Squires, Phil Hanawalt, Dave Perkins, and Ann Stapleton for helpful comments, Jessica Ruvinsky for instructions on photosynthesis measurements, and John Fernandes for searching GenBank for maize ribosomal protein cDNAs. P.C. is a member of the Research Career of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET).
This work was supported by the National Science Foundation (grant no. IBN 98–72657) and the U.S. Department of Agriculture (grant no. 2003–00745). P.C. was a postdoctoral fellow of Fundación Antorchas.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.047043.
References
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Freeling M (1990) Hypoxic stress-induced changes in ribosomes of maize seedling roots. Plant Physiol 94: 1237–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Rousseaux MC, Searles PS, Zaller JG, Giordano CV, Robson TM, Caldwell MM, Sala OE, Scopel AL (2001) Impacts of solar ultraviolet-B radiation on terrestrial ecosystems of Tierra del Fuego (southern Argentina): an overview of recent progress. J Photochem Photobiol B 62: 67–77 [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2002) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Barakat A, Szick-Miranda K, Chang I-F, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127: 398–415 [PMC free article] [PubMed] [Google Scholar]
- Bawden FC, Kleczkowski A (1959) Photoreactivation of nucleic acid from tobacco mosaic virus. Nature 183: 503–504 [DOI] [PubMed] [Google Scholar]
- Bawden FC, Sinha RC (1961) Effects of ultraviolet radiation on infection by intact and phenol-disrupted red clover mottle virus. Virology 14: 198–206 [DOI] [PubMed] [Google Scholar]
- Beltrán-Peña E, Aguilar R, Ortiz-Lopez A, Dinkova TD, Sanchez de Jimenez ES (2002) Auxin stimulates S6 ribosomal protein phosphorylation in maize thereby affecting protein synthesis regulation. Physiol Plant 115: 291–297 [DOI] [PubMed] [Google Scholar]
- Brenneisen P, Wenk J, Wlaschek M, Krieg T, Scharffetter-Kochanek K (2000) Activation of p70 ribosomal protein S6 kinase is an essential step in the DNA damage-dependent signaling pathway responsible for the ultraviolet B-mediated increase in interstitial collagenase (MMP-1) and stromelysin-1 (MMP-3) protein levels in human dermal fibroblasts. J Biol Chem 275: 4336–4344 [DOI] [PubMed] [Google Scholar]
- Brimacombe R, Greuer B, Gulle H, Kosack M, Mitchell P, Oßwald M, Stade K, Stiege W (1990) New techniques for the analysis of intra-RNA and RNA protein cross-linking data from ribosomes. In G Spedding, ed, Ribosomes and Protein Synthesis. A Practical Approach. IRL Press, Oxford, pp 131–159
- Britt AB (1996) DNA damage and repair in plants. Annu Rev Plant Physiol Plant Mol Biol 4: 75–100 [DOI] [PubMed] [Google Scholar]
- Burnette WN (1981) Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112: 195–203 [DOI] [PubMed] [Google Scholar]
- Caldwell MM (1971) Solar UV irradiation and the growth and development of higher plants. Photophysiology 6: 131–177 [Google Scholar]
- Casati P, Walbot V (2003) Gene expression profiling in response to ultraviolet radiation in Zea mays genotypes with varying flavonoid content. Plant Physiol 132: 1739–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V (2004) Rapid transcriptome responses of maize to UV-B in irradiated and shielded tissues. Genome Biol 5: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz HR, Aguilar R, de Jimenez ES (2004) Functional characterization of a maize ribosomal S6 protein kinase (ZmS6K), a plant ortholog of metazoan p70(S6K). Biochemistry 43: 533–539 [DOI] [PubMed] [Google Scholar]
- Fewell SW, Woolford JL (1999) Ribosomal protein s14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B Pre-mRNA and to 18S rRNA. Mol Cell Biol 19: 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt KE, Wilson MI, Greenberg BM (1999) Tryptophan photolysis leads to a UVB-induced 66 kDa photoproduct of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in vitro and in vivo. Photochem Photobiol 70: 49–56 [Google Scholar]
- Greuer B, Osswald M, Brimacombe R, Stoffler G (1987) RNA-protein cross-linking in Escherichia coli 30S ribosomal-subunits. Determination of sites on 16S RNA that are cross-linked to proteins S3, S4, S7, S9, S10, S11, S17, S18 and S21 by treatment with BIS-(2-chloroethyl)-methylamine. Nucleic Acids Res 15: 3241–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell J, Smith LH, Wilkins MB, Jenkins GI, Nimmo HG (1996) Higher plant phosphoenolpyruvate carboxylase kinase is regulated at the level of translatable mRNA in response to light or a circadian rhythm. Plant J 10: 1071–1078 [Google Scholar]
- Iordanov MS, Pribnow P, Magun JL, Dinh T-H, Pearson JA, Chen SL-Y, Magun BE (1997) Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol 17: 3373–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanov MS, Pribnow D, Magun JL, Dinh T-H, Pearson JA, Magun BE (1998) Ultraviolet radiation triggers the ribotoxic stress response in mammalian cells. J Biol Chem 273: 15794–15803 [DOI] [PubMed] [Google Scholar]
- Jäckle H, Kalthoff K (1978) Photoreactivation of RNA in UV-irradiated insect eggs (Smittia SP chironomidae diptera). 1. Photosensitized production and light-dependent disappearance of pyrimidine dimers. Photochem Photobiol 27: 309–315 [DOI] [PubMed] [Google Scholar]
- Jäckle H, Kalthoff K (1980) Photoreversible UV-inactivation of messenger-RNA in an insect embryo (Smittia spec chironomidae, diptera). Photochem Photobiol 32: 749–761 [DOI] [PubMed] [Google Scholar]
- Kakegawa T, Ito M, Hayakawa A, Matsuda M, Tamura S, Saito H, Kaspar RL, Kobayashi H (2002) Rapamycin induces binding activity to the terminal oligopyrimidine tract of ribosomal protein mRNA in rats. Arch Biochem Biophys 402: 77–83 [DOI] [PubMed] [Google Scholar]
- Kalthoff K, Urban K, Jäckle H (1978) Photoreactivation of RNA in UV-irradiated insect eggs (Smittia SP chironomidae diptera). 2. Evidence for heterogeneous light-dependent repair activities. Photochem Photobiol 27: 317–322 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kozlov IA, Nielsen PE, Orgel LE (1998) A method for the 32P labeling of peptides or peptide nucleic acid oligomers. Bioconjug Chem 9: 415–417 [DOI] [PubMed] [Google Scholar]
- Levy S, Avni D, Hariharan N, Perry RP, Mayuhas O (1991) Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci USA 88: 3319–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BJ, Vilardell J, Warner JR (1996) An RNA structure involved in feedback regulation of splicing and of translation is critical for biological fitness. Proc Natl Acad Sci USA 93: 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh KB, Bonham-Smith PC (2001) Establishment of Arabidopsis thaliana ribosomal protein RPL23A-1 as a functional homologue of Saccharomyces cerevisiae ribosomal protein L25. Plant Mol Biol 46: 673–682 [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Avni D, Biberman Y, Shama S, Ostareck DH, Ostareck Lederer A, Hentze MW (1997) Growth-dependent translational control of TOP mRNAs. FASEB J 11: 2500 [Google Scholar]
- Murphy TM, Kuhn DN, Murphy JB (1973) Ultraviolet irradiation of the components of the wheat embryo in vitro protein synthesizing system. Biochemistry 12: 1782–1789 [DOI] [PubMed] [Google Scholar]
- Murphy TM, Wright LA Jr, Murphy JB (1974) Inhibition of protein synthesis in cultured tobacco cells by ultraviolet radiation. Photochem Photobiol 21: 219–225 [DOI] [PubMed] [Google Scholar]
- Noah JW, Shapkina T, Wollenzien P (2000) UV-induced crosslinks in the 16S rRNAs of Escherichia coli, Bacillus subtilis and Thermus aquaticus and their implications for ribosome structure and photochemistry. Nucleic Acids Res 28: 3785–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Kaji A, He ZW, Ma WY, Miyamoto K, Yang CS, Dong ZG (2001) Inhibitory mechanisms of tea polyphenols on the ultraviolet B-activated phosphatidylinositol 3-kinase-dependent pathway. J Biol Chem 276: 46624–46631 [DOI] [PubMed] [Google Scholar]
- Osswald M, Greuer M, Brimacombe R (1990) Localization of a series of RNA-protein cross-link sites in the 23S and 5S ribosomal-RNA from Escherichia coli, induced by treatment of 50S subunits with 3 different bifuntional reagents. Nucleic Acids Res 18: 6755–6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul ND, Gwynn-Jones D (2003) Ecological roles of solar UV radiation: towards an integrated approach. Trends Ecol Evol 18: 48–55 [Google Scholar]
- Polidoros AN, Scandalios JG (1998) Circadian expression of the maize catalase Cat3 gene is highly conserved among diverse maize genotypes with structurally different promoters. Genetics 149: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Masson J, Koncz C, Afsar K, Jakovleva L, Paszkowski J (1999) Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J 18: 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles PS, Flint SD, Caldwell MM (2001) A meta analysis of plant field studies simulating stratospheric ozone depletion. Oecologia 127: 1–10 [DOI] [PubMed] [Google Scholar]
- Sesto A, Navarro M, Burslem F, Lorcano JL (2002) Analysis of ultraviolet B response in primary human keratinocytes using oligonucleotide arrays. Proc Natl Acad Sci USA 99: 2965–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Nover L (1982) Heat-shock-induced alterations of ribosomal protein phosphorylation in plant cell cultures. Cell 30: 427–437 [DOI] [PubMed] [Google Scholar]
- Stapleton AE, Walbot V (1994) Flavonoids can protect maize DNA from the induction of ultraviolet-radiation damage. Plant Physiol 105: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiege W, Atmadja J, Zobawa M, Brimacombe R (1986) Investigation of the tertiary folding of Escherichia coli ribosomal RNA by intra-RNA cross-linking in vivo. J Mol Biol 191: 135–138 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Wool IG (1993) The primary structure of rat ribosomal protein L23a. The application of homology search to the identification of genes for mammalian and yeast ribosomal proteins and a correlation of rat and yeast ribosomal proteins. J Biol Chem 268: 2755–2761 [PubMed] [Google Scholar]
- Thomas G (2002) The S6 kinase signaling pathway in the control of development and growth. Biol Res 35: 305–313 [DOI] [PubMed] [Google Scholar]
- Turck F, Kozma SC, Thomas G, Nagy F (1998) A heat-sensitive Arabidopsis thaliana kinase substitutes for human p70–S6k function in vivo. Mol Cell Biol 18: 2038–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F (2004) Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol 134: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valery C, Grob JJ, Verrando P (2001) Identification by cDNA microarray technology of genes modulated by artificial ultraviolet radiation in normal human melanocytes: relation to melanocarcinogenesis. J Invest Dermatol 117: 1471–1482 [DOI] [PubMed] [Google Scholar]
- Wacker A (1961) Strahlenchemische veranderungen von pyrimiden invivo und invitro. J Chim Phys 58: 1040–1045 [Google Scholar]
- Williams AJ, Werner-Fraczek J, Chang IF, Bailey-Serres J (2003) Regulated phosphorylation of 40S ribosomal protein S6 in root tips of maize. Plant Physiol 132: 2086–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans JFGH, de Mots A (1965) Spectrophotometric characteristics of the chlorophylls and their pheophytins in ethanol. Biochim Biophys Acta 109: 448–455 [DOI] [PubMed] [Google Scholar]
- Wool IG, Chan YL, Gluck A (1995) Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol 73: 933–947 [DOI] [PubMed] [Google Scholar]
- Wower I, Wower J, Meinke M, Brimacombe R (1981) The use of 2-iminothiolane as an RNA-protein cross-linking agent in Escherichia coli ribosomes, and the localization on 23S RNA of sites cross-linked to proteins L4, L6, L21, L23, L27 and L29. Nucleic Acids Res 9: 4285–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Hu YY, Wang JL, Chatterjee M, Shi YG, Kaufman RJ (2002) Ultraviolet light inhibits translation through activation of the unfolded protein response kinase PERK in the lumen of the endoplasmic reticulum. J Biol Chem 277: 18077–18083 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Spirin AS (1988) Hot tritium bombardment technique for ribosome surface-topography. Methods Enzymol 164: 426–439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.